Abstract
Huntington disease (HD) is a neurodegenerative disorder that is caused by a CAG repeat expansion in HTT. The length of this repeat, however, only explains a proportion of the variability in age of onset in patients. Genome-wide association studies have identified modifiers that contribute toward a proportion of the observed variance. By incorporating tissue-specific transcriptomic information with these results, additional modifiers can be identified. We performed a transcriptome-wide association study assessing heritable differences in genetically determined expression in diverse tissues, with genome-wide data from over 4000 patients. Functional validation of prioritized genes was undertaken in isogenic HD stem cells and patient brains. Enrichment analyses were performed with biologically relevant gene sets to identify the core pathways. HD-associated gene coexpression modules were assessed for associations with neurological phenotypes in an independent cohort and to guide drug repurposing analyses. Transcriptomic analyses identified genes that were associated with age of HD onset and displayed colocalization with gene expression signals in brain tissue (FAN1, GPR161, PMS2, SUMF2), with supporting evidence from functional experiments. This included genes involved in DNA repair, as well as novel-candidate modifier genes that have been associated with other neurological conditions. Further, cortical coexpression modules were also associated with cognitive decline and HD-related traits in a longitudinal cohort. In summary, the combination of population-scale gene expression information with HD patient genomic data identified novel modifier genes for the disorder. Further, these analyses expanded the pathways potentially involved in modifying HD onset and prioritized candidate therapeutics for future study.
Graphical Abstract
Graphical Abstract.
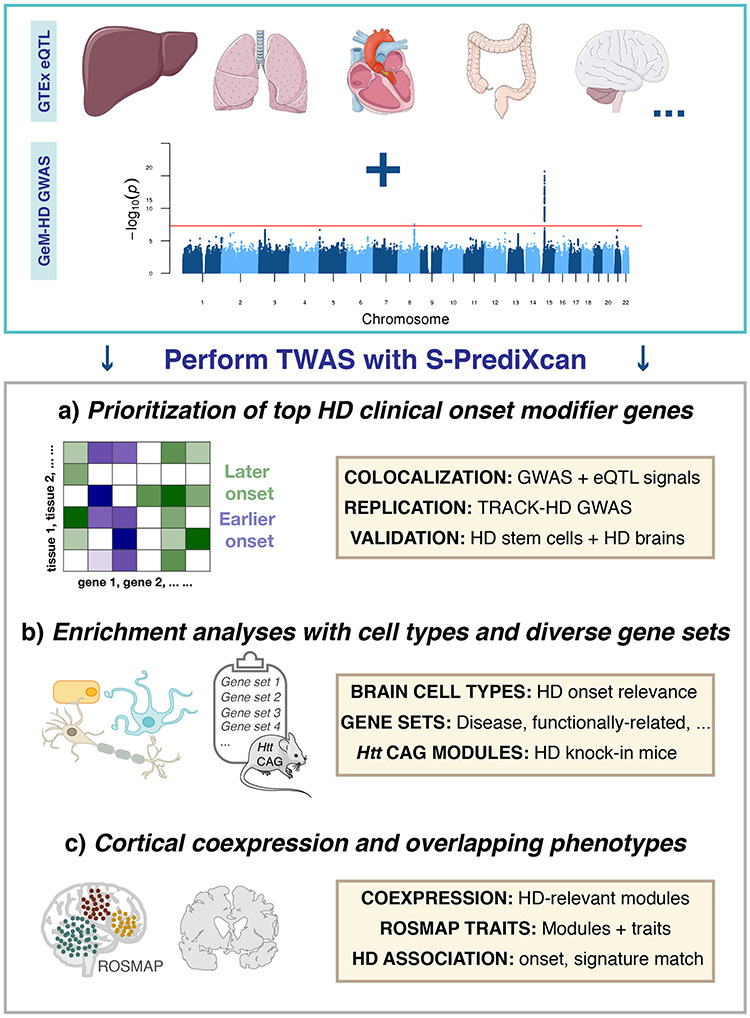
Schematic representation of methodology employed to identify transcriptomic modifiers of Huntington disease (HD). Transcriptome-wide association analyses facilitated (A) the identification of top transcriptomic HD onset modifier genes, (B) relevant biologically diverse gene sets that are enriched for this trait and (C) human cortical coexpression modules that are relevant for HD onset and related phenotypes in an aging cohort, along with signature matching of HD signals for drug repurposing.
Introduction
Although genomic studies have made significant progress in identifying genetic variants associated with human disease, there has been less focus on the study of genomic modifiers of disease to date (1,2). Such research is of great importance since it can be used to complement the development of genetic prediction models, provide novel therapeutic approaches and improve genetic counseling strategies. Huntington disease (HD) is an autosomal dominant neurodegenerative disorder that is caused by an expanded CAG tract in the HTT gene (3). Individuals with repeat lengths of 40 CAGs and greater display full penetrance, and there is an inverse relationship between repeat length and clinical age of onset of the disorder (4). However, the length of this repeat only explains approximately 67% of the variability in age of onset observed between affected individuals, and a large proportion of the residual clinical differences in age of onset observed between patients is likely to be heritable (5,6). Identifying this residual component is important since HD is the most prevalent monogenic neurological condition in the developed world (3). Moreover, understanding this genetic component will help guide the selection of participants and provide objective measures for clinical trials evaluating experimental therapeutics.
Recent studies have started to make progress in identifying modifier genetic variants, both for HD (7–11) and other neurological disorders (12–14). For example, genetic variants in the interrupting sequence between the pathogenic CAG repeat and the polymorphic CCG repeat have been shown to influence age of onset of HD patients and are particularly relevant for patients who carry reduced penetrance alleles (10,11). Additionally, genome-wide association studies (GWAS) have identified trans-modifiers of clinical age of onset in HD (7,8) and have highlighted the important role of DNA repair genes in modulating age of onset, potentially through altering the somatic instability of the CAG repeat.
Interestingly, a subset of these HD modifiers has been shown to modulate the age of onset of spinocerebellar ataxias, which are also caused by pathogenic CAG repeats (15), indicating the potential transferability of these genomic findings in HD to related disorders. Although these studies have made significant advances in our understanding of the genetic modulation of clinical onset in HD, they only explain a limited proportion of the residual variance observed between HD patients. Further, since the signals derived from GWAS are typically in non-coding regions, prioritized regions still need to be refined through the incorporation of functional genomic information into analyses (16). This is pertinent, since it is well documented that changes in gene expression are a hallmark of HD pathology (17,18), and HTT is expressed ubiquitously across tissues (19).
Examination of heritable gene expression profiles has recently been made possible through the development of transcriptome-wide association study (TWAS)-based approaches. These enhance GWAS-derived information through leveraging population-scale expression quantitative trait locus (eQTL) data derived from diverse tissues (20–22). These analyses are used to assess the relevance of gene expression with regard to the GWAS trait, returning gene-level association results. Because of a reduced multiple testing burden compared with GWAS, these transcriptome-inferred analyses have more power to prioritize additional candidate regions, which can in turn be used to inform future in vitro and in vivo genetic modifier validation studies. Further, gene-level association results can also be used in enrichment analyses to determine whether particular pathways or gene sets are relevant to the trait of interest.
We therefore assessed whether heritable variation in gene expression was associated with the clinical onset of HD by analyzing GWAS data for this phenotype from over 4000 patients in combination with transcriptomic information from 48 tissues (summarized in the Graphical Abstract). The key aims of this study were to identify top candidate modifier genes and to prioritize core biologically relevant gene sets involved in modifying HD onset. The expression of prioritized genes was further evaluated in an allelic series of isogenic HD human pluripotent stem cells (hPSCs), as well as through an orthogonal analysis of proteins in pathologically relevant brain regions in HD patient samples. Building on these findings, we showed an enrichment for associations in disease-relevant gene sets, as well as in genes belonging to coexpression modules that are either huntingtin CAG dependent or associated with disease-relevant traits in the human brain. Finally, we performed HD TWAS signature matching to inform future studies of drug repurposing.
Results
Transcriptomic analyses prioritize novel genomic modifiers for the clinical onset of HD
We performed an extensive TWAS using eQTL data from 48 tissues from the GTEx Project (23). In the discovery analyses, HD age of clinical onset GWAS summary statistics from 4082 patients, recruited by the GeM-HD Consortium, were used (8). Since mutant HTT is expressed throughout the body and induces pathology in numerous tissues (3), we chose to perform this agnostic approach to tissue selection, using all information available from the GTEx Project. This approach is also recommended by the S-PrediXcan developers to maximize discovery power (20). Loci prioritized from the TWAS were further refined using Bayesian colocalization analysis, which assessed the probability that at a given locus, the GWAS and eQTL signals share a common causal variant.
Combining these eQTL data with the HD clinical onset GWAS, TWAS in the discovery cohort identified 15 candidate genes located in eight genomic regions (Table 1, Fig. 1, Supplementary Material, Fig. S1 and Supplementary Material, Table S1) where the genetic component of expression was predicted to significantly associated with HD clinical onset. Further, a supplementary analysis of two large cortical gene expression data sets from CommonMind Consortium and ROSMAP did not identify any additional genes not captured in GTEx analysis.
Table 1.
Summary of the 15 prioritized transcriptomic modifier genes where imputed expression is associated with the age of clinical onset of Huntington disease in the GeM-HD cohort
| Gene | Start position (location) | Top P-value | Top Z-score | Striatal CAG modulea | Colocalized (any tissue) | Colocalized (brain) | Description |
|---|---|---|---|---|---|---|---|
| GPR161 | 1:168053997 (1q24) | 4.8 × 10−5 | 4.06 | No | Yes | Yes | G-protein–coupled receptor involved in neuronal tube formation. |
| PMS2 | 7:6012870 (7p22) | 5.1 × 10−5 | 4.05 | No | Yes | Yes | DNA repair pathway, involved in mismatch repair. |
| EIF2AK1 | 7:6061881 (7p22) | 1.1 × 10−5 | 4.40 | Yes (M1) | Yes | Yes | Eukaryotic translation initiation factor. Potential genetic modifier of multiple sclerosis. |
| SUMF2 | 7:56131695 (7p11) | 3.9 × 10−6 | 4.62 | Yes (M20) | Yes | Yes | Sulfatase-modifying factor gene. |
| CHCHD2 | 7:56169262 (7p11) | 5.1 × 10−6 | --4.56 | No | Yes | No | Neurodegenerative disorder risk gene. Localized to mitochondria and involved in mitochondrial respiration. |
| RRM2B | 8:103216730 (8q22) | 6.3 × 10−7 | --4.98 | Yes (M20) | Yes | No | Ribonucleotide reductase, involved in DNA repair and mitochondrial phenotypes. |
| FXYD2 | 11:117671559 (11q23) | 3.9 × 10−5 | --4.11 | No | Yes | No | Subunit of the sodium/potassium-transporting ATPase. Located in the mitochondria. |
| CHRFAM7A | 15:30653443 (15q13) | 2.2 × 10−7 | 5.18 | NA | No | No | Nicotinic acetylcholine receptor fusion protein. |
| GOLGA8H | 15:30896329 (15q13) | 8.7 × 10−8 | 5.35 | No | No | No | Golgin protein gene. |
| ARHGAP11B | 15:30916697 (15q13) | 8.6 × 10−6 | --4.45 | NA | No | No | Human-specific gene, involved in cortical folding. |
| FAN1 | 15:31196055 (15q13) | 4.2 × 10−11 | 6.60 | No | Yes | Yes | Fanconi anemia–associated gene (DNA repair pathway). |
| MTMR10 | 15:31231144 (15q13) | 5.8 × 10−7 | 5.00 | Yes (M7) | No | No | Myotubularin-related protein. |
| KLF13 | 15:31619058 (15q13) | 4.5 × 10−5 | --4.08 | No | No | No | Transcription factor. |
| HMHA1 | 19:1065922 (19p13) | 9.8 × 10−5 | --3.90 | No | No | No | Minor histocompatibility protein, located adjacent to DNA-directed RNA polymerase, POLR2E, as well as ABCA7. |
| ATP4A | 19:36040945 (19q13) | 3.7 × 10−5 | 4.13 | Yes (M20) | No | No | P-type cation-transporting ATPase. Located downstream of a number of FXYD genes. |
Genes are annotated for chromosomal location, whether the expression and GWAS-based signals colocalize and membership to striatal coexpression modules where expression is influenced by Htt CAG repeat lengths. NA, not applicable. Bold text represents that this criteria has been met.
aCAG-dependent coexpression modules from Langfelder et al.
Figure 1.
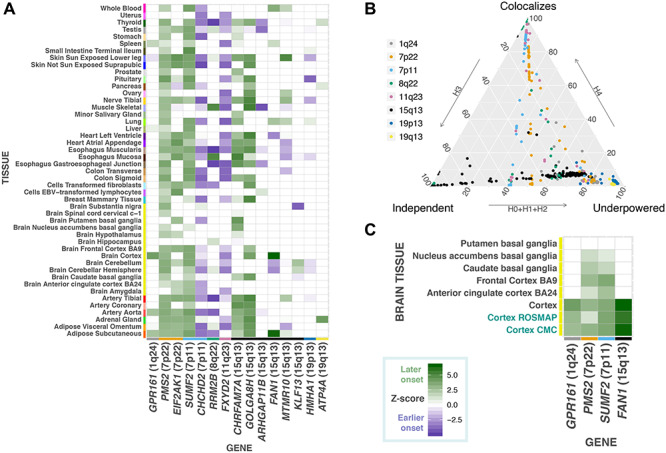
Transcriptome-wide association analysis of Huntington disease clinical onset prioritizes novel candidate modifier genes. (A) Gene-level Z-scores of top modifier genes across tissues in the GeM-HD discovery cohort. Positive Z-scores indicate that increased gene expression is associated with a later age of clinical onset in patients, whereas negative Z-scores are associated with earlier onset. (B) Ternary plot of COLOC posterior probabilities colored by chromosomal location displaying TWAS regions that displayed evidence for colocalization (i.e. COLOC PP4 > 0.5) versus those regions that were either underpowered or where eQTL and GWAS signals represent independent associations. (C) Gene-level Z-scores from TWAS analyses in brain-related tissues for genes that showed evidence for colocalization in these regions in GTEx, as well as the CommonMind Consortium and ROSMAP (colored). These genes showed consistent associations in cortical tissues displaying that increased expression was associated with delayed HD onset.
In these discovery analyses, the most significant TWAS association was for FAN1 in the cortex (Z-score 6.60, P = 4.25 × 10−11), a signal that appears to be driven by eQTLs linked to the one of the chromosome 15 signals detected in the GeM-HD GWAS (top GWAS variant: rs2140734, delays onset by 1.4 years, GTEx eQTL cortex: normalized effect size 0.38, P = 3.2 × 10−7). FAN1 showed consistent associations across the cortical TWAS models from GTEx, CommonMind Consortium and ROSMAP (P < 1.0 × 10−8, Fig. 1), as well as colocalization in the large eQTL meta-analysis (24) from brain regions (PP.H4 = 0.89). In addition to FAN1, three other genes, GPR161, PMS2 and SUMF2, showed colocalization in a brain tissue (Fig. 1 and Table 1).
Four novel prioritized genes, at two loci (chromosome 7p22: PMS2 P = 5.1 × 10−5 and EIF2AK1 P = 1.1 × 10−5; chromosome 7p11: SUMF2 P = 3.9 × 10−6 and CHCHD2 P = 5.1 × 10−6), showed independent evidence for colocalization between the GWAS and expression association signals (i.e. PP4 > 0.5, Fig. 1), as well as nominal independent replication in the TRACK-HD analyses (Supplementary Material, Table S2). Of note, protein truncating mutations in the mismatch repair gene, PMS2, have been shown to cause cancers such as Lynch syndrome (25). Increased expression of PMS2 (Z-score = 4.05) was associated with later age of HD onset and could potentially play a role in mediating the somatic instability of the CAG repeat. The EIF2AK1 and CHCHD2 genes are relevant for neurological phenotypes, since genetic variants have been, respectively, associated with modulating multiple sclerosis progression (12), as well as risk for Parkinson disease and Lewy body disorders, respectively (26).
The association of multiple genes at the same locus, supported by colocalization analyses, could be the result of co-regulation. Colocalization analyses employ a Bayesian statistical test to assess whether two genomic association signals share a common causal variant. They therefore serve as a post-filtering step to prioritize signals where there is a high probability that the GWAS and eQTLs association results arise from a single causal variant, as opposed to S-PrediXcan associations that may have resulted simply from linkage disequilibrium (Fig. 1 and Supplementary Material, Table S2). The value of these colocalization analyses is indicated by the clear clustering of GTEx colocalized modifier genes into biological networks, including an enrichment for DNA repair genes with ATPase activity (Supplementary Material, Fig. S2). It should, however, be noted that non-colocalization could also have been observed for some loci due to limited power or multiple causal variants at a chromosomal region.
Examination of the remaining TWAS signals uncovered additional noteworthy genes. These included RRM2B, a p53-inducible ribonucleotide reductase, and GPR161, a cilium-related G-protein–coupled receptor that has been implicated in neural tube development via the sonic hedgehog pathway (27), both of which displayed evidence of colocalization. The Rho GTPase activating protein gene, HMHA1, was prioritized on chromosome 19p13; however, this gene is located directly downstream of a notable Alzheimer disease gene (28), ABCA7. Although ABCA7 was not prioritized after accounting for multiple testing across tissues, over 60% of ABCA7–tissue combinations displayed nominally significant P-values, indicating that variants related to this gene may have caused this signal. Finally, the mismatch repair gene, MSH3 gene, which was prioritized in the TRACK-HD GWAS (7) used in our replication analyses, also displayed a number of highly-ranked associations in the discovery TWAS (top Z-score = −3.75, P = 1.77 × 10−4).
Further assessment of prioritized transcriptomic HD modifier genes in model systems and HD patient brains
Functional confirmation of prioritized TWAS genes was performed in order to provide orthogonal validation of these findings. In this regard, we assessed alterations of top candidate transcriptomic modifiers at the gene expression level in an allelic series of isogenic HD hPSCs and at the protein level in cortical and striatal HD patient and age-matched control brains.
Using isogenic HD hPSCs as an in vitro model of HD, it was found that DNA repair genes (i.e. FAN1, PMS2 and MSH3) displayed increased expression levels at longer CAG repeat lengths in pluripotent stem cells (Fig. 2). Cells in this early developmental stage display increased activity of DNA repair systems (29), and the higher expression levels at longer CAG lengths could be the result of preventative mechanisms relating to the repeat expansion. Conversely, protein levels for these genes were decreased in HD patient brains compared with controls (Fig. 2 and Supplementary Material, Table S3; FAN1 cortex P = 0.0004; PMS2 cortex P = 0.017; MSH3 caudate nucleus P = 0.04). This may therefore reflect deficiencies in DNA repair in the final stages of the disease. Alternatively, negative correlations between RNA and protein levels have been previously observed for a subset of genes in knock-in mHtt mice, which may reflect the consequences of impaired posttranscriptional regulation as a result of mutant huntingtin (17).
Figure 2.
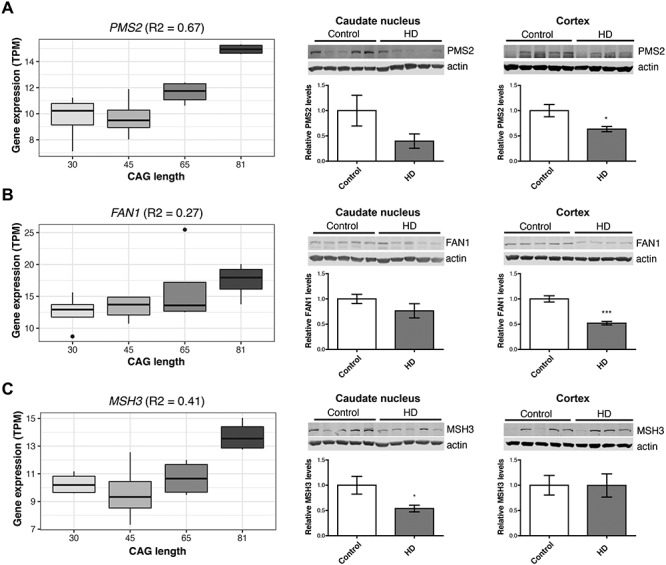
DNA repair genes are dysregulated in HD stem cell lines, as well as patient brains, but show differing directions of effect at the transcript and protein level. Gene expression of DNA repair genes, (A) PMS2, (B) FAN1 and (C) MSH3, was elevated at longer CAG lengths in human pluripotent stems cells (hPSCs, left), but relative protein levels are decreased in HD patient brains compared with matched controls (middle and right). Asterisks indicate P < 0.05.
Additional supporting evidence was also obtained in vitro for the GPR161 gene, since expression levels were decreased at high CAG lengths in allelic neural progenitor cells (R2 = 0.43, P = 0.006). Further, GPR161 protein levels were also lower in the caudate nucleus of HD patients compared with age-matched controls (P = 0.005, Fig. 3), with a trend toward significance in the same direction in the cortex in these patients (Supplementary Material, Table S3). The most significant TWAS association for GPR161 was in the cortex, indicating that increased gene expression is associated with a later age of onset in HD patients. This influence on HD clinical onset is in line with previous experiments that have observed increased genomic instability of medulloblastomas in conditional knockout Gpr161 mice (27). Further, the recent GeM-HD GWAS study (11) has implicated genetic variation near another G-protein–coupled receptor gene, GPR151, in modifying clinical onset in HD (top variant: rs79727797). Network-based analyses of GPR161 and GPR151 suggest the importance of serotonin signaling through shared protein domains of the receptors (Supplementary Material, Fig. S3).
Figure 3.
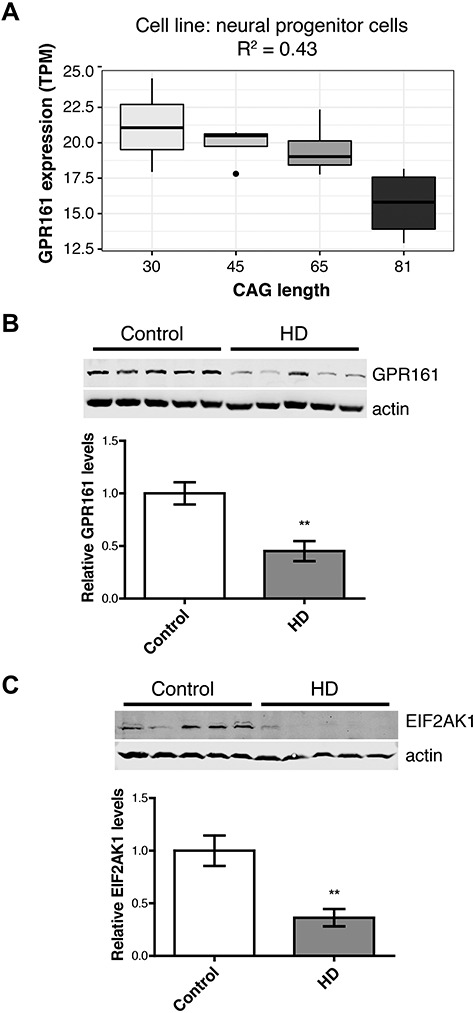
Orthogonal validation of key transcriptomic modifier genes confirms the dysregulation of candidate HD modifiers in humans. The ciliary G-protein–coupled receptor, GPR161, was (A) down-regulated at increasing CAG lengths in neural progenitor cells and (B) decreased relative protein levels in the caudate nucleus in HD patients compared with controls. (C) Another candidate HD modifier protein, EIF2AK1, was also dysregulated in this striatal region in patients. Asterisks indicate P < 0.05.
HD transcriptomic modifier associations are enriched in relevant cell types and gene sets
Next, we searched for HD clinical onset–relevant cell types in the brain using cortical single cell sequencing data. This was performed with RolyPoly (30), which uses a regression-based polygenic model to determine which cell types in particular tissue are the most relevant to a trait of interest. These analyses detected a significant relationship with brain microvascular endothelial cells (BMECs) and disease onset (P < 0.05, Fig. 4). BMECs constitute the major structural components of the blood–brain barrier, and recent studies of iPSC-derived versions of this cell type have shown that HD BMECs display both angiogenic and blood–brain barrier impairments in HD patients compared with control lines (31). An analysis of the prioritized transcriptomic HD modifier genes in this published gene expression data set (31) revealed that the two colocalized and replicated chromosome 7p11 genes were dysregulated in BMECs derived from HD patients (i.e. CHCHD2 P = 4.4 × 10−9, fold change −1.9 and SUMF2 P = 0.0016, fold change 1.1). We also chose to assess the prioritized genes at the protein level using patient-derived BMEC cell lines. In these analyses, MSH3, RRM2B and EIF2AK1 were dysregulated in our HD BMEC cell lines (Supplementary Material, Fig. S4 and Supplementary Material, Table S4), indicating the potential importance of these proteins in this cell type in HD.
Figure 4.
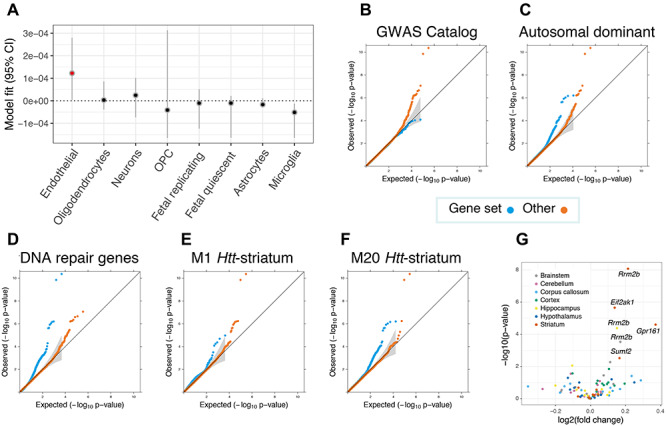
Enrichment analyses revealed brain cell types, striatal coexpression modules and gene sets that are most relevant for modifying the clinical onset of Huntington disease (HD). (A) Inferring relevant brain cell types from the GWAS data supplemented with cortical single cell sequencing information suggests that brain microvascular endothelial cells (annotated in red) could be relevant for the trait (P < 0.05). Cell types are ranked according to parameter estimates. Representative Q–Q plots for significantly enriched or depleted S-PrediXcan modifier associations for genes belonging to various gene sets (B–D), as well as coexpression networks that are related to murine Htt-CAG size over time (E and F). M20Htt-striatum genes are involved in p53 signaling and Brca1 in DNA damage response, whereas M1Htt-striatum genes are implicated in protein ubiquitination, tRNA charging and HD signaling. (G) Volcano plot for prioritized transcriptomic modifier genes in in Q175 mice versus wild type at 6 months of age for six brain-related tissues. Significantly dysregulated genes after correcting for multiple testing corrected are annotated, with all genes differentially expressed in the brain were up-regulated in mice with expanded Htt CAGs.
In an attempt to identify groups of genes that are important for modifying HD clinical onset, we evaluated 17 diverse gene sets for enrichment with regard to TWAS associations (Fig. 4 and Supplementary Material, Table S5), based on their mean S-PrediXcan Z2-scores. There was a significant underrepresentation of GWAS catalogue genes, which reflects the fact that the majority of GWAS conducted to date focused on complex diseases and that clinical modifiers of a Mendelian disease represents a unique phenotype. Disease-related genes with annotations in ClinVar (32) were significantly enriched for HD-related associations, particularly those causing autosomal dominant disorders and ones that result from haploinsufficiency. Autosomal recessive genes, however, were not significantly enriched for HD clinical onset associations. These findings indicate that more subtle changes in expression of genes that are intolerant to loss-of-function variants may play an important role in modulating clinical age of onset in HD. Associations for DNA repair genes were also significantly enriched, adding confirmatory evidence for somatic instability as a modifier for HD clinical onset (7,23). Of potential translational relevance, drug target genes (33) were also prioritized in the analyses. These gene set analyses therefore provide an avenue to identify the biological mechanisms that underlie the differences in age of onset observed between HD patients.
Gene coexpression modules that display a positive correlation to huntingtin–CAG length are enriched for HD genomic modifier associations
In order to determine whether groups of genes whose expression is mediated by mutant huntingtin were enriched in the HD modifier association results (mean S-PrediXcan Z2-scores from the TWAS), we analyzed transcriptomic coexpression networks from a large time course experiment from HD knock-in mice with increasing Htt CAG lengths (17). An analysis of the 13 striatal and 5 cortical repeat length−dependent modules from that study (17) revealed that two striatal modules, M1Htt-striatum and M20Htt-striatum, were significantly enriched for S-PrediXcan TWAS associations (Supplementary Material, Table S6). Both of these are positively correlated with Htt CAG length. Interestingly, M20Htt-striatum displayed the most significant association of these up-regulated modules in the original study (17).
Analyses of gene expression profiles in the aging human brain identify HD-relevant coexpression modules and overlapping phenotypes
Next, we assessed previously identified gene coexpression modules (n = 47) detected in the human cortex from a longitudinal aging cohort, ROSMAP (34), for enrichment in the HD clinical onset TWAS results (mean S-PrediXcan Z2-scores). This was performed in order to identify groups of coexpressed cortical genes that could be particularly important for modulating HD onset. Fifteen modules showed significant differences in mean Z2-scores between gene sets belonging to the cortical modules, with seven modules displaying a significant enrichment for HD-related phenotype associations compared with other genes (Fig. 5 and Supplementary Material, Tables S7 and S8). Notably, the top m109human-cortex module, that has been associated with Alzheimer disease and cognitive decline (34), was statistically underrepresented in HD-related associations (P = 1.1 × 10−11). Conversely, the mitochondrial module, m131human-cortex, which was also implicated in cognition in that previous study, had significantly enriched HD-related scores when compared with other genes (P = 4.1 × 10−6).
Figure 5.
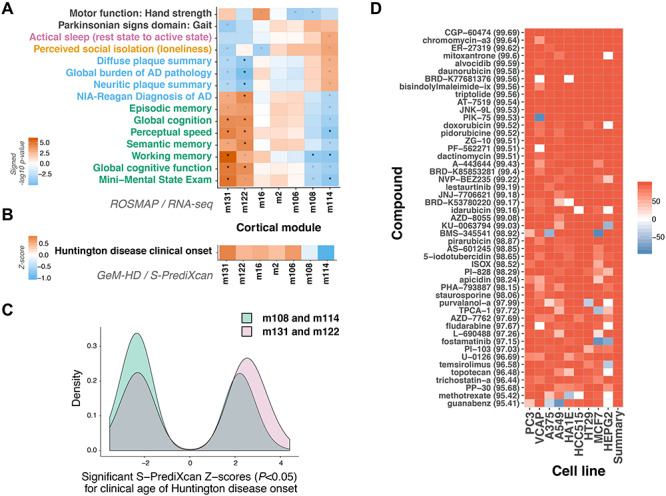
Expression of a subset of cortical gene coexpression modules influences the clinical onset of Huntington disease (HD) as well as related phenotypes. (A) Cortical coexpression modules that show enrichments for TWAS results for HD clinical age of onset (n = 7) are also associated with relevant phenotypes in an independent longitudinal aging cohort. Increased expression of mitochondrial-related modules m122 and m131 is associated with improved performance for cognitive-related traits, whereas the opposite effect was observed for m108 (cholesterol and hedgehog signaling-related) and m114 (cadherin binding-related) expression. Within-category Bonferroni significant traits are displayed, with asterisks denoting significant associations after Bonferroni correction for multiple testing. Trait categories are annotated (black, motor and gait; pink, sleep and circadian rhythms; orange, lifestyle/personality; blue, pathology; green, cognitive decline). Black asterisks: significant after correcting for all comparisons, grey asterisks: significant after adjusting for within-trait comparisons. (B) A similar effect pattern was observed for these modules with regard to their influence on age of clinical onset in HD. When assessing significant S-PrediXcan Z-scores (i.e. P < 0.05), on average, increased expression of genes belonging to modules m108 and m114 is more likely to lead to an earlier age of HD onset, whereas the opposite pattern (i.e. increased expression and later onset) is observed for the other modules. (C) Collectively, significant HD clinical onset S-PrediXcan Z-scores for the top phenotype-related coexpression cortical modules also display differences in distributions. There were significantly more negative Z-scores detected for genes belonging to modules m108 and m114 compared with m131 and m122 (P = 2.0 × 10−6). (D) A screen of over 27 000 perturbagens in nine cell lines identified highly similar (>95%) perturbagens in the Broad Connectivity Map database (median Connectivity Map/tau scores indicated in parentheses). Similar compounds, with potential use for drug repurposing in HD, are shown.
The ROSMAP study longitudinally also collected various phenotypic measures for participants. This allowed us to assess which of these traits the seven HD onset–related coexpression modules are associated with in aging individuals in the ROSMAP cohort, to inform what other biological processes these groups of genes are potentially important for. In these analyses, increased expression of m122human-cortex and m131human-cortex was associated with improved performance for cognitive-related traits, whereas the opposite effect was observed for m108human-cortex and m114human-cortex expression (Fig. 5). Beneficial cortical modules were enriched for mitochondrial (m106, m122 and m131) and neuronal/synaptic (m16) gene ontology terms, whereas those deleterious ones were involved in cholesterol and hedgehog signaling pathways (m108), as well as cadherin binding (m114) (34).
Subsequently, we wanted to investigate the effect of increased expression of the genes from HD-enriched cortical modules from the ROSMAP analyses on clinical onset. Therefore, to investigate whether a consistent direction of effect was observed in HD patients, we assessed the mean direction of significant Z-scores for these modules in the HD clinical onset TWAS results. Mirroring the ROSMAP effects, increased expression of m108human-cortex and m114 human-cortex was correlated with an earlier age of HD onset, whereas increased expression of m122human-cortex and m131human-cortex correlated with later age of HD onset (Fig. 5). Although only passing within-trait correction for multiple testing, these modules were also associated with other phenotypes relating to HD (e.g. m131human-cortex and gait disturbances P = 1.20 × 10−4). In fact, m131human-cortex is the most significantly associated module with regard to gait out of all 47 ROSMAP cortical coexpression modules. We also studied whether expression patterns for the individual, prioritized HD transcriptomic modifier genes were associated with available phenotypes in the aging cohort. These analyses revealed that MTMR10 and PMS2 were associated with relevant traits after correcting for multiple testing, with greatest statistical significance being achieved for working memory and global cognitive function decline (Supplementary Material, Fig. S5 and Supplementary Material, Table S9).
Perturbagens that give rise to gene expression profiles that are similar to beneficial TWAS signatures can be prioritized for drug repurposing. Once again, in these analyses, we selected genes from the seven HD-enriched cortical ROSMAP modules that displayed significant (P < 0.05) brain-derived TWAS associations (n = 77). We performed signature matching of these TWAS signals in the CMAP Database using their Query Tool (https://clue.io/) (35). The CMAP database contains gene expression signatures for over 27 000 perturbagens (i.e. compounds, as well as genetic knockdown and overexpression). In this regard, signature matching was used to identify highly similar (connectivity scores >95%) and dissimilar (connectivity scores <95%) perturbagens (Supplementary Material, Table S10 and Fig. 5). The gene sets used in these analyses included 3 of the 15 prioritized TWAS genes (i.e. m122 human-cortex PMS2 and EIF2AK1, as well as m2 human-cortex SUMF2). In these analyses, top perturbational classes displaying similar signatures included inhibitors of topoisomerase, PI3K, MTOR, CDK and FLT3. Conversely, the most dissimilar compound signature was obtained for kinetin riboside (−99.23). Top gene perturbagens with similar signatures included knockdown of the mitochondrial transporter, SLC25A28, as well as the NF-kappa-B inhibitor beta gene, NFKBIB.
Discussion
Genomic approaches in HD can assist in understanding differences in clinical onset observed between patients, as well as aid in identifying novel biomarkers and therapeutic strategies to improve disease management. To our knowledge, the current study provides the most extensive TWAS of the modification of the age onset in a Mendelian disease to date and has generated a number of novel candidate regions, as well as neurobiological pathways, for future study.
These analyses provide additional support for the role of DNA repair in disease onset, pointing toward somatic expansion of CAG tracts as one of the potential mechanisms behind this trait. By employing an integrated omic approach, we present evidence for another mismatch repair gene, PMS2, being involved in modifying the clinical onset of HD. Additionally, we show genes dysregulated in HD are enriched for disease modification signals, along with other important disease- and drug-related gene sets. Further, we identified cortical gene coexpression modules that are associated with cognitive decline as well as HD clinical onset that can also be targeted in future studies with important implications for aging populations. These findings indicate the powerful capabilities of indirectly assessing gene expression through TWAS-driven incorporation of large-scale transcriptomic information into the study of genomic modifiers of disease.
Assisted by colocalization-based post-filtering, our TWAS analyses identified a number of novel modifier regions. Colocalized genes were more likely to be replicated (Table 1), forming a core list of candidates for future study. The top candidates include the four genes that colocalized in brain tissues (i.e. GPR161, PMS2, SUMF2 and FAN1) and showed consistent evidence for up-regulation being associated with delayed onset in cortical tissues. Further, regions where more than one colocalized gene is located, and coregulation is therefore suspected, would benefit from interrogation with functional experiments such as targeted genome editing in future studies. This is important since the current study does not provide direct causal evidence for these genes with regard to HD onset.
The potential role of G-protein–coupled receptors (GPCRs) in modifying HD clinical onset provides an additional interesting line for further research. GPR161 can potentially be linked to DNA repair–related processes since conditional Gpr161 knockout mice have been shown to exhibit increased DNA damage (27). With regard to GPR151, biobank-scale genomic studies have revealed that protein-truncating variants are protective against obesity (36) and metabolic dysfunction has been observed in HD (2). Furthermore, functional characterization of neuronal Gpr151 suggested that the gene is important in the paraventricular nucleus and could therefore be potentially implicated in food- and drug-seeking behaviors (37). Future studies should therefore aim at delineating the potential role of these two orphan GPCRs with regard to, in relation to modifying age of onset of the disorder.
The TWAS also detected a number of associations with genes involved in ATP- and mitochondrial-related processes, an important finding since mitochondrial impairment has been suggested to contribute to the complex pathogenesis of HD (3,19). Although previous studies in isogenic HD cell lines have provided evidence for the dysregulation of the mitochondrial-related neurodegenerative disorder risk gene, CHCHD2 (38), our study provides the first evidence for the gene being involved with clinical onset in HD patients. The other TWAS gene previously implicated in neurodegenerative disorders is EIF2AK1 (12), although the HD signal at this locus may primarily be driven by the expression of the mismatch repair gene, PMS2. However, immunoblotting of EIF2AK1 in patient brains and BMECs indicated that this protein could be dysregulated in disease. Further, aberrant mRNA translation initiation and elongation have been implicated in neurodegeneration and altered in nucleotide repeat expansion disorders (39). The relevance of EIF2AK1 therefore cannot be excluded.
A subset of the transcriptomic modifier genes was orthogonally validated at the protein level by evaluating cortical and caudate nucleus lysates from HD patients compared with matched controls. Although not a direct comparison with regard to the age of clinical onset phenotype, we hypothesized that relevant modifier genes may also be dysregulated in the disease state. Our findings that CAG-dependent coexpression modules are enriched in TWAS associations further support this assumption. These HD clinical onset associated murine modules can be prioritized for future study to improve our biological understanding of the trait and to potentially develop therapeutic modifiers.
In addition to the Htt CAG coexpression module and gene set analyses, examination of human cortical coexpression information was highly informative, indicating the mechanistic insights that can be uncovered through TWAS-driven approaches. Notably, the enriched HD-associated ROSMAP cortical modules showed shifts in their HD onset association distributions that were correlated with their phenotypic effects in the aging individuals. The beneficial cortical modules identified here may have broader implications beyond HD, since they may be involved in processes pertaining to general brain health. The GeM-HD GWAS showed that there is a correlation between motor onset and cognitive impairment and psychiatric symptoms and that the main HD genetic modifiers are also associated with one or more of these traits (8). Cognitive impairments present a significant burden to the quality of life of HD patients and are often observed prior to the onset of motor symptoms (40). Our findings begin to provide further insights into the neurodegenerative processes that lead to cognitive deficits in HD and how they are distinct from Alzheimer disease. However, although ROSMAP participants had no dementia at enrolment, concomitant Alzheimer disease may have partially influenced these results.
Signature matching of gene expression profiles to large databases of perturbagens is a novel way to repurpose drugs to treat different diseases (41). Employing this approach, informed by genetic data generated from numerous HD patients, yielded a number of leads for future research. These include chromomycin, which has been proposed as a potential therapeutic target for neurological disorders caused by repeat expansions (42), as well as mitoxantrone and guanabenz, which have been implicated as potential medications for multiple sclerosis (43,44). Additionally, anthracyclines have been shown to correct gene expression perturbations in HD mice (45), whereas BMS-345541 (46) and the histone deacetylase inhibitor, trichostatin-a (47), have been shown to restore HD-related deficits in vitro. Further, knockdown with antisense oligonucleotides of genes that yielded similar expression profiles could also be explored in the future in model systems. Conversely, kinetin riboside, an apoptosis inducer that has been shown to cause ATP depletion and genotoxicity (48), displayed the most dissimilar gene expression signature when compared with the beneficial HD modifier TWAS signals. ATP deficits are a hallmark of HD, and, whereas the precursor of kinetin riboside, kinetin (also known as N6-furfuryladenine), has recently been proposed as a novel therapeutic molecule for HD (49,50), kinetin riboside may cause deleterious gene expression changes through alterations in biological feedback loops leading to DNA repair processes.
The current study is not without limitations. TWAS approaches are useful for identifying trait-related genomic regions of interest but should not be viewed as providing causal evidence with regard to highlighted genes (51). Therefore, future mechanistic studies are therefore warranted for the top HD modifier genes. These include the novel modifier genes with the most supporting evidence, such as GPR161, due to colocalization of signals in the cortex; PMS2, since the gene is a key member of the mismatch repair system, and the neurological disorder risk genes, CHCHD2 and EIF2AK1. Additionally, although agnostic approaches to tissue selection have been recommended for TWAS (20), the sample size of the GTEx brain tissues is relatively limited, and future HD genomic modifier studies would benefit from the inclusion of additional striatal and cortical samples. A recent, more targeted HD modifier study compared with the current investigation, eQTL information from the CommonMind Consortium only provided additional evidence for the FAN1 gene (52). Although the HD patients in the replication study were extensively phenotyped for disease progression (7), the sample size of this cohort also limited our power to confirm associations. Finally, the applicability of TWAS is inherently limited with regard to the importance of steady-state gene expression on the phenotype being studied. Our gene expression–informed analyses identified a number of novel findings, but further in-depth analyses of the role of deleterious coding variation are also required.
HD is characterized by a spectrum of symptoms that include motor, psychiatric and cognitive impairments (53). More work is required to elucidate whether any of the individual genomic HD modifier genes alter clinical severity and progression, in addition to age of clinical onset. Previous research has shown that the FAN1 and RRM2B HD modifiers already act in the prediagnosis phase and that they are associated with distinct endophenotypes at this stage (54). Future studies should therefore determine whether there is a pleiotropic effect between variants and genes that influence motor onset, and the severity of psychiatric and cognitive symptoms.
In conclusion, by jointly modeling HD clinical onset GWAS summary statistics with population-scale tissue-specific eQTL information, we expanded the number of candidate modifier genes and gained increased insights into HD disease mechanisms. Transcriptomic imputation can therefore be seen as a complementary approach to GWAS, as it can reveal novel information not previously detected with traditional methodologies. Significant progress can be made if these findings help inform clinical translation by paving the way forward for comprehensive drug repurposing. Further, expanding future TWAS to incorporate modifiers of other neurodegenerative disorders will aid in identifying common pathways across these diseases, as well to ascertain those that are unique to each disorder.
Materials and Methods
TWAS and Bayesian colocalization analyses
S-PrediXcan (20) was used to perform a TWAS by integrating gene expression prediction models (http://predictdb.org/) generated from the Genotype–Tissue Expression (GTEx) Consortium (v7 release) (23) with summary statistics from two GWAS of the genetic modifiers of HD. These models were built using eQTL information from 48 tissues, which includes 13 brain regions. GWAS data from 4082 European HD patients (CAG ranges: 40–55) generated by the Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium (8) was used in the discovery analysis. Analyses were restricted to protein coding genes, and a false discovery rate (FDR) of 0.1 for each tissue was used to determine statistical significance. Since eQTL data can display correlation across tissues (23), we selected this threshold in an attempt to balance minimizing detecting false-positive associations, while maximizing discovery power. We also further assessed prioritized genes in dorsolateral prefrontal cortex TWAS models from two large consortia, the CommonMind Consortium (n = 538) (55) and ROSMAP (n = 411) (34).
To validate prioritized genes further, filtering was performed employing Bayesian colocalization analyses with COLOC (56). Non-ambiguous variants in the GeM-HD GWAS data were interrogated for colocalization in relation to the eQTL association results from the GTEx Consortium (23) as well as eQTLs from a large meta-analysis of brain region samples from GTEx, CommonMind and ROSMAP (n = 1194) (24). COLOC produces posterior probabilities for four hypotheses: H0, no causal variant; H1, causal variant for trait 1 only; H2, causal variant for trait 2 only; H3, two distinct causal variants; H4, one common causal variant. In order to be defined as colocalized in a tissue, regions were required to show strong evidence for one common causal variant (i.e. PP4 > 0.5), without substantial evidence for the PrediXcan associations occurring as a result of linkage disequilibrium contamination.
Regions that were significantly associated in the discovery analysis and were validated through Bayesian colocalization analyses were further examined for replication. We assessed replication in S-PrediXcan gene–level associations generated using GWAS of disease progression in 216 European HD patients recruited by the TRACK-HD Consortium (7). We considered a gene to have suggestive evidence for replication at a nominal P < 0.05 when the association effect was in the same direction in significant discovery and replication tissues.
The biomaRt package in R was used to access Ensembl database (grch37.ensembl.org) to obtain relevant genomic coordinates of genes of interest as well as mouse gene homologues. Networks of physical and other interactions based on diverse genomic and proteomic data with gene sets of interest were visualized using GeneMANIA (57).
Orthogonal validation of prioritized transcriptomic modifier genes using data from HD allelic hPSCs, HD knock-in mice and human HD patient brains
Gene expression of prioritized genes was assessed using RNA-seq data from isogenic allelic hPSC lines for four different HTT CAG lengths (CAG lengths: 30, 45, 65, 81; n = 4 per length) that had been differentiated into five cell line types: pluripotent stem cells, hepatocytes, neural progenitor cells, post-mitotic neurons and skeletal muscle cells. Further details are described in Ooi et al. (58). The expression for each gene (transcripts per kilobase million) and cell line combination was assessed as a continuous trait with linear regression for different CAG lengths.
HD knock-in mouse RNA-seq data from Langfelder et al. (17) were used to determine whether prioritized transcriptomic HD modifier gene was differentially expressed in Q175 HD mice (n = 8) versus Q20 wild-type mice (n = 8) at 6 months for 12 tissues, including 6 brain-related regions.
Protein levels for a subset of the prioritized genes (i.e. CHCHD2, EIF2AK1, FAN1, GPR161, MSH3, MTMR10, PMS2, RRM2B and SUMF2) were assessed using western blot analysis in HD patient brains (n = 10) compared with age-matched controls (n = 9) in both the striatum (caudate nucleus) and cortex. These samples were obtained from de-identified archived brain tissue samples from the Huntington Disease Biobank at the University of British Columbia. All samples were collected, stored and accessed with informed consent and approval of the University of British Columbia/Children’s and Women’s Health Centre of British Columbia Research Ethics Board (UBC C&W REB H06-70467 and H06-70410).
For immunoblotting, a small piece (2 mm × 2 mm) of frozen cortex or caudate nucleus was cut using a clean razor blade on dry ice. Each brain section was lysed, and 75 μg of total protein was resolved on a 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel. Proteins were transferred to 0.2 μm nitrocellulose membranes that were blotted for GPR161 (1:250, Abcam, ab 83007), EIF2AK1 (1:500, Abcam, ab154076), PMS2 (1:500, Abcam, ab110638), MTMR10 (1:250, Abcam, ab179756), CHCHD2 (1:500, Abcam, ab220688), MSH3 (1:250, Abcam, ab154486), FAN1 (1:1000, Abcam, ab185554), SUMF2 (1:250, Thermo Fisher Scientific, PA5-54961), RRM2B (p53R2, 1:1000, Abcam, ab154194) and actin (1:5000, Sigma-Aldrich, A2228). Proteins were detected with IR dye 800CW goat anti-mouse-labeled (1:250, Rockland 610-131-007, Gilbertsville, PA) and AlexaFluor 680 goat anti-rabbit (1:250, Molecular Probes A21076, Eugene, OR)-labeled secondary antibodies and the LiCor Odyssey Infrared Imaging system.
Assessing important cell types for HD clinical onset using single cell sequencing information
Relevant cell types for modifying HD clinical onset were inferred using RolyPoly (30), which employs a regression-based polygenic model to detect enrichment of GWAS signal based on gene expression information for different cell populations. Developer-recommended criteria were implemented using publically available single cell sequencing data from human cortex (59). These data had been clustered into six major brain cell types: astrocytes, oligodendrocytes, oligodendrocyte precursor cells, neurons, microglia and vascular cells, as well as replicating and quiescent fetal neuronal populations (59). Significant differences in expression between HD patients and controls for prioritized transcriptomic HD modifier genes in BMECs were assessed using RNA-seq information from a previous study, as described by the authors (31).
Brain microvascular endothelial cell differentiation
GM09197 HD, C1- and C2-induced hPSC lines were generated as described (38). GM03621 HD fibroblasts were obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research. Induced hPSCs from this line were generated using an episomal reprogramming strategy as described (60) and characterized for pluripotency and karyotype. Differentiation to BMECs was performed using a protocol modified from Lippmann et al. (61) Briefly, iPSCs were grown in mTeSR1 (STEMCELL Technology cat# 85850) and were passaged as aggregates onto Matrigel-coated plates (Corning cat# 354230). Cells were grown to 40% confluency before changing media to DMEM/F12 (Gibco cat# 11320033) supplemented with 20% KnockOut serum replacement (Gibco cat # 10828028), 1× MEM Non-Essential Amino Acids Solution (Gibco cat# 11140050), 1× GlutaMAX-I (Gibco cat # 35050061) and 0.1 mm beta-mercaptoethanol. DMEM/F12 complete media were changed daily for 5 days. On day 6, media were changed to human endothelial media (Gibco cat# 11111044) supplemented with 20 ng/ml bFGF (ProSpec cat # CYT-218) and 1% platelet poor-derived serum (Sigma cat# P2918). Endothelial media were changed daily for 2 days. On day 8, cells were dissociated with Accutase and seeded at 25000 cells/cm2 on collagen IV-coated (400 μg/ml; Sigma C5533) and fibronectin (100 μg/ml; Sigma F4759)-coated plates. Endothelial media were changed daily for 4 days, and confluent cells were harvested for immunoblotting.
Enrichment of transcriptomic gene–level association results for HD onset in gene sets
We performed TWAS association enrichment analyses (mean S-PrediXcan Z2-score for all gene–tissue pairs, as previously described) (20) with regard to diverse gene sets in the GeM-HD cohort. Bonferroni correction for multiple testing was applied to determine statistically significant enrichment of gene sets. Seventeen gene sets (https://github.com/macarthur-lab/gene_lists; downloaded November 2017, n = 17) from a repository, which includes annotations for genes grouped according to biological function, involvement in specific inheritance models of diseases and known drug targets (complete descriptions can be found in Supplementary Material, Table S4), were assessed. Further, gene sets belonging to coexpression modules identified through RNA-seq analysis in brain regions most relevant to HD (i.e. cortex and striatum) (3) were also assessed for enrichment in the TWAS data. These gene sets included striatal and cortical coexpression modules that are mediated by murine Htt CAG length and age (six CAG lengths and three time points, n = 144 mice per tissue) (17) and coexpression modules identified in the human cortex (n = 478) (34).
Relevance of genetic modifiers of HD age of onset to related traits in the aging brain
The human cortical samples (n = 478) used to identify coexpression modules relevant to modifying HD clinical onset were collected from two prospective cohorts (ROSMAP), which extensively characterized the study participants for cognitive measures for up to 20 years and performed pathological characterization postmortem (https://www.radc.rush.edu/documentation.htm) (34). This allowed for the opportunity to determine the relevance of the genetic HD modifiers to related phenotypes observed in the aging brain. Cortical coexpression modules (n = 47) that displayed significant enrichments for S-PrediXcan associations with HD clinical age of onset were examined with regard to clinical phenotypes in an independent longitudinal aging cohort. In these analyses, the expression of significantly enriched HD modifier modules (n = 7) was assessed for association with clinical and pathological traits (n = 83), including those relevant to HD, in the ROSMAP cohort. Modules that displayed significant associations with at least one ROSMAP trait were examined with regard to the direction of significant Z-scores (P < 0.05) in the HD clinical age of onset TWAS. Cortically expressed prioritized HD transcriptomic modifier genes, as well as MSH3 and HTT, were also assessed with regard to associations with these ROSMAP phenotypes. Individual gene expression levels were only available for a subset of the phenotypes present in the ROSMAP cohort examined in Mostafavi et al. (34) (n = 13), thus gene-level analyses were performed in this subset of traits.
Association between each module and each trait was conducted by first representing each module by the mean gene expression level of the module’s gene members (applied to standardized gene expression data), and then using Spearman correlation to associate module means with observed phenotypic traits. Bonferroni correction for multiple testing was used to determine statistical significance with traits and genes (significance thresholds: module–trait analyses P < 8.6 × 10−5 and gene–multi-trait analyses P < 3.2 × 10−4; nominal significance for gene–individual–trait analyses P < 0.004).
Perturbagen signature matching for potential drug repurposing in HD
HD modifier TWAS signals matched to gene expression profiles from over 27 000 perturbagens in nine cell lines in Connectivity Map (CMap) database (35) (https://clue.io/). TWAS Z-scores were used as a proxy for gene expression (i.e. positive Z-scores as up-regulated genes and negative Z-scores as down-regulated ones), with perturbagens giving rise to highly similar median CMap connectivity score profiles (i.e. >95%) being considered potential targets for drug repurposing. These analyses were restricted to genes that fulfilled all of the following criteria: (i) members of one of the seven cortical modules from ROSMAP that displayed enrichment for HD onset associations, (ii) displaying at least suggestive TWAS significance (P < 0.05) in at least one GTEx brain tissue and (iii) the direction of these TWAS Z-scores should match the direction of the predicted mean influence on HD onset of the coexpression module (i.e. positive TWAS Z-scores for m2, m16, m106, m122 and m131, and negative TWAS Z-scores for m108 and m114).
All downstream statistical analyses were conducted using R version 3.4.4 software (The R Foundation).
Supplementary Material
Acknowledgements
We would like to acknowledge the GeM-HD, TRACK-HD, ROSMAP and CommonMind consortia for providing access to their summary statistic information. The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of Interest statement. The authors declare no conflicts interests.
Contributor Information
Galen E B Wright, Centre for Molecular Medicine and Therapeutics, Vancouver, British Columbia V5Z 4H4, Canada; Department of Medical Genetics, University of British Columbia, Vancouver, British Columbia V6H 3N1, Canada; BC Children’s Hospital Research Institute, Vancouver, British Columbia V5Z 4H4, Canada.
Nicholas S Caron, Centre for Molecular Medicine and Therapeutics, Vancouver, British Columbia V5Z 4H4, Canada; Department of Medical Genetics, University of British Columbia, Vancouver, British Columbia V6H 3N1, Canada; BC Children’s Hospital Research Institute, Vancouver, British Columbia V5Z 4H4, Canada.
Bernard Ng, Centre for Molecular Medicine and Therapeutics, Vancouver, British Columbia V5Z 4H4, Canada; Department of Medical Genetics, University of British Columbia, Vancouver, British Columbia V6H 3N1, Canada; Department of Statistics, University of British Columbia, Vancouver, British Columbia V6T 1Z4, Canada.
Lorenzo Casal, Centre for Molecular Medicine and Therapeutics, Vancouver, British Columbia V5Z 4H4, Canada; Department of Medical Genetics, University of British Columbia, Vancouver, British Columbia V6H 3N1, Canada; BC Children’s Hospital Research Institute, Vancouver, British Columbia V5Z 4H4, Canada.
William Casazza, Centre for Molecular Medicine and Therapeutics, Vancouver, British Columbia V5Z 4H4, Canada; Department of Medical Genetics, University of British Columbia, Vancouver, British Columbia V6H 3N1, Canada; Department of Statistics, University of British Columbia, Vancouver, British Columbia V6T 1Z4, Canada.
Xiaohong Xu, Translational Laboratory in Genetic Medicine (TLGM), Agency for Science, Technology and Research (A*STAR), Singapore 138648, Singapore.
Jolene Ooi, Translational Laboratory in Genetic Medicine (TLGM), Agency for Science, Technology and Research (A*STAR), Singapore 138648, Singapore.
Mahmoud A Pouladi, Translational Laboratory in Genetic Medicine (TLGM), Agency for Science, Technology and Research (A*STAR), Singapore 138648, Singapore; Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117597, Singapore; Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117597, Singapore.
Sara Mostafavi, Centre for Molecular Medicine and Therapeutics, Vancouver, British Columbia V5Z 4H4, Canada; Department of Medical Genetics, University of British Columbia, Vancouver, British Columbia V6H 3N1, Canada; Department of Statistics, University of British Columbia, Vancouver, British Columbia V6T 1Z4, Canada.
Colin J D Ross, BC Children’s Hospital Research Institute, Vancouver, British Columbia V5Z 4H4, Canada; Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117597, Singapore; Faculty of Pharmaceutical Sciences, University of British Columbia, Vancouver, British Columbia V6T 1Z3, Canada.
Michael R Hayden, Centre for Molecular Medicine and Therapeutics, Vancouver, British Columbia V5Z 4H4, Canada; Department of Medical Genetics, University of British Columbia, Vancouver, British Columbia V6H 3N1, Canada; BC Children’s Hospital Research Institute, Vancouver, British Columbia V5Z 4H4, Canada.
Funding
Canadian Institutes of Health Research (Foundation Grant ERT 155723 to M.R.H.).
References
- 1. Harper A.R., Nayee S. and Topol E.J. (2015) Protective alleles and modifier variants in human health and disease. Nat. Rev. Genet., 16, 689–701. [DOI] [PubMed] [Google Scholar]
- 2. Caron N.S., Wright G.E.B. and Hayden M.R. (2020) In Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K. and Amemiya A. (eds), GeneReviews®. The National Center for Biotechnology Information, Seattle, WA. [Google Scholar]
- 3. Bates G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R., Nance M., Ross C.A., Scahill R.I., Wetzel R. et al. (2015) Huntington disease. Nat. Rev. Dis. Primers, 1, 15005. [DOI] [PubMed] [Google Scholar]
- 4. Keum J.W., Shin A., Gillis T., Mysore J.S., Abu Elneel K., Lucente D., Hadzi T., Holmans P., Jones L., Orth M. et al. (2016) The HTT CAG-expansion mutation determines age at death but not disease duration in Huntington disease. Am. J. Hum. Genet., 98, 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gusella J.F. and MacDonald M.E. (2009) Huntington's disease: the case for genetic modifiers. Genome Med., 1, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rosenblatt A., Brinkman R.R., Liang K.Y., Almqvist E.W., Margolis R.L., Huang C.Y., Sherr M., Franz M.L., Abbott M.H., Hayden M.R. et al. (2001) Familial influence on age of onset among siblings with Huntington disease. Am. J. Med. Genet., 105, 399–403. [PubMed] [Google Scholar]
- 7. Hensman Moss D.J., Pardinas A.F., Langbehn D., Lo K., Leavitt B.R., Roos R., Durr A., Mead S., TRACK-HD investigators, REGISTRY investigators et al. (2017) Identification of genetic variants associated with Huntington's disease progression: a genome-wide association study. Lancet Neurol., 16, 701–711. [DOI] [PubMed] [Google Scholar]
- 8. Genetic Modifiers of Huntington's Disease Consortium (2015) Identification of genetic factors that modify clinical onset of Huntington's disease. Cell, 162, 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Becanovic K., Norremolle A., Neal S.J., Kay C., Collins J.A., Arenillas D., Lilja T., Gaudenzi G., Manoharan S., Doty C.N. et al. (2015) A SNP in the HTT promoter alters NF-kappaB binding and is a bidirectional genetic modifier of Huntington disease. Nat. Neurosci., 18, 807–816. [DOI] [PubMed] [Google Scholar]
- 10. Wright G.E.B., Collins J.A., Kay C., McDonald C., Dolzhenko E., Xia Q., Becanovic K., Drogemoller B.I., Semaka A., Nguyen C.M. et al. (2019) Length of uninterrupted CAG, independent of polyglutamine size, results in increased somatic instability, hastening onset of Huntington disease. Am. J. Hum. Genet., 104, 1116–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Genetic Modifiers of Huntington's Disease Consortium (2019) CAG repeat not Polyglutamine length determines timing of Huntington's disease onset. Cell, 178, 887–900 e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sadovnick A.D., Traboulsee A.L., Zhao Y., Bernales C.Q., Encarnacion M., Ross J.P., Yee I.M., Criscuoli M.G. and Vilarino-Guell C. (2017) Genetic modifiers of multiple sclerosis progression, severity and onset. Clin. Immunol., 180, 100–105. [DOI] [PubMed] [Google Scholar]
- 13. Blitterswijk M., Mullen B., Nicholson A.M., Bieniek K.F., Heckman M.G., Baker M.C., DeJesus-Hernandez M., Finch N.A., Brown P.H., Murray M.E. et al. (2014) TMEM106B protects C9ORF72 expansion carriers against frontotemporal dementia. Acta Neuropathol., 127, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deming Y., Li Z., Kapoor M., Harari O., Del-Aguila J.L., Black K., Carrell D., Cai Y., Fernandez M.V., Budde J. et al. (2017) Genome-wide association study identifies four novel loci associated with Alzheimer's endophenotypes and disease modifiers. Acta Neuropathol., 133, 839–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bettencourt C., Hensman-Moss D., Flower M., Wiethoff S., Brice A., Goizet C., Stevanin G., Koutsis G., Karadima G., Panas M. et al. (2016) DNA repair pathways underlie a common genetic mechanism modulating onset in polyglutamine diseases. Ann. Neurol., 79, 983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hauberg M.E., Zhang W., Giambartolomei C., Franzen O., Morris D.L., Vyse T.J., Ruusalepp A., CommonMind C., Sklar P., Schadt E.E. et al. (2017) Large-scale identification of common trait and disease variants affecting gene expression. Am. J. Hum. Genet., 100, 885–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Langfelder P., Cantle J.P., Chatzopoulou D., Wang N., Gao F., Al-Ramahi I., Lu X.H., Ramos E.M., El-Zein K., Zhao Y. et al. (2016) Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat. Neurosci., 19, 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ament S.A., Pearl J.R., Cantle J.P., Bragg R.M., Skene P.J., Coffey S.R., Bergey D.E., Wheeler V.C., MacDonald M.E., Baliga N.S. et al. (2018) Transcriptional regulatory networks underlying gene expression changes in Huntington's disease. Mol. Syst. Biol., 14, e7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saudou F. and Humbert S. (2016) The biology of huntingtin. Neuron, 89, 910–926. [DOI] [PubMed] [Google Scholar]
- 20. Barbeira A.N., Dickinson S.P., Bonazzola R., Zheng J., Wheeler H.E., Torres J.M., Torstenson E.S., Shah K.P., Garcia T., Edwards T.L. et al. (2018) Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun., 9, 1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W., Jansen R., Geus E.J., Boomsma D.I., Wright F.A. et al. (2016) Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet., 48, 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M. et al. (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet., 48, 481–487. [DOI] [PubMed] [Google Scholar]
- 23. GTEx Consortium (2017) Genetic effects on gene expression across human tissues. Nature, 550, 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qi T., Wu Y., Zeng J., Zhang F., Xue A., Jiang L., Zhu Z., Kemper K., Yengo L., Zheng Z. et al. (2018) Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat. Commun., 9, 2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Broeke S.W., Brohet R.M., Tops C.M., Klift H.M., Velthuizen M.E., Bernstein I., Capella Munar G., Gomez Garcia E., Hoogerbrugge N., Letteboer T.G. et al. (2015) Lynch syndrome caused by germline PMS2 mutations: delineating the cancer risk. J. Clin. Oncol., 33, 319–325. [DOI] [PubMed] [Google Scholar]
- 26. Ogaki K., Koga S., Heckman M.G., Fiesel F.C., Ando M., Labbe C., Lorenzo-Betancor O., Moussaud-Lamodiere E.L., Soto-Ortolaza A.I., Walton R.L. et al. (2015) Mitochondrial targeting sequence variants of the CHCHD2 gene are a risk for Lewy body disorders. Neurology, 85, 2016–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimada I.S., Hwang S.H., Somatilaka B.N., Wang X., Skowron P., Kim J., Kim M., Shelton J.M., Rajaram V., Xuan Z. et al. (2018) Basal suppression of the sonic hedgehog pathway by the G-protein-coupled receptor Gpr161 restricts medulloblastoma pathogenesis. Cell Rep., 22, 1169–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Steinberg S., Stefansson H., Jonsson T., Johannsdottir H., Ingason A., Helgason H., Sulem P., Magnusson O.T., Gudjonsson S.A., Unnsteinsdottir U. et al. (2015) Loss-of-function variants in ABCA7 confer risk of Alzheimer's disease. Nat. Genet., 47, 445–447. [DOI] [PubMed] [Google Scholar]
- 29. Wiatr K., Szlachcic W.J., Trzeciak M., Figlerowicz M. and Figiel M. (2018) Huntington disease as a neurodevelopmental disorder and early signs of the disease in stem cells. Mol. Neurobiol., 55, 3351–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Calderon D., Bhaskar A., Knowles D.A., Golan D., Raj T., Fu A.Q. and Pritchard J.K. (2017) Inferring relevant cell types for complex traits by using single-cell gene expression. Am. J. Hum. Genet., 101, 686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lim R.G., Quan C., Reyes-Ortiz A.M., Lutz S.E., Kedaigle A.J., Gipson T.A., Wu J., Vatine G.D., Stocksdale J., Casale M.S. et al. (2017) Huntington's disease iPSC-derived brain microvascular endothelial cells reveal WNT-mediated angiogenic and blood-brain barrier deficits. Cell Rep., 19, 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M. and Maglott D.R. (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res., 42, D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nelson M.R., Wegmann D., Ehm M.G., Kessner D., St Jean P., Verzilli C., Shen J., Tang Z., Bacanu S.A., Fraser D. et al. (2012) An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science, 337, 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mostafavi S., Gaiteri C., Sullivan S.E., White C.C., Tasaki S., Xu J., Taga M., Klein H.U., Patrick E., Komashko V. et al. (2018) A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer's disease. Nat. Neurosci., 21, 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., Gould J., Davis J.F., Tubelli A.A., Asiedu J.K. et al. (2017) A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell, 171, 1437–1452 e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emdin C.A., Khera A.V., Chaffin M., Klarin D., Natarajan P., Aragam K., Haas M., Bick A., Zekavat S.M., Nomura A. et al. (2018) Analysis of predicted loss-of-function variants in UK Biobank identifies variants protective for disease. Nat. Commun., 9, 1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Broms J., Grahm M., Haugegaard L., Blom T., Meletis K. and Tingstrom A. (2017) Monosynaptic retrograde tracing of neurons expressing the G-protein coupled receptor Gpr151 in the mouse brain. J. Comp. Neurol., 525, 3227–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu X., Tay Y., Sim B., Yoon S.I., Huang Y., Ooi J., Utami K.H., Ziaei A., Ng B., Radulescu C. et al. (2017) Reversal of phenotypic abnormalities by CRISPR/Cas9-mediated gene correction in Huntington disease patient-derived induced pluripotent stem cells. Stem Cell Reports, 8, 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kapur M., Monaghan C.E. and Ackerman S.L. (2017) Regulation of mRNA translation in neurons-a matter of life and death. Neuron, 96, 616–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paulsen J.S., Smith M.M., Long J.D., PREDICT HD investigators and Coordinators of the Huntington Study Group (2013) Cognitive decline in prodromal Huntington disease: implications for clinical trials. J. Neurol. Neurosurg. Psychiatry, 84, 1233–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C. et al. (2019) Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov., 18, 41–58. [DOI] [PubMed] [Google Scholar]
- 42. Chen Y.W., Satange R., Wu P.C., Jhan C.R., Chang C.K., Chung K.R., Waring M.J., Lin S.W., Hsieh L.C. and Hou M.H. (2018) Co(II)(Chromomycin)(2) complex induces a conformational change of CCG repeats from i-motif to base-extruded DNA duplex. Int. J. Mol. Sci., 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martinelli Boneschi F., Vacchi L., Rovaris M., Capra R. and Comi G. (2013) Mitoxantrone for multiple sclerosis. Cochrane Database Syst. Rev., CD002127. [DOI] [PubMed] [Google Scholar]
- 44. Way S.W., Podojil J.R., Clayton B.L., Zaremba A., Collins T.L., Kunjamma R.B., Robinson A.P., Brugarolas P., Miller R.H., Miller S.D. et al. (2015) Pharmaceutical integrated stress response enhancement protects oligodendrocytes and provides a potential multiple sclerosis therapeutic. Nat. Commun., 6, 6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stack E.C., Del Signore S.J., Luthi-Carter R., Soh B.Y., Goldstein D.R., Matson S., Goodrich S., Markey A.L., Cormier K., Hagerty S.W. et al. (2007) Modulation of nucleosome dynamics in Huntington's disease. Hum. Mol. Genet., 16, 1164–1175. [DOI] [PubMed] [Google Scholar]
- 46. Caron N.S., Desmond C.R., Xia J. and Truant R. (2013) Polyglutamine domain flexibility mediates the proximity between flanking sequences in huntingtin. Proc. Natl. Acad. Sci., 110, 14610–14615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Naia L., Cunha-Oliveira T., Rodrigues J., Rosenstock T.R., Oliveira A., Ribeiro M., Carmo C., Oliveira-Sousa S.I., Duarte A.I., Hayden M.R. et al. (2017) Histone deacetylase inhibitors protect against pyruvate dehydrogenase dysfunction in Huntington's disease. J. Neurosci., 37, 2776–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cabello C.M., Bair W.B. 3rd, Ley S., Lamore S.D., Azimian S. and Wondrak G.T. (2009) The experimental chemotherapeutic N6-furfuryladenosine (kinetin-riboside) induces rapid ATP depletion, genotoxic stress, and CDKN1A(p21) upregulation in human cancer cell lines. Biochem. Pharmacol., 77, 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bowie L.E., Maiuri T., Alpaugh M., Gabriel M., Arbez N., Galleguillos D., Hung C.L.K., Patel S., Xia J., Hertz N.T. et al. (2018) N6-Furfuryladenine is protective in Huntington's disease models by signaling huntingtin phosphorylation. Proc. Natl. Acad. Sci., 115, E7081–E7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Maiuri T., Bowie L.E. and Truant R. (2019) DNA repair Signaling of huntingtin: the next link between late-onset neurodegenerative disease and oxidative DNA damage. DNA Cell Biol., 38, 1–6. [DOI] [PubMed] [Google Scholar]
- 51. Wainberg M., Sinnott-Armstrong N., Mancuso N., Barbeira A.N., Knowles D.A., Golan D., Ermel R., Ruusalepp A., Quertermous T., Hao K. et al. (2019) Opportunities and challenges for transcriptome-wide association studies. Nat. Genet., 51, 592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goold R., Flower M., Moss D.H., Medway C., Wood-Kaczmar A., Andre R., Farshim P., Bates G.P., Holmans P., Jones L. et al. (2019) FAN1 modifies Huntington's disease progression by stabilising the expanded HTT CAG repeat. Hum. Mol. Genet., 28, 650–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McAllister B., Gusella J.F., Landwehrmeyer G.B., Lee J.-M., MacDonald M.E., Orth M., Rosser A.E., Williams N.M., Holmans P., Jones L. et al. (2020) The onset and prevalence of motor and psychiatric symptoms in Huntington’s disease. bioRxiv. doi: 10.1101/2020.05.26.116798. [DOI] [Google Scholar]
- 54. Long J.D., Lee J.-M., Aylward E.H., Gillis T., Mysore J.S., Abu Elneel K., Chao M.J., Paulsen J.S., MacDonald M.E. and Gusella J.F. (2018) Genetic modification of Huntington disease acts early in the prediagnosis phase. Am. J. Hum. Genet., 103, 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huckins L.M., Dobbyn A., Ruderfer D.M., Hoffman G., Wang W., Pardinas A.F., Rajagopal V.M., Als T.D., Huckins T.N., Girdhar K. et al. (2019) Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat. Genet., 51, 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C. and Plagnol V. (2014) Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet., 10, e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Franz M., Rodriguez H., Lopes C., Zuberi K., Montojo J., Bader G.D. and Morris Q. (2018) GeneMANIA update 2018. Nucleic Acids Res., 46, W60–W64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ooi J., Langley S.R., Xu X., Utami K.H., Sim B., Huang Y., Harmston N.P., Tay Y.L., Ziaei A., Zeng R. et al. (2019) Unbiased profiling of isogenic Huntington disease hPSC-derived CNS and peripheral cells reveals strong cell-type specificity of CAG length effects. Cell Rep., 26, 2494–2508 e2497. [DOI] [PubMed] [Google Scholar]
- 59. Darmanis S., Sloan S.A., Zhang Y., Enge M., Caneda C., Shuer L.M., Hayden Gephart M.G., Barres B.A. and Quake S.R. (2015) A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci., 112, 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. et al. (2011) A more efficient method to generate integration-free human iPS cells. Nat. Methods, 8, 409–412. [DOI] [PubMed] [Google Scholar]
- 61. Lippmann E.S., Azarin S.M., Kay J.E., Nessler R.A., Wilson H.K., Al-Ahmad A., Palecek S.P. and Shusta E.V. (2012) Derivation of blood-brain barrier endothelial cells from human pluripotent stem cells. Nat. Biotechnol., 30, 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


