Abstract
Background & objectives:
Since the beginning of the year 2020, the pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) impacted humankind adversely in almost all spheres of life. The virus belongs to the genus Betacoronavirus of the family Coronaviridae. SARS-CoV-2 causes the disease known as coronavirus disease 2019 (COVID-19) with mild-to-severe respiratory illness. The currently available diagnostic tools for the diagnosis of COVID-19 are mainly based on molecular assays. Real-time reverse transcription-polymerase chain reaction is the only diagnostic method currently recommended by the World Health Organization for COVID-19. With the rapid spread of SARS-CoV-2, it is necessary to utilize other tests, which would determine the burden of the disease as well as the spread of the outbreak. Considering the need for the development of such a screening test, an attempt was made to develop and evaluate an IgG-based ELISA for COVID-19.
Methods:
A total of 513 blood samples (131 positive, 382 negative for SARS-CoV-2) were collected and tested by microneutralization test (MNT). Antigen stock of SARS-CoV-2 was prepared by propagating the virus in Vero CCL-81 cells. An IgG capture ELISA was developed for serological detection of anti-SARS-CoV-2 IgG in serum samples. The end point cut-off values were determined by using receiver operating characteristic (ROC) curve. Inter-assay variability was determined.
Results:
The developed ELISA was found to be 92.37 per cent sensitive, 97.9 per cent specific, robust and reproducible. The positive and negative predictive values were 94.44 and 98.14 per cent, respectively.
Interpretation & conclusions:
This indigenously developed IgG ELISA was found to be sensitive and specific for the detection of anti-SARS-CoV-2 IgG in human serum samples. This assay may be used for determining seroprevalence of SARS-CoV-2 in a population exposed to the virus.
Keywords: COVID-19, diagnosis, ELISA, human, IgG antibodies, microneutralization test, SARS-CoV-2, standardization
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was first reported from China during December 20191. Within a short period of time, the virus has spread across different continents affecting almost 215 countries and territories. Till date, SARS-CoV-2 has infected about 5.4 million people and led to the death of approximately 0.34 million people across the globe2. Coronaviruses are a large family of viruses within the family Coronaviridae, and the order Nidovirales3. They cause diseases in a variety of domestic and wild animals as well as in humans. Human coronoviruses (HCoVs)-229E and OC43 were the first HCoVs discovered in the 1960s4. Although these viruses caused self-limiting respiratory illnesses, the first HCoV that caused SARS epidemic emerged in China during 20035. A second outbreak of severe respiratory illness called the Middle East respiratory syndrome (MERS) occurred in Saudi Arabia during 20126.
The COVID-19 infection is known to spread from one person to others via droplets produced while coughing and sneezing. The available data suggest an incubation period between 2 and 14 days, with an average of five days. Approximately 80 per cent of the infected cases had mild illness; 14-15 per cent experienced severe illness with pneumonia and five per cent were seriously ill, mainly older individuals and people with other medical conditions such as asthma, hypertension, diabetes or heart disease7. However, some COVID-positive individuals found to be asymptomatic could act as carriers of the infection8. One of the main challenges in containing the spread of SARS-CoV-2 is the diagnosis of a large number of infected individuals within a population. Majority of the asymptomatic cases may remain undetected due to the limited resources for diagnosis.
As approved by the World Health Organization (WHO), molecular assays are currently the mainstay of diagnosis of COVID-199. Serological assays have also been used as a screening tool for the surveillance of the emerging and re-emerging diseases. However, serological assays are not currently recommended for case detection and are not included in the WHO laboratory testing guidelines for COVID-19. The actual burden of infection and its spread can be determined using detection of IgM and IgG antibodies against SARS-CoV-2 by serological assays such as ELISA10,11,12,13,14. Haveri et al15 reported the presence of serum IgM and IgG antibodies against SARS-CoV-2 using immunofluorescence assays. Commercial and non-commercial IgM/IgG ELISA for COVID-19 (n=61) have been certified for commercial purpose/research by different certifying agencies16. About 16 ELISA kits are still in the stage of development. These assays would be easy to use and be cost-effective tools for the detection and surveillance of COVID-19 in resource-limited settings14,15. Here, we report the standardization of an indigenous IgG-ELISA for the detection of anti-COVID-19 IgG antibodies at the Indian Council of Medical Research-National Institute of Virology (ICMR-NIV), Pune, India.
Material & Methods
Human clinical specimens: A total of 513 blood samples were included for assay validation. Among them, 150 serum samples were from real-time reverse transcription-polymerase chain reaction (real-time RT-PCR)-confirmed COVID-19 patients after the second week of the onset of the disease. Those who were real-time RT-PCR positive for influenza A (H1N1) pdm09 (n=13), influenza A (H3N2) (n=5), human coronavirus OC43 (n=5), rhinovirus (n=2), respiratory syncytial virus (n=2), influenza B (n=3), parainfluenza type 4 (n=3), hepatitis B virus (n=5), hepatitis C virus (n=5) and dengue IgG and chikungunya IgG positive samples (n=10 each) were used as a panel to test cross-reactivity. Two hundred and seventy nine serum samples were COVID-19 real-time RT-PCR negative from healthy blood donors collected prior to the SARS-COV-2 outbreak. The remaining 21 serum samples were real-time RT-PCR negative for COVID-19, which were collected during the current pandemic. All these serum samples had been tested using microneutralization test (MNT) for SARS-CoV-217. All samples were stored at −20°C until further use. Institutional Human Ethics Committee approved this study. A written informed consent was obtained from all the participants.
Cell and antigen preparation
Virus propagation and titration: SARS-CoV-2 was isolated from throat/nasal swab specimen (NIV-2020-770) in Vero CCL-81 cells at the containment facility of ICMR-NIV, Pune18. SARS-CoV-2 virus stock was prepared by propagating the virus in Vero CCL-81 cells. The cytopathic effect started at the post-infection day (PID) 2, and the culture supernatant was harvested at the PID 3. Virus titrations were performed in Vero CCL-81 cells using tissue culture infectious dose 50% (TCID50) assay. Virus titre (TCID50/ml) was calculated by the Reed-Muench method19 and found to be 106.5 TCID50/ml.
Gamma inactivation of the virus: Gamma irradiation of the virus stock was performed using Co-60 source (24 kGy) of GC-5000 Gamma chamber (BRIT, Mumbai). This irradiated stock was again inoculated in Vero CCL-81 twice for five days to confirm the inactivation of the virus.
Concentration of gamma-inactivated antigen: Gamma-irradiated SARS-CoV-2-infected tissue culture fluid was concentrated using 30 kDa filters (Millipore, Germany) and further passed through 0.2 μm filters, aliquoted and stored at −80°C. Concentrated viral antigen was also aliquoted in 1 and 2 ml volumes in frosted glass bottles and further lyophilized. The lyophilized vials were stored at −20°C to be used as antigen in ELISA tests.
Microneutralization test (MNT): All serum samples were tested for neutralization against SARS-CoV-2 (ID NIV2020-770) in the Biosafety Level 3 (BSL-3) laboratory of ICMR-NIV, Pune. The assay was performed as described by Manenti et al17 with a few modifications. Briefly, two-fold (1:10 to 1:1280), serially diluted heat-inactivated serum samples (50 μl) in minimal essential medium (Gibco, Thermo Fisher Scientific, USA) supplemented with two per cent foetal bovine serum (Gibco, Thermo Fisher Scientific, USA) were prepared. In the next step, 50 μl virus suspension of 2 log tissue culture infective dose of previously titered virus stock was added to each well of ELISA plate and incubated at 37°C for one hour. The mixture was then added on to the preformed monolayers of Vero CCL-81 cells in 96-well plates (Nunc, Thermo Fisher Scientific, USA) and incubated at 37°C with five per cent CO2. After incubation for 72 h, the medium was decanted and washed with saline and the plates were stained with one per cent amido black. The serum neutralization titre is the reciprocal of the highest dilution resulting in an infection reduction of >50 per cent. A cut-off of neutralization titre >20 was considered as positive.
Anti-SARS-CoV-2 human IgG ELISA: An IgG capture ELISA was developed for serological diagnosis of SARS-CoV-2 infection from patients' serum. Briefly, 96-well polystyrene microtitre ELISA plates (Nunc, Thermo Fisher Scientific, USA) were coated with inactivated SARS-CoV-2 antigen (1:10 diluted, 100 μl/well) in 1x phosphate-buffered saline (PBS) (pH 7.2, 1 M). The antigen-coated plates were kept overnight at 4°C. These wells were blocked with a Liquid Plate Sealer (CANDOR Bioscience GmbH, Germany) for two hours at room temperature (25-30°C). The plates were washed three times with 10 mM PBS, pH 7.4 with 0.1 per cent Tween-20 (PBST) (Sigma-Aldrich, USA). To the coated and blocked wells, 100 μl of 1:100 diluted human serum samples were added and incubated at 37°C for one hour. These wells were washed five times using 1x PBST. Following this, anti-human IgG horseradish peroxidase (HRP) was diluted in StabilZyme (SurModics, Inc., USA) and added 100 μl/well; and plates were incubated for one hour at 37°C. After incubation, the plates were washed as described above. Further, 100 μl of 3,3', 5, 5'-tetramethylbenzidine (TMB) substrate was added and incubated for 10 min. The reaction was stopped by 1 N sulphuric acid (H2SO4), and the absorbance values were measured at 450 nm using an ELISA reader. Positive and negative controls were prepared as described below and were included in the respective wells in the test.
Using virus neutralization test as the gold standard, the sensitivity and specificity of IgG ELISA was determined.
Serum controls for anti- SARS-CoV-2 human IgG ELISA: Human serum samples tested and confirmed by real-time RT-PCR and MNT were selected for the preparation of positive and negative controls. High positive control serum was prepared by pooling 15 serum samples collected from COVID-19 survivors infected during the current SARS-COV-2 pandemic. Negative control serum was prepared by pooling samples from 10 healthy individuals, whose samples were collected and stored before this pandemic.
Determination of cut-off values and assay validation: A total of 513 serum samples, including the 131 positive and 382 negative samples as confirmed by the MNT, were tested by ELISA. The end point cut-off was determined by the analysis of a receiver operating characteristic (ROC) curve based on positive divided by negative (P/N) values. The panels of serum samples described above were selected to validate cross-reactivity. The inter-assay variability was assessed by testing two negative and two positive control serum samples in 15 different work sessions, and four replicates within each plate were used to determine the coefficient of variation (CV).
Results
Among the 150 real-time RT-PCR-confirmed COVID-19 serum samples, 131 were positive by MNT and 19 were negative. In addition, among 21 serum samples collected during this pandemic which were negative by real-time RT-PCR, remained negative when tested by MNT.
Determination of cut-off value and assay performance: After the ROC analysis, the area under the curve (AUC) was found to be 0.986. The cut-off threshold for the assay was set at 0.200, which gave a sensitivity of 92.37 per cent and a specificity of 97.9 per cent (Fig. 1). The calculated threshold cut-off for the ELISA at optical density (OD) at 450 nm was more than the average OD of negative control +0.2 and the sample to negative control a ratio >1.5, differentiating between the presence and absence of IgG antibodies in the samples. The assay was found to have a negative predictive value of 98.14 per cent and a positive predictive value of 94.44 per cent.
Fig. 1.
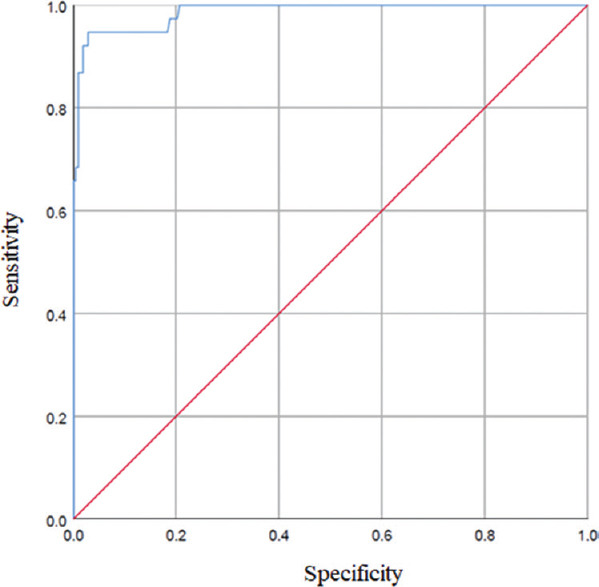
Receiver operating characteristic (ROC) analysis showing sensitivity versus specificity for discrimination of positive and negative serum samples, which were derived for in-house IgG ELISA compared to microneutralization test results (ROC area under the curve: 0.986).
Of the 131 MNT-positive serum samples tested by the in-house IgG ELISA, four were found to be negative and 127 were positive. However, one serum sample which was IgG positive by the in-house ELISA was negative by MNT. The inter-assay CV was nine per cent for the positive control serum and 9.4 per cent for the negative control. The intra-assay CV of four replicates within each plate for positive control was 3.5 per cent and for negative control it was 7.2 per cent.
Inter-laboratory evaluation of the assay: Inter-laboratory comparison of indigenously developed anti-SARS human IgG ELISA was performed at two centres. A panel of 150 coded (75 positive and 75 negative) human serum samples (including the panel to test cross-reactivity) were provided along with four ready-to-use SARS-COV-2 IgG ELISA kits. The concordance obtained was 99.33 and 100 per cent at both the centres when compared with the results of ICMR-NIV, Pune. A correlation analysis was performed to analyze the inter-laboratory performance between the different laboratories. The results indicated a positive correlation between the reference laboratory and the other laboratories (Lab 1: r=0.9153, P <0.001 with r2 value 0.8387; Lab 2: r=0.9523, P <0.001 with r2 value of 0.9068) (Fig. 2).
Fig. 2.
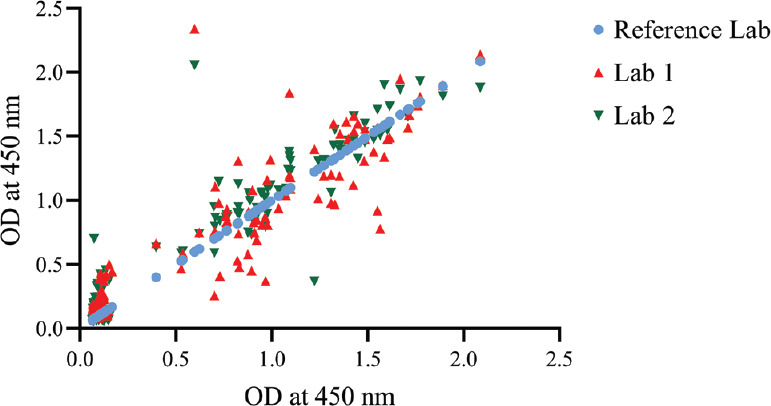
Inter-laboratory correlation analysis: A panel of 150 serum samples tested at two external laboratories and compared with the ICMR-National Institute of Virology reference laboratory indicated a positive correlation between the reference laboratory and the other laboratories.
Correlation between anti-SARS-CoV-2 human IgG ELISA and MNT: The correlation between the in-house IgG ELISA against the neutralization titres indicated a strong positive correlation (r=0.7836, P<0.001) (Fig. 3).
Fig. 3.
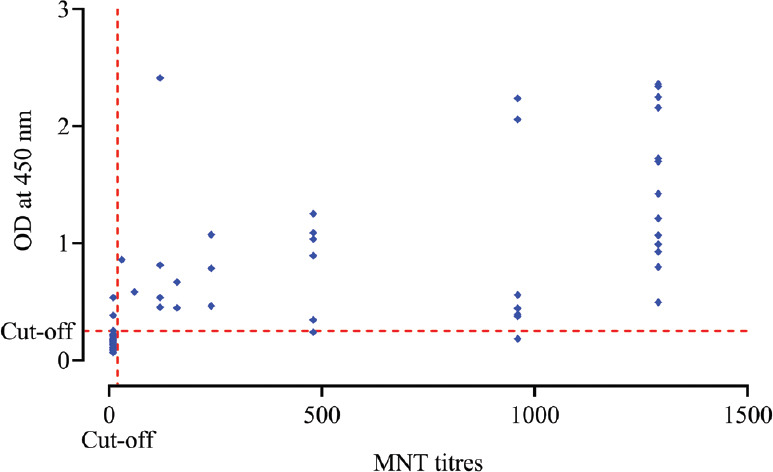
Correlation analysis of SARS-CoV-2 antibody titre of microneutralization test (MNT) compared with optical densities of the in-house SARS-CoV-2 IgG ELISA (r=0.7836, P<0.001).
Discussion
The U.S. Food and Drug Administration (FDA) has granted 31 molecular diagnostic kits (real-time RT-PCR, RT-LAMP and point-of-care test) for emergency use authorization. For surveillance of the emerging and re-emerging viruses, serological assays have been widely recommended. Several IgM/IgG ELISA kits for COVID-19 are in the stage of development. However, validated and approved SARS-CoV-2 serological assays are lacking for case detection17 and are not included in laboratory testing guidelines for COVID-19 of the WHO9. Due to the non-availability of an indigenous, approved and cost-effective kit, an in-house ELISA was developed and validated for the detection of anti-SARS-CoV-2 human IgG antibodies. The virus neutralization is regarded as the gold standard method for a serological assay20. The in-house ELISA results correlated strongly with neutralization assay, indicating that the assay was sensitive and specific for the detection of anti-SARS-CoV-2 IgG. The assay performance was not different with the use of serum or plasma. No cross-reactivity with other respiratory viruses was detected. This kit may be used to detect exposed immune-protected individuals. This ELISA will be useful in screening healthcare workers, industry workers, etc. The use of whole-cell antigen instead of recombinant nucleocapsid antigen or spike protein antigen provided a broad sensitivity to the assay. However, the assay performance needs to be assessed in a large cohort of related human/zoonotic coronaviruses. The development of a sensitive and specific assay based on whole-virus antigen that could be produced at lower manufacturing cost will provide easy-to-use and affordable kits to resource-limited settings.
In conclusion, our findings suggested that this indigenous anti-SARS-CoV-2 human IgG-ELISA was sensitive and specific for the detection of IgG antibodies among individuals who have been exposed to SARS-CoV-2. The ELISA results correlated strongly with virus neutralizing antibodies. This assay may be used for ascertaining the seroprevalence against SARS-CoV-2 in a population and for epidemiological studies.
Footnotes
Financial support & sponsorship: None.
Conflicts of Interest: None.
References
- 1.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–3. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. WHO coronavirus disease (COVID-19) Dashboard. [accessed on May 27, 2020]. Available from: https://covid19whoint/">https://covid19whoint/
- 3.Holmes KV. Coronaviruses (Coronaviridae) Encyclopedia Virol. 1999:291–8. [Google Scholar]
- 4.Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121:190–3. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 5.Liang G, Chen Q, Xu J, Liu Y, Lim W, Peiris JS, et al. Laboratory diagnosis of four recent sporadic cases of community-acquired SARS, Guangdong Province, China. Emerg Infect Dis. 2004;10:1774–81. doi: 10.3201/eid1010.040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Severe respiratory illness associated with a novel coronavirus - Saudi Arabia and Qatar, 2012. MMWR Morb Mortal Wkly Rep. 2012;61:820. [PubMed] [Google Scholar]
- 7.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72-314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. doi: 101001/jama20202648. [DOI] [PubMed] [Google Scholar]
- 8.Lai CC, Liu YH, Wang CY, Wang YH, Hsueh SC, Yen MY, et al. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARSCoV-2): Facts and myths. J Microbiol Immunol Infect. 2020 doi: 10.1016/j.jmii.2020.02.012. doi: 101016/jjmii202002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: Interim guidance. Geneva: WHO; 2020. Mar 2, [Google Scholar]
- 10.Ye F, Xu S, Rong Z, Xu R, Liu X, Deng P, et al. Delivery of infection from asymptomatic carriers of COVID-19 in a familial cluster. Int J Infect Dis. 2020;94:133–8. doi: 10.1016/j.ijid.2020.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai SL, Wang JY, Zhou YQ, Yu DS, Gao XM, Li LL, et al. [Analysis of the first cluster of cases in a family of novel coronavirus pneumonia in Gansu Province] Zhonghua Yu Fang Yi Xue Za Zhi. 2020;54:E005. doi: 10.3760/cma.j.issn.0253-9624.2020.0005. [DOI] [PubMed] [Google Scholar]
- 12.Xiao SY, Wu Y, Li J. Evolving status of the 2019 novel coronavirus infections: Proposal of conventional serologic assays for disease diagnostics and infection monitoring 2020. J Med Virol. 2020;92:464–7. doi: 10.1002/jmv.25702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long QX, Deng HJ, Chen J, Hu J, Liu BZ, Liao P, et al. Antibody responses to SARS-CoV-2 in COVID-19 patients: The perspective application of serological tests in clinical practice. medRxiv. 2020 doi: https://doiorg/101101/2020031820038018. [Google Scholar]
- 14.Lee NY, Li CW, Tsai HP, Chen PL, Syue LS, Li MC, et al. A case of COVID-19 and pneumonia returning from Macau in Taiwan: Clinical course and anti-SARS-CoV-2 IgG dynamic. J Microbiol Immunol Infect. 2020:S1684. doi: 10.1016/j.jmii.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haveri A, Smura T, Kuivanen S, Österlund P, Hepojoki J, Ikonen N, et al. Serological and molecular findings during SARS-CoV-2 infection: The first case study in Finland, January to February 2020. Euro Surveill. 2020;25:2000266. doi: 10.2807/1560-7917.ES.2020.25.11.2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, et al. SARS-CoV-2 specific antibody responses in COVID-19 patients. Emerg Infect Dis. 2020:26. doi: 10.3201/eid2607.200841. doiorg/103201/eid2607200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manenti A, Maggetti M, Casa E, Martinuzzi D, Torelli A, Trombetta CM, et al. Evaluation of SARS-CoV-2 neutralizing antibodies using a CPE-based colorimetric live virus micro-neutralization assay in human serum samples. J Med Virol. 2020 doi: 10.1002/jmv.25986. doi: 101002/jmv25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkale P, Patil S, Yadav PD, Nyayanit DA, Sapkal G, Baradkar S, et al. First isolation of SARS-CoV-2 from clinical samples in India. Indian J Med Res. 2020;151:244–50. doi: 10.4103/ijmr.IJMR_1029_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Epidemiol. 1938;27:493–7. [Google Scholar]
- 20.Rapid / CLIA / ELISA Kits approved for testing of Covid-19 with the conditions. [accessed on May 27, 2020]. Available from: https://cdscogovin/opencms/resources/UploadCDSCOWeb/2018/UploadPublic_NoticesFiles/Rapiddetection%20Kit-16-04-2020pdf,">https://cdscogovin/opencms/resources/UploadCDSCOWeb/2018/UploadPublic_NoticesFiles/Rapiddetection%20Kit-16-04-2020pdf .


