Abstract
The ‘discovery’ stage of genome-wide association studies required amassing large, homogeneous cohorts. In order to attain clinically useful insights, we must now consider the presentation of disease within our clinics and, by extension, within our medical records. Large-scale use of electronic health record (EHR) data can help to understand phenotypes in a scalable manner, incorporating lifelong and whole-phenome context.
However, extending analyses to incorporate EHR and biobank-based analyses will require careful consideration of phenotype definition. Judgements and clinical decisions that occur ‘outside’ the system inevitably contain some degree of bias and become encoded in EHR data. Any algorithmic approach to phenotypic characterization that assumes non-biased variables will generate compounded biased conclusions. Here, we discuss and illustrate potential biases inherent within EHR analyses, how these may be compounded across time and suggest frameworks for large-scale phenotypic analysis to minimize and uncover encoded bias.
Introduction
The application of large-scale genome-wide association studies (GWAS) has yielded significant and important insights into the genetic architecture of complex traits and diseases. Global collaboration and data sharing approaches have enabled highly standardized sample collection, genotyping, analysis and replication. For example, efforts from the Psychiatric Genomics Consortium have led to collection of >250 000 individuals with 10 psychiatric traits and disorders, and consequent identification of >200 disease-associated loci (1). By necessity, this ‘discovery’ stage of GWAS required the collection of large, homogeneous cohorts. However, biological insight into disease aetiopathology from these studies has been limited. In addition, GWAS analyses to date have focussed almost exclusively on White- and European-descent populations (2–5), significantly limiting potential biological insights, reducing replicability and applicability of biomarkers, and potentially leading to more deeply stratified healthcare, with precision medicine approaches available only for a subset of patients (2,5). In order to attain clinically useful insights, we must now consider the presentation of disease within our clinics and our communities rather than within a narrowly defined cohort including only White cases and controls. By extension, we must look within our medical records to uncover the next stage of genetic associations with complex traits. Such large-scale use of electronic health record (EHR) data can help to understand phenotypes in a scalable manner, incorporating lifelong and whole-phenome context (6–11).
Much has been written regarding the importance of properly accounting for race and ancestry in genetic studies, both in large-scale GWAS (2–4,12–14) and in EHR analyses. However, biases inherent in diagnostic practices may pose an even greater threat to cross-ancestry studies. It is neither appropriate to assume that EHR phenotypes are ‘ground truth’, nor that ‘clinician validation’ or text notes are a ‘gold standard’ for understanding diagnoses; rather, EHR represent complex, multi-level human decisions with potential for bias, compounded across legal, medical and diagnostic systems.
Here, we define EHR ‘bias’ as influences on a variable related to the individual or to the system assigning that variable rather than a true description of the variable itself. Studies that fail to account for these biases may falsely interpret their results to infer differences between groups that are a result of structured bias rather than biological truth; genetic analyses that do not examine critically the potential differences in phenotypic and diagnostic accuracy between groups will be highly vulnerable to spurious findings or underpowered analyses.
Implicit Bias of Hidden Variables: The Anatomy of Diagnostic Bias in Structured and Unstructured Data
Bias is a dynamic, multi-level process occurring across individuals, local and global systems (Fig. 1). Although certain levels of bias may be encoded directly within EHR, system- and community-level bias influence data in EHR without mapping to a specific variable within the dataset. For example, physician bias may be encoded in specific variables such as diagnosis, medications and treatment; hospital bias may be encoded in types of insurance or lengths of stay. In comparison, community-level bias may not impact a specific variable, but rather serves to influence hospital and physician practices. Notably, each type of variable can have bias from multiple sources. For example, the decision to involuntarily commit an individual is encoded in the system as a given legal status such as ‘9.39’, which is influenced by multiple sources of bias including state law, hospital policy, judicial systems and physician discretion. Frameworks for system-level bias can serve to identify direct and indirect sources of bias encoded in EHR data, improving both within-EHR analysis and harmonizing studies across different EHRs.
Figure 1.
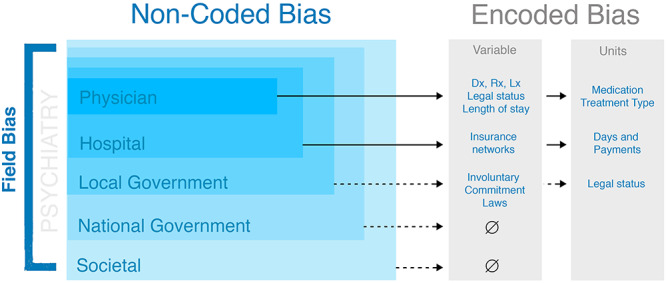
Multi-level biases are inherent in EHR data. These may be explicitly encoded and detectable (e.g. length of stay) or implicit (e.g. societal bias).
Understanding structured bias requires evaluation of bias in data from the level of individual diagnosis to systemic practices. For example, bias occurs in clinical practice to varying degrees, with increased risk of bias where objective clinical markers are unavailable. In particular, the field of psychiatry is notable for its lack of objective markers to guide decision-making processes, and is thus an ideal use case for discussing how clinical bias can become encoded in EHR data. Psychiatric diagnostic criteria are codified in the Diagnostic and Statistical Manual (‘DSM’) (15). Although updated periodically (16), these criteria include field-level biases and potentially outdated diagnostic criteria or classifications, which are based on observed behaviours, either directly or via collateral. For example, the Diagnostic and Statistical Manual version 5 (DSM-5) criteria for any given disease requires meeting a set of symptoms for a specified duration of time. The development of disease definition and assessment of each individual diagnostic criterion are subject to unique biases. Thus, the problem of bias in psychiatric phenotypes encoded in EHR data is that both the assessment of disease and the disease definition itself inherently lack ground truth or gold standard.
Such biases pose particular problems for EHR–genetic studies that compare automatically inferred cases versus controls. Although minor uncertainties or algorithmic inaccuracies may be acceptable in return for the substantial increase in sample size offered by these approaches, introduction of systematic bias or bias occurring primarily in one group of patients poses a substantial risk to the accuracy and interpretability of these studies. For example, diagnostic biases in the psychiatric field may lead to systematic differences in diagnoses across racial and ethnic groups or between genders. Consequently, genetic association analyses may be less well powered among these groups or may produce spurious results. In Figure 2, we consider the impact of diagnostic biases on case–control definitions, and association statistics, for a polygenic trait [e.g. schizophrenia (SCZ) (17) or major depressive disorder (18)]. If case–control definitions are applied accurately, we partition our population into cases and controls (Fig. 2Ai), and will identify a small number of significantly associated variants (Fig. 2Bi). If instead some bias systematically increases diagnosis of a disorder among some patients, our ‘cases’ will now include many true controls (Fig. 2Aii). As these biases may fluctuate across clinics, hospitals or due to other unknown factors (Fig. 1), assignment of individuals presenting with some subset of disease symptoms or endophenotypes [i.e. those individuals falling in the middle of our liability threshold model (18,19), Fig. 2Aii] to ‘case’ or ‘control’ groups will be essentially biologically random, driven by circumstances of bias or access to the healthcare system rather than relating to disease presentation. Genetic analyses and estimate of heritability based on such inferences will therefore be substantially underpowered (20) (Fig. 2Bii). Conversely, bias that systematically decreases likelihood of diagnosis [e.g. due to race- or gender-specific stereotyping in disease definition (21–26) or due to bias that reduces lack of access to healthcare (27–34)] may mean that only ‘extreme’ presentations of a disease are identified and included as ‘cases’ (Fig. 2Aiii). In these instances, ‘control’ populations will likely also include a number of ‘true cases’ (Fig. 2Aiii); consequently, association statistics may be inflated (Fig. 2Biii).
Figure 2.
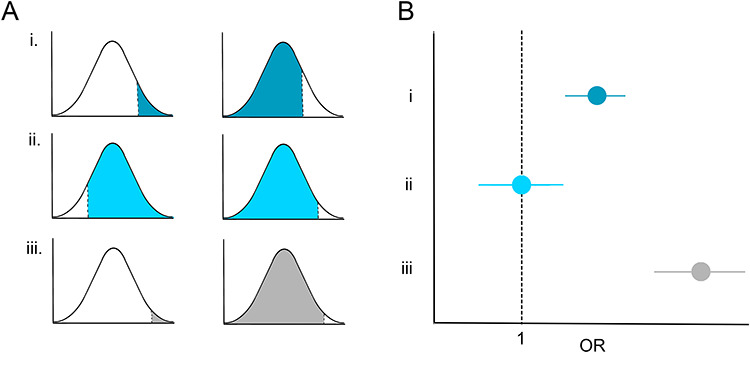
Impact of case–control definition bias on genetic studies. (A) Case (left) and control (right) definitions shown on a liability threshold model when (i) individuals are diagnosed accurately and without bias; (ii) where bias leads to over-diagnosis; and (iii) where bias leads to under-diagnosis. (B) Genetic associations arising from the situations described in (A). (i) A variant is identified as associated with a given trait; (ii) over-diagnosis leads to underpowered studies; true associations may be missed; and (iii) under-diagnosis leads to spurious results; associations may be inflated.
Brief History of Bias in Psychiatry: Past Evidence and Ongoing Examples
Diagnostic biases have been well documented in psychiatric research and include both patterns of under- and over-diagnosis. For example, numerous studies have found that Black patients are disproportionately more likely to be diagnosed with SCZ than White patients (35–37). A meta-analysis of 41 studies found Black patients to be preferentially diagnosed with SCZ by an odds ratio (OR) of 2.43 (38). White patients, on the other hand, are more likely to be diagnosed with an affective disorder (39). This divergence has been attributed to epigenetic differences (40), with little to no empirical support, and differences in socioeconomic status (41), but the diagnostic discrepancy remains present even after controlling for age, sex, income, site and education (42), suggesting that physician bias or systemic bias presenting across various levels of healthcare and legislative processes (Fig. 1) may present the most notable factor. In addition, biased racial expectations by physicians towards patients may lead to inflated rates of SCZ diagnoses in Black patients. One such expectation is that of dishonesty. A study by Eack et al. (43), for instance, found that Black patients were perceived as less honest by interviewers, potentially due to misattribution of cultural guardedness as dishonesty, and this perception predicted over-diagnosis of SCZ. Another study determined that presence of diagnostic criteria are differentially weighed in diagnosis of SCZ for Black and White patients (44). The presence of negative symptoms especially disproportionately corresponded to SCZ diagnosis in Black patients, whereas presence of the same symptoms did not correspond to a diagnosis of SCZ in non-Black patients. Such over-diagnosis leads to disproportionately high prescription of antipsychotic medications (45), long-acting antipsychotic injections (46) and hospitalizations in Black patients (45), contributing to worse outcomes in Black patients 1 year after first episode psychosis (47), and may lead to underpowered genetic association analyses unless diagnostic biases are addressed and corrected for in EHR–GWAS (Fig. 2).
We observed these same patterns of diagnostic bias in our own EHR data (12) (C. Seah, H.R. Dueñas and L.M. Huckins, manuscript in preparation; Fig. 3). Analysis of EHR data of 722 patients with psychosis (defined by International Statistical Classification of Diseases codes F2-, F31.5, F31.64, F32.3, F33.3) enrolled in Mount Sinai BioMe aligned with previous observations of overrepresentation of Black patients with psychosis (OR = 1.7) and underrepresentation of White patients with psychosis (OR = 0.29). Furthermore, Black patients were more likely to be diagnosed with SCZ (OR = 1.92) or schizoaffective disorder (OR = 1.83) as opposed to depression with psychosis (OR = 1.19). Black patients with psychosis were more likely to be prescribed intramuscular (IM) Haldol injections (OR = 2.05) than their White peers (OR = 0.23), a medication commonly indicated for agitation. When controlled for racial distributions of patients with psychosis, Black patients were still preferentially given Haldol injections (OR = 1.08) compared with their White peers (OR = 0.72). In fact, we found that Black patients were prescribed psychiatric medications almost twice as often (1.92×) as White patients.
Figure 3.
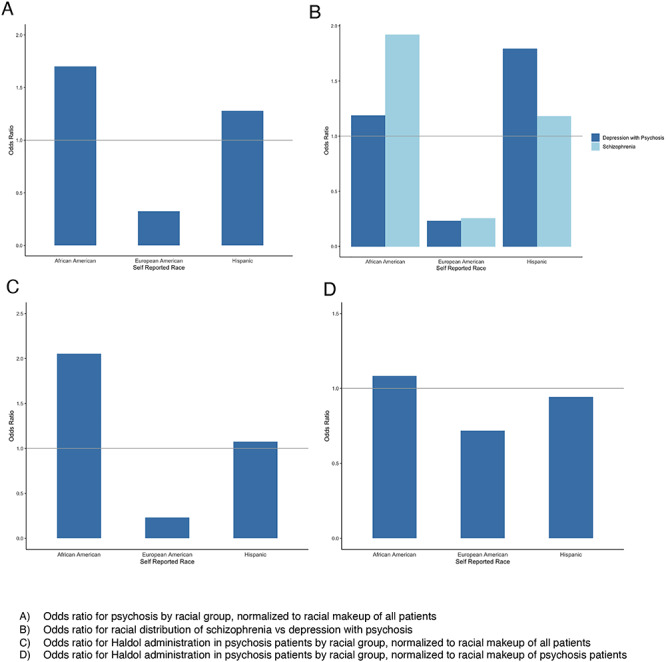
Systemic differences in psychosis diagnosis and treatment in EHR. (A) Odds ratio for psychosis by racial group, normalized to racial makeup of all patients. (B) Odds ratio for racial distribution of schizophrenia versus depression with psychosis. (C) Odds ratio for Haldol administration in psychosis patients by racial group, normalized to racial makeup of all patients. (D) Odds ratio for Haldol administration in psychosis patients by racial group, normalized to racial makeup of psychosis patients.
Further analysis reveals that White patients are less likely than Black patients to be placed on ‘violence precautions’, (OR = 0.58 and OR = 1.05, respectively); a marker in the EHR that indicates to physicians they should have a higher caution for violence and a lower threshold to take measures such as IM medications for agitation. These precautions are a result of patient history in the EHR as well as outside the EHR, through collateral such as law enforcement or family. The discrepancies in assigning ‘violence precautions’ contributing to potential increased use of IM Haldol among Black patients illustrates how system-level bias that exists outside the hospital system may contribute to encoded biases with multiple, complex origins.
However, diagnostic bias does not always lead to over-diagnosis. For example, current estimates of individuals with autism spectrum disorders (ASD) systematically underestimate numbers of women with the disease, overrepresenting men on an order of 4:1 (48). This skewed sex ratio present in ASD is largely due to under-diagnosis of girls with the disorder. Although this may stem in part from fundamental sex-based aetiological differences (49), the attribution of the presentation in men as the norm likely contributes significantly to this skew. This male norm has historically been upheld by significant gender biases in research study recruitment (50), with brain volumetric studies (51) and task functional magnetic resonance imaging studies (52) overrepresenting men by a ratio of 8:1 and 15:1, respectively. In addition, bias encoded in screening questionnaires preferentially identify and diagnose boys, leaving autism undiagnosed in girls. One example is the Autism Diagnosis Interview-Revised, which preferentially scores symptoms commonly expressed by boys and algorithmically predicts autism in boys at a higher rate than girls (53). For example, it, alongside the Autism Diagnostic Observation Schedule-Generic, underscores and downplays sensorimotor symptoms, which have been shown to be more prevalent in girls (53). Other symptoms more present in girls, such as more imaginative and pretend play at a young age (23) and restricted interests related to people and animals as opposed to inanimate objects (25), are unaccounted for. Individual items on another diagnostic tool, the Autism Spectrum Quotient (AQ-10), have also been shown to preferentially underestimate autistic traits in girls and cannot be used individually (26). When girls are diagnosed, they are diagnosed later than boys, according to a study of 2275 individuals with autism (54). Later diagnosis of women in adulthood adds to practical difficulties for clinicians, as it is more challenging to obtain an accurate developmental history (55), leading to compounded diagnostic error (Fig. 4). Late diagnosis and misdiagnosis leads to higher rates of anxiety, as well as self-reported exhaustion and confusion about one’s identity in women with ASD (56).
Figure 4.
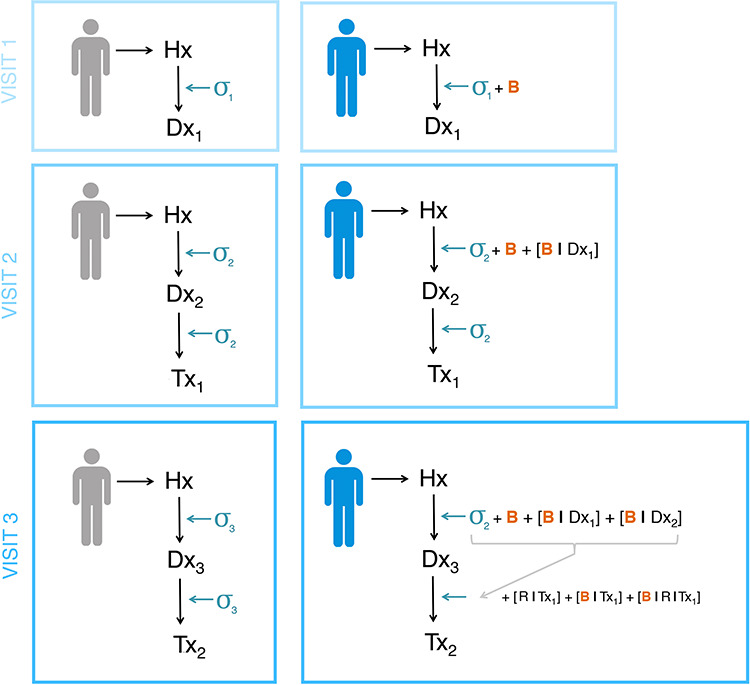
Diagnostic journeys for two individuals through a healthcare system. Left: an ‘ideal’ non-biased diagnostic journey. Visit 1 produces a preliminary diagnosis (Dx1), with some uncertainty (σ1); a second visit refines this diagnosis (Dx2), resulting in reduced diagnostic uncertainty (σ2 < σ1) and treatment (Tx2). Visit 3 results in an increasingly appropriate diagnosis and treatment, and uncertainty continues to reduce (σ3 < < σ1). Right: modelling potential compound bias throughout a diagnostic journey. Here, an initial visit results in a diagnosis (Dx1) influenced by both uncertainty (σ1) and bias (B). Bias compounds across visits; Dx2 is influenced by both current physician bias (B) and bias relating to the previous diagnosis (B | Dx1). Similarly, treatments assigned at visit 3 will be influenced by compound bias based on knowledge of previous treatment options (B | Tx1) and response to those treatments (B | R | Tx1). Hx, hospital system; Dx, diagnosis; Tx, treatment; σ, uncertainty; B, bias; B | Dx, bias regarding previous diagnosis Dx; B | Tx, bias regarding previous treatment Tx; R | Tx, response to treatment Tx.
Sex-based differences in disease presentation also contribute to misdiagnosis and under-diagnosis in eating disorders. Here, however, diagnostic standards are determined based on a female stereotype of disease (57). Women have a 4.2-fold greater lifetime prevalence of eating disorders than men (58). Presentations of eating disorders vary significantly by gender (59), with women more likely to endorse loss of control while eating, and men more likely to overeat (60), as well as women more likely to desire thinness, and men equally likely to desire gaining or losing weight (61). Diagnostic criteria for eating disorders focus on female manifestations of the disease, for instance, up until the DSM-5, amenorrhoea was listed as diagnostic criteria for anorexia nervosa (15,16,62), a criterion that further biases physicians’ diagnostic patterns to recognize women with the disorder. Societal stigma contributes to later recognition of eating disorders in men, with a study noting that 30% of men surveyed had been misdiagnosed due to not fulfilling the gendered stereotype of patients with eating disorders (57). Outcomes are even worse in transgender patients, who describe being misgendered or withholding gender information from physicians due to worries about stigma, and therefore receiving substandard care (63).
Compounded Bias, False Conclusions, Incorrect Care
Importantly, biases within EHR data do not occur as the result of a single interaction or societal factor. Rather, we expect that repeated, longitudinal interactions with healthcare systems will result in compound biases. Specifically, any given psychiatric diagnosis or treatment is non-independent of previous diagnosis or treatments, and therefore, biases present in diagnosis or treatment are amplified over time. Initial bias is likely to be compounded if the original diagnosis was biased, generating biased treatments, poor patient outcomes and misguided understanding of underlying biology.
For example, in Figure 4, we compare diagnostic journeys for two individuals through a healthcare system. First, we describe the assumed, ‘ideal’ diagnostic journey of an individual (Fig. 4; left). An initial appointment produces a preliminary diagnosis (Dx1), with some uncertainty (σ1); a second visit refines this diagnosis (Dx2), resulting in reduced diagnostic uncertainty (such that σ2 < σ1), and perhaps including an appropriate treatment (Tx2). Subsequent visits result in further refinement of diagnoses and identification of increasingly appropriate treatments, and uncertainty continues to reduce (σ3 < < σ1). However, this scenario does not accurately reflect the impact of compound bias throughout the system (Fig. 4; right). In this scenario, bias (B) is introduced when assigning diagnoses and treatments and is compounded across visits. Rather than arriving at a refined diagnosis with low uncertainty, this patient may instead face iterative and compound diagnostic biases (B, B | Dx1, B | Dx2), as well as iterative and compound biases relating to treatment (B | Tx1) and treatment response (B | R | Tx1). As each subsequent diagnosis and treatment is related to previous ones, compounded bias occurs as an individual further interacts with the healthcare system. Given that both unbiased and biased data appear identical in an EHR, we cannot, for example, differentiate true psychotic disease with resistance from inaccurate diagnosis resulting in non-response. Accordingly, as we develop definitions of phenotypes based on algorithmic approaches to large datasets, we must find a way to differentiate between phenotypes characterized by variables encoded in the EHR and those that are a result of variables within the variables, such as bias, that will generate precisely false conclusions.
Frameworks for Uncovering Implicit Bias in Variables: Proposed Analytic Approaches
Creating automated algorithms to identify patterns and infer phenotypes in EHR is attractive in its simplicity and potential power. Simple definitions of case counts and specific treatments may rapidly and easily expand case and control counts for GWAS or other studies that require amassing very large case and control numbers to maximize power. However, we urge caution in the development and application of these algorithms. We have demonstrated that, far from being gold standard, validated phenotyping tools, EHR may encode several levels of systemic, compound bias in arriving at treatments and diagnosis. Put simply, we expect that a person’s race, gender identity, sexuality and many other intersectional factors will impact their experiences of our healthcare systems. Consequently, algorithms and tools that process EHR data without adjustment risk encoding, and potentially reinforcing, such bias in downstream analyses.
Here, we outline analytical approaches that may be adopted to characterize and minimize diagnostic bias in EHR analyses. First, researchers should seek to minimize phenome-wide heterogeneity among cases and controls as defined in their studies. Broadly, we expect that patterns of comorbidities and known risk factors should present at relatively stable levels across all case or controls groups within the sample (e.g. when partitioning samples according to race or gender), unless there is strong biological reason to expect otherwise. Although we expect natural fluctuations in comorbidities between cases, we do not expect systematic differences in comorbidity profiles between case groups. For example, consider a diagnosis of SCZ applied in three groups of patients (White, Black and Hispanic), as in our earlier example. We have shown that both the initial diagnosis and downstream treatments for the diagnosis are significantly biased between these three groups. Consequently, we might also expect different comorbidity profiles or treatment histories among these three groups; most obviously, different medications prescribed and taken (as we have shown), but also differential rates of other, similar diagnoses (e.g. depression with psychosis versus SCZ). Such phenome-wide differences may be present across a broad range of diagnoses, are indicative of biased case/control assignments and will confound downstream genetic associations. Many well-established methods exist for researchers to probe comorbidity profile and phenome-wide associations. For example, phenome-wide association studies (9–11,64–68) or lab-wide association studies (69) systematically probe associations across the full EHR, whereas phenotype risk score approaches (70) identify significant phenome-wide risk factors and predictors of disease outcomes and trajectories.
Second, where possible, researchers should consider order and convergence of diagnoses across the lifespan. Approaching EHR analyses as retrospective ‘snapshots’ risks wrongly conflating unrelated symptoms occurring years or even decades apart. For example, reasonable EHR approaches to automatically identifying individuals with treatment-resistant depression may select individuals with a history of multiple depression diagnoses and multiple prescriptions of antidepressants. Failure to account properly for order and times between these events may conflate separate, unrelated episodes of depression or treatments, reducing case-definition specificity and resulting in underpowered studies. In Figure 5A, we illustrate three individuals with various lifetime diagnoses and treatments. From a ‘snapshot’ perspective, all individuals have identical phenome-wide profiles. However, sampling throughout the life course will produce radically different phenomes for each individual. Furthermore, sequential diagnoses, versus simultaneous diagnoses or treatments, may allow for introduction of different biases, which should be carefully considered. For example, historical biases as diagnostic boundaries shift and fluctuate, or compound biases across iterations of hospital visits (Fig. 4).
Figure 5.
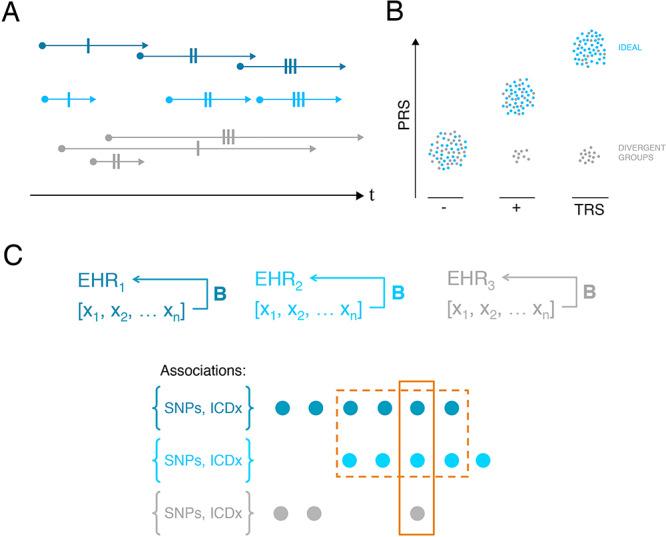
Approaches to detect biases in EHR. (A) Consider order and convergence of diagnoses across the lifespan. Here, we consider three individuals (dark blue, light blue and grey), with identical variables within the EHR (single-dashed lines represent identical diagnoses, etc.). From a retrospective, ‘snapshot’ perspective, all individuals appear identical; however, order and persistence of diagnoses clearly differ. (B) Leverage known biology. Here, we illustrate how polygenic risk scores might be applied to test accuracy of phenotype definitions. In this case, we consider controls (‘-’), schizophrenia cases (‘+’) and individuals with ‘TRS’. If these definitions are accurately assigned, we expect increased PRS in cases versus controls and in TRS versus others (blue). If however some bias affects phenotype assignment, we may identify a group of individuals with divergent PRS (grey). (C) Here, we illustrate three EHR (EHR1, EHR2 and EHR3), with overlapping but not identical sets of biases B and phenotypes [x1…xN]. Genotypic and phenotypic associations {SNPs, ICDx}, which are present across multiple different EHR are more likely to represent true biological signal rather than representing biased inferences.
Third, researchers should leverage known biology to probe the accuracy of diagnostic classifications. Although insufficient for clinical predictions when used alone, polygenic risk scores (PRS) or known genetic correlations between traits may be used to provide additional evidence for diagnostic classifications. In Figure 5B, we take treatment-resistant schizophrenia (TRS) in an EHR as an example. ‘Case’ status in these studies may be ascertained through clinician interview and validation (in best case scenarios) or by automated mining of antipsychotic prescription history to identify increasing dosages and/or large numbers of different antipsychotic drug prescriptions. We suggest that the latter approach may identify two distinct groups: first, individuals truly suffering from TRS; second, individuals with some separate, distinct disorder. Antipsychotic treatment in these instances will be ineffective (treating not a core symptom, but rather a misdiagnosis). In order to disentangle these groups, we can leverage known biology. For example, previous work has demonstrated that individuals with TRS have significantly increased SCZ–PRS compared with both controls and SCZ cases. Modelling SCZ–PRS among controls, SCZ cases and TRS ‘cases’ may validate diagnosis (individuals shown in blue, Fig. 5B) or conversely may indicate misdiagnosis (individuals shown in black, Fig. 5B).
To date, validation in genetic association studies has relied upon replication in an independent dataset. For researchers using EHR data, we caution that systemic biases will differ across healthcare systems, likely in a non-random fashion. Studies at each EHR site must be carefully tailored to characterize and address specific local data biases; this may mean development of individual algorithms and approaches separately within each healthcare system. Here, we propose that the same theoretical framework underpinning trans-ancestral fine-mapping of genomic loci may be useful to researchers. That is, associations that are present across multiple different EHR are more likely to represent true biological signal rather than representing biased inferences (Fig. 5C). In order to infer potential sources of shared and distinct biases across healthcare systems and EHR, researchers should consider carefully the various systemic biases that may be inherent in each different cohort in their studies, considering, for example, different local, governmental and legal structures in place at each site.
Any algorithmic approach built on bias will perpetuate bias and false conclusions. Without knowledge of the exact nature of the bias itself, it is possible to design algorithmic approaches to large structured datasets that may leverage this uncertainty to identify the impact of implicit bias on variables. Overall, the development of analytical frameworks to address bias in large datasets has the potential to both elucidate biology of disease and characterize implicit bias encoded in systems.
Funding
L.M.H. acknowledges funding from the Seaver Foundation through a Seaver Faculty Foundation Award and from National Institute of Mental Health R01MH121923.
Conflict of Interest statement. None declared.
References
- 1. Sullivan P.F., Agrawal A., Bulik C.M., Andreassen O.A., Borglum A.D., Breen G., Cichon S., Edenberg H.J., Faraone S.V., Gelernter J. et al. (2018) Psychiatric genomics: an update and an agenda. Am. J. Psychiatry, 175, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Popejoy A. and Fullerton S. (2016) Genomics is failing on diversity. Nature, 538, 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peterson R.E., Kuchenbaecker K., Walters R.K., Chen C.-Y., Popejoy A.B., Periyasamy S., Lam M., Iyegbe C., Strawbridge R.J., Brick L. et al. (2019) Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell, 179, 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wojcik G.L., Graff M., Nishimura K.K., Tao R., Haessler J., Gignoux C.R., Highland H.M., Patel Y.M., Sorokin E.P., Avery C.L. et al. (2019) Genetic analyses of diverse populations improves discovery for complex traits. Nature, 570, 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bustamante C.D., De La Vega F.M. and Burchard E.G. (2011) Genomics for the world. Nature, 475, 163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glicksberg B.S., Johnson K.W. and Dudley J.T. (2018) The next generation of precision medicine: observational studies, electronic health records, biobanks and continuous monitoring. Hum. Mol. Genet., 27, R56–R62. [DOI] [PubMed] [Google Scholar]
- 7. Wolford B.N., Willer C.J. and Surakka I. (2018) Electronic health records: the next wave of complex disease genetics. Hum. Mol. Genet., 27, R14–R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Robinson J.R., Denny J.C., Roden D.M. and Driest S.L.V. (2018) Genome-wide and phenome-wide approaches to understand variable drug actions in electronic health records. Clin. Transl. Sci., 11, 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pendergrass S.A., Brown-Gentry K., Dudek S.M., Torstenson E.S., Ambite J.L., Avery C.L., Buyske S., Cai C., Fesinmeyer M.D., Haiman C. et al. (2011) The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genet. Epidemiol., 35, 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Denny J.C., Ritchie M.D., Basford M.A., Pulley J.M., Bastarache L., Brown-Gentry K., Wang D., Masys D.R., Roden D.M. and Crawford D.C. (2010) PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinforma. Oxf. Engl., 26, 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheutlin A.B., Dennis J., Karlsson Linnér R., Moscati A., Restrepo N., Straub P., Ruderfer D., Castro V.M., Chen C.-Y., Ge T. et al. (2019) Penetrance and pleiotropy of polygenic risk scores for schizophrenia in 106,160 patients across four health care systems. Am. J. Psychiatry, 176, 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Belbin G.M., Odgis J., Sorokin E.P., Yee M.-C., Kohli S., Glicksberg B.S., Gignoux C.R., Wojcik G.L., Van Vleck T., Jeff J.M. et al. (2017) Genetic identification of a common collagen disease in Puerto Ricans via identity-by-descent mapping in a health system. Elife, 6, e25060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duncan L., Shen H., Gelaye B., Meijsen J., Ressler K., Feldman M., Peterson R. and Domingue B. (2019) Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun., 10, 3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bigdeli T., Ripke S., Peterson R., Trzaskowski M., Bacanu S.-A., Abdellaoui A., Andlauer T., Beekman A., Berger K., Blackwood D. et al. (2017) Genetic effects influencing risk for major depressive disorder in China and Europe. Transl. Psychiatry, 7, e1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. American Psychiatric Association (2013) Diagnostic and Statistical Manual of Mental Disorders (DSM-5®). American Psychiatric Pub, Arlington, VA. [Google Scholar]
- 16. American Psychiatric Association (2013) Highlights of Changes from DSM-IV-TR to DSM-5. American Psychiatric Association, Arlington, VA, pp. 1–19. [Google Scholar]
- 17. Wray N.R. and Visscher P.M. (2010) Narrowing the boundaries of the genetic architecture of schizophrenia. Schizophr. Bull., 36, 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Corfield E.C., Yang Y., Martin N.G. and Nyholt D.R. (2017) A continuum of genetic liability for minor and major depression. Transl. Psychiatry, 7, e1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruderfer D.M., Ripke S., McQuillin A., Boocock J., Stahl E.A., Pavlides J.M.W., Mullins N., Charney A.W., Ori A.P.S., Loohuis L.M.O. et al. (2018) Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell, 173, 1705–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wray N.R., Lee S.H., Mehta D., Vinkhuyzen A.A.E., Dudbridge F. and Middeldorp C.M. (2014) Research review: polygenic methods and their application to psychiatric traits. J. Child Psychol. Psychiatry, 55, 1068–1087. [DOI] [PubMed] [Google Scholar]
- 21. Manzato E., Gualandi M., Tarabbia C., Romano D., Pascoli L.D. and Scanelli G. (2017) Anorexia nervosa: an update on genetic, biological and clinical aspects in males. Ital. J. Gend.-Specif. Med., 3, 59–70. [Google Scholar]
- 22. Feldman M.B. and Meyer I.H. (2007) Eating disorders in diverse lesbian, gay, and bisexual populations. Int. J. Eat. Disord., 40, 218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kreiser N.L. and White S.W. (2014) ASD in females: are we overstating the gender difference in diagnosis? Clin. Child Fam. Psychol. Rev., 17, 67–84. [DOI] [PubMed] [Google Scholar]
- 24. Anderson C.B. and Bulik C.M. (2004) Gender differences in compensatory behaviors, weight and shape salience, and drive for thinness. Eat. Behav., 5, 1–11. [DOI] [PubMed] [Google Scholar]
- 25. Lai M.-C. and Baron-Cohen S. (2015) Identifying the lost generation of adults with autism spectrum conditions. Lancet Psychiatry, 2, 1013–1027. [DOI] [PubMed] [Google Scholar]
- 26. Murray A.L., Allison C., Smith P.L., Baron-Cohen S., Booth T. and Auyeung B. (2017) Investigating diagnostic bias in autism spectrum conditions: an item response theory analysis of sex bias in the AQ-10. Autism Res. Off. J. Int. Soc. Autism Res., 10, 790–800. [DOI] [PubMed] [Google Scholar]
- 27. Marques L., Alegria M., Becker A.E., Chen C.-N., Fang A., Chosak A. and Diniz J.B. (2011) Comparative prevalence, correlates of impairment, and service utilization for eating disorders across US ethnic groups: implications for reducing ethnic disparities in health care access for eating disorders. Int. J. Eat. Disord., 44, 412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coffino J.A., Udo T. and Grilo C.M. (2019) Rates of help-seeking in US adults with lifetime DSM-5 eating disorders: prevalence across diagnoses and differences by sex and ethnicity/race. Mayo Clin. Proc., 94, 1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee-Winn A., Mendelson T. and Mojtabai R. (2014) Racial/ethnic disparities in binge eating: disorder prevalence, symptom presentation, and help-seeking among Asian Americans and non-Latino Whites. Am. J. Public Health, 104, 1263–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Merikangas K.R., He J., Burstein M., Swendsen J., Avenevoli S., Case B., Georgiades K., Heaton L., Swanson S. and Olfson M. (2011) Service utilization for lifetime mental disorders in U.S. adolescents: results of the National Comorbidity Survey–Adolescent Supplement (NCS-A). J. Am. Acad. Child Adolesc. Psychiatry, 50, 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Richman L.S., Kohn-Wood L.P. and Williams D.R. (2007) The role of discrimination and racial identity for mental health service utilization. J. Soc. Clin. Psychol., 26, 960–981. [Google Scholar]
- 32. Rawal P., Romansky J., Jenuwine M. and Lyons J.S. (2004) Racial differences in the mental health needs and service utilization of youth in the juvenile justice system. J. Behav. Health Serv. Res., 31, 242–254. [DOI] [PubMed] [Google Scholar]
- 33. Mandell D.S., Listerud J., Levy S.E. and Pinto-martin J.A. (2002) Race differences in the age at diagnosis among Medicaid-eligible children with autism. J. Am. Acad. Child Adolesc. Psychiatry, 41, 1447–1453. [DOI] [PubMed] [Google Scholar]
- 34. Magaña S., Parish S.L., Rose R.A., Timberlake M. and Swaine J.G. (2012) Racial and ethnic disparities in quality of health care among children with autism and other developmental disabilities. Intellect. Dev. Disabil., 50, 287–299. [DOI] [PubMed] [Google Scholar]
- 35. Minsky S., Vega W., Miskimen T., Gara M. and Escobar J. (2003) Diagnostic patterns in Latino, African American, and European American psychiatric patients. Arch. Gen. Psychiatry, 60, 637–644. [DOI] [PubMed] [Google Scholar]
- 36. Strakowski S.M., Flaum M., Amador X., Bracha H.S., Pandurangi A.K., Robinson D. and Tohen M. (1996) Racial differences in the diagnosis of psychosis. Schizophr. Res., 21, 117–124. [DOI] [PubMed] [Google Scholar]
- 37. Schwartz R.C. and Blankenship D.M. (2014) Racial disparities in psychotic disorder diagnosis: a review of empirical literature. World J. Psychiatry, 4, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Olbert C.M., Nagendra A. and Buck B. (2018) Meta-analysis of Black vs. White racial disparity in schizophrenia diagnosis in the United States: do structured assessments attenuate racial disparities? J. Abnorm. Psychol., 127, 104–115. [DOI] [PubMed] [Google Scholar]
- 39. Strakowski S.M., Keck P.E.J., Arnold L.M., Collins J., Wilson R.M., Fleck D.E., Corey K.B., Amicone J. and Adebimpe V.R. (2003) Ethnicity and diagnosis in patients with affective disorders. J. Clin. Psychiatry, 64, 747–754. [DOI] [PubMed] [Google Scholar]
- 40. Schwartz E.K., Docherty N.M., Najolia G.M. and Cohen A.S. (2019) Exploring the racial diagnostic bias of schizophrenia using behavioral and clinical-based measures. J. Abnorm. Psychol., 128, 263–271. [DOI] [PubMed] [Google Scholar]
- 41. Ojeda V.D. and Bergstresser S.M. (2008) Gender, race-ethnicity, and psychosocial barriers to mental health care: an examination of perceptions and attitudes among adults reporting unmet need. J. Health Soc. Behav., 49, 317–334. [DOI] [PubMed] [Google Scholar]
- 42. Gara M.A., Vega W.A., Arndt S., Escamilla M., Fleck D.E., Lawson W.B., Lesser I., Neighbors H.W., Wilson D.R., Arnold L.M. et al. (2012) Influence of patient race and ethnicity on clinical assessment in patients with affective disorders. Arch. Gen. Psychiatry, 69, 593–600. [DOI] [PubMed] [Google Scholar]
- 43. Eack S.M., Bahorik A.L., Newhill C.E., Neighbors H.W. and Davis L.E. (2012) Interviewer-perceived honesty as a mediator of racial disparities in the diagnosis of schizophrenia. Psychiatr. Serv. Wash. DC, 63, 875–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trierweiler S.J., Neighbors H.W., Munday C., Thompson E.E., Binion V.J. and Gomez J.P. (2000) Clinician attributions associated with the diagnosis of schizophrenia in African American and non-African American patients. J. Consult. Clin. Psychol., 68, 171–175. [DOI] [PubMed] [Google Scholar]
- 45. Rost K., Hsieh Y.-P., Xu S., Menachemi N. and Young A.S. (2011) Potential disparities in the management of schizophrenia in the United States. Psychiatr. Serv. Wash. DC, 62, 613–618. [DOI] [PubMed] [Google Scholar]
- 46. Aggarwal N.K., Rosenheck R.A., Woods S.W. and Sernyak M.J. (2012) Race and long-acting antipsychotic prescription at a community mental health center: a retrospective chart review. J. Clin. Psychiatry, 73, 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li H., Eack S.M., Montrose D.M., Miewald J.M. and Keshavan M. (2011) Longitudinal treatment outcome of African American and Caucasian patients with first episode psychosis. Asian J. Psychiatr., 4, 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fombonne E. (2003) Epidemiological surveys of autism and other pervasive developmental disorders: an update. J. Autism Dev. Disord., 33, 365–382. [DOI] [PubMed] [Google Scholar]
- 49. Schaafsma S.M. and Pfaff D.W. (2014) Etiologies underlying sex differences in autism spectrum disorders. Front. Neuroendocrinol., 35, 255–271. [DOI] [PubMed] [Google Scholar]
- 50. Lai M.-C., Lerch J.P., Floris D.L., Ruigrok A.N.V., Pohl A., Lombardo M.V. and Baron-Cohen S. (2017) Imaging sex/gender and autism in the brain: etiological implications. J. Neurosci. Res., 95, 380–397. [DOI] [PubMed] [Google Scholar]
- 51. Lange, N., Travers, B.G., Bigler, E.D., Prigge, M.B., Froehlich, A.L., Nielsen, J.A., Cariello, A.N., Zielinski, B.A., Anderson, J.S., Fletcher, P.T., Alexander, A.A. and Lainhart, J.E. (2015) Longitudinal volumetric brain changes in autism spectrum disorder ages 6–35 years. Autism research : official journal of the International Society for Autism Research, 8, 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Philip R.C.M., Dauvermann M.R., Whalley H.C., Baynham K., Lawrie S.M. and Stanfield A.C. (2012) A systematic review and meta-analysis of the fMRI investigation of autism spectrum disorders. Neurosci. Biobehav. Rev., 36, 901–942. [DOI] [PubMed] [Google Scholar]
- 53. Beggiato A., Peyre H., Maruani A., Scheid I., Rastam M., Amsellem F., Gillberg C.I., Leboyer M., Bourgeron T., Gillberg C. et al. (2017) Gender differences in autism spectrum disorders: divergence among specific core symptoms. Autism Res. Off. J. Int. Soc. Autism Res., 10, 680–689. [DOI] [PubMed] [Google Scholar]
- 54. Begeer S., Mandell D., Wijnker-Holmes B., Venderbosch S., Rem D., Stekelenburg F. and Koot H.M. (2013) Sex differences in the timing of identification among children and adults with autism spectrum disorders. J. Autism Dev. Disord., 43, 1151–1156. [DOI] [PubMed] [Google Scholar]
- 55. Green R.M., Travers A.M., Howe Y. and McDougle C.J. (2019) Women and autism spectrum disorder: diagnosis and implications for treatment of adolescents and adults. Curr. Psychiatry Rep., 21, 22. [DOI] [PubMed] [Google Scholar]
- 56. Bargiela S., Steward R. and Mandy W. (2016) The experiences of late-diagnosed women with autism spectrum conditions: an investigation of the female autism phenotype. J. Autism Dev. Disord., 46, 3281–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Räisänen U. and Hunt K. (2014) The role of gendered constructions of eating disorders in delayed help-seeking in men: a qualitative interview study. BMJ Open, 4, e004342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qian J., Hu Q., Wan Y., Li T., Wu M., Ren Z. and Yu D. (2013) Prevalence of eating disorders in the general population: a systematic review. Shanghai Arch. Psychiatry, 25, 212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kinasz K., Accurso E.C., Kass A.E. and Le Grange D. (2016) Does sex matter in the clinical presentation of eating disorders in youth? J. Adolesc. Health Off. Publ. Soc. Adolesc. Med., 58, 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Striegel-Moore R.H., Rosselli F., Perrin N., DeBar L., Wilson G.T., May A. and Kraemer H.C. (2009) Gender difference in the prevalence of eating disorder symptoms. Int. J. Eat. Disord., 42, 471–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Strother E., Lemberg R., Stanford S.C. and Turberville D. (2012) Eating disorders in men: underdiagnosed, undertreated, and misunderstood. Eat. Disord., 20, 346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bell C.C. (1994) DSM-IV: diagnostic and statistical manual of mental disorders. JAMA, 272, 828–829. [Google Scholar]
- 63. Thapliyal P., Hay P. and Conti J. (2018) Role of gender in the treatment experiences of people with an eating disorder: a metasynthesis. J. Eat. Disord., 6, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hall M.A., Verma A., Brown-Gentry K.D., Goodloe R., Boston J., Wilson S., McClellan B., Sutcliffe C., Dilks H.H., Gillani N.B. et al. (2014) Detection of pleiotropy through a phenome-wide association study (PheWAS) of epidemiologic data as part of the Environmental Architecture for Genes Linked to Environment (EAGLE) study. PLoS Genet., 10, e1004678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pendergrass S.A., Brown-Gentry K., Dudek S., Frase A., Torstenson E.S., Goodloe R., Ambite J.L., Avery C.L., Buyske S., Bůžková P. et al. (2013) Phenome-wide association study (PheWAS) for detection of pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network. PLoS Genet., 9, e1003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ye Z., Mayer J., Ivacic L., Zhou Z., He M., Schrodi S.J., Page D., Brilliant M.H. and Hebbring S.J. (2015) Phenome-wide association studies (PheWASs) for functional variants. Eur. J. Hum. Genet. EJHG, 23, 523–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ritchie M.D., Denny J.C., Crawford D.C., Ramirez A.H., Weiner J.B., Pulley J.M., Basford M.A., Brown-Gentry K., Balser J.R., Masys D.R. et al. (2010) Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am. J. Hum. Genet., 86, 560–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wei W.-Q., Teixeira P.L., Mo H., Cronin R.M., Warner J.L. and Denny J.C. (2016) Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J. Am. Med. Inform. Assoc. JAMIA, 23, e20–e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dennis J.K., Sealock J.M., Straub P., Hucks D., Actkins K., Faucon A., Goleva S.B., Nirachou M., Singh K., Morley T. et al. (2020) Lab-wide association scan of polygenic scores identifies biomarkers of complex disease. medRxiv. doi: 10.1101/2020.01.24.20018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bastarache L., Hughey J.J., Hebbring S., Marlo J., Zhao W., Ho W.T., Van Driest S.L., McGregor T.L., Mosley J.D., Wells Q.S. et al. (2018) Phenotype risk scores identify patients with unrecognized Mendelian disease patterns. Science, 359, 1233–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]


