Sir,
An outbreak of pneumonia was reported from Wuhan, People's Republic of China, in December 2019 that was linked to a novel coronavirus designated as SARS-CoV-21. On January 31, 2020, the International Health Regulations Emergency Committee of the World Health Organization (WHO) declared the COVID-19 outbreaks as a Public Health Emergency of International Concern (PHEIC)2. In this communication, we report the early response to SARS-CoV-2 outbreak in India. All the overseas travellers reported to seven major airports (Delhi, Mumbai, Kolkata and later Chennai, Hyderabad, Bengaluru and Cochin were added). The initial COVID-19 testing strategy included individuals who had international travel history with symptoms, symptomatic contacts of laboratory-confirmed COVID-19 patients and symptomatic healthcare workers managing respiratory distress/severe acute respiratory illness (SARI)3. Throat and nasal swabs collected from the travellers were tested for influenza, other common respiratory viruses [parainfluenza viruses (PIV), Adeno, Rhino, respiratory syncytial virus (RSV) and human metapneumovirus (hMPV)] along with known human coronaviruses (229E, Oc43, HKU 1 and NL 63)4 in addition to the SARS-CoV-2 as per WHO guidelines5. A total of 362 clinical samples were received during January 22 to February 29, 2020 collected at the airport screening centre and a few samples of the SARS-CoV-2 suspected overseas returnees developing signs and symptoms were referred by the State Health department from 24 States. All the specimens were tested by real-time reverse-transcription-polymerase chain reaction (qRT-PCR) for the following viruses: influenza A [A(H1N1) pdm09 and A(H3N2)], influenza B [B/Yamagata and B/Victoria] along with housekeeping RNaseP gene (CDC, WHO) and RSV A&B, hMPV, PIV 1, 2, 3, 4, rhinovirus and adenovirus including 229E, OC43, HKU1 and NL63 using a protocol described earlier4. RNA was extracted using MagMax-96 Viral RNA Isolation kit as per manufacturer's protocol (Thermo Fisher Scientific, Lithuania). Nucleic acid amplification was performed using one-step qRT-PCR (SuperScript™ III kit, Invitrogen, USA). A 25 μl PCR reaction mixture comprised 10 μmol of each forward and reverse primers, 5 μmol of TaqMan probe, 12.5 μl 2× buffer, 0.5 μl SuperScript™ III enzyme and 5 μl nucleic acid templates. Thermal cycling conditions were as follows: 50°C for 30 min for reverse transcription, initial denaturation at 94°C for five minutes, 45 cycles of three steps, 15 sec at 94°C, 15 sec at 50°C and 30 sec at 55°C incubation step during which fluorescence data were collected. The WHO qRT-PCR protocol for screening (E gene assay) and confirmation (RdRp, N and ORF 1b genes) assay was used to detect SARS-CoV-2 (protocols of Charité Laboratories, Berlin, and Hong Kong University, respectively)5. Appropriate permission was obtained from the institutional ethics committee to publish the data.
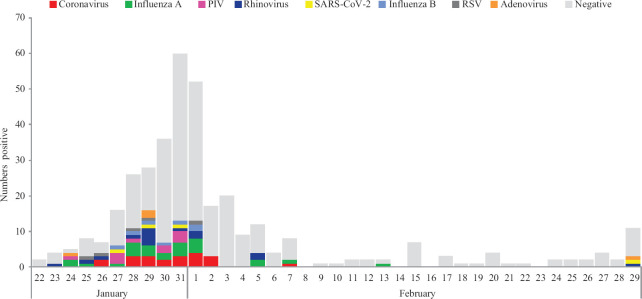
In view of the increasing number of cases with the novel virus globally, the ICMR-National Institute of Virology, Pune, swiftly established screening as well as confirmatory assays for SARS-CoV-2. Of the 362 clinical samples, 250 (68.9%) were male and 109 (30.1%) were female. Majority of the suspected cases [258 (71.8%)] were between 21 and 40 yr of age, as most of them were either medical students or working professionals (Table I). Of the 362 clinical specimens tested, 84 (23.2%) were positive for one or more respiratory viruses (10 with co-infections) and 278 were negative. Viral positivity was highest in the 41-50 yr age group (Table II) and SARS-CoV-2 was detected only among four (1.1%) cases, all of them had a travel history to China or to the countries affected with COVID-19 outbreak; the first three SARS-CoV-2-positive cases were from Kerala and the fourth one from Delhi. With the establishment of SARS-CoV-2 testing facility at the Virus Research and Diagnostic Laboratories (VRDLs) across India6, there was an incremental decline in the number of samples referred to the NIV from the first week of February onwards. Among other respiratory viruses, Influenza A viruses were the most commonly detected [25 (6.8%); 95% confidence interval: 14.5-16.2] viruses followed by human coronaviruses (5.7%) (Figure). Among Influenza A viruses, majority were Influenza A (H1N1) pdm09 (n=17) followed by A (H3N2) (n=8) and Influenza B among seven (1.9%) cases. Among human coronaviruses (n=21), OC43 was most commonly detected (n=15) followed by HKU (n=4), 229E (n=2) and NL63 (n=1) (one had co-infection). Rhinovirus was detected in 15 cases followed by parainfluenza virus (n=10). Day-wise distribution is shown in the Figure. Among the positive cases, cough, 57 (67.9%) was the predominant symptom followed by fever, 48 (57%), sore throat, 42 (50%) and nasal discharge 29 (34.5%). Only seven (8.3%) positive cases had a comorbid condition. Twelve (14.3%) positive cases received antivirals and 31 (36.9%) received antibiotics. Only one case needed mechanical ventilation. During the study period, January 22 to February 29, 2020, respiratory virus detection rate was 23 per cent and with circulation of one or more respiratory viruses. Only four imported cases were confirmed for SARS-CoV-2. Thus, this study highlights the importance of screening for other respiratory viruses in case of patients presenting with respiratory symptoms. All the SARS-CoV-2-infected persons had an associated overseas travel history. The first three cases were detected in the end of January 2020 who had a travel history of Wuhan, China. After that for almost a month, no SARS-CoV-2-positive case was detected. The fourth case confirmed at NIV, Pune, had a travel history of Milan, Italy. Other than influenza, 5.8 (n=21) per cent of human coronaviruses were reported, and the most common human coronavirus was OC-43 (4.1%). An earlier study from ICMR-NIV, Pune showed that the frequency of human coronavirus was 1-3 per cent, with OC-43 being the predominant virus in circulation (unpublished data). Utilizing the capability of VRDL network, early detection of cases have been facilitated.
Table I.
Age-wise distribution of per cent positivity in those tested
| Age group (yr) | Total | Negative | Positive | Per cent positivity |
|---|---|---|---|---|
| 0-10 | 16 | 11 | 5 | 31.3 |
| 11-20 | 38 | 28 | 10 | 26.3 |
| 21-30 | 159 | 127 | 32 | 20.1 |
| 31-40 | 99 | 75 | 24 | 24.2 |
| 41-50 | 30 | 19 | 11 | 36.7 |
| 51-60 | 12 | 10 | 2 | 16.7 |
| 61-70 | 5 | 5 | 0 | 0.0 |
| >70 | 3 | 3 | 0 | 0.0 |
| Total | 362 | 278 | 84 | 23.2 |
Table II.
Age-wise detection of viral pathogens in 362 travellers
| Age group (yr) | Influenza A | Human coronavirus | Rhinovirus | PIV | Influenza B | RSV | SARS-CoV-2 | Adenovirus | HMPV |
|---|---|---|---|---|---|---|---|---|---|
| 0-10 | 5 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 1 |
| 11-20 | 1 | 2 | 2 | 1 | 3 | 1 | 1 | 0 | 0 |
| 21-30 | 10 | 7 | 3 | 6 | 3 | 0 | 2 | 2 | 1 |
| 31-40 | 5 | 7 | 7 | 2 | 1 | 3 | 0 | 1 | 0 |
| 41-50 | 3 | 2 | 3 | 1 | 0 | 0 | 1 | 1 | 0 |
| 51-60 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 61-70 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| >70 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 25 | 21 | 15 | 10 | 7 | 6 | 4 | 4 | 2 |
PIV, parainfluenza virus; RSV, respiratory syncytial virus; HMPV, human metapneumovirus
Figure.
Day-wise distribution of respiratory viruses from January 22 to February 29, 2020. PIV, parainfluenza virus; RSV, respiratory syncytial virus.
Acknowledgment
The authors gratefully acknowledge Prof. Balram Bhargava, the Secretary, Department of Health Research (DHR) & Director General, ICMR, for his guidance and support, Drs R.R. Ganagakhedkar, N. Gupta, S. Giri, I. Praharj, Harmeet Kaur, N. Vijay and N. Agrawal for coordination of samples. Shri. Jadhav from NIV, Pune for data analysis. The authors thank Maximum Containment Facility staff Ms T.D. Mujumdar, Shrimati S. Patil, Shri H. Dighe, Shrimati A. Waghmare, Shri S Baradkar and Ms K Kalale for extending their support. We are also grateful to Integrated Disease Surveillance Programme (IDSP) and Emergency Medical Relief (EMR) Cell of the Ministry of Health & Family Welfare, New Delhi.
Footnotes
Financial support & sponsorship: Financial support was provided by the Indian Council of Medical Research, Department of Health Research, Ministry of Health & Family Welfare, New Delhi to the ICMR-National Institute of Virology, Pune, Maharashtra, India.
Conflicts of Interest: None.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV) Geneva, Switzerland: WHO; 2020. [Google Scholar]
- 3.Gupta N, Praharaj I, Bhatnagar T, Vivian Thangaraj JW, Giri S, Chauhan H, et al. Severe acute respiratory illness surveillance for coronavirus disease 2019, India, 2020. IndianJ Med Res. 2020;151:236–40. doi: 10.4103/ijmr.IJMR_1035_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koul PA, Mir H, Saha S, Chadha MS, Potdar V, Widdowson MA, et al. Respiratory viruses in returning Hajj & Umrah pilgrims with acute respiratory illness in 2014-2015. Indian J Med Res. 2018;148:329–33. doi: 10.4103/ijmr.IJMR_890_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corman V, Bleicker T, Brünink S, Drosten C, Landt O, Koopmans M, et al. Diagnostic detection of Wuhan coronavirus 2019 by real-time RT-PCR. Geneva: World Health Organization; 2020. [accessed on January 16, 2020]. Available from: https://wwwwhoint/docs/default-source/ coronaviruse/wuhan-virus-assay-v1991527e5122341d99287a1b17c111902pdf,">https://wwwwhoint/docs/default-source/ coronaviruse/wuhan-virus-assay-v1991527e5122341d99287a1b17c111902pdf . [Google Scholar]
- 6.Gupta N, Potdar V, Praharaj I, Giri S, Sapkal G, Yadav P, et al. Laboratory preparedness for SARS-CoV-2 testing in India: Harnessing a network of Virus Research & Diagnostic Laboratories. Indian J Med Res. 2020;151:216–25. doi: 10.4103/ijmr.IJMR_594_20. [DOI] [PMC free article] [PubMed] [Google Scholar]