Abstract
Cancer is caused by genetic alterations that affect cellular fitness, and multicellular organisms have evolved mechanisms to suppress cancer such as cell cycle checkpoints and apoptosis. These pathways may be enhanced by the addition of tumor suppressor gene paralogs or deletion of oncogenes. To provide insights to the evolution of cancer suppression across the mammalian radiation, we estimated copy numbers for 548 human tumor suppressor gene and oncogene homologs in 63 mammalian genome assemblies. The naked mole rat contained the most cancer gene copies, consistent with the extremely low rates of cancer found in this species. We found a positive correlation between a species’ cancer gene copy number and its longevity, but not body size, contrary to predictions from Peto’s Paradox. Extremely long-lived mammals also contained more copies of caretaker genes in their genomes, suggesting that the maintenance of genome integrity is an essential form of cancer prevention in long-lived species. We found the strongest association between longevity and copy numbers of genes that are both germline and somatic tumor suppressor genes, suggesting that selection has acted to suppress both hereditary and sporadic cancers. We also found a strong relationship between the number of tumor suppressor genes and the number of oncogenes in mammalian genomes, suggesting that complex regulatory networks mediate the balance between cell proliferation and checks on tumor progression. This study is the first to investigate cancer gene expansions across the mammalian radiation and provides a springboard for potential human therapies based on evolutionary medicine.
Keywords: mammals, genome, tumor suppressor genes, oncogenes, cancer, life history
Introduction
Cancer is a genetic disease arising from the accumulation of mutations that affect cellular survival and division. These mutations, which confer fitness benefits to cellular lineages in the form of greater survival, proliferative potential, and immortality (Hanahan and Weinberg 2011), either arise during somatic evolution within the lifetime of a single individual or are inherited through the germline and thus responsible for genetic predispositions to cancer (Bodmer and Tomlinson 2010). According to current estimates, between ∼1% and ∼3.5% of human genes are mutated in cancers (Futreal et al. 2004; Sondka et al. 2018), with ∼90% of known cancer-associated mutations occurring somatically, ∼20% occurring in the germline, and ∼10% occurring as both somatic and germline mutations (Sondka et al. 2018). The types of mutations leading to cancer vary widely, from single point mutations to translocations, but also copy number variations (Stratton et al. 2009) that can include the amplification of oncogenes and/or the deletion of tumor suppressor genes (Nunney 1999; Vogelstein and Kinzler 2004).
Although cancer progresses as a consequence of natural selection acting on mutations in cellular lineages, there has also been selection acting at the organismal level to suppress tumor growth, leading to conserved mechanisms such as cell cycle checkpoints and apoptosis (Leroi et al. 2003). Thus, in both the cellular and organismal contexts, cancer is a phenotype that has evolved under multiple levels of selection since the origins of multicellularity (Domazet-Lošo and Tautz 2010; Aktipis et al. 2015). Tumor suppression may be especially challenging in organisms with large bodies and longer lifespans, since cancer is an age- and body size-related disease (Nunney 2018; Seluanov et al. 2018). In humans, longer average leg length results in a higher risk of nonsmoking-related cancers (Albanes 1998), and greater than average height has been associated with a higher lifetime risk of melanoma (Lahmann et al. 2016). The probability of getting cancer increases with age in humans (Frank 2007), and >50% of cancers in the United States are diagnosed in patients who are 65 years and older (White et al. 2014), making advancing age the single biggest risk factor for cancer.
Since the number of potentially carcinogenic somatic mutations increases as a function of the number of dividing cells throughout an individual’s lifetime, large multicellular animals with long lifespans theoretically face a higher lifetime risk of cancer than do smaller and shorter lived ones (Peto et al. 1975; Peto 1977). However, across mammals (Abegglen et al. 2015), cancer mortality risk has not been found to be associated with body size or life span: In fact, although the cancer mortality rate in humans is estimated to be between 11% and 25% (Ferlay et al. 2015), it may be as low as 5% in elephants (Abegglen et al. 2015). Thus, the data support Peto’s Paradox: Larger and long-lived animals actually get less cancer than humans, even though greater size and longevity requires more complex genetic controls over cell growth (Nunney 1999). This suggests that certain animal lineages may have evolved enhanced cancer suppression mechanisms in order to offset tradeoffs associated with large bodies and long lifespans (Roche et al. 2012).
The genetic controls of cancer suppression in nature’s giants has been the focus of much research (Abegglen et al. 2015; Caulin et al. 2015; Sulak et al. 2016; Vazquez et al. 2018; Tollis et al. 2019), as well as in mammals with pronounced longevity such as naked mole rats (Heterocephalus glaber) (Seluanov et al. 2009; Tian et al. 2013). Low cancer mortality rates in elephants may be related to the redundancy provided by as many as 20 genomic copies of the tumor suppressor gene TP53 (Abegglen et al. 2015; Caulin et al. 2015; Sulak et al. 2016), which is responsible for apoptosis, senescence, and cell cycle arrest in the presence of damaged DNA (Kumari et al. 2014). Elephant cells have a higher apoptotic response to DNA damage compared with humans (Abegglen et al. 2015; Sulak et al. 2016), suggesting programed cell death rather than the repair of damaged DNA is the mechanism of cancer suppression in elephants. Thus, gene duplication can be a major mechanism for the evolution of new traits (Ohta 1989), and redundancies of tumor suppressor genes in large and long-lived animals, such as TP53 in elephants, could effectively require more somatic mutations to trigger malignancies and thus protection from tumors (Nunney 1999).
If the evolution of large bodies and long lifespans was accompanied by compensatory and independent anticancer adaptations such as tumor suppressor gene duplications, then we should be able to predict lineage-specific tumor suppressor gene expansions in certain lineages. However, to date, the genomic investigations into any link between tumor suppressor gene duplications and the evolution of large bodies or long lifespans have been limited to either a single taxon (Seluanov et al. 2007; Abegglen et al. 2015; Vazquez et al. 2018) or a small number of genomes (Caulin et al. 2015). The recent availability of a large number of diverse mammalian genome assemblies (Koepfli et al. 2015; Lewin et al. 2018) provides an unsourced opportunity to learn about potential genetic mechanisms underlying cancer suppression.
Here, we provide a comparative analysis that examines the relationship between life history traits and the number of cancer gene duplications in a species’ genome. By leveraging the Catalogue of Somatic Mutations in Cancer (COSMIC; Sondka et al. 2018), we estimated the copy numbers of over 500 human cancer gene homologs in 63 complete genomes from across the mammalian radiation. We then performed phylogenetically informed statistical tests to determine the effects of life history traits such as body mass and longevity on cancer gene copy numbers in mammals. Our study has three goals: 1) to identify mammalian species with the largest numbers of cancer gene paralogs, 2) to determine which life history traits are predictive of cancer gene copy numbers in mammals, and 3) to identify specific mammalian species, or genes, that should be further studied to understand mechanisms of cancer resistance. Combining genomics and trait data, we uncover unique aspects of certain species’ biology and find evidence for multiple, independent mechanisms of cancer suppression across the tree of life, opening up possibilities for future human cancer therapies.
Results
Human Cancer Gene Duplications in Mammalian Genomes
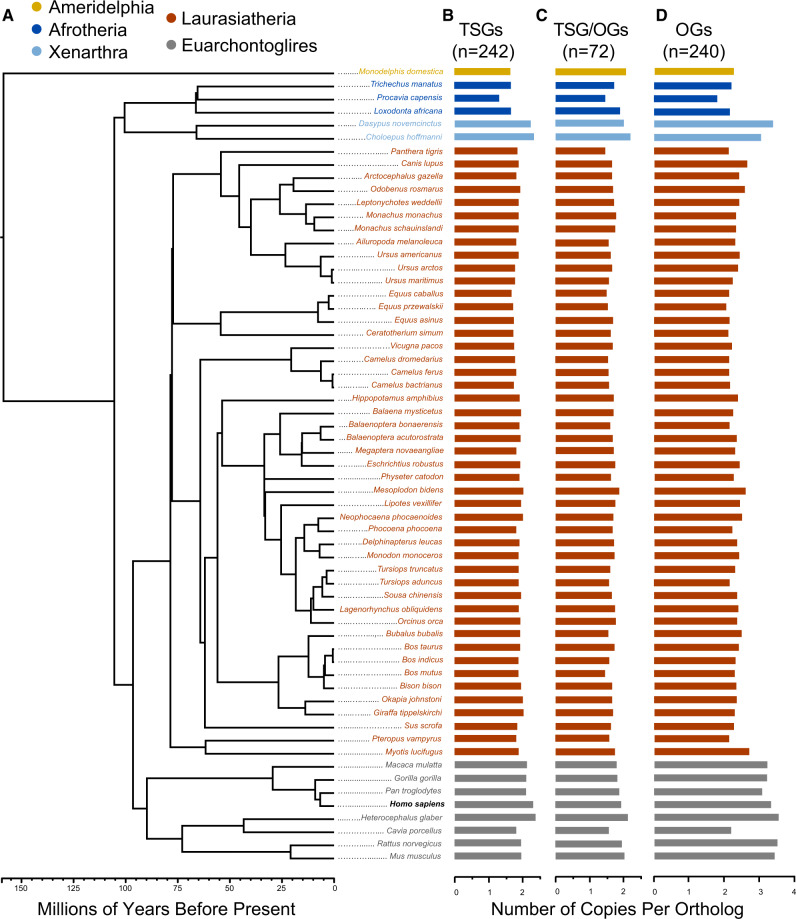
We queried 63 mammalian genome assemblies representing the eutherian superorders, Afrotheria, Xenarthra, Euarchontoglires, and Laurasiatheria, and one marsupial (fig. 1A and supplementary table 1, Supplementary Material online) for 548 human cancer genes, including 242 tumor suppressor genes and 240 oncogenes (as well as 72 classified as both tumor suppressor and oncogenes) that are causally linked to cancer according to the Cancer Gene Census (Sondka et al. 2018) from the COSMIC database (Tate et al. 2019). Within the tumor suppressor genes, 143 genes contained somatic mutations linked to cancer, 35 contained germline mutations, and 43 contained both somatic and germline mutations. To collect homologs for each human cancer gene in mammalian genomes, we performed a protein BLAT search (Kent 2002) using the human peptide, followed by a nucleotide BLAT of the top hit and putative ortholog within the novel genome, and a BlastX search of each putative paralog to the NCBI human peptide database to ensure a reciprocal match (see Materials and Methods for more details). We then counted the total number of human cancer gene homologs collected for each species, as well as the number of paralogs per species per gene.
Fig. 1.
Cancer gene duplications in mammalian genomes. (A) Phylogeny of 63 mammalian genomes used in this study, from Fritz et al. (2009). The tip label for humans (Homo sapiens) is in bold. (B) Copy numbers of tumor suppressor genes (TSGs) in each mammalian genome, normalized as the total number of TSG copies divided by the total number of TSG orthologs detected in a genome. (C) Normalized copy numbers for genes that are both tumor suppressor genes and oncogenes (TSGs/OGs) in each genome. (D) The normalized copy numbers of OGs. TSG and OG classifications were taken from the Cancer Gene Census (Sondka et al. 2018).'
Of the 546 successfully validated human cancer gene orthologs, we found that 476 (87.34%) had at least a 1:many relationship (i.e., there are at least two copies of the gene in at least one mammalian genome). We also found that 293 (53.76%) had three copies in at least one mammalian genome, 176 (36.33%) had four copies in at least one mammalian genome, 124 (24.95%) had five copies in at least one mammalian genome, and 45 (8.26%) genes had ten copies in at least one mammalian genome. We did not find an effect of genome assembly length (phylogenetic generalized least squares or PGLS regression, supplementary fig. 1, Supplementary Material online), or assembly completeness (PGLS, supplementary fig. 2, Supplementary Material online), on estimated cancer gene copy number. To test for a systematic bias in our ability to identify human cancer genes in nonhuman genomes due to the degree of divergence between humans and the other mammals, we tested for but found no relationship between estimated cancer gene copy number in a mammalian genome and its time to most recent common ancestor (TMRCA) with human as estimated from TimeTree (www.timetree.org, last accessed November 2019) (Kumar et al. 2017).
In order to assess the evolutionary conservation of each human cancer gene ortholog across mammals, we collected 100-way vertebrate phastCons scores (Siepel et al. 2005) for each gene using the UCSC Human Genome Browser (Kent et al. 2002) and validated the presence of each ortholog in the human and either opossum, elephant, or armadillo genome using OrthoDB v9 (Zdobnov et al. 2017). Overall, human cancer genes are not conserved at the sequence level and ranged from 0.001 to 0.9 with a mean of 0.2 (supplementary fig. 3, Supplementary Material online). We found that only 58 human cancer genes had average phastCons scores above a cutoff of 0.3 which was previously determined to be evolutionarily conserved (Siepel et al. 2005), and that 30 of these (79%) were oncogenes. We found that cancer gene phastCons scores had only a weak effect on their copy number estimates in the human genome (P = 0.005, R2 = 0.014) (supplementary fig. 4, Supplementary Material online). All queried human cancer genes contained orthologs in human and either opossum, elephant, and armadillo. Therefore, although human cancer genes are generally not conserved at the nucleotide level, our results suggest that they were present in the common ancestor of eutherians. There were only two human cancer orthologs missing from opossum based on OrthoDB, and we could not detect these genes in the opossum assembly using our BLAT/BLAST search either, suggesting that these orthologs are missing from the opossum assembly.
Nearly Cancer Free: Naked Mole Rats Have More Paralogous Cancer Gene Copies in Their Genomes
Although we found that many human cancer genes are duplicated in mammalian genomes, the number of predicted paralogous gene copies varied widely per species. For instance, we detected 13 copies of TP53 in the African savannah elephant (Loxodonta africana), which is similar to the number of annotated TP53 homologs for this species on Ensembl (Caulin et al. 2015) and what has been reported in other species of elephant (Abegglen et al. 2015; Sulak et al. 2016), whereas we annotated only a single copy in human. However, the elephant was in the bottom one-fifth for total number of tumor suppressor gene duplications overall (fig. 1B), with an average of 1.65 gene copies per ortholog. The naked mole rat (Heterocephalus glaber) contained the most tumor suppressor gene duplications of the queried mammalian genomes, with an average of 2.39 copies per ortholog, followed by two-toed sloth (Choloepus hoffmanni; 2.33), human (2.30), and nine-banded armadillo (Dasypus novemcinctus; 2.16) (fig. 1B). The naked mole rat also contained the most oncogene duplications (3.40 copies per ortholog), followed by brown rat (Rattus norvegicus; 3.36), house mouse (Mus musculus; 3.29), and nine-banded armadillo (3.23) (fig. 1C).
We further divided the tumor suppressor genes by comparing levels of gene duplication in caretaker or gatekeeper genes, according to Caulin et al. (2015). Caretaker genes are responsible for the inhibition of DNA damage as well as the mediation and control of DNA repair, whereas gatekeeper genes are responsible for the control of cell cycle checkpoints and proliferation (Kinzler and Vogelstein 1997). The mammalian genome with the highest number of caretaker gene duplications was the naked mole rat (2.03), followed by two other euarchontoglires: human (1.61) and brown rat (1.57). The two-toed sloth had the most predicted gatekeeper gene duplications (2.54), followed by naked mole rat (2.42), and human (2.39).
Because natural selection may act differently on somatic mutations compared with germline mutations (Vicens and Posada 2018), we considered an alternative subdivision of tumor suppressor genes depending on whether the cancer-associated mutations only occurred somatically, only in the germline, or both. When we compared human cancer gene duplications across the eutherian superorders, we found that the genomes of euarchontoglires (n = 8) contained a higher median oncogene and germline tumor suppressor gene duplication, whereas xenarthran genomes (n = 2) contained a higher median duplication of genes that are both tumor suppressors and oncogenes, as well as a higher median duplication when all tumor suppressor genes were classified together (supplementary fig. 5, Supplementary Material online). In contrast, afrotherian (n = 3) and laurasiatherian (n = 47) genomes contained lower median duplications of all types of human cancer genes. We found no relationship between cancer gene copy number and TMRCA with humans; therefore, the higher cancer gene copy numbers in rodents are likely due to the fact that euarchontoglires have accumulated more gene expansions compared with other mammals (Demuth et al. 2006) and not related to potential biases stemming from our use of human peptides as query sequences.
Cannot Have One without the Other: The Balance between Tumor Suppressor Genes and Oncogenes
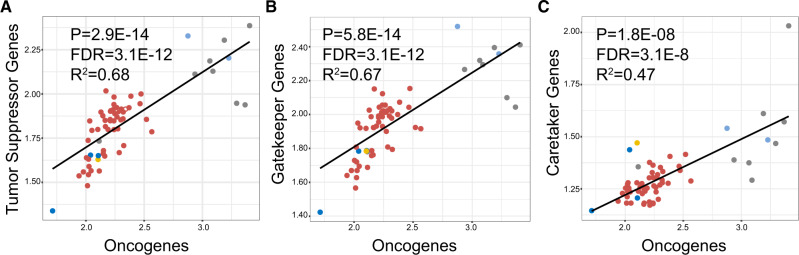
We found a strong correlation between the number of tumor suppressor genes and the number of oncogenes in mammalian genomes (P = 2.95e-14, false discovery rate or FDR = 3.08e-12, R2 = 0.68), which includes a strong correlation between the number of gatekeeper genes and the number of oncogenes (P = 5.81e-14, FDR = 3.08e-12, R2 = 0.67), as well as a positive relationship between the number of caretaker genes and the number of oncogenes (P = 1.77e-08, FDR = 3.13e-08, R2 = 0.47) (fig. 2).
Fig. 2.
Positive correlations between tumor suppressor gene and oncogene copy numbers in 63 mammalian genomes. (A) PGLS regression between copy numbers of all tumor suppressor genes and copy numbers of oncogenes in mammalian genomes. (B) PGLS between copy numbers of caretaker genes and copy numbers of oncogenes. (C) PGLS between copy numbers of gatekeeper genes and copy numbers of oncogenes. Colors represent mammalian clades following figure 1A. All y axes are given in terms of normalized cancer gene copy number (the total number of cancer gene homologs divided by the number of found cancer gene orthologs). FDR: false discovery rate.
Longevity Matters More than Just Size
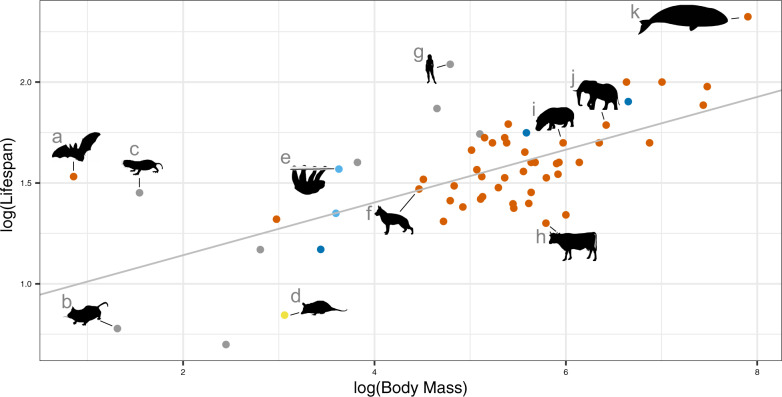
Mammalian megafauna such as elephants and whales are made of, respectively, 100× and 1,000× more cells than smaller human-sized mammals. Such a large number of cells may confer a greater risk of cancer-causing mutations during somatic evolution (Caulin et al. 2015). Therefore, large-bodied mammals may have evolved compensatory adaptations for extra cancer defenses to ensure survival through reproductive age (Peto 1977; Caulin and Maley 2011; Abegglen et al. 2015). We hypothesized that a potential genetic mechanism for cancer suppression in large mammals could be a redundancy of tumor suppressor genes that would effectively require more “hits” to trigger carcinogenesis (Nunney 1999; Caulin and Maley 2011). Conversely, we expected to find a paucity of oncogenes in the genomes of larger mammals relative the genomes of smaller mammals. To test this, we collected body mass (in grams) and lifespan (in years) data for the 63 mammalian species in this study (fig. 3).
Fig. 3.
Relationship between body mass (g) and lifespan data (years) for 63 mammals. The gray line represents a linear regression between log10 body mass and log10 maximum lifespan (slope = 0.13, intercept = 0.88). Colors represent mammalian clades following figure 1A. Silhouettes from PhyloPic.org represent key lineages (animal figures not drawn to scale): (a) little brown bat (Myotis lucifugus), (b) house mouse (Mus musculus), (c) naked mole rat (Heterocephalus glaber), (d) gray opossum (Monodelphis domestica, by Sarah Werning https://creativecommons.org/licenses/by/3.0/), (e) two-toed sloth (Choleopus hoffmanni), (f) dog (Canis lupus familiaris), (g) human (Homo sapiens), (h) cow (Bos taurus), (i) hippopotamus (Hippopotamus amphibius), (j) African bush elephant (Loxodonta africana, by Jan A. Venter, Herbert H.T. Prins, David A. Balfour, and Rob Slotow, vectorized by T. Michael Keesey, https://creativecommons.org/licenses/by/3.0/), and (k) bowhead whale (Balaena mysticetus, by Chris Huh, https://creativecommons.org/licenses/by-sa/3.0/).
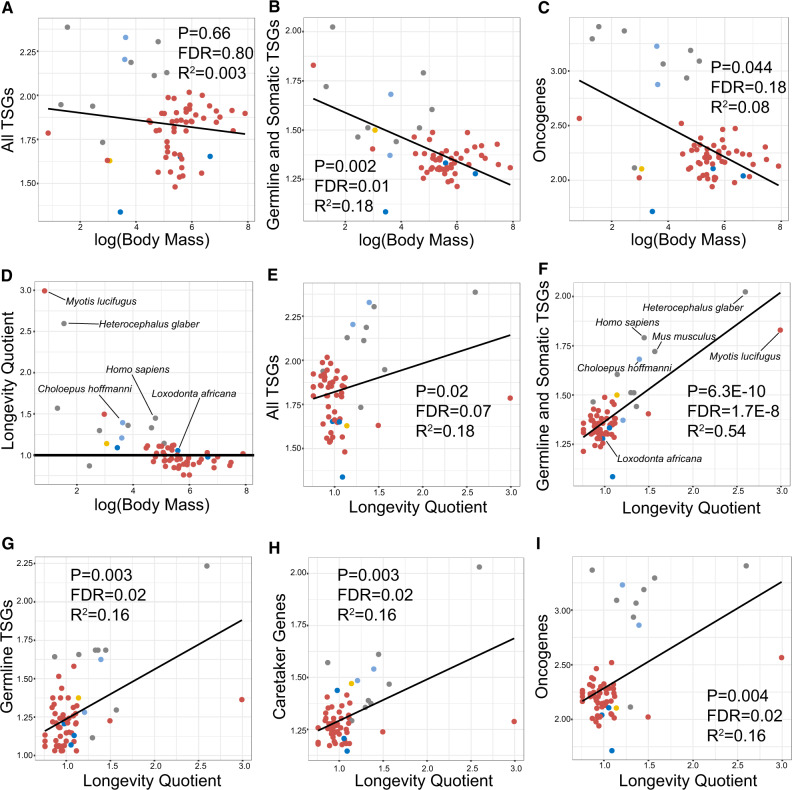
We found no relationship between the number of tumor suppressor gene duplications and body mass across mammals when we considered all tumor suppressor genes together (fig. 4A); however, we did find that, surprisingly, copy numbers of tumor suppressor genes which are mutated in both germline and soma are negatively correlated with body mass (P = 0.002, FDR = 0.01, R2 = 0.18, fig. 4B). We also found a negative relationship between body size and oncogene copy numbers (P = 0.04, R2 = 0.08, fig. 4C). However, these results did not pass significance criteria after applying the FDR to account for multiple testing (see Materials and Methods), or when taking phylogeny into account using a PGLS regression. Therefore, larger mammals did have fewer oncogenes in their genomes than smaller mammals, albeit not to a statistically significant degree. We found no evidence that larger mammals contain more tumor suppressor genes.
Fig. 4.
Cancer gene duplications are positively associated with longevity, and not body mass, in mammalian genomes. (A) There is no association between tumor suppressor gene (TSG) duplications and body mass (given in log10) when we grouped all TSG types together. (B) The only subset of TSGs whose copy numbers in mammalian genomes are significantly associated with body mass are those which are mutated in both human germline and soma. This relationship is slightly negative and is of moderate significance (FDR = 0.01). (C) Larger mammals have fewer oncogene duplications; however, this relationship in nonsignificant when taking into account both phylogeny and multiple testing. (D) LQs versus body mass for 62 mammals included in this study. Key species relevant to this study are labeled. Black line indicates LQ = 1.0, where the observed longevity is equal to the expected longevity. (E) There is a positive correlation between TSG duplications and LQ when we grouped all TSG types together; however, this relationship is nonsignificant when taking into account multiple testing (FDR = 0.07). (F) The strongest correlation (R2 = 0.54) was found between duplications in TSGs mutated in both human germline and soma and longevity. LQ is also correlated with duplications in TSGs with only germline mutations (G), caretaker genes (H), and oncogenes (I). Colors represent mammalian clades following figure 1A. All y axes are given in terms of normalized cancer gene copy number (the total number of cancer gene homologs divided by the number of found cancer gene orthologs).
As cancer is a disease of not just body size but also longevity, we tested for a relationship between human cancer gene copy numbers and longevity in 63 mammals (see Materials and Methods). Briefly, we collected maximum body mass and lifespan data for 2,549 mammals in order to calculate longevity quotients (LQs) for each species. LQ gives an indication of how long a species’ lifespan is compared with other species of similar size, where LQ = observed longevity/expected longevity (Austad and Fischer 1991). We calculated the expected longevity for each species by fitting a linear regression to log10(maximum longevity) and log10(body mass) using nonflying eutherian mammals, following Austad and Fischer (1991) and Foley et al. (2018), obtaining a slope of 0.27 and an intercept of 0.14 (R2 = 0.51) (fig. 4D). We were unable to obtain lifespan data for Sowerby’s beaked whale (Mesoplodon bidens); therefore, we did not calculate LQ for this species.
Across mammals, we found a positive relationship between copy numbers of all tumor suppressor genes and LQ (fig. 4E), although this result did not pass significance criteria after correcting for multiple testing. We found the strongest (and most statistically significant) positive relationship between LQ and copy numbers of tumor suppressor genes mutated in both germline and soma (P = 6.26e-10, FDR = 1.7e-8, R2 = 0.54) (fig. 4F). We also found significant relationships between LQ and copy numbers of germline tumor suppressor genes (P = 0.003, FDR = 0.02, R2 = 0.16, fig. 4G), caretaker genes (P = 0.003, FDR = 0.02, R2 = 0.16, fig. 4H), and oncogenes (P = 0.004, FDR = 0.02, R2 = 0.17, fig. 4I). All of these tests passed the criteria for significance after correcting for multiple testing, and results did not change when we tested these models on subsets and random samples from the 63 species. We found no effect of basal metabolic rate (BMR), diet, placental invasiveness, mean annual temperature, or biome on cancer gene copy number (supplementary fig. 6, Supplementary Material online).
In order to determine whether these patterns were unique to cancer genes, we also estimated copy numbers for curated sets of 87 housekeeping genes and 214 nervous system genes from Dorus et al. (2004) (supplementary figs. 7 and 8, Supplementary Material online). We found no relationship between any of the life history variables and housekeeping gene copy number. We found a weak effect of body mass (P = 0.02, R2 = 0.11) and LQ (P = 0.02, R2 = 0.10) on nervous system gene copy number; however, these results did not pass significance criteria after correction for multiple testing. Therefore, according to our results, longevity had a striking and unique effect on the number of cancer gene copies in a mammalian genome, and the strongest predictor of cancer gene copy number in a mammalian genome was its lifespan relative to other mammals of similar size.
Discussion
The evolutionary relatedness of other mammals to humans has led to the realization that patterns of tumor suppressor gene duplication in mammalian genomes may shed light on cancer resistance mechanisms and lead to novel therapies (Caulin and Maley 2011; Abegglen et al. 2015; Caulin et al. 2015; Keane et al. 2015; Tollis et al. 2017; Vazquez et al. 2018). However, to date, the few studies that have queried mammalian genomes for tumor suppressor gene duplications have mostly focused on few taxa (Caulin et al. 2015; Keane et al. 2015) or a small number of genes (Abegglen et al. 2015; Sulak et al. 2016; Vazquez et al. 2018). Here, we have provided the most comprehensive survey of cancer gene duplications across the mammalian radiation to date, incorporating 548 known human cancer genes and 63 mammalian genomes. Our results show that ∼87% of human cancer genes are duplicated in at least one mammalian genome, and that there are many lineage-specific cancer gene expansions.
A caveat of this study is that it is impossible to identify true biological gene deletions in draft genomes which may harbor assembly errors and varying levels of incompleteness. Nonetheless, we found no relationship between assembly length or predicted assembly completeness and cancer gene copy number. Another potential problem in our analysis is the possibility of an ascertainment bias that stems from using human peptides as query sequences; evolutionarily distant mammalian genomes could contain undetected homologs due to high levels of sequence divergence. Several results suggest that this was not a problem for our study: 1) We found no relationship between the TMRCA with human and cancer gene copy number in our mammalian genome assemblies, 2) all queried human cancer genes shared orthologous matches in every tested mammal, with only two exceptions in the marsupial, suggesting widespread orthology, and 3) we found only a weak relationship between evolutionary sequence conservation and cancer gene copy number. All these results suggest that our ability to detect cancer gene copies in a given mammalian genome did not rely on evolutionary distance from humans. Other caveats are the lack of experimental evidence that the human cancer genes in this study share the same function in other species, and that there may be species-specific pathways for tumor suppression in addition to those causally linked to human cancers. Thus, our results must be taken with some caution until these are experimentally verified.
Our finding that the naked mole rat has the most cancer gene duplications is consistent with previous studies suggesting that naked mole rats have very strong cancer defenses and antiaging adaptations (Buffenstein 2005, 2008; Seluanov et al. 2009; Tian et al. 2013), and that until recently (Delaney et al. 2016) there have been no reported cases of cancer in the species. Surprisingly, even though murine rodents such as brown rats and mice are extremely cancer-prone (Andervont and Dunn 1962), we found that rodents including naked mole rat, brown rat, and house mouse contained more copies of every category of cancer gene than other mammals, except for the two-toed sloth which had the most gatekeepers, somatic tumor suppressor genes, and genes that are both tumor suppressors and oncogenes. We also found that the brown rat and mouse contained the most housekeeping genes, and that the naked mole rat, brown rat, and mouse contained the most nervous system genes. This is consistent with the observation that the rodent lineage has experienced more gene family expansions than other mammals (Demuth et al. 2006). Rodents contain a diversity of lifespan phenotypes that range from 4 years in mice to over 30 years in naked mole rats. The repression of telomerase activity, which is a well-studied anticancer mechanism, was found to be related to body mass rather than lifespan in rodents (Seluanov et al. 2007), suggesting that larger species must mitigate the risk of tumorigenesis that stems from cell proliferation. Although we found support for a positive relationship between tumor suppressor gene copy number and body mass and lifespan in euarchontoglires (P = 0.02, R2 = 0.69) (supplementary fig. 5, Supplementary Material online), these tests did not pass significance criteria after correction for multiple testing. Still, the naked mole rat had both the highest LQ and the most cancer gene duplications, meaning our results make it difficult to reject a correlation between body mass, lifespan, and anticancer adaptations in rodents.
The two-toed sloth and nine-banded armadillo also contained a comparatively large number of cancer gene duplications. These species are xenarthran mammals which diverged from other eutherians during the early Cenozoic (Meredith et al. 2011), making them only very distantly related to rodents and primates. To our knowledge, there has not been any cancer reported in sloths; however, sloth fur carries an anticancer fungus (Higginbotham et al. 2014). There have been reports of cancer in armadillos (Pence et al. 1983; Lee et al. 2015). Neither of these species are particularly large-bodied, nor do they exhibit remarkable longevity according to our estimates. Therefore, it is not clear why these xenarthrans would have special anticancer defenses. Among mammals, xenarthrans are thought to have the lowest BMR (McNab 1985; Pauli et al. 2016), which may be a potential anticancer mechanism for cancer-resistant species (Caulin and Maley 2011) due to reduced oxidative damage to DNA which could otherwise trigger tumor formation (Adelman et al. 1988; Xie et al. 2004). However, we found no relationship between BMR and cancer gene copy number. We suggest that sloths and armadillos should be the focus of future studies investigating cancer resistance.
Bats (order Chiroptera), on the other hand, have both high metabolic rates and exceptional longevity (Munshi-South and Wilkinson 2010) (fig. 4D). We found that the little brown bat (Myotis lucifugus) was an outlier compared with other laurasiatherians with high cancer gene copy numbers, in particular for tumor suppressor genes with both germline and somatic mutations (1.83 copies per ortholog vs. 1.42 average for laurasiatherians). Recent comparative studies have suggested that bats in the long-lived genus Myotis do not display telomere shortening with age (Foley et al. 2018), showed evidence of positive selection in genes that control DNA damage (Foley et al. 2018), and have unique gene expression patterns in pathways controlling DNA repair and tumor suppression that increase with age (Huang et al. 2019). Similarly, a previous study found accelerated protein evolution in DNA damage and response pathways that was unique to long-lived mammals (Li and de Magalhães 2013). Meanwhile, we found that copy numbers of caretaker genes were significantly correlated with longevity across mammals. It was recently suggested that the number of DNA repair genes in a species’ genome is related to its rate of diversification, and that “living fossils” contain fewer DNA repair genes due to a lower mutational load (Voskarides et al. 2019). We tested for this by calculating the evolutionary distinctiveness (ED; Faith et al. 2007) for each species in our study and found no relationship between ED and cancer gene copy number (data not shown). Altogether, these results are consistent with other studies (MacRae et al. 2015) and suggest that the maintenance of genome integrity may be an essential form of cancer prevention in long-lived species.
Copy numbers of cancer genes bearing both germline and somatic mutations according to COSMIC yielded the strongest positive correlation with longevity, followed by germline-only-mutated cancer genes. This suggests that pathways associated with hereditary cancers, which are caused by germline mutations, may face different constraints than those associated with sporadic cancers. Vicens and Posada (2018) analyzed ratios of the proportion of nonsynonymous substitutions per site to the proportion of synonymous substitutions per site in human cancer genes across mammals and found evidence for relaxed selective constraint in cancer genes with germline mutations relative to those with somatic mutations. Thus, there may be stronger purifying selection acting on pathways associated with highly deleterious sporadic cancers, which would reduce the fixation rate of copy number variants related to those pathways. Still, hereditary cancers are likely strongly selected against in long-lived species due to the fact that in general cancer is a late-onset disease. Our results suggest that additional copies of genes with both germline and somatic mutations linked to cancer may provide an extra safeguard against both hereditary and sporadic cancers in long-lived species.
Interestingly, we found that copy numbers of both tumor suppressor genes and oncogenes were positively correlated with longevity across mammals. At the same time, the number of tumor suppressor genes in a genome was strongly correlated with the number of oncogenes. Caulin et al. (2015) found a positive correlation between the number of oncogenes and the number of gatekeepers, whereas we found the positive correlation also holds for caretaker genes. It is possible that Caulin at al. (2015) may not have had the statistical power to detect this relationship. Their study lacked several mammalian genomes included here, such as the naked mole rat which, according to our results, contains the most caretaker genes. It seems logical that long-lived species would have more copies of tumor suppressor genes, which control the cell cycle and programed cell death, thus inhibiting tumor growth (Vogelstein and Kinzler 2004). Meanwhile, oncogenes are genes that, when amplified, activate and promote cell growth and proliferation. It seems paradoxical, therefore, that there would also be oncogene expansions in long-lived species. However, longer-lived species may have more complex regulatory networks that mediate the balance between cellular proliferative potential and necessary checks on tumor progression. In the human and mouse genomes, oncogenes without a tumor suppressor gene at a nearby genomic location were found more likely to become amplified (Wu et al. 2017). Oncogenes and tumor suppressor genes were found to be among the evolutionarily oldest sets of genes in eukaryotes, associated with the timing of the origins of both multicellular and vertebrate life (Makashov et al. 2019); this is consistent with our observation that oncogenes are highly conserved at the sequence level. The coamplification of oncogene and tumor suppressor gene families may be the result of a positive feedback system in which the addition of tumor suppressor genes is an inhibitory response to the expansion of oncogenes, or vice-versa.
Our study demonstrates that species with high LQs contain a particularly high number of cancer genes in their genomes, and we suggest this may be a mechanism for cancer defenses in long-lived species. However, although longevity is related to body size in general (see fig. 3), we find that cancer gene copy number actually does not increase with body size. This is slightly counterintuitive, especially when considering that even though cancer risk increases with body size (Nunney 2018), there is no relationship between body mass and cancer risk across species (Abegglen et al. 2015). Thus, although Peto’s Paradox appears to be real, our data suggest that extra tumor suppressor genes are not contributing to cancer suppression in many large mammals (with some notable exceptions such as TP53 in elephants). However, tumor suppressor gene redundancy is not the only potential mechanism of cancer resistance. Other solutions to Peto’s Paradox might include lower somatic mutation rates, highly efficient apoptosis, increased sensitivity to contact inhibition, or lower metabolic rates in larger organisms (Caulin and Maley 2011).
In conclusion, our study has shed light on many aspects of the evolution of cancer suppression in mammals. The naked mole rat genome contains more cancer gene copies than all of the other mammals we investigated, even after correcting for phylogeny and ascertainment biases, which is consistent with the very low cancer rates found in this species. We also found that although euarchontoglires (the mammalian superorder that include primates and rodents) have the most cancer gene copies, there have also been significant cancer gene duplications in xenarthrans (sloths and armadillos), and that laurasiatherians contain comparatively much fewer cancer gene copies with the little brown bat as an outlier. We suggest that these species should be the focus of future functional assays that investigate potential cancer defense mechanisms. We found that contrary to predictions stemming from Peto’s Paradox, body size does not explain much of the variance in mammalian cancer gene copy number, whereas longevity does. Tumor suppressor genes that are mutated in both the germline and the soma had significantly more duplications in the genomes of mammals with pronounced longevity, including the little brown bat and the naked mole rat, suggesting extra cancer defenses against both heritable and spontaneous cancers in long-lived species. Our study adds to the growing body of evidence that adaptations for cancer defense mechanisms have occurred across the mammalian tree of life, and that mammals with exceptional longevity relative to their body size may hold genetic keys to cancer prevention.
Materials and Methods
Collection of Orthologs
A list of human cancer genes, including tumor suppressor genes and oncogenes, was downloaded from COSMIC (last accessed December 2018). For each cancer gene, we retrieved 1) gene symbols, 2) gene names, and 3) whether cancer-related mutations were germline, somatic, or both. Each tumor suppressor gene was given the classification of being a gatekeeper gene or a caretaker gene according to the 59 gatekeepers and caretakers reported in Caulin et al. (2015); we classified the remaining tumor suppressor genes as either gatekeepers or caretakers using the descriptions for each gene available from GeneCards (Stelzer et al. 2016) (Supplementary Material online). We then gathered human (GRCh38.p13) protein sequences for each human cancer gene ortholog from Ensembl v98 using BioMart (Kinsella et al. 2011). The complete genome assemblies of 63 mammals were collected from NCBI (O’Leary et al. 2016) and The Bowhead Whale Genome Resource (Keane et al. 2015). Each genome assembly was assessed for completeness by calculating the proportion of complete universal single copy orthologs from mammalia_odb10 with BUSCO v4.0 (Simão et al. 2015).
To collect human cancer gene orthologs in mammalian genomes, we used the human peptide in a protein BLAT search with minscore = 55 and minidentity = 60. To collect cancer gene paralogs, we used the DNA sequence of the top scoring hit from the protein BLAT in a second BLAT search of the same genome. We then used each putative paralogous hit at least 33.3% of the length of the coding sequence of the human ortholog as a query in a BlastX (Boratyn et al. 2012) of the RefSeq human protein database (taxid:9606), keeping only copies that returned a top hit of the original human peptide with ≥65% protein identity. This approach was more conservative than using the BLAT parameters on the UCSC Genome Browser web server; however, using web BLAT settings resulted in a large number of false positives (i.e., the reciprocal BlastX returned the wrong human ortholog). Whenever the first search resulted in either missing homologs or a large number of false positives, we reran each analysis with either more relaxed BLAT search parameters and/or a slightly more conservative protein identity cutoff for BlastX (≥70%).
Despite relaxing the parameters, two cancer genes (PRDM1 and QKI) consistently returned zero copies. We therefore performed the bioinformatic search steps for these genes manually using web BLAT on 33 mammalian genomes hosted on the UCSC Genome Browser, and ultimately obtained verified homologs for both genes. To account for potential missing orthologs due to incomplete genome sequencing and assembly, we normalized the cancer gene copy number estimates for each mammalian genome to equal the total number of cancer gene homologs divided by the number of found cancer gene orthologs. To estimate evolutionary conservation in human cancer genes, we averaged the per-site phastCons scores (phastCons100way, Siepel et al. 2005) for the Ensembl whole gene annotations of every human cancer gene ortholog using the UCSC Table Browser (Karolchik et al. 2004).
Dorus et al. (2004) provided a list of 94 housekeeping genes, seven of which were also found in COSMIC and were thus removed from analysis, as well as 214 nervous system genes. The human peptides for these genes were used as queries in a bioinformatic search as described above using the 63 mammalian genomes.
Collection of Trait Data
We collected maximum body mass and lifespan data for the 63 mammals whose genomes were analyzed using the Amniote Life History Database (Myhrvold et al. 2015). Additionally, we collected mean BMR (ml O2/h) for 25 of these species, 19 of which were previously reported (Wilson Sayres et al. 2011). We were able to obtain BMR from six more of our studied species from additional sources (Gumal and Irwan 1998; Jones et al. 2009; Sieg et al. 2009). We collected additional data for each species including biome (tropical or nontropical), habitat (aquatic or terrestrial), and diet (carnivore, omnivore, or herbivore) from Animal Diversity Web (Meyers et al. 2020). Information on the mean annual temperature for 23 species was available from PanTHERIA (Jones et al., 2009). We obtained placentation data (hemochorial, endotheliochorial, or epitheliochorial) for 31 mammals from Comparative Placentation (http://placentation.ucsd.edu/homefs.html) and Mossman (1987). We pruned a mammalian supertree phylogeny (Fritz et al. 2009) to only include the 63 mammals whose genomes were queried in this study, and tested for potential sampling bias in this pruned phylogeny at http://www.evolution.rdg.ac.uk/pe/index.html (last accessed May 2020), finding no node density artifact (Venditti et al., 2006). We calculated the ED for each species using ed.calc from the package caper v1.0.1 (Orme et al. 2018) in R v3.6.1 (R Core Team 2018). The life history data as well as the cancer gene copy number estimates for all 63 mammals are in supplementary appendix A, Supplementary Material online.
LQ Calculations
To increase the number of data points compared with previous studies of LQ (Austad and Fischer 1991; Foley et al. 2018), we accessed the Amniote Life History Database and collected maximum body mass and lifespan data for 2,549 mammalian species including 2,060 nonflying eutherians, 260 chiropterans, and 299 marsupials. We calculated the expected longevity of each species by fitting a linear regression to log10(maximum longevity) and log10(body mass) using the nonflying eutherian mammal data as in Austad and Fischer (1991) and Foley et al. (2018). Calculations were performed in R v3.5.1, and we accounted for phylogenetic dependence in the data using PGLS in caper v1.0.1 using the mammalian supertree phylogeny.
Phylogenetically Informed Regressions of Cancer Gene Copy Numbers and Trait Data
We tested the relationships between normalized human cancer gene copy number (for total tumor suppressor genes, germline tumor suppressor genes, somatic tumor suppressor genes, gatekeepers, caretakers, oncogenes, and genes that are both tumor suppressors and oncogenes) and the trait data. In total, we completed 106 PGLS regressions using caper v1.0.1, available in supplementary appendix B, Supplementary Material online. We applied a FDR where q = 0.05 in order to account for multiple testing (Mcdonald 2014). We also performed the PGLS on subsets of mammals with different body masses relative to the average for humans (62 kg): equal to or less than human body mass, equal to or less than half of human body mass, equal to or less than one-tenth of human body mass, equal to or greater than human body mass, and equal to or greater than double human body mass. Finally, we reran all PGLS models on smaller random samples (ten each at n = 15, n = 32).
Data Availability
All sequence data and genome coordinates from this study are made publicly available at the Harvard Dataverse (https://doi.org/10.7910/DVN/3CVMQI). The automated bioinformatic pipeline is made available at https://github.com/bhanratt/orthologcollector.
Supplementary Material
Acknowledgments
This work was supported in part by NIH (Grant Nos. U54 CA217376, U2C CA233254, P01 CA91955, R01 CA149566, R01 CA170595, R01 CA185138, and R01 CA140657) as well as CDMRP Breast Cancer Research Program (Award No. BC132057) and the Arizona Biomedical Research Commission (Grant No. ADHS18-198847 to C.C.M.). We would like to thank Brian Hanratty from the Bioinformatics Core at Arizona State University and Michael I. Byars from Northern Arizona University. We also extend gratitude toward Dr Melissa Wilson (Arizona State University) for valuable comments during early manuscript preparations.
References
- Abegglen LM, Caulin AF, Chan A, Lee K, Robinson R, Campbell MS, Kiso WK, Schmitt DL, Waddell PJ, Bhaskara S, et al. 2015. Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA 314(17):1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman R, Saul RL, Ames BN.. 1988. Oxidative damage to DNA: relation to species metabolic rate and life span. Proc Natl Acad Sci U S A. 85(8):2706–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktipis CA, Boddy AM, Jansen G, Hibner U, Hochberg ME, Maley CC, Wilkinson GS.. 2015. Cancer across the tree of life: cooperation and cheating in multicellularity. Philos Trans R Soc Lond B Biol Sci. 370(1673):20140219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanes D. 1998. Height, early energy intake, and cancer: evidence mounts for the relation of energy intake to adult malignancies. BMJ 317(7169):1331–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andervont HB, Dunn TB.. 1962. Occurrence of tumors in wild house mice. J Natl Cancer Inst. 28:1153–1163. [PubMed] [Google Scholar]
- Austad SN, Fischer KE.. 1991. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J Gerontol. 46(2):B47–B53. [DOI] [PubMed] [Google Scholar]
- Bodmer W, Tomlinson I.. 2010. Rare genetic variants and the risk of cancer. Curr Opin Genet Dev. 20(3):262–267. [DOI] [PubMed] [Google Scholar]
- Boratyn GM, Schäffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL.. 2012. Domain enhanced lookup time accelerated BLAST. Biol Direct 7(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffenstein R. 2005. The naked mole-rat: a new long-living model for human aging research. J Gerontol A Biol Sci Med Sci. 60(11):1369–1377. [DOI] [PubMed] [Google Scholar]
- Buffenstein R. 2008. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol B 178(4):439–445. [DOI] [PubMed] [Google Scholar]
- Caulin AF, Graham TA, Wang L-S, Maley CC.. 2015. Solutions to Peto’s paradox revealed by mathematical modelling and cross-species cancer gene analysis. Philos Trans R Soc Lond B Biol Sci. 370(1673):20140222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulin AF, Maley CC.. 2011. Peto’s Paradox: evolution’s prescription for cancer prevention. Trends Ecol Evol. 26(4):175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney MA, Ward JM, Walsh TF, Chinnadurai SK, Kerns K, Kinsel MJ, Treuting PM.. 2016. Initial case reports of cancer in naked mole-rats (Heterocephalus glaber). Vet Pathol. 53(3):691–696. [DOI] [PubMed] [Google Scholar]
- Demuth JP, Bie TD, Stajich JE, Cristianini N, Hahn MW.. 2006. The evolution of mammalian gene families. PLoS One 1(1):e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Lošo T, Tautz D.. 2010. Phylostratigraphic tracking of cancer genes suggests a link to the emergence of multicellularity in metazoa. BMC Biol. 8(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM, Lahn BT.. 2004. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell 119(7):1027–1040. [DOI] [PubMed] [Google Scholar]
- Faith DP, Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie J.. 2007. Mammals in the edge: conservation priorities based on threat and phylogeny. PLoS One 2(3):e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F.. 2015. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136(5):E359–E386. [DOI] [PubMed] [Google Scholar]
- Foley NM, Hughes GM, Huang Z, Clarke M, Jebb D, Whelan CV, Petit EJ, Touzalin F, Farcy O, Jones G, et al. 2018. Growing old, yet staying young: the role of telomeres in bats’ exceptional longevity. Sci Adv. 4(2):eaao0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SA. 2007. Dynamics of cancer: incidence, inheritance, and evolution. Princeton (NJ: ): Princeton University Press. [PubMed] [Google Scholar]
- Fritz SA, Bininda-Emonds ORP, Purvis A.. 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol Lett. 12(6):538–549. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR.. 2004. A census of human cancer genes. Nat Rev Cancer 4(3):177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumal MT, Irwan M.. 1998. The ecology and role of the large flying fox (Pteropus vampyrus) in Sarawakian rain forrests—keluang embet embet. Hornbill 2:82–95. [Google Scholar]
- Hanahan D, Weinberg RA.. 2011. Hallmarks of cancer: the next generation. Cell 144(5):646–674. [DOI] [PubMed] [Google Scholar]
- Higginbotham S, Wong WR, Linington RG, Spadafora C, Iturrado L, Arnold AE.. 2014. Sloth hair as a novel source of fungi with potent anti-parasitic, anti-cancer and anti-bacterial bioactivity. PLoS One 9(1):e84549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Whelan CV, Foley NM, Jebb D, Touzalin F, Petit EJ, Puechmaille SJ, Teeling EC.. 2019. Longitudinal comparative transcriptomics reveals unique mechanisms underlying extended healthspan in bats. Nat Ecol Evol. 3(7):1110–1120. [DOI] [PubMed] [Google Scholar]
- Jones KE, Bielby J, Cardillo M, Fritz SA, O’Dell J, Orme CDL, Safi K, Sechrest W, Boakes EH, Carbone C, et al. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90(9):2648–2648. [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ.. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32(Database issue):D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane M, Semeiks J, Webb AE, Li YI, Quesada V, Craig T, Madsen LB, van Dam S, Brawand D, Marques PI, et al. 2015. Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 10(1):112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12(4):656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D.. 2002. The human genome browser at UCSC. Genome Res. 12(6):996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. 2011. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database 2011(0):bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B.. 1997. Gatekeepers and caretakers. Nature 386(6627):761–763. [DOI] [PubMed] [Google Scholar]
- Koepfli K-P, Paten BGenome 10K Community of ScientistsO’Brien SJ.. 2015. The Genome 10K Project: a way forward. Annu Rev Anim Biosci. 3(1):57–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Kumari R, Kohli S, Das S.. 2014. p53 regulation upon genotoxic stress: intricacies and complexities. Mol Cell Oncol. 1(3):e969653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmann PH, Hughes MCB, Williams GM, Green AC.. 2016. A prospective study of measured body size and height and risk of keratinocyte cancers and melanoma. Cancer Epidemiol. 40:119–125. [DOI] [PubMed] [Google Scholar]
- Lee B, Oh S, Lee S, Kim Y, Youn S, Kim Y, Kwon S, Kim D-Y.. 2015. Squamous cell carcinoma in a nine-banded armadillo (Dasypus novemcinctus). J Zoo Wildl Med. 46(2):333–334. [DOI] [PubMed] [Google Scholar]
- Leroi AM, Koufopanou V, Burt A.. 2003. Cancer selection. Nat Rev Cancer 3(3):226–231. [DOI] [PubMed] [Google Scholar]
- Lewin HA, Robinson GE, Kress WJ, Baker WJ, Coddington J, Crandall KA, Durbin R, Edwards SV, Forest F, Gilbert MTP, et al. 2018. Earth BioGenome Project: sequencing life for the future of life. Proc Natl Acad Sci U S A. 115(17):4325–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, de Magalhães JP.. 2013. Accelerated protein evolution analysis reveals genes and pathways associated with the evolution of mammalian longevity. Age 35(2):301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae SL, Zhang Q, Lemetre C, Seim I, Calder RB, Hoeijmakers J, Suh Y, Gladyshev VN, Seluanov A, Gorbunova V, et al. 2015. Comparative analysis of genome maintenance genes in naked mole rat, mouse, and human. Aging Cell 14(2):288–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makashov AA, Malov SV, Kozlov AP.. 2019. Oncogenes, tumor suppressor and differentiation genes represent the oldest human gene classes and evolve concurrently. Sci Rep. 9(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdonald JH. 2014. Handbook of biological statistics. Baltimore (MD: ): Sparky House Publishing. [Google Scholar]
- McNab BK. 1985. Energetics, population biology, and distribution of xenarthrans, living and extinct In: Montgomery GG, editor. The evolution and ecology of armadillos, sloths and vermilinguas. Washington/London: Smithsonian Institution Press. [Google Scholar]
- Meredith RW, Janečka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simão TLL, Stadler T, et al. 2011. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334(6055):521–524. [DOI] [PubMed] [Google Scholar]
- Meyers PR, Espinosa CS, Parr T, Jones GS, Dewey TA.. 2020. The Animal Diversity Web [online]. Available from: https://animaldiversity.org. Accessed November 2019.
- Mossman H. 1987. Vertebrate fetal membranes: comparative ontogeny and morphology; evolution; phylogenetic significance; basic functions; research opportunities. New Brunswick (NJ: ): Rutgers University Press. [Google Scholar]
- Munshi-South J, Wilkinson GS.. 2010. Bats and birds: exceptional longevity despite high metabolic rates. Ageing Res Rev. 9(1):12–19. [DOI] [PubMed] [Google Scholar]
- Myhrvold NP, Baldridge E, Chan B, Sivam D, Freeman DL, Ernest S.. 2015. An amniote life-history database to perform comparative analyses with birds, mammals, and reptiles. Ecology 96(11):3109–3109. [Google Scholar]
- Nunney L. 1999. Lineage selection and the evolution of multistage carcinogenesis. Proc R Soc Lond B 266(1418):493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunney L. 2018. Size matters: height, cell number and a person’s risk of cancer. Proc R Soc B 285(1889):20181743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. 1989. Role of gene duplication in evolution. Genome 31(1):304–310. [DOI] [PubMed] [Google Scholar]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44(D1):D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme D, Freckleton R, Thomas G, Petzoldt T, Fritz S, Isaac N, Pearse W.. 2018. caper: comparative analyses of phylogenetics and evolution in R. R package version 1.0.1. Available from: https://CRAN/R-project.org/package=caper. Accessed November 2019.
- Pauli JN, Peery MZ, Fountain ED, Karasov WH.. 2016. Arboreal folivores limit their energetic output, all the way to slothfulness. Am Nat. 188(2):196–204. [DOI] [PubMed] [Google Scholar]
- Pence DB, Tran RM, Bishop ML, Foster SH.. 1983. Fibroma in a nine-banded armadillo (Dasypus novemcingtus). J Comp Pathol. 93(2):179–184. [DOI] [PubMed] [Google Scholar]
- Peto R. 1977. Epidemiology, multistage models, and short-term mutagenicity tests In: Hiatt HH, Watson JD, Winsten JA, editors. The origins of human cancer. New York: Cold Spring Harbor Laboratory; p. 403–1428. [Google Scholar]
- Peto R, Roe FJ, Lee PN, Levy L, Clack J.. 1975. Cancer and ageing in mice and men. Br J Cancer 32(4):411–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. Available from: https://www.R-project.org/. Accessed November 2019.
- Roche B, Hochberg ME, Caulin AF, Maley CC, Gatenby RA, Misse D, Thomas F.. 2012. Natural resistance to cancers: a Darwinian hypothesis to explain Peto’s paradox. BMC Cancer 12(1):387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Chen Z, Hine C, Sasahara THC, Ribeiro A, Catania KC, Presgraves DC, Gorbunova V.. 2007. Telomerase activity coevolves with body mass not lifespan. Aging Cell 6(1):45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Gladyshev VN, Vijg J, Gorbunova V.. 2018. Mechanisms of cancer resistance in long-lived mammals. Nat Rev Cancer 18(7):433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Hine C, Azpurua J, Feigenson M, Bozzella M, Mao Z, Catania KC, Gorbunova V.. 2009. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc Natl Acad Sci U S A. 106(46):19352–19357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieg AE, O’Connor MP, McNair JN, Grant BW, Agosta SJ, Dunham AE.. 2009. Mammalian metabolic allometry: do intraspecific variation, phylogeny, and regression models matter? Am Nat. 174(5):720–733. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15(8):1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA.. 2018. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 18(11):696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et al. 2016. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinf. 54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA.. 2009. The cancer genome. Nature 458(7239):719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulak M, Fong L, Mika K, Chigurupati S, Yon L, Mongan NP, Emes RD, Lynch VJ.. 2016. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife 5:1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. 2019. COSMIC: the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 47(D1):D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Azpurua J, Hine C, Vaidya A, Myakishev-Rempel M, Ablaeva J, Mao Z, Nevo E, Gorbunova V, Seluanov A.. 2013. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499(7458):346–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Robbins J, Webb AE, Kuderna LFK, Caulin AF, Garcia JD, Bèrubè M, Pourmand N, Marques-Bonet T, O’Connell MJ, et al. 2019. Return to the sea, get huge, beat cancer: an analysis of cetacean genomes including an assembly for the humpback whale (Megaptera novaeangliae). Mol Biol Evol. 314:1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Schiffman JD, Boddy AM.. 2017. Evolution of cancer suppression as revealed by mammalian comparative genomics. Curr Opin Genet Dev. 42:40–47. [DOI] [PubMed] [Google Scholar]
- Vazquez JM, Sulak M, Chigurupati S, Lynch VJ.. 2018. A zombie LIF gene in elephants is upregulated by TP53 to induce apoptosis in response to DNA damage. Cell Rep. 24(7):1765–1776. [DOI] [PubMed] [Google Scholar]
- Venditti C, Meade A, Pagel M.. 2006. Detecting the node-density artifact in phylogeny reconstruction. Syst Biol. 55(4):637–643. [DOI] [PubMed] [Google Scholar]
- Vicens A, Posada D.. 2018. Selective pressures on human cancer genes along the evolution of mammals. Genes 9(12):582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW.. 2004. Cancer genes and the pathways they control. Nat Med. 10(8):789–799. [DOI] [PubMed] [Google Scholar]
- Voskarides K, Dweep H, Chrysostomou C.. 2019. Evidence the DNA repair genes, a family of tumor suppressor genes, are associated with evolution rate and size of genomes. Hum Genomics. 13(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MC, Holman DM, Boehm JE, Peipins LA, Grossman M, Henley SJ.. 2014. Age and cancer risk. Am J Prev Med. 46(3):S7–S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Sayres MA, Venditti C, Pagel M, Makova KD.. 2011. Do variations in substitution rates and male mutation bias correlate with life-history traits? A study of 32 mammalian genomes. Evolution 65(10):2800–2815. [DOI] [PubMed] [Google Scholar]
- Wu WKK, Li X, Wang X, Dai RZW, Cheng ASL, Wang MHT, Kwong T, Chow TC, Yu J, Chan MTV, et al. 2017. Oncogenes without a neighboring tumor-suppressor gene are more prone to amplification. Mol Biol Evol. 34(4):903–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Yang H, Cunanan C, Okamoto K, Shibata D, Pan J, Barnes DE, Lindahl T, McIlhatton M, Fishel R, et al. 2004. Deficiencies in mouse myh and ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-Ras oncogene in lung tumors. Cancer Res. 64(9):3096–3102. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Tegenfeldt F, Kuznetsov D, Waterhouse RM, Simão FA, Ioannidis P, Seppey M, Loetscher A, Kriventseva EV.. 2017. OrthoDB v9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 45(D1):D744–D749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data and genome coordinates from this study are made publicly available at the Harvard Dataverse (https://doi.org/10.7910/DVN/3CVMQI). The automated bioinformatic pipeline is made available at https://github.com/bhanratt/orthologcollector.