Abstract
Our understanding of gut functioning and pathophysiology has grown considerably in the past decades, and advancing technologies enable us to deepen this understanding. Single-cell RNA sequencing (scRNA-seq) has opened a new realm of cellular diversity and transcriptional variation in the human gut at a high, single-cell resolution. ScRNA-seq has pushed the science of the digestive system forward by characterizing the function of distinct cell types within complex intestinal cellular environments, by illuminating the heterogeneity within specific cell populations and by identifying novel cell types in the human gut that could contribute to a variety of intestinal diseases. In this review, we highlight recent discoveries made with scRNA-seq that significantly advance our understanding of the human gut both in health and across the spectrum of gut diseases, including inflammatory bowel disease, colorectal carcinoma and celiac disease.
Introduction
Since its early scientific investigations in the 1960s, we have accumulated a vast amount of knowledge on gut physiology and gastrointestinal diseases (1). The multifunctional nature of the human intestine, illustrated by its key role in food digestion, nutrient absorption and transportation, in immune response to pathogens and in forming a physical defense barrier, implies an exceptional biological complexity. Although tremendous scientific effort has been applied to grasp this complexity, it was not until recent technological advances like single-cell RNA sequencing (scRNA-seq) analysis that the cellular landscape of the human gut could be assessed at a high resolution. Single-cell transcriptomics has unveiled remarkable heterogeneity within major cell types and has identified new cell subpopulations that contribute to the complex intestinal cellular composition. Moreover, scRNA-seq has offered an unprecedented view of human disease by deconvoluting cellular interactions and pathway crosstalk that underlie disease pathophysiology (2). In this review, we discuss the findings of signature studies that employed scRNA-seq to profile cell types in normal gut mucosa (3) and in mucosa of patients with celiac disease (CeD) (4), inflammatory bowel disease (IBD) (5), including both Crohn’s disease (CD) (6,7) and ulcerative colitis (UC) (8–10), and colorectal carcinoma (CRC) (11–13), as detailed in Table 1. Since corresponding human data is as of yet unavailable, we also discuss a study of the mouse small intestinal epithelium that identified the cellular response to bacterial and helminth infections (14).
Table 1.
Single-cell transcriptomic studies in human gut
| Reference | Sample | Target cell population | Number of generated cells, sample size and identified cell types |
|---|---|---|---|
| Healthy gut | |||
| Wang et al. 2019 (3) | Healthy donors, | Epithelial cells | • 14.537 cells—6 donors |
| Mucosal biopsies of ileum, colon and rectum | • 7 epithelial cell subsets | ||
| Celiac disease (CeD) | |||
| Atlasy et al. 2019 (4) preprint | Healthy controls and CeD | Immune cells | • 3.994 cells—6 donors and 8 CeD patients |
| Mucosal biopsies of duodenum | • 5 main immune cell lineages (7 subsets of T cells, 1 subset of B cells, 2 subsets of plasma cells, 7 subsets of myeloid cells, 4 subsets of mast cells) | ||
| Inflammatory bowel disease (IBD) | |||
| Huang et al. 2019 (5) | Healthy donors and IBD (UC, CD, IBD-U), all pediatric | Epithelial, stromal and immune cells | • 73.165 cells—pediatric: 6 donors, 6 IBD-U (colitis), 2 UC, 3 CD patients • 9 major cell types: epithelial cells (10 clusters), stromal cells (8 clusters), and 7 immune cell lineages (10 myeloid cell clusters, 7 B cell subsets, 2 plasma cell subsets, and 16 subsets of T and NK cells) |
| Mucosal biopsies of colon | |||
| Martin et al. 2019 (6) | CD | Stromal and immune cells | • 82.417 cells—11 CD patients |
| Mucosal biopsies of ileum (matched inflamed and non-inflamed); peripheral blood | • 47 (33 if combining shared annotations) cell subsets: 8 stromal cell subsets, 25 immune cell subsets (from 7 distinct lineages) | ||
| Uniken Venema et al. 2019 (7) | CD | Immune cells | • 5.292 T cells—3 CD patients |
| Mucosal biopsies of inflamed ileum and peripheral blood | • 6 distinct T cell subsets | ||
| Parikh et al. 2019 (8) | Healthy donors and UC | Epithelial cells | • 11.175 cells—3 donors, 3 UC patients |
| Mucosal biopsies of colon (matched inflamed and non-inflamed UC mucosa) | • 10 epithelial cell subsets in healthy colon, 12 cell subsets in inflamed UC colon | ||
| Kinchen et al. 2018 (9) | Healthy donors and UC | Stromal cells | • 9.591 cells from 5 donors, 5 UC patients |
| Mucosal biopsies of colon (matched inflamed and non-inflamed UC mucosa) | • 11 stromal cell subsets in healthy colon, 12 subsets in UC colon | ||
| colonic organoids | |||
| Smillie et al. 2019 (10) | Healthy donors and UC | Epithelial, stromal and immune cells | • 360.650 cells—12 donors, 18 UC patients |
| Mucosal biopsies of colon (matched inflamed and non-inflamed UC mucosa) | • 51 cell subsets: 15 epithelial cell subsets; 13 stromal cell subsets, 23 immune cell subsets | ||
| Colorectal carcinoma (CRC) | |||
| Li et al. 2017 (11) | CRC | Epithelial, stromal and immune cells | • 969 cells—resected primary tumors of CRC patients and 622 cells—the nearby normal mucosa of 7 of these patients |
| Tumor tissue and matched adjacent normal mucosa of colon, rectum or caecum | • 7 distinct cells types: epithelial cells (9 clusters), stromal cells (3 subsets of fibroblasts, endothelial cells), immune cells (T cells, B cells, mast cells and myeloid cells) | ||
| Uhlitz et al. 2020 (12) preprint | CRC | Epithelial, stromal and immune cells | • ~50.000 cells—8 CRC patients |
| Tumor tissue and matched adjacent normal mucosa of colon, rectum or caecum; matching CRC organoids | • 7 main types of epithelial cells, 5 tumor-specific epithelial cell subsets; stromal cells (pericytes, glial cells, endothelial cells and 5 subsets of fibroblasts); immune cell lineages (26 subsets in total assigned over T cells, B cells, plasma cells, myeloid cells and mast cells) | ||
| Zhang et al. 2018 (13) | CRC | Immune cells | • 11.138 T cells—12 CRC patients |
| Tumor tissue and matched adjacent normal mucosa of colon and rectum; peripheral blood | • 20 T cell subsets | ||
| Infectious disease (in mouse intestine) | |||
| Haber et al. 2017 (14) | Healthy mice and mice infected with Salmonella enterica or Heligmosomoides polygyrus | Epithelial cells | • 53.193 cells—small intestine and organoids of 2–4 mice per group • 15 epithelial cell subsets |
| Dissociated cells from small intestine and epithelial organoids | |||
UC, ulcerative colitis; CD, Crohn’s disease; IBD-U, inflammatory bowel disease unclassified
This review (1) describes the key scRNA-seq findings in the three main cellular compartments of the intestinal mucosa—epithelial, stromal and immune—and (2) highlights cellular remodeling and cell–cell interactions in gut disease.
Epithelial cell compartment
The intestinal epithelium lines the luminal surface of the gut mucosa and carries out a diversity of vital functions: it maintains a physical barrier, shielding the interior intestinal milieu from luminal content and pathogens, executes absorptive and metabolic tasks, controls bacterial growth and actively contributes to immune responses (15). Conventionally, we recognize undifferentiated intestinal stem cells, positioned at the crypt base, which via transit-amplifying (TA) cells give rise to the specialized intestinal cell lineages. These include absorptive enterocytes/colonocytes, enteroendocrine cells, goblet cells, Paneth cells and, less known, tuft cell-expressing receptors to sense luminal pathogens (16,17) and microfold cells (M-cells) guiding transport of luminal antigens to the lamina propria (18). Structural deviations in the epithelial compartment can cause intestinal barrier dysfunction that marks many intestinal disorders, including infectious diseases, inflammatory bowel disease (IBD), celiac disease (CeD) and colorectal carcinoma (CRC) (19,20). The scRNA-seq studies (1) identified a novel (BEST4 expressing) absorptive cell type regulating pH balance (5,8,10), (2) showed the existence of Paneth-like cells in the colon (3,8), (3) distinguished an inflammation-associated subset of goblet cells (8), (4) highlighted the role of M-cells in disease (10) and (5) reported specific responses of epithelial cells to intestinal infection (14).
BEST4 expressing absorptive cells
This newly identified distinct subpopulation of intestinal absorptive cells highly expresses the calcium-sensitive chloride channel bestrophin-4 (BEST4) and the pH detecting proton channel otopetrin 2 (OTOP2) and is therefore predicted to transport salt, ions and metals (8,10). By maintaining luminal pH, BEST4/OTOP2 cells are thought to support optimal microbial growth, marking a novel component in the host–microorganism interaction. Moreover, BEST4+ cells are a previously unknown source of the paracrine hormone uroguanylin, which regulates intestinal electrolyte homeostasis by binding to the guanylyl cyclase C (GC-C) receptor and, thereby, increases intracellular levels of cyclic guanosine monophosphate (cGMP) (21,22). Dysfunctional cGMP/GC-C signaling has been implicated in compromised epithelial barrier function, increased intestinal inflammation and tumor growth (23), accelerating the progression of gastrointestinal disorders such as IBD and colon carcinoma (24,25). Single-cell profiling showed that both IBD (8,10) and CRC (11,12) are marked by the loss of BEST4/OTOP2 cells, supporting the role of cGMP/GC-C dysregulation in these gut diseases.
Paneth-like cells in the colon
Paneth cells, found in the crypt base in the small intestine, form a secretory lineage that is crucial for epithelial barrier function and epithelial cell renewal (26,27). These cells secrete antimicrobial peptides and factors that support intestinal stem cells. In contrast to the small intestine, healthy colonic crypts do not harbor Paneth cells and, therefore, rely on other sources for these factors. Colonic Paneth-like cells (PLCs) have been identified in mice but remained obscure in humans (28,29). Following up on a scRNA-seq study that describes a population of PLCs in the human colon (30), Wang et al. indeed verified the existence of PLCs in adult colon and showed that these cells, much like ileal Paneth cells, express genes involved in bacterial defense and genes that encode factors to sustain intestinal stem cells (3). Moreover, another scRNA-seq study detected a subset of crypt-base goblet cells that highly express the antimicrobial peptide lysozyme (LYZ) in inflamed colon and which most likely act as PLCs (8). While impaired Paneth cell function has been shown to contribute to the pathogenesis of ileal CD and CeD (31–33), the involvement of colonic PLCs in gut diseases is yet to be elucidated.
Inflammation-associated goblet cells
Luminal secretion of mucins by goblet cells is critical for the establishment of a chemical and physical barrier as a frontline of innate host defense (34). Dysregulated goblet cell function contributes to barrier breakdown in UC (35) and CeD (36); however the pathways that underlie this breakdown are still unknown. ScRNA-seq studies mapping the cells of colonic epithelia reveal an exceptional goblet cell diversity, distinguishing several subsets of varying maturity and localization within the intestinal crypts (8). There appears to be a positional remodeling of goblet cells in IBD, along with the emergence of a disease-associated subset of goblet cells in inflamed colon. Moreover, the goblet-cell-secreted antibacterial defense factor WFDC2 is lost in active UC, suggesting a novel functional role of this factor in the maintenance of the mucosal barrier.
The role of M-cells in disease
M-cells contribute to the adaptive immunity in the gut by delivering luminal antigens to the underlying mucosal lymphoid tissues (37). While M-cells normally reside in the follicle-associated epithelia of the small intestine and are rarely found in healthy colon, scRNA-seq shows that M-cells markedly expand in the inflamed colon of UC patients (10). Activated M-cells highly express chemokines recruiting immune cells to the site of inflammation. These specialized epithelial cells highly express a large number of genes known to be associated with IBD susceptibility, pinpointing M-cells as a central node in the cell–cell interaction network during IBD inflammation (10). Besides inflammation, infectious conditions have been shown to ectopically induce M-cells, where they act as a portal for pathogen invasion in the mucosa (38). The only available scRNA-seq study that investigated responses of epithelial cells during intestinal infection was limited to mice and could not detect M-cells at the resolution of their data, and therefore, this study was unable to report infection-induced changes in M-cells (14).
Epithelial response to the intestinal infection
ScRNA-seq reveals that the restructuring of the epithelial barrier, involving shifts in cell proportions and cell-intrinsic programs, is specific to the identity of the pathogen (14). For instance, goblet and tuft cells—secretory cells that are known to respond to parasites—accumulate in mouse small intestine during helminth infection, whereas the proportions of absorptive enterocytes and Paneth cells increase in response to Salmonella infection. The question whether these findings translate to the human gut warrants further investigation.
Lastly, single-cell profiling of tumors and matched normal tissues provides a unique opportunity to identify changes in the epithelial cell compartment in CRC. Two scRNA-seq studies describe a pronounced expansion of undifferentiated stem-/TA-like cells within tumors, comprising more than 90% of all tumor epithelial cells (11,12). While stem cells are essential for tissue homeostasis and regeneration, they also drive therapy resistance in cancer. Tumor-specific stem-/TA-like cells show higher expression of bottom-crypt markers than cells in the normal colon epithelium, have high proliferative activity and express genes linked to oncogenic processes (11,12). These scRNA-seq findings imply that epithelial cells in CRC display considerable cell plasticity and have multilineage differentiation capacity.
Stromal cell compartment
Residing within the intestinal lamina propria, stromal cells such as fibroblasts, myofibroblasts, pericytes and endothelial cells provide a supportive matrix for the epithelium. Stromal cells dynamically interact with both epithelial and immune cells, playing crucial roles in regulating epithelial barrier homeostasis, gut innate immunity, tissue repair and tumor development (39,40). Recently, scRNA-seq studies profiling gut mucosal cells revealed previously unknown heterogeneity within the stromal compartment. In addition, these studies identified new and distinct intestine-specific mesenchymal subsets and uncovered their functional role to maintain and regenerate the intestinal epithelium in health and disease.
Among stromal transcriptomes, most studies distinguish the following distinct fibroblast subsets along the crypt–villus axis of the human gut: myofibroblasts, lamina propria fibroblasts, SOX6+ (upper crypt) fibroblasts, RSPO3+ (crypt base) fibroblasts and disease-associated subsets of fibroblasts (Table 2). These fibroblast subtypes show transcriptional, spatial and functional diversity. Lamina propria fibroblasts were shown to diffusely populate the mucosal connective tissue and express non-fibrillar collagens and elastic fibers. In turn, fibroblasts that characteristically express transcription factor SOX6 and Wnt ligands WNT5A and WNT5B reside in close proximity to the epithelial monolayer, suggesting their role in epithelial cell proliferation and differentiation and, hence, in epithelial barrier maintenance. Another fibroblast subset is defined by the expression of RSPO3, WNT2B and TNFRSF13B and spatial proximity to the crypt base, regulating the survival of intestinal stem cells. Single-cell studies show that the abovementioned fibroblasts can be detected in normal gut mucosa as well as in inflamed mucosa of IBD patients (5,9,10) and in tumors of CRC patients (12). Two specific disease-associated fibroblast types have been identified: inflammation-associated fibroblasts (IAFs) in IBD, which are almost exclusively present in inflamed mucosa and appear to play an important role in recruiting immune cells to the gut mucosa, and cancer-associated fibroblasts (CAFs) that generally seem to play a tumor-promoting role producing pro-oncogenic growth factors.
Table 2.
Fibroblast subtypes in the human gut identified by single-cell transcriptomics
| Subset | Gene markers* | Location/function | Reference (subset annotation)** |
|---|---|---|---|
| 1. Myofibroblasts | MYH11, ACTG2, DES | Distributed throughout the lamina propria Express contractile genes Relatively unchanged in inflammation |
(5) Huang et al. (6) Martin et al. (smooth muscle cells) (9) Kinchen et al. (10) Smillie et al. (12) Uhlitz et al. |
| 2. Lamina propria fibroblasts | CCL2, CCL8, CCL11, CCL13, CXCL1, APOE, ADAMDEC1 | Distributed throughout the lamina propria; Involved in structural organization of extracellular matrix |
(5) Huang et al. (6) Martin et al. (fibroblasts) (9) Kinchen et al. (S1) (10) Smillie et al. (WNT2B+ Foshi/lo fibroblasts) (12) Uhlitz et al. (fibroblasts) |
| 3. SOX6+ fibroblasts | SOX6, F3, WNT5A, WNT5B, BMP2, BMP4, FRZB | Reside in a close proximity to epithelial cells (near the villus) Regulate epithelial regeneration |
(5) Huang et al. (epithelial proximal fibroblasts) (9) Kinchen et al. (S2) (10) Smillie et al. (WNT5B+ fibroblasts) (12) Uhlitz et al. (upper crypt fibroblasts) |
| 4. RSPO3+ fibroblasts | RSPO3, S3, S7, WNT2B, TNFRSF13B | Decrease upon inflammation Reside near the crypt Regulate the survival of intestinal stem cells |
(5) Huang et al. (WNT2Bhi and TNFRSF13B+ fibroblasts) (9) Kinchen et al. (S3) (10) Smillie et al. (12) Uhlitz et al. (crypt-base fibroblasts) |
| 5a. Inflammation-associated fibroblasts (IAFs) | IL6, IL11, CXCL3, CXCL5, CXCL6, MMP3, MMP10, CHI3L1, OSMR | Almost exclusive for inflamed mucosa (in IBD) Mobilize the immune response |
(5) Huang et al. (inflammatory fibroblasts) (6) Martin et al. (activated fibroblasts) (9) Kinchen et al. (S4)# (10) Smillie et al. |
| 5b. Cancer-associated fibroblasts (CAFs) | TGFB1, TGFB3, MMP2, MMP3, MMP11 | Exclusive for tumor tissue (in CRC) Produce multiple pro-oncogenic growth factors Mediate paracrine responses in tumors |
(11) Li et al. (12) Uhlitz et al. |
*selected subset-defining gene markers that overlap in the studies listed under ‘Reference’.
**indicated only if subset annotation differs from the one indicated under ‘Subset’.
#IAFs described by Kinchen et al. had a mixed gene expression signature of RSPO3+ fibroblasts and IAFs when compared to the clusters described by Smillie et al. and Huang et al.
5a and 5b divide a category of disease-associated subsets of fibroblasts: IAFs in IBD and CAFs in CRC.
GO, gene ontology; IBD, inflammatory bowel disease; CRC, colorectal cancer.
Immune cell compartment
The gut is the largest immune organ in the human body, and the mucosal immune system is crucial in health and disease, as it guards the barrier between the body’s internal milieu and the microbiome in the gut lumen (41). Although many mucosal immune cells are gut-resident and their main role is maintaining homeostasis, scRNA-seq studies provide additional evidence for their active involvement in inflammation and carcinogenesis.
ScRNA-seq highlights T cells as the most functionally diverse and flexible immune cells in the human gut. Instead of the classic denomination based on surface markers (i.e. CD4-CD8), scRNA-seq differentiates cells based on their gene expression, classifying T cells based on their origin, spatial localization and function. Under homeostatic circumstances, the gut mucosa harbors a vast reservoir of naive, central memory and resident memory T cells. In disease, the number of specific T cell subsets expands, bearing out the fluidity and the functional diversity of the compartment (6,10). Studies that employ scRNA-seq to profile human gut cells in CRC show similar inflammatory responses as have been observed in IBD (11–13). In active IBD, tissue-resident T cells fulfill a multitude of different functions: pro-inflammatory—through cytotoxic (TNF, IFNG) or antimicrobial (IL22, IL17A) pathways, and anti-inflammatory—through suppressive pathways (IL10, TIGIT). Still, separate populations of cytotoxic T cells and regulatory T cells (Treg) are clearly present in IBD. While classically cytotoxic T cells are CD8+ T cells, scRNA-seq reveals that on gene expression level, cytotoxic T cell subset consists of both CD4+ and CD8+ T cells (7,10). Regulatory T cells, as characterized by the expression of IL10 and CTLA4, are present in the healthy gut and expand during inflammation.
Furthermore, scRNA-seq has provided new insight into IL17 expressing cells, which are known to play a central role in chronic inflammation in IBD (42). Although these cells are classically identified as one Th17 cell population, scRNA-seq provided evidence for the existence of a much wider array of Th17 cell subtypes (6,7,10). Thus, Th17 cell subtypes are ranging from classic Th17 CD4+ T cells, which have an inflammation-modulating phenotype and appear to share a lineage with Treg cells, to the Th17-like cells, with a cytotoxic phenotype, on the other end of the spectrum. The latter are a mixed population of CD4+ and CD8+ T cells. ScRNA-seq detected the marked expansion of this population in the gut mucosa in both IBD and CRC, and it seems to play an important role in aggravating tissue damage and subsequent cancer progression (10,12,13).
Along with the T cells, B cells are a very abundant immune population in the gut mucosa, which further increases in numbers upon active inflammation. Moreover, in active IBD many B cells evolve into plasma cells, favoring IgG producing phenotype over IgA (5,10), which is consistent with the immunoglobulin class switching known in IBD.
ScRNA-seq characterized myeloid cell populations and demonstrated that myeloid cells exist on a scale of active development from monocytes to dendritic cells (DCs) and macrophages (Mfs). DCs survey the mucosa by sampling antigen, and scRNA-seq shows that monocyte-derived DCs form a stable population in the human intestine under homeostasis (43). Once activated, DCs migrate to the lymph nodes to interact with T and B cells. ScRNA-seq reveals that activated DCs, as characterized by the expression of NFκB-inducing cytokines and lymph-attracting chemokines, are more numerous in the mucosa of patients with IBD than in healthy controls (6). Likewise, gut-resident Mfs represent the most abundant mononuclear phagocytes in the body under physiological conditions, and activated pro-inflammatory Mfs are overrepresented in the gut mucosa of IBD patients (6,10,43). These activated DCs and pro-inflammatory Mfs have a central role in IBD, perpetuating disease activity independently of the adaptive immune inflammatory mechanisms targeted by anti-TNFα therapy (6).
Functional networks
In healthy gut mucosa, cells from the different compartments interact to maintain gut barrier function. For instance, together Paneth(like) cells, BEST4+ cells and lamina propria (myo)fibroblasts stimulate epithelial cell renewal, while gut-resident DCs surveil the epithelium for invading antigens, and Mfs and T cells maintain the immune barrier. In disease, this well-orchestrated functional homeostasis is disturbed and remodeled.
One of the strengths of scRNA-seq is that it enables the construction of functional cellular networks in health and in disease while pinpointing the central network hubs. Figure 1 outlines major disease-associated changes in cell composition of the intestinal mucosa and maps cell–cell interaction formed in human gut disease. ScRNA-seq nominates disease-associated cell subsets, such as IAFs/CAFs, M-cells, activated endothelial cells, activated Mfs, activated DCs and inflammatory T cells, as central hubs in the cross-lineage network that drive epithelial barrier breakdown and aggravate disease progression.
Figure 1.
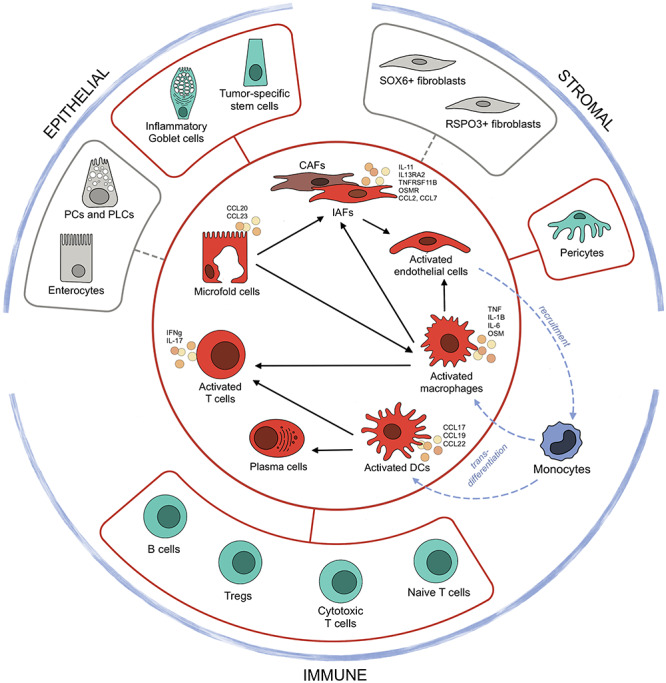
Gut disease-specific features identified by single-cell transcriptomics. The inner circle includes the cell types (in red color) that expand in disease and form key network hubs, mediating inter-lineage crosstalk. The outer circle highlights changes in the proportions of cell subtypes in each compartment that don’t directly contribute to the disease-associated cell–cell network but yet have detrimental effects for intestinal barrier homeostasis. Cell types in gray are depleted in active disease, while cell types in green considerably expand. Cell–cell interactions and their direction are marked by black arrows. Blue dashed arrows delineate the recruitment of circulating classical monocytes by activated endothelial cells, which in turn differentiate into pathogenic activated macrophages and DCs. Annotations: IAFs, inflammation-associated fibroblasts; CAFs, cancer-associated fibroblasts; DCs, dendritic cells; PCs, Paneth cells; PLCs, Paneth-like cells.
Furthermore, scRNA-seq defines the cellular remodeling in the three main intestinal cell compartments (epithelial, stromal and immune) during disease. Enterocytes and Paneth(like) cells in the epithelium, and SOX6+ and RSPO3+ subsets of fibroblasts in the stroma, whose functioning is essential for intestinal homeostasis, have been found to be depleted in inflamed mucosa, reflecting reduced compartmentalization in the diseased gut. On the other hand, scRNA-seq detected the expansion of pericytes, inflammatory goblet cells in IBD and tumor-specific stem cells in CRC. Even more pronounced changes have been described for the immune compartment, where naive T cells, cytotoxic T cells, Tregs and B cells largely contribute to the increased pool of immune cells at the site of inflammation.
Discussion
Single-cell studies have shown that there is a remarkable cellular diversity between patients with similar phenotypes: single-cell transcriptome signatures stratify CRC tumors into subgroups with distinct patient survival (11) and stratify CD patients with ileal inflammation into subgroups with distinct response to anti-TNFa therapy (6). Molecular phenotyping will thus become a crucial step in personalized medicine, and further exploration of pathophysiological diversity in diseases of the gut will greatly improve our ability to realize this personalized medicine. At the same time, single-cell techniques are evolving further, first of all, allowing for higher throughput and lower cost per sample (44). Other new developments in high-resolution transcriptome-wide technologies are capable to infer the spatial localization of the cells of which gene expression is measured, shedding more light on the functioning of the gut mucosa as an organ (45,46). Single-cell technologies revolutionized the way we approach human biology, culminating in an exciting effort to map all human cells as championed by the Human Cell Atlas (https://www.humancellatlas.org). Consequently, defining human gut at single-cell resolution will continue to reshape our understanding of gastrointestinal health and disease.
Conflict of Interest statement. None declared.
Funding
R.K. Weersma is supported by a Diagnostics Grant from the Dutch Digestive Foundation (D16–14). E.A.M. Festen is supported by a MLDS Career Development grant (CDG 14–04).
References
- 1. Drossman D.A. (2016) Functional gastrointestinal disorders: history, pathophysiology, clinical features, and Rome IV. Gastroenterology, 150, 1262–1279. [DOI] [PubMed] [Google Scholar]
- 2. Zeng T. and Dai H. (2019) Single-cell RNA sequencing-based computational analysis to describe disease heterogeneity. Front. Genet., 10, 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Y., Song W., Wang J., Wang T., Xiong X., Qi Z., Fu W., Yang X. and Chen Y.-G. (2020) Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J. Exp. Med., 217, e20191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atlasy N., Bujko A., Brazda P.B., Janssen-Megens E., Bækkevold E.S., Jahnsen J., Jahnsen F.L. and Stunnenberg H.G. (2019) Single cell transcriptome atlas of immune cells in human small intestine and in celiac disease. bioRxiv, 721258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang B., Chen Z., Geng L., Wang J., Liang H., Cao Y., Chen H., Huang W., Su M., Wang H. et al. (2019) Mucosal profiling of pediatric-onset colitis and IBD reveals common Pathogenics and therapeutic pathways. Cell, 179, 1160–1176. [DOI] [PubMed] [Google Scholar]
- 6. Martin J.C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J.A., Gettler K., Chuang L.-S., Nayar S., Greenstein A.J. et al. (2019) Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell, 178, 1493–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Uniken Venema W.T., Voskuil M.D., Vila A.V., van der Vries G., Jansen B.H., Jabri B., Faber K.N., Dijkstra G., Xavier R.J., Wijmenga C. et al. (2019) Single-cell RNA sequencing of blood and Ileal T cells from patients with Crohn’s disease reveals tissue-specific characteristics and drug targets. Gastroenterology, 156, 812–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parikh K., Antanaviciute A., Fawkner-Corbett D., Jagielowicz M., Aulicino A., Lagerholm C., Davis S., Kinchen J., Chen H.H., Alham N.K. et al. (2019) Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature, 567, 49–55. [DOI] [PubMed] [Google Scholar]
- 9. Kinchen J., Chen H.H., Parikh K., Antanaviciute A., Jagielowicz M., Fawkner-Corbett D., Ashley N., Cubitt L., Mellado-Gomez E., Attar M. et al. (2018) Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell, 175, 372–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smillie C.S., Biton M., Ordovas-Montanes J., Sullivan K.M., Burgin G., Graham D.B., Herbst R.H., Rogel N., Slyper M., Waldman J. et al. (2019) Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell, 178, 714–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li H., Courtois E.T., Sengupta D., Tan Y., Chen K.H., Goh J.J.L., Kong S.L., Chua C., Hon L.K., Tan W.S. et al. (2017) Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet., 49, 708–718. [DOI] [PubMed] [Google Scholar]
- 12. Uhlitz F., Bischoff P., Sieber A., Obermayer B., Blanc E., Lüthen M., Sawitzki B., Kamphues C., Beule D., Sers C. et al. (2020) A census of cell types and paracrine interactions in colorectal cancer. bioRxiv, 2020.01.10.901579. [Google Scholar]
- 13. Zhang L., Yu X., Zheng L., Zhang Y., Li Y., Fang Q., Gao R., Kang B., Zhang Q., Huang J.Y. et al. (2018) Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature, 564, 268–272. [DOI] [PubMed] [Google Scholar]
- 14. Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y. et al. (2017) A single-cell survey of the small intestinal epithelium. Nature, 551, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peterson L.W. and Artis D. (2014) Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol., 14, 141–153. [DOI] [PubMed] [Google Scholar]
- 16. Harris N. (2016) The enigmatic tuft cell in immunity. Science (80), 351, 1264–1265. [DOI] [PubMed] [Google Scholar]
- 17. Schneider C., O’Leary C.E. and Locksley R.M. (2019) Regulation of immune responses by tuft cells. Nat. Rev. Immunol., 19, 584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dillon A. and Lo D.D. (2019) M cells: intelligent engineering of mucosal immune surveillance. Front. Immunol., 10, 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vancamelbeke M. and Vermeire S. (2017) The intestinal barrier: a fundamental role in health and disease. Expert Rev. Gastroenterol. Hepatol., 11, 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Camilleri M., Madsen K., Spiller R., Van Meerveld B.G. and Verne G.N. (2012) Intestinal barrier function in health and gastrointestinal disease. Neurogastroenterol. Motil., 24, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brenna Ø., Bruland T., Furnes M.W., van Beelen Granlund A., Drozdov I., Emgård J., Brønstad G., Kidd M., Sandvik A.K. and Gustafsson B.I. (2015) The guanylate cyclase-C signaling pathway is down-regulated in inflammatory bowel disease. Scand. J. Gastroenterol., 50, 1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rahbi H., Narayan H., Jones D.J.L. and Ng L.L. (2012) The uroguanylin system and human disease. Clin. Sci., 123, 659–668. [DOI] [PubMed] [Google Scholar]
- 23. Camilleri M. (2015) Guanylate Cyclase C agonists: emerging gastrointestinal therapies and actions. Gastroenterology, 148, 483–487. [DOI] [PubMed] [Google Scholar]
- 24. Lan D., Niu J., Miao J., Dong X., Wang H., Yang G., Wang K. and Miaob Y. (2016) Expression of guanylate cyclase-C, guanylin and uroguanylin is downregulated proportionally to the ulcerative colitis disease activity index. Sci. Rep., 6, 25034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waldman S.A. and Camilleri M. (2018) Guanylate cyclase-C as a therapeutic target in gastrointestinal disorders. Gut, 67, 1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Porter E.M., Bevins C.L., Ghosh D. and Ganz T. (2002) The multifaceted Paneth cell. Cell. Mol. Life Sci., 59, 156–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clevers H.C. and Bevins C.L. (2013) Paneth cells: maestros of the small intestinal crypts. Annu. Rev. Physiol., 75, 289–311. [DOI] [PubMed] [Google Scholar]
- 28. Rothenberg M.E., Nusse Y., Kalisky T., Lee J.J., Dalerba P., Scheeren F., Lobo N., Kulkarni S., Sim S., Qian D. et al. (2012) Identification of a cKit+ colonic Crypt Base secretory cell that supports Lgr5+ stem cells in mice. Gastroenterology, 142, 1195–1205.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sasaki N., Sachs N., Wiebrands K., Ellenbroek S.I.J., Fumagalli A., Lyubimova A., Begthel H., van den Born M., van Es J.H., Karthaus W.R. et al. (2016) Reg4 + deep crypt secretory cells function as epithelial niche for Lgr5 + stem cells in colon. Proc. Natl. Acad. Sci., 113, E5399–E5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao S., Yan L., Wang R., Li J., Yong J., Zhou X., Wei Y., Wu X., Wang X., Fan X. et al. (2018) Tracing the temporal-spatial transcriptome landscapes of the human fetal digestive tract using single-cell RNA-sequencing. Nat. Cell Biol., 20, 721–734. [DOI] [PubMed] [Google Scholar]
- 31. Jäger S., Stange E.F. and Wehkamp J. (2013) Inflammatory bowel disease: an impaired barrier disease. Langenbeck's Arch. Surg., 398, 1–12. [DOI] [PubMed] [Google Scholar]
- 32. Liu T.-C., Gurram B., Baldridge M.T., Head R., Lam V., Luo C., Cao Y., Simpson P., Hayward M., Holtz M.L. et al. (2016) Paneth cell defects in Crohn’s disease patients promote dysbiosis. JCI Insight, 1, e86907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schumann M., Siegmund B., Schulzke J.D. and Fromm M. (2017) Celiac disease: role of the epithelial barrier. Cell. Mol. Gastroenterol. Hepatol., 3, 150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim Y.S. and Ho S.B. (2010) Intestinal goblet cells and Mucins in health and disease: recent insights and progress. Curr. Gastroenterol. Rep., 12, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gersemann M., Becker S., Kübler I., Koslowski M., Wang G., Herrlinger K.R., Griger J., Fritz P., Fellermann K., Schwab M. et al. (2009) Differences in goblet cell differentiation between Crohn’s disease and ulcerative colitis. Differentiation, 77, 84–94. [DOI] [PubMed] [Google Scholar]
- 36. Capuano M., Iaffaldano L., Tinto N., Montanaro D., Capobianco V., Izzo V., Tucci F., Troncone G., Greco L. and Sacchetti L. (2011) MicroRNA-449a overexpression, reduced NOTCH1 signals and scarce goblet cells characterize the small intestine of celiac patients. PLoS One, 6, e29094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mabbott N.A., Donaldson D.S., Ohno H., Williams I.R. and Mahajan A. (2013) Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol., 6, 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kobayashi N., Takahashi D., Takano S., Kimura S. and Hase K. (2019) The roles of Peyer’s patches and microfold cells in the gut immune system: relevance to autoimmune diseases. Front. Immunol., 10, 2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Owens B.M.J. and Simmons A. (2013) Intestinal stromal cells in mucosal immunity and homeostasis. Mucosal Immunol., 6, 224–234. [DOI] [PubMed] [Google Scholar]
- 40. Nowarski R., Jackson R. and Flavell R.A. (2017) The stromal intervention: regulation of immunity and inflammation at the epithelial-Mesenchymal barrier. Cell, 168, 362–375. [DOI] [PubMed] [Google Scholar]
- 41. Turner J.R. (2009) Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol., 9, 799–809. [DOI] [PubMed] [Google Scholar]
- 42. Srenathan U., Steel K. and Taams L.S. (2016) IL-17+ CD8+ T cells: differentiation, phenotype and role in inflammatory disease. Immunol. Lett., 178, 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cerovic V., Bain C.C., Mowat A.M. and Milling S.W.F. (2014) Intestinal macrophages and dendritic cells: what’s the difference? Trends Immunol., 35, 270–277. [DOI] [PubMed] [Google Scholar]
- 44. Hess J.F., Kohl T.A., Kotrová M., Rönsch K., Paprotka T., Mohr V., Hutzenlaub T., Brüggemann M., Zengerle R., Niemann S. and Paust N. (2020) Library preparation for next generation sequencing: a review of automation strategies. Biotechnol. Adv., 41, 107537. [DOI] [PubMed] [Google Scholar]
- 45. Eng C.-H.L., Lawson M., Zhu Q., Dries R., Koulena N., Takei Y., Yun J., Cronin C., Karp C., Yuan G.-C. and Cai L. (2019) Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature, 568, 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R., Welch J., Chen L.M., Chen F. and Macosko E.Z. (2019) Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science (80), 363, 1463–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]


