Abstract
We investigated how the two rounds of whole-genome duplication that occurred at the base of the vertebrate lineage have impacted ancient microsyntenic associations involving developmental regulators (known as genomic regulatory blocks, GRBs). We showed that the majority of GRBs identified in the last common ancestor of chordates have been maintained as a single copy in humans. We found evidence that dismantling of the duplicated GRB copies occurred early in vertebrate evolution often through the differential retention of the regulatory gene but loss of the bystander gene’s exonic sequences. Despite the large evolutionary scale, the presence of duplicated highly conserved noncoding regions provided unambiguous proof for this scenario for multiple ancient GRBs. Remarkably, the dismantling of ancient GRB duplicates has contributed to the creation of large gene deserts associated with regulatory genes in vertebrates, providing a potentially widespread mechanism for the origin of these enigmatic genomic traits.
Keywords: gene deserts, whole-genome duplications, origin of vertebrates, genomic regulatory block
Complex spatiotemporal regulation of transcription is crucial for animal embryonic development. This regulation relies heavily on long-range distal enhancers, which are a hallmark of metazoans (Irimia et al. 2013; Sebe-Pedros et al. 2016) and are particularly widespread in vertebrates (Marlétaz et al. 2018). Long-range enhancers engage in precise physical interactions with their target promoters, which can be located hundreds of kbs away. These enhancers are particularly prevalent in the case of developmental transcription factors (hereafter trans-dev genes; Woolfe et al. 2005), and create complex cis-regulatory landscapes that leave distinctive signatures on how the genome is organized around these genes. Among these signatures, the massive size of the intergenic regions associated with trans-dev genes is probably the most conspicuous (Nelson et al. 2004). In the most extreme cases, these gene-free regions can be longer than 1 Mb, and are commonly known as gene deserts (Nobrega et al. 2003). These regulatory-rich gene deserts associated with trans-dev genes constitute ∼30% of all human gene deserts (Ovcharenko et al. 2005). Moreover, whereas the majority of gene deserts have a higher proportion of repetitive regions and are of relatively recent evolutionary origin, gene deserts associated with trans-dev genes are much more ancient and stable, containing numerous transcriptional enhancers and a high density of highly conserved noncoding regions (HCNRs) shared across bony vertebrates (Ovcharenko et al. 2005). However, the origin of vertebrate regulatory gene deserts remains a mystery. One of the challenges of studying the evolution of gene desserts is the difficulty to discriminate homologous gene deserts from those that could have independently evolved around the same genes in different lineages. Thus, the presence of conserved syntenic reference points such as HCNRs and/or conserved neighboring genes is necessary to unambiguously establish gene desert homology, something that becomes increasingly challenging with evolutionary distance. Therefore, given the nearly complete absence of noncoding sequences conserved between vertebrates and nonvertebrates (Royo et al. 2011; Clarke et al. 2012), not much can be said about the origin of regulatory gene deserts except that many of them date back to at least the last common ancestor of vertebrates.
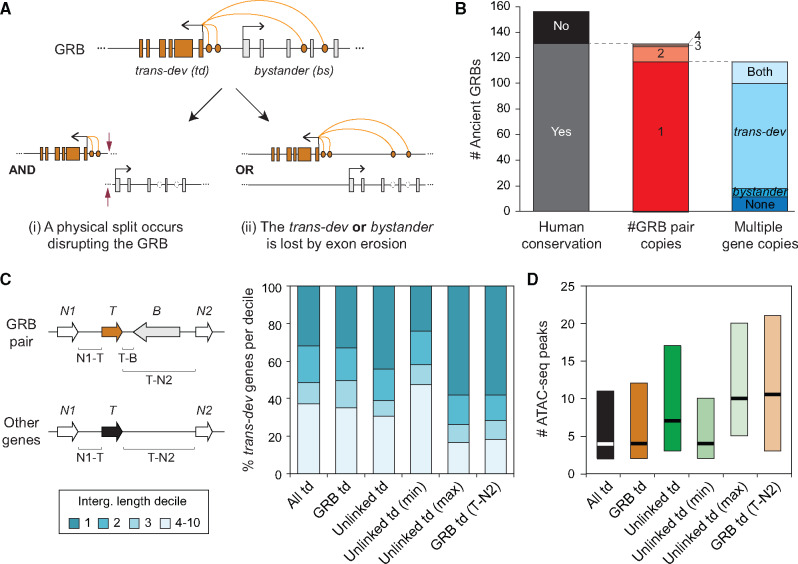
In addition to intergenic regions, distal enhancers from trans-dev genes can also constrain genomic organization when they are located within the introns of neighboring genes. This creates microsyntenic associations between trans-dev and non-trans-dev (bystander) genes, known as genomic regulatory blocks (GRBs; fig. 1A) (Kikuta et al. 2007). GRBs are thus the expanded regulatory landscapes of the trans-dev genes, and closely coincide with topological-associated domains (TADs) (Harmston et al. 2017). Importantly, as the disruption of GRBs would separate enhancers from their target trans-dev genes, these microsyntenic associations are often highly conserved in evolution (Engstrom et al. 2007; Kikuta et al. 2007; Dong et al. 2009; Irimia, Tena et al. 2012). Furthermore, double-strand breaks have been shown to occur preferentially at TAD borders in human and mouse, reinforcing the idea that TADs (and GRBs) are particularly resistant to genomic rearrangements (Canela et al. 2017). In fact, many of these microsyntenic pairs are among the most ancient features of animal genomes, dating back at least to the last common ancestor of bilaterian animals (Irimia, Tena et al. 2012; Simakov et al. 2013). This is especially important because, with a few notable exceptions (Royo et al. 2011; Clarke et al. 2012), orthologous cis-regulatory elements have not been identified across phyla. Thus, ancient GRBs are often the only source of information to gain insight into the evolution of homologous gene regulatory landscapes in deep evolutionary times. However, despite their remarkable evolutionary conservation, GRBs can also be modified under certain circumstances. In particular, the genetic redundancy created by the whole-genome duplication (WGD) of teleosts was shown to have triggered a wholesale dismantling of GRB duplicates (Kikuta et al. 2007; Dong et al. 2009). In this lineage, the most common outcome of the dismantling was the preservation of the trans-dev gene and associated enhancers accompanied by the loss of the bystander copy’s exonic sequences (fig. 1A, scenario [ii]).
Fig. 1.
Fates of ancient GRBs after the two WGDs in vertebrates. (A) Genetic redundancy allows the dismantling of GRBs after WGD. Two scenarios are depicted: (i) The GRB microsyntenic association is disrupted by a break point, in which both the trans-dev and the bystander genes may be maintained, but in different genomic locations; This scenario would impact the regulation of the trans-dev gene. (ii) The GRB microsyntenic association is dismantled by the differential loss of either the trans-dev or the bystander gene. The loss can occur by a large deletion of the genomic locus or by pseudogenization and “exon erosion.” Although the former would impact the regulation of the trans-dev when the bystander is lost, the latter would not. (B) Summary of the fates of the 156 studied ancient GRB pairs present by the last common ancestor of chordates. Human conservation, whether the human genome has conserved at least one linked copy or not of the ancient GRB associations; #GRB pair copies, for those conserved, the number of copies of GRB pairs maintained in human (1–4); Multiple gene copies, for those GRB pairs in single copy, in how many cases, there are multiple ohnologs for both the trans-dev and bystander genes (i.e., not linked), only for the trans-dev or bystander gene, or for none. (C) Percent of trans-dev (T) genes of different types that have at least one intergenic region (N1-T and/or T-B [for ancient GRB pairs] or T-N2 [for other trans-dev genes]) within the first, second, third, or another decile of intergenic region lengths genome-wide (i.e., trans-dev genes in decile 1 have at least one intergenic region whose length is among the top 10% of all intergenic regions). All td, all trans-dev genes linked to at least one non-trans-dev gene (n = 745); GRB td, trans-dev genes that are part of conserved ancient GRB pairs (n = 103); Unlinked td, trans-dev ohnologs from ancient GRBs that are not linked to the ancient bystander gene (n = 171); Unlinked td (max/min), the unlinked trans-dev ohnolog with the largest/smallest intergenic region (n = 107); GRB td (T-N2), for these cases, the distance from the trans-dev gene in a conserved ancient GRB pair to the gene after the bystander (gene N2) is considered as the only intergenic distance (n = 103). Genes are only counted once in each category and cases for which the two downstream neighbors (N1 and N2) are not present are not considered. All lengths for each category are provided in supplementary table S3, Supplementary Material online. The use of N1 and N2 to label gene neighbors does not imply that these genes are the ohnologs of the neighboring genes flanking a trans-dev gene before WGDs. (D) For each type of trans-dev gene, number of ATAC-seq peaks found in the intergenic region with the highest number of peaks, except for “GRB td (T-N2),” where only the number of peaks between the trans-dev and the gene N2 is considered (as in C; n = 655, 82, 131, 78, 78, and 82, respectively).
In the case of the two rounds of WGD that occurred at the base of vertebrates (Dehal and Boore 2005; Putnam et al. 2008), it is unknown how they have impacted the evolution of ancient GRBs. Furthermore, except in the case of a few isolated loci (Irimia, Royo et al. 2012; Maeso et al. 2012; Acemel et al. 2016), the mechanisms leading to the dismantling of ancient GRB are poorly understood. Here, we identified GRBs that were present in the last common ancestor of chordates and investigated their fates after the vertebrate WGDs. We found that the microsyntenic associations of most ancient GRBs have been maintained in single copy, and that the majority of additional duplicate copies dismantled these associations very early in vertebrate evolution, likely by the differential loss of the bystanders and retention of the trans-dev genes. Interestingly, we observed that such loss of bystanders have substantially contributed to create multiple human gene deserts, and propose that this has been an important source for regulatory gene desert formation at the origin of vertebrates.
Results
Evolutionary History of Ancient GRBs in the Human Genome
To investigate gene desert and GRB evolution before the origin of vertebrates and their subsequent fate after the vertebrate WGDs, we first defined a list of 745 developmental transcription factor (trans-dev) genes in the human genome belonging to 363 families that were already present in the last common ancestor of chordates (see Materials and Methods for details). Of these 745 trans-dev genes, 236 (31.7%) are flanked by at least one intergenic region that falls within the 10% largest intergenic regions of the human genome, or 143 (19.2%) and 94 (12.6%) if the top 5% or 3% intergenic regions are used to define gene deserts, respectively. Next, we assessed which of these trans-dev genes were in GRBs that were likely present in the last common ancestor of chordates. For this purpose, we compiled a list of previously reported microsyntenic nonparalogous gene pairs detected using 13 metazoan species dating back to the last common ancestor of Eumetazoans (Irimia, Tena et al. 2012), and also performed a de novo search for gene pairs with conserved microsynteny employing more recently published genomes from slow-evolving nonvertebrate species (see Materials and Methods for details; both analyses included the human genome). From the union of these ancient microsyntenic associations, we then identified those gene pairs containing a trans-dev gene and a non-trans-dev (bystander) gene. This resulted in a list of 116 ancient putative GRBs, thus involving 32.0% of the 363 ancient trans-dev families. The majority of these GRBs (85/116, 73.3%) contained only one microsyntenic association between a trans-dev and a bystander gene, but in the remaining 31 cases the trans-dev gene was linked to two or more bystander genes (supplementary table S1, Supplementary Material online). Therefore, these 116 ancient GRBs corresponded to a total of 156 unique trans-dev-bystander ancient syntenic pairs, hereafter GRB pairs. For simplicity, we here studied the evolution of these GRB pairs independently. For specific comparisons, we also compiled a list of 52 ancient human non-trans-dev gene pairs whose expression is highly correlated across thousands of transcriptomic data sets and that are in head-to-head orientation (likely sharing a bidirectional promoter) (Irimia, Tena et al. 2012) (supplementary table S2, Supplementary Material online).
The human genome has retained at least one copy of 131 out of 156 (84.0%) ancient GRB pairs (fig. 1B and supplementary table S1, Supplementary Material online). However, the microsyntenic association between trans-dev and bystander was maintained in more than one copy for only 14/131 (10.7%) of those ancestral GRB pairs (e.g., ONECUT1/WDR72 on chromosome 15 and ONECUT2/WDR7 on chromosome 18), whereas the vast majority of ancestral syntenic pairs were present in a single copy (117 out of 131) (fig. 1B and supplementary table S1, Supplementary Material online). A similar percentage of the ancient gene pairs with coregulated expression (3/52, 5.8%) were also maintained in multiple copies (P = 0.41, two-sided Fisher’s exact test between GRB and coregulated pairs) (supplementary table S2, Supplementary Material online). The large fraction of single-copy associations after WGD could be due to several reasons. First, even if multiple ohnologs (i.e., paralogs derived from WGDs) are maintained for both the trans-dev and the bystander genes, their microsyntenic association may be lost by genomic rearrangements (fig. 1A, scenario [i]). Second, trans-dev and non-trans-dev genes are known to be retained at different rates after WGD (Putnam et al. 2008; Cañestro et al. 2013), which could also result in the loss of the GRB syntenic association (fig. 1A, scenario [ii]). Consistent with the second scenario, only 17/117 (14.5%) of single-copy GRB pairs have retained multiple copies of both the trans-dev and the bystander genes (fig. 1B and supplementary table S1, Supplementary Material online). In the vast majority of cases, only the trans-dev gene has been retained in multiple copies (82/117, 70.1%), which is significantly more than the seven cases (6.0%) observed for the bystander gene (P = 8.57e-12, proportion test). These differences are due to higher retention rates of the trans-dev genes, because the bystander genes were retained in multiple copies at rates similar to those of genes in coregulated pairs (24/117 [20.5%] vs. 26/104 [25%]; P = 0.54, two-sided Fisher’s exact test). The differential loss of bystander genes is illustrated by the cases of ancestral multibystander GRBs in which different trans-dev ohnologs have remained associated with only one of the original bystander families (supplementary figs. S1–S3 and table S1, Supplementary Material online). In these cases, it is likely that specific bystanders were differentially lost in each vertebrate GRB copy, similar to what has been reported in teleosts (Kikuta et al. 2007; Dong et al. 2009).
To approximate the relative timing at which the loss of the GRB syntenic associations occurred, we next studied ancestral GRB pair evolution in two slow-evolving basal-branching vertebrate species, the elephant shark Callorhinchus milii and the spotted gar Lepisosteus oculatus, and in chicken. We could find evidence of linkage for additional pairs of trans-dev and bystander ohnologs that are not linked in the human genome for only 2/131 (1.5%) ancestral GRB pairs: FOXP3/GPR173 in L. oculatus, and PBX4/MVB12A in L. oculatus, C. milii as well as in chicken (supplementary table S1, Supplementary Material online). A similar pattern was found for the 25 ancestral GRB pairs that have not been conserved in human, for which only two (8%) gene pairs were linked in any of the other vertebrate genomes: MKL2/DCUN1D3 in C. milii, and KDM1A/TMEM30B in C. milii and L. oculatus. Therefore, these data indicate that the majority of losses of GRB syntenic associations likely occurred before the last common ancestor of gnathostomes, soon after the two rounds of WGD.
Dismantlement of Ancient GRB Pairs Often Contributes to Regulatory Gene Deserts in the Human Genome
Our results thus far show that the majority of ancient GRB pair associations have been conserved in a single copy and that most losses seemingly occurred early in vertebrate evolution, likely involving the differential loss of the bystander copies and preservation of trans-dev ohnologs (scenario [ii] in fig. 1A). The differential loss of the bystander gene could occur by a large deletion of the locus or by pseudogenization of the exonic sequence (exon erosion), with dramatically different effects on the conservation of the regulatory landscape of the trans-dev gene: whereas a large deletion would remove putative regulatory elements of the trans-dev gene, erosion of the bystander exons would allow essential regulatory elements to be maintained. Interestingly, considering that many bystanders are unusually large genes with very long introns (Kikuta et al. 2007; Irimia, Tena et al. 2012), the latter could result in the creation of a large gene-free region in the formerly bystander locus, that is, a “gene desertification” of the GRB.
To evaluate the predictions made by this hypothesis, we first calculated the length of the intergenic regions around different sets of the 745 ancient trans-dev genes in the human genome (supplementary table S3, Supplementary Material online). As expected for genes with complex regulation (Nelson et al. 2004), trans-dev genes that are part of GRBs with conserved ancient synteny (GRB td) were enriched for large intergenic regions, but to an extent similar to that of the whole set of ancient human trans-dev genes associated with at least one non-trans-dev gene (fig. 1C; 33.0% of “GRB td” have at least one intergenic region that is among the 10% largest [decile 1] intergenic regions genome-wide [>229 kb], compared with 31.7% for all ancient trans-dev genes [All td]; P = 0.822, two-sided Fisher’s exact test). The fraction of genes with at least one intergenic region in the top decile was significantly increased for the 171 trans-dev ohnologs from ancient GRB pairs that are no longer linked to the original bystander genes (Unlinked td; 44.4%, P = 0.0017, two-sided Fisher’s exact test compared with all trans-dev genes). Interestingly, this enrichment was much stronger when only the unlinked trans-dev ohnolog with the largest intergenic region was considered (Unlinked td (max), 57.9%, P = 2.31e-07, two-sided Fisher’s exact test). This is consistent with the asymmetric evolution of gene regulatory landscapes reported for vertebrate ohnologs, in which some ohnologous copies have much larger intergenic regions than others (Marlétaz et al. 2018). Furthermore, these patterns were stronger in the case of GRBs ancestrally associated with a single bystander compared with those GRB pairs from multibystander GRBs (supplementary fig. S5, Supplementary Material online), in line with the differential bystander retention observed for the latter set (supplementary fig. S1, Supplementary Material online). Remarkably, the distribution of deciles for the ohnologs of unlinked trans-dev genes with the largest intergenic regions (Unlinked td (max)) closely matched that of the trans-dev genes in conserved ancestral GRB pairs when the distance to the neighboring gene after the bystander (i.e., including also the gene body of the bystander) is used as its intergenic distance (GRB td (T-N2); fig. 1C). In fact, most (58.3%) of these distances including the gene body of the bystander are longer than 229 kb, which is the lower size limit of the top 10% largest intergenic distances in human. This means that, in all these cases, the erosion of the bystander genes would immediately qualify the resultant intergenic regions as gene deserts. Importantly, these patterns, especially the enrichment on decile 1 intergenic lengths, were not observed for a set of ancestral microsyntenic pairs of nondevelopmental genes (supplementary fig. S4 and table S3, Supplementary Material online).
Similar patterns were observed when comparing the complexity of regulatory landscapes among sets of trans-dev genes, measured as the number of ATAC-seq peaks found in the different intergenic regions (fig. 1D). For this, we first obtained the corresponding mouse orthologous regions, and then used ATAC-seq data for multiple stages from 12 developing mouse tissues from the ENCODE project to count the number of significant ATAC-seq peaks detected in at least two tissues (see Materials and Methods for details). Consistent with the differences in intergenic region lengths, unlinked trans-dev genes from conserved ancestral GRB pairs have higher numbers of associated ATAC-seq peaks compared with linked (and all) trans-dev genes (fig. 1D). This is even higher for the unlinked ohnolog with the largest number of peaks (Unlinked td (max)), whose distribution is again similar to that observed for linked trans-dev genes when the bystander loci are considered as part of the trans-dev gene’s intergenic region (GRB td (T-N2)). In addition, by using single-cell RNA-seq data from mouse E9.5 embryos, we did not observe any significant difference in the number of cell types in which linked and unlinked trans-dev genes are expressed (supplementary fig. S6A, Supplementary Material online), or in the similarity of expression patterns between ohnologs from ancient GRBs versus the rest of trans-dev genes (supplementary fig. S6B and C, Supplementary Material online), suggesting that retention or loss of bystander genes does not seem to play a major role in the evolution of gene expression after WGDs.
In summary, the unlinked ohnologs we identified correspond to 76/236 (32.2%) of the ancient human trans-dev genes that have at least one intergenic region that falls within the 10% largest of the human genome. Moreover, this fraction increases up to 36.3% (52/143) or 44.7% (42/94) if the top 5% or 3% intergenic regions are used to define gene deserts, respectively, suggesting that bystander erosion could have contributed to the origin of gene deserts in a substantial number of cases. However, the erosion of bystander exons in duplicated GRBs can only be unequivocally demonstrated by detecting bystander pseudogenic exon remnants or by the presence of duplicated HCNRs that were originally located within the introns of the bystander gene before the two WGDs (McEwen et al. 2006; Wang et al. 2007; Maeso et al. 2012; Acemel et al. 2016). To investigate these possibilities, we used a set of duplicated HCNRs in which one of the copies is present within the intron of a bystander gene of an ancient GRB pair and the other in a gene-free region surrounding the unlinked paralogous trans-dev gene, which we compiled from a previous report (McEwen et al. 2006) and from a de novo search using mouse embryonic ATAC-seq peaks (see Materials and Methods). In total, we identified 23 pairs of HCNRs that were ancestrally located in bystander introns associated with 16/99 (16.2%) ancestral GRB pairs in which at least one linked and one unlinked trans-dev genes have been retained in human (supplementary table S4, Supplementary Material online). In addition, we performed a systematic search for exon remnants from bystanders in the intergenic regions of unlinked trans-dev genes, which resulted in the discovery of a high-confidence exon remnant from a paralog of FAM172A associated with NR2F2 (supplementary table S4, Supplementary Material online).
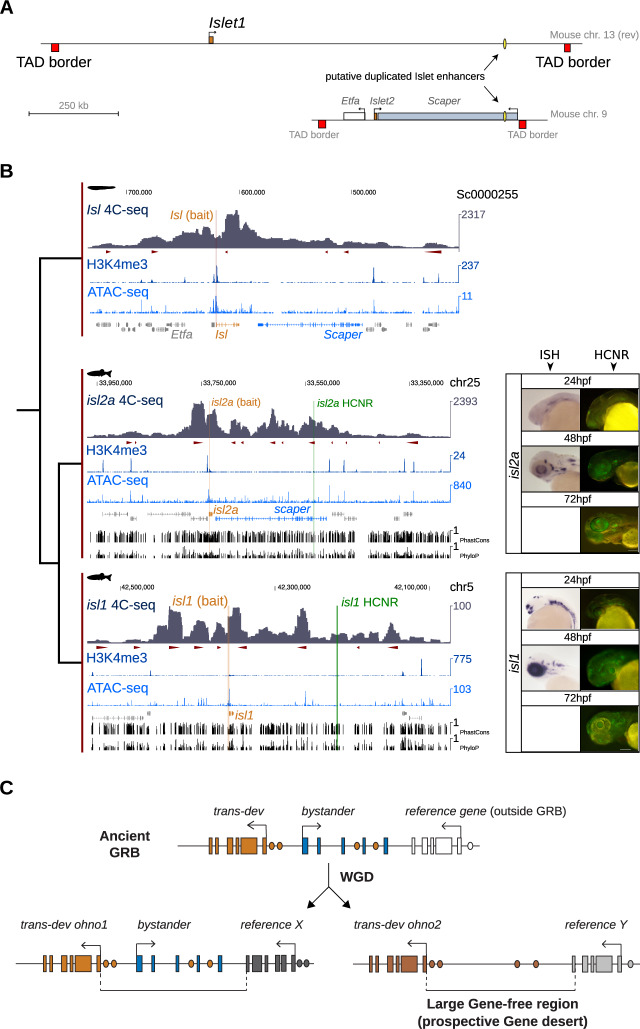
One example of bystander erosion is provided by the GRB pair formed by Islet/Scaper, conserved from human to sponges (Irimia, Tena et al. 2012; Wong et al. 2019). McEwen et al. identified a duplicated HCNR in mammals, with one copy in the intron of Scaper, near Isl2, and another in a gene desert of 1.4 Mb near Isl1 (McEwen et al. 2006). In both cases, Isl1 and Isl2, along with the conserved duplicated elements, were located within the borders of single mouse TADs (fig. 2A). These two sequences were conserved across multiple vertebrates, including zebrafish (fig. 2B). To assess if the duplicated HCNRs interacted with the respective Islet promoters, we generated circular chromosome conformation capture sequencing (4C-seq) using zebrafish isl1 and isl2a promoters as viewpoints. In both cases, the pattern of interactions indicates that the HCNRs are included within the gene regulatory landscape of the respective Islet promoters (fig. 2B and supplementary fig. S7, Supplementary Material online). The same situation was observed in mouse, where both duplicated HCNRs interact with the promoters of their corresponding Isl paralogs as shown with our 4C-seq data and available HiC experiments (Bonev et al. 2017) (supplementary figs. S8 and S9, Supplementary Material online). Furthermore, we generated 4C-seq data in amphioxus embryos using the promoter of its single Isl gene as viewpoint, and also found it to interact with the orthologous Scaper locus (fig. 2B and supplementary fig. S7, Supplementary Material online). Next, to probe the potential enhancer activity of these elements, we generated stable zebrafish lines using the ZED vector (Bessa et al. 2009). Both HCNRs drove expression in the nervous system in overlapping, but also distinct domains, consistent with the expression of their target genes (fig. 2B and C). A similar scenario was observed for other trans-dev genes with large gene deserts that were originally involved in ancestral GRBs, such as Otx1/2-Ehbp1, for which we could also show interaction between duplicated HCNRs and their respective promoters in zebrafish and mouse and between Otx and the orthologous Ehbp1 locus in amphioxus (supplementary figs. S10–S13, Supplementary Material online).
Fig. 2.
The evolution of the Islet-Scaper GRB pair exemplifies the contribution of bystander erosion to the origin of gene deserts. (A) Schematic representation of Isl1 and Isl2 gene regulatory landscapes in the mouse genome (regions contained between the TAD borders identified by Dixon et al. [2012]). Scaper, the bystander associated with Isl2, is depicted, as well as the pair of ohnologous HCNRs identified by McEwen et al. (2006). (B) 4C-seq signal using Isl promoters as viewpoints (orange) and H3K4me3 and ATAC-seq signal from 24 hpf zebrafish or 15 hpf amphioxus embryos, and conservation tracks from the UCSC Genome Browser (PhastCons and PhyloP, for zebrafish only); ohnologous HCNRs are highlighted in green. Red arrowheads indicate significant interactions with either Islet promoter. Right: In situ hybridization of isl1 and isl2a from Thisse et al. (2004) and GFP expression driven by the HCNRs associated with isl1 and isl2a at different timepoints using the ZED vector. During the first few days of development, the isl1 enhancer showed dynamic expression. At 24 hpf, the reporter shows expression in the anterior telencephalon, and a subset of retina and ventral hindbrain cells. At 48 and 72 h, it maintains expression in a sparse subset of neural cells that may overlap with small, localized regions of isl1 expression. In the case of isl2a, the enhancer found within scaper drove consistent expression in a diffuse anterior domain at 24 hpf, as well as in the pineal gland and ventral hindbrain. The expression becomes more restricted at 48 and 72 hpf; to the pineal gland, subsets of the retinal and otic cells, and faintly in the diencephalon. Scale bar: 100 μm. (C) Model of presumptive gene desert formation by GRB dismantling through bystander exon erosion.
Discussion
We found that most ancient GRB pairs have been retained in a single copy in the vertebrate lineage after two rounds of WGD. Only a small subset of GRB pairs have been maintained in multiple copies, a fraction similar to that observed for ancient pairs of coregulated genes in a head-to-head orientation, suggesting no bias for or against retention of GRB syntenic associations in multiple copies after WGD. Moreover, data from slow-evolving, basal-branching gnathostomes suggest that the dismantlement of most extra duplicates likely occurred shortly after the two WGDs (although it should be noted that the exact fraction of inferred early losses might decrease by looking at a larger number of vertebrate species). Although it is difficult to assess it confidently, given the amount of evolutionary time involved and the scarcity of duplicated HCNRs present in vertebrate genomes (McEwen et al. 2006), we also found evidence that a substantial fraction of the dismantled GRB syntenic associations happened by differential loss of the bystander gene by pseudogenization (bystander exon erosion). This pattern is similar to that reported for the more recent teleost-specific WGD (Kikuta et al. 2007; Dong et al. 2009), for which a larger number of HCNRs within bystander genes could be used to unequivocally prove this fate. In the case of the vertebrate WGDs, a few preduplicative HCNRs provided such proof for ∼16% of the ancient GRB pairs. Given that sequence conservation of ancient HCNRs is extremely rare (Holland et al. 2008; Royo et al. 2011), it is possible that similar processes have occurred with many other ancient GRBs in vertebrates despite the lack of conservation of duplicated and ancient HCNRs, as suggested by the analysis of intergenic length distributions. Remarkably, for many cases in which a trans-dev ohnolog has been retained without the association with a bystander copy, we could observe a large associated gene-free region, including for those with HCNR-based evidence for bystander erosion. This highlights a mechanism for gene desert creation after WGD (fig. 2C), by which the already-large intergenic regions of trans-dev genes can be massively increased by the addition of the genomic sequences corresponding to the bystander loci. This is in contrast to a gradual increase of gene-free regions through evolutionary time as the major path for gene desert formation, although both scenarios are certainly not mutually exclusive and likely occur together.
In summary, we show that in 32.2–44.7% of human trans-dev genes associated with gene deserts, erosion of ancient bystanders could have contributed to the formation of their gene deserts. Importantly, these numbers are based only on the data from GRB pairs present in the last common ancestor of chordates, and it should be noted that we do not know the actual repertoire of GRB syntenic associations present immediately before the vertebrate WGDs. Therefore, desertification of preduplicative GRBs may have contributed to origin of many or even most vertebrate regulatory gene deserts. Whatever the extent, erosion of bystanders from duplicated GRBs certainly contributed to explain the origin of this enigmatic genomic trait, which is particularly prevalent in vertebrates.
Materials and Methods
We assembled a comprehensive catalog of ancient GRBs (present in the last common ancestor of chordates) using three main sources: 1) gene pairs with ancient microsyntenic associations identified by comparing 13 metazoan genomes (Irimia, Tena et al. 2012) (595 pairs); 2) gene pairs containing duplicated HCNRs identified by McEwen et al. (2006) (18 pairs); and 3) a de novo search for ancient microsyntenic associations (1,538 pairs; supplementary table S5, Supplementary Material online; see supplementary materials and methods, Supplementary Material online, for details). Gene pairs from the three sources were then combined into a nonredundant set of pairs, and we defined ancient GRB pairs as gene pairs formed by a trans-dev and a non-trans-dev gene. trans-dev genes were defined as “transcription factors involved in the regulation of developmental processes” based on Gene Ontology information (see supplementary methods, Supplementary Material online, for details).
To investigate the evolution of GRB pairs after WGDs, we first defined a consensus set of ohnologs based on different sources (Makino and McLysaght 2010; Singh et al. 2015; Ensembl Paralogs) (supplementary table S6, Supplementary Material online). Based on this ohnology information, for each tested ancestral GRB, we counted the number of trans-dev and bystander ohnologs conserved in the human genome and reassessed which pairwise combinations were linked together and separated by no more than two intervening genes. Genes that had no ohnolog were assumed to have gone back to single copy after the two WGDs.
To obtain pairs of ohnologous regulatory elements that were differentially associated with ohnologous trans-dev genes (one within a bystander and another one in an intergenic region), we collected putative ancient duplicated enhancers from two sources: 1) duplicated HCNRs identified by McEwen et al. (2006) and reported to be within a neighboring gene and an intergenic region; and 2) a de novo search for duplicated ATAC-seq-defined regulatory elements. For the latter, we downloaded 132 ATAC-seq experiments corresponding to several stages from 12 mouse developing tissues (biosamples) from the ENCODE portal (supplementary table S7, Supplementary Material online; see supplementary materials and methods, Supplementary Material online, for details).
The length of the intergenic region between each gene pair was calculated for the human hg38 genome, using only one representative transcript per gene, and considering only protein-coding genes that were included in the OrthoFinder homology clusters. Telomere and centromere regions (downloaded from the UCSC Table function) were discarded. Intergenic regions were then ranked and deciles were made (the top decile corresponded to regions of at least 228,558 bp, and the first three deciles to 54,957 bp; the median intergenic region was 19,949 bp).
4C-seq experiments were performed and analyzed as described earlier (Acemel et al. 2016) (primers provided in supplementary table S8, Supplementary Material online). Significant 4C-seq contacts were defined as those regions displaying interaction frequencies higher than the expected interaction frequency obtained by fitting a monotonic regression to each individual 4C-seq experiment as proposed before (de Wit et al. 2015; further details provided in supplementary materials and methods, Supplementary Material online, code available in GitLab: https://gitlab.com/rdacemel/grb_4c-seq). To probe the conservation of HCNRs associated with isl2a and isl1 in zebrafish, and the open chromatin region located between hey2 and hddc2 (hey2-E1), the sequences were cloned into the Zebrafish Enhancer Detection (ZED) vector and transgenic lines were generated and screened as previously described (Bessa et al. 2009). Single-cell RNA-seq data for mouse E9.5 embryos were obtained from Cao et al. (2019); normalized expression values for the studied trans-dev ohnologs are provided in supplementary table S9, Supplementary Material online.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The research has been funded by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (ERC-StG-LS2-637591 to M.I. and ERC-AdG-LS8-740041 to J.L.G.-S.), the Spanish Ministry of Science and Innovation (BFU2017-89201-P to M.I., RYC-2016-20089 and PGC2018-099392-A-I00 to I.M., BFU2016-74961-P to J.L.G.-S., and BFU2016-81887-REDT/AEI to J.L.G.-S. and M.I.), the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007-2013) under REA grant agreement 608959, the “Centro de Excelencia Severo Ochoa 2013-2017” (SEV-2012-0208), and the “Unidad de Excelencia María de Maetzu 2017-2021” (MDM-2016-0687). We acknowledge the support of the CERCA Programme/Generalitat de Catalunya and of the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) to the EMBL partnership.
Author Contributions
M.T.-S., R.D.A., P.N.F., I.M., and M.I. performed bioinformatic analyses. E.M.K., R.D.A., M.S.M., S.N., and J.J.T. performed molecular biology and transgenesis experiments. J.L.G.-S., I.M., and M.I. conceived, designed, and coordinated the study. E.M.K., I.M., and M.I. wrote the article with the input of the other authors.
References
- Acemel RD, Tena JJ, Irastorza-Azcarate I, Marletaz F, Gomez-Marin C, de la Calle-Mustienes E, Bertrand S, Diaz SG, Aldea D, Aury JM, et al. 2016. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat Genet. 48:336–341. [DOI] [PubMed] [Google Scholar]
- Bessa J, Tena JJ, de la Calle-Mustienes E, Fernández-Miñán A, Naranjo S, Fernández A, Montoliu L, Akalin A, Lenhard B, Casares F, et al. 2009. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn. 238:2409–2417. [DOI] [PubMed] [Google Scholar]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot JP, Tanay A, et al. 2017. Multiscale 3D genome rewiring during mouse neural development. Cell 171:557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A, Maman Y, Jung S, Wong N, Callen E, Day A, Kieffer-Kwon KR, Pekowska A, Zhang H, Rao SSP, et al. 2017. Genome organization drives chromosome fragility. Cell 170:507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañestro C, Albalat R, Irimia M, Garcia-Fernàndez J.. 2013. Impact of gene gains, losses and duplication modes on the origin and diversification of vertebrates. Semin Cell Dev Biol. 24:83–94. [DOI] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. 2019. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SL, VanderMeer JE, Wenger AM, Schaar BT, Ahituv N, Bejerano G.. 2012. Human developmental enhancers conserved between deuterostomes and protostomes. PLoS Genet. 8:e1002852.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Vos ES, Holwerda SJ, Valdes-Quezada C, Verstegen MJ, Teunissen H, Splinter E, Wijchers PJ, Krijger PH, de Laat W.. 2015. CTCF binding polarity determines chromatin looping. Mol Cell. 60:676–684. [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL.. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3:e314.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B.. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Fredman D, Lenhard B.. 2009. Synorth: exploring the evolution of synteny and long-range regulatory interactions in vertebrate genomes. Genome Biol. 10:R86.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B.. 2007. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 17:1898–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmston N, Ing-Simmons E, Tan G, Perry M, Merkenschlager M, Lenhard B.. 2017. Topologically associating domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. Nat Commun. 8:441.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland LZ, Satoh N, Azumi K, Benito-Gutiérrez È, Bronner-Fraser M, Brunet F, Butts T, Candiani S, Dishaw LD, Ferrier DEK, et al. 2008. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 18:1100–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Maeso I, Roy SW, Fraser HB.. 2013. Ancient cis-regulatory constraints and the evolution of genome architecture. Trends Genet. 29:521–528. [DOI] [PubMed] [Google Scholar]
- Irimia M, Royo JL, Burguera D, Maeso I, Gómez-Skarmeta JL, Garcia-Fernandez J.. 2012. Comparative genomics of the Hedgehog loci in chordates and the origins of Shh regulatory novelties. Sci Rep. 2:433.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Tena JJ, Alexis MS, Fernandez-Miñan A, Maeso I, Bogdanovic O, de la Calle-Mustienes E, Roy SW, Gómez-Skarmeta JL, Fraser HB.. 2012. Extensive conservation of ancient microsynteny across metazoans due to cis-regulatory constraints. Genome Res. 22:2356–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engstrom PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, et al. 2007. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 17:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeso I, Irimia M, Tena JJ, González-Pérez E, Tran D, Ravi V, Venkatesh B, Campuzano S, Gómez-Skarmeta JL, Garcia-Fernàndez J.. 2012. An ancient genomic regulatory block conserved across bilaterians and its dismantling in tetrapods by retrogene replacement. Genome Res. 22:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T, McLysaght A.. 2010. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci U S A. 107:9270–9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlétaz F, Firbas PN, Maeso I, Tena JJ, Bogdanovic O, Perry M, Wyatt CD, de la Calle-Mustienes E, Bertrand S, Burguera D, et al. 2018. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 564:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen GK, Woolfe A, Goode D, Vavouri T, Callaway H, Elgar G.. 2006. Ancient duplicated conserved noncoding elements in vertebrates: a genomic and functional analysis. Genome Res. 16:451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C, Hersh B, Carroll S.. 2004. The regulatory content of intergenic DNA shapes genome architecture. Genome Biol. 5(4):R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM.. 2003. Scanning human gene deserts for long-range enhancers. Science 302:413.. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L.. 2005. Evolution and functional classification of vertebrate gene deserts. Genome Res. 15:137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam N, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453:1064–1071. [DOI] [PubMed] [Google Scholar]
- Royo JL, Maeso I, Irimia M, Gao F, Peter IS, Lopes CS, D’Aniello S, Casares F, Davidson EH, Garcia-Fernández J, et al. 2011. Transphyletic conservation of developmental regulatory state in animal evolution. Proc Natl Acad Sci U S A. 108:14186–14191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe-Pedros A, Ballare C, Parra-Acero H, Chiva C, Tena JJ, Sabido E, Gomez-Skarmeta JL, Di Croce L, Ruiz-Trillo I.. 2016. The dynamic regulatory genome of Capsaspora and the origin of animal multicellularity. Cell 165:1224–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simakov O, Marletaz F, Cho SJ, Edsinger-Gonzales E, Havlak P, Hellsten U, Kuo DH, Larsson T, Lv J, Arendt D, et al. 2013. Insights into bilaterian evolution from three spiralian genomes. Nature 493:526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PP, Arora J, Isambert H.. 2015. Identification of ohnolog genes originating from whole genome duplication in early vertebrates, based on synteny comparison across multiple genomes. PLoS Comput Biol. 11(7):e1004394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill JP, Thisse C.. 2004. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 77:505–519. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhong J, Su B, Zhou Y, Wang YQ.. 2007. Comparison of Pax1/9 locus reveals 500-myr-old syntenic block and evolutionary conserved noncoding regions. Mol Biol Evol. 24:784–791. [DOI] [PubMed] [Google Scholar]
- Wong ES, Tan SZ, Garside V, Vanwalleghem G, Gaiti F, Scott E, McGlinn E, Francois M, Degnan BM.. 2019. Early origin and deep conservation of enhancers in animals. bioRxiv. 633651.
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3:e7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.