Abstract
The southern African indigenous Khoe-San populations harbor the most divergent lineages of all living peoples. Exploring their genomes is key to understanding deep human history. We sequenced 25 full genomes from five Khoe-San populations, revealing many novel variants, that 25% of variants are unique to the Khoe-San, and that the Khoe-San group harbors the greatest level of diversity across the globe. In line with previous studies, we found several gene regions with extreme values in genome-wide scans for selection, potentially caused by natural selection in the lineage leading to Homo sapiens and more recent in time. These gene regions included immunity-, sperm-, brain-, diet-, and muscle-related genes. When accounting for recent admixture, all Khoe-San groups display genetic diversity approaching the levels in other African groups and a reduction in effective population size starting around 100,000 years ago. Hence, all human groups show a reduction in effective population size commencing around the time of the Out-of-Africa migrations, which coincides with changes in the paleoclimate records, changes that potentially impacted all humans at the time.
Keywords: Khoe-San, southern Africa, population structure
Introduction
Genetics has played an increasingly important role in revealing human evolutionary history, by demonstrating that Homo sapiens emerged from Africa (Cann et al. 1987; Ramachandran et al. 2005), with some groups outside Africa admixing with archaic humans (Meyer et al. 2012; Prüfer et al. 2014). Our deepest roots include indigenous groups of current-day southern Africa, with modern-day Khoe-San representing one branch in the earliest population divergence in Homo sapiens, and all other Africans and non-Africans representing the other branch (Gronau et al. 2011; Veeramah et al. 2012; Schlebusch et al. 2012, 2017; Schlebusch and Jakobsson 2018). Southern African hunter-gatherers (San) and herders (Khoekhoe) are collectively referred to as Khoe-San (Schlebusch 2010). Khoe-San people speak Khoisan languages, a group of languages that rely heavily on “click” sounds. Three out of the five major Khoisan language families are spoken in southern Africa, namely, Kx’a (formerly called Northern Khoisan), Tuu (formerly Southern Khoisan), and Khoe-Kwadi (formerly Central Khoisan). These three language families show no linguistic relatedness to each other (Güldemann 2014). A few complete genomes from Khoe-San individuals have been investigated with poor representation among the different groups (Meyer et al. 2012; Kim et al. 2014; Mallick et al. 2016). As the Khoe-San represents one of two branches of the deepest population divergence within Homo sapiens, it is crucial to reveal their evolutionary history and their genetic diversity in order to understand the early evolutionary history of our species.
We sequenced and analyzed 25 complete high-coverage genomes from five different Khoe-San groups, representing the three main Khoisan linguistic phyla, across an extensive geographic area. These genomes were placed into a global context by jointly investigating 11 previously published genomes from the HGDP panel, sequenced on the same platform and subjected to similar single nucleotide polymorphism (SNP) calling procedures (Meyer et al. 2012; Raghavan et al. 2014), and another 67 genomes sequenced on the Complete Genomics platform (Drmanac et al. 2010; Lachance et al. 2012; 1000 Genomes Project Consortium 2015). Using these data sets, we characterized genome variation across the world and inferred past population history, where Khoe-San groups showed greater genetic diversity than any other group, but still revealed a reduction in effective population size coinciding with the Out-of-Africa migrations and bottleneck. We further discovered a number of selection targets in the Khoe-San and other groups, and within our common ancestors of >300,000 years ago. These results shed new light on Pleistocene human demographic history and evolution.
Results and Discussion
Among the genomes of 25 individuals (mean coverage 53.4× after mapping and quality filtering; supplementary sections 1–3, Supplementary Material online and table 1), we called 20,020,719 autosomal SNPs (table 1 and supplementary table S5.1, Supplementary Material online). After group-wide quality filtering (supplementary sections 1–3, Supplementary Material online), 18,637,959 autosomal biallelic SNPs remained (table 1), 1,960,665 (10.5%) of which were novel (compared with dbSNP build 151). The two southern Khoe-San groups (Nama and Karretjie People) presented the most novel variants (table 1 and supplementary fig. S5.4, Supplementary Material online). Although many novel variants were singletons (supplementary fig. S5.3A, Supplementary Material online and table 1), 3.2% of them were both novel and present in more than one copy; demonstrating that many variants common among the Khoe-San have not been reported yet. Of the 5,101,560 variants present in all five Khoe-San groups, 24,517 were novel (supplementary fig. S5.4, Supplementary Material online). These variants, common among Khoe-San groups but absent in other populations, have not been previously characterized.
Table 1.
Summary of Genomic (autosomal) Variation in Five Individuals each from Five Khoe-San Groups.
| Category | Total | Karretjie | Nama | |Gui ‖Gana | Ju|’hoansi | !Xun |
|---|---|---|---|---|---|---|
| Dinucleotide SNPs (filtered) | 18,637,959 | 10,555,587 | 10,514,246 | 10,649,570 | 10,429,573 | 10,676,563 |
| Exonic SNPs (%) | 0.653 | 0.597 | 0.598 | 0.596 | 0.599 | 0.598 |
| Novel variants versus dbSNP built 151 (% of variants) | 1,960,665 (10.5%) | 578,935 (5.5%) | 632,492 (6.0%) | 491,543 (4.6%) | 548,528 (5.3%) | 477,360 (4.5%) |
| Singletons (% of variants) | 5,403,107 (29.0%) | 4,547,752 (43.1) | 4,504,833 (42.8) | 4,639,215 (43.6) | 4,315,691 (41.4) | 4,660,181 (43.6) |
| Non-singleton novel variants (% of variants) | 602,402 (3.2%) | 108,129 (1.0%) | 95,736 (0.9%) | 97,282 (0.9%) | 106,546 (1.0%) | 91,365 (0.9%) |
| Mean Depth per Individual, duplicates excluded (all positions in ref genome) | 53.4 (45.1–59.8) | 52.5 (45.1–56.9) | 55.3 (53.7–56.4) | 51.7 (48.2–54.8) | 52.8 (48.0–59.8) | 54.7 (51.7–57.1) |
| Mean heterozygosity (genomic) | 0.001274 | 0.001273 | 0.001266 | 0.001275 | 0.001263 | 0.001291 |
| Heterozygosity variable sites (Called+Filtered Variants) | 0.183249 | 0.183205 | 0.182149 | 0.183438 | 0.181665 | 0.18579 |
| Tajima’s D | −0.7827 | −0.3349 | −0.3200 | −0.3609 | −0.2830 | −0.3639 |
| Tajima’s D (exonic) | −1.1412 | −0.5198 | −0.4844 | −0.5477 | −0.4731 | −0.5446 |
| Mean DAF | 0.1746052 | 0.2746496 | 0.2751872 | 0.2727002 | 0.2770476 | 0.2724151 |
| Mean DAF exonic | 0.1607157 | 0.2650433 | 0.2663403 | 0.2632778 | 0.2680222 | 0.2638948 |
| Total indels (VQSRed) | 2,176,524 | 1,441,604 | 1,458,457 | 1,433,307 | 1,439,949 | 1,461,609 |
| Deletions | 1,267,661 | 802,507 | 799,461 | 812,743 | 795,648 | 815,037 |
| Insertions | 908,863 | 634,150 | 634,609 | 640,933 | 631,300 | 642,247 |
| Complex indels | 527,796 | 513,400 | 512,696 | 514,178 | 512,684 | 514,099 |
| Structural variants | 4,452 | 1,979 | 2,030 | 2,419 | 2,139 | 2,362 |
| Proportion Structural variants with genes |
0.378 | 0.39 | 0.37 | 0.379 | 0.377 | 0.374 |
The Khoe-San exhibited the greatest genetic diversity (mean heterozygosity per individual: 1.154 × 10−3; fig. 1C and supplementary figs. S5.1 and S5.5, Supplementary Material online), compared with other African genomes (mean heterozygosity: 1.079 × 10−3, Mbuti, Mandenka, Yoruba, and Dinka). However, modern-day Khoe-San groups received 10–30% admixture from a mixed eastern African-Eurasian group ∼1,500 years ago (Schlebusch et al. 2017; Skoglund et al. 2017). When genomic material attributed to recent admixture was masked out, the genetic diversity of the Khoe-San (mean heterozygosity after masking: 1.106 × 10−3) decreased and approached that of other African groups (supplementary fig. S8.1, Supplementary Material online), but still remained significantly greater (P = 0.013, Wilcoxon test). Among the Khoe-San, the !Xun had the highest heterozygosity and allelic diversity (table 1 and supplementary figs. S5.1 and S5.5, Supplementary Material online), also sharing the most alleles with all other African groups (fig. 2B), pointing to the highest amount of admixture into the !Xun from non-Khoe-San African groups among the Khoe-San.
Fig. 1.
Sample locations and genetic diversity in the Khoe-San. (A) Sample locations across the world. Colors depict the various data sets included in the study and sample sizes are indicated after the population code. CG, Complete Genomics diversity set (Drmanac et al. 2010); HGDP, HGDP data (Meyer et al. 2012); KGP, 1000 Genomes typed on Complete Genomics platform (1000 Genomes Project Consortium 2015); KSP, this study; LC, Lachance et al. (2012); SGDP, Simons Genome Diversity Project (Mallick et al. 2016); BBA, Ballito Bay A (Schlebusch et al. 2017). The locations chosen for the CEU, GIH, and MXL reflect the ancestry of the population (not the sampling location). (B) Sample locations across Africa. Populations in boldface display newly sequenced individuals. (C) Genetic (autosomal) variation for three population groups: Khoe-San, other sub-Saharan Africans, and non-Africans. The summary statistics were calculated on the joint KSP and HGDP group called data set to avoid biases. The KSP and HGDP data sets were both sequenced on Illumina platforms. Note that the HGDP San individual was not included in the metrics shown here. Heterozygosity was computed from the number of variable positions divided by number of sequenced positions, and averaged across individuals. Mean total runs of homozygosity (ROH) displays the sum over the lengths 0.2–1 Mb. Average (across the genome) number of distinct alleles (allelic richness) and average number of alleles are unique to a single population (private allelic richness) in a sample of eight haploid genomes per variable site. Standard errors were calculated. For heterozygosity, it is the standard error of the mean per individual, averaged across individuals. For ROH, it is the standard error of the mean of individuals. Standard errors for heterozygosity and for allelic richness were very small (<0.08%, see supplementary sections 5.2, 5.3, and 5.5, Supplementary Material online, for details). (D) Private allelic richness (per variable site) of alleles shared by pairwise combinations of the five Khoe-San populations. We distinguish three groups: northern San (Ju|’hoansi and !Xun), central San (|Gui and ‖Gana), and southern San (Nama and Karretjie). (E) Venn diagram summarizing private and shared variants in the Khoe-San versus other Africans versus non-Africans.
Fig. 2.
Grouped bar-plots summarizing private allele sharing as a fraction of the total number of variant sites in the data set: (A) Privately shared alleles of various Khoe-San groups with comparative groups. (B) Privately shared alleles of comparative groups.
In a set of 99 sequenced individuals (from 31 populations), we inferred population stratification across the globe (∼27 million variants; supplementary figs. S6.1, S6.4, and S6.6, Supplementary Material online). The first two principal components (PCs) (supplementary fig. S6.1, Supplementary Material online) explained 7.5% of the global genetic variation and roughly divided it into three groups: non-Africans, Khoe-San, and other Africans. Subsequent PCs summarized variation in other African hunter-gatherer groups (eastern- and western-rainforest hunter-gatherers and Hadza), as well as variation within the Khoe-San (northern, southern, and central) (supplementary fig. S6.1, Supplementary Material online). Variation among non-Africans first became visible at PC20 (we note, however, that the African data set was larger than the non-African data set, 60 vs. 39 individuals). This PCA—based on a globally representative, whole-genome data set—illustrates the extent of African diversity and is a reflection of global genetic diversity, in contrast to inferences based on SNP genotypes, where non-African variation is magnified through ascertainment bias and sample bias (supplementary section 6, Supplementary Material online).
We found a distinct signal of eastern African/non-African affinity and shared private variants among the Khoe-San, particularly for the Nama (supplementary section 6.7, Supplementary Material online, fig. 2, and supplementary figs. S6.1–S6.7 and S6.16–S6.19, Supplementary Material online). This outcome is consistent with recent migration of mixed (eastern African-Eurasian) herding groups to southern Africa, and potentially long-term gene flow between eastern African hunter-gatherers (e.g., Hadza) and Khoe-San (Pickrell et al. 2012, 2014; Schlebusch et al. 2012, 2017; Breton et al. 2014; Macholdt et al. 2014; Skoglund et al. 2017). This pattern can also be seen in mtDNA and Y chromosome data (supplementary section 5.10, Supplementary Material online) (Naidoo et al. 2020), with haplogroup sharing detected between the Ju|’hoansi and Hadza.
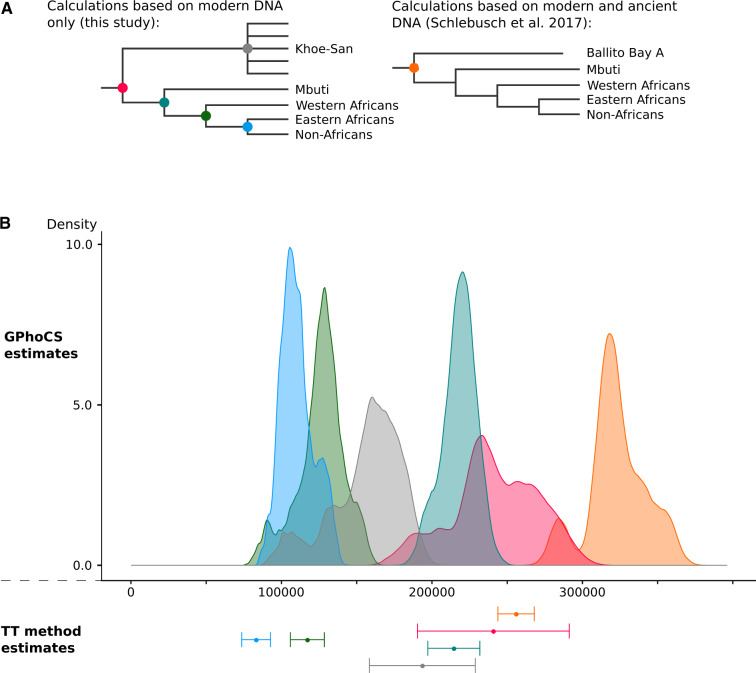
We estimated population divergence between the Khoe-San and various other groups using different and complementary approaches (Gronau et al. 2011; Schlebusch et al. 2017). We applied a mutation rate of 1.25 × 10−8 per base pair per generation and a generation time of 30 years to convert estimates to years ago in the past (unscaled estimates, means, medians, and standard deviations are available in supplementary tables S7.1 and S7.2, Supplementary Material online). Consistent with previous studies (Gronau et al. 2011; Veeramah et al. 2012; Schlebusch et al. 2012, 2017; Schlebusch and Jakobsson 2018), the deepest divergences included the Khoe-San populations (fig. 3 and supplementary tables S7.1 and S7.2 and figs. S7.1, S7.2, and S7.6, Supplementary Material online); a result probably not caused by “archaic admixture” into the Khoe-San (supplementary section 10 and fig. S10.1, Supplementary Material online). Modern-day Khoe-San have, however, >10% of their genetic material tracing to a recent admixture with external groups (Schlebusch et al. 2017; Skoglund et al. 2017). By sequencing the genome of the Stone Age boy from Ballito Bay (BBA), South Africa, the deepest population divergence in Homo sapiens was estimated to 350,000–260,000 years ago (Schlebusch et al. 2017). Consistent with the recent admixture into all modern-day Khoe-San groups, which reduces population divergence time estimates (Schlebusch et al. 2017) (supplementary section 8 and figs. S7.2, S7.4, and S8.2, Supplementary Material online), we found the mean divergence time of all Khoe-San populations from all other groups to be within the 200–300 ka range (supplementary tables S7.2 and S7.2, Supplementary Material online, and fig. 3). These dates correlate well with previous estimates (Gronau et al. 2011; Veeramah et al. 2012) that also fall within the 200–300 ka (kiloannum: thousand years ago) range when applying the mutation rate used here. The Ju|’hoansi (with the lowest level of recent admixture) had a point estimate of ∼270 ka (∼9,000 generations), SD 20 ka (GphoCS method; TT method: ∼260 ka, SD 12 ka), whereas the Nama (with the greatest level of recent admixture) had a point estimate of ∼210 ka, SD 30 ka (TT method: ∼210 ka, SD 30 ka; supplementary tables S7.1 and S7.2, Supplementary Material online). The Mbuti then diverged around ∼220 ka, SD 10 ka (TT method: 215 ka, SD 9 ka), with the other population divergences occurring subsequently. We inferred a mean divergence time of ∼160 ka, SD 20 ka (TT method: ∼190 ka, SD 20 ka) among the different San groups, consistent with previous estimates (Schlebusch et al. 2017).
Fig. 3.
Population divergence estimates. (A) Schematic overview of the estimated population divergences. The colored nodes correspond to the population divergences that were estimated with the TT method and GPhoCS, and the estimates are presented in (B). (B) Distribution of divergence time estimates based on GPhoCS (unscaled estimates, means, and medians available in supplementary table S7.1, Supplementary Material online) and mean ± standard error of the divergence time estimated with the TT method (supplementary table S7.2, Supplementary Material online).
We note that the population history of humans may not always be well represented by divergence models, as gene flow often occurs among human groups, and isolation-by-distance models may sometimes be better descriptions (Vicente et al. 2019). For instance, there is distinct sharing of private alleles between the !Xun/Ju|’hoansi (who traditionally live in the northwestern part of southern Africa) and Mbuti central African rainforest foragers, indicating gene-flow across south-central Africa (fig. 2). The indigenous southern African hunter-gatherer genetic component, might thus have extended far beyond southern Africa in the past (Skoglund et al. 2017; Henn et al. 2018; Scerri et al. 2018, 2019; Schlebusch and Jakobsson 2018; Vicente et al. 2019). A likely consequence is that all population divergence estimates should be interpreted as lower bounds and that the actual population structure could be much older.
The effective ancestral population size (Ne) of currently living individuals can be estimated from genome data (Li and Durbin 2011), and the resolution for certain time periods can be affected by evaluating different numbers of genomes, with increasing numbers improving resolution closer to the present day (Schiffels and Durbin 2014). All human groups were inferred to have had an Ne of ∼30,000 about 300 ka, with a reduction in estimated effective size starting around 150–100 ka (assuming a mutation rate of 1.25 × 10−8 per base pair per generation and a generation time of 30 years; fig. 4 and supplementary fig. S7.11, Supplementary Material online). Non-African populations reached a lowest level (Ne ∼2,000) in the bottleneck around 80 ka, coinciding with the Homo sapiens Out-of-Africa migration event (Nielsen et al. 2017). Surprisingly, most African populations also showed a reduction in estimated Ne during this period, reaching ∼1/3 of the previous Ne (fig. 4 and supplementary figs. S7.10 and S7.11, Supplementary Material online). The decline in effective population sizes appears to be the largest among eastern African populations, followed by western Africans, and subsequently by the rainforest hunter-gatherer populations. Khoe-San groups seem to be the least affected; however, the genome of the 2,000-year-old Ballito Bay boy (unaffected by recent admixture into Khoe-San groups) also showed a reduction in effective population size (fig. 3A;Schlebusch et al. 2017).
Fig. 4.
Estimates of effective population size across time. (A) Effective population sizes estimated for autosomal data from single individuals (i.e., two chromosomes) for the Khoe-San (average over the five individuals in each population), the HGDP individuals, and the Stone Age southern African Ballito Bay A boy (BBA; Schlebusch et al. 2017). (B) African temperature variation estimated from the reconstruction of sea surface temperature in the southwestern Indian Ocean (Caley et al. 2018). (C) Khoe-San effective population sizes estimated from single individuals (“two chromosomes,” solid gray), pairs of individuals (“four chromosomes,” solid colored lines), and five individuals (“ten chromosomes,” colored dotted lines). The curves are averaged over all MSMC runs for all different combinations of individuals (respectively, five, ten, and one).
If we jointly analyze two individuals (four haploid genomes) instead of one, it should provide more resolution on the timing of the bottleneck (Schiffels and Durbin 2014), because the mean time to first coalescence for four haploid genomes is 85 ka (assuming an average ancestral Ne of 17,000 and a generation time of 30 years). For this analysis, we found that all Khoe-San groups showed a reduction to 1/3 of the previous Ne between 100 and 20 ka (fig. 4C and supplementary fig. S7.12, Supplementary Material online). The same pattern was also observed with samples of five individuals (ten haploid genomes), though it could not be detected with samples of one single modern-day Khoe-San individual (fig. 4 and supplementary fig. S7.12, Supplementary Material online). We simulated data under a bottleneck model and ran MSMC on samples of one, two, four, and five individuals under a range of varying conditions of bottleneck strength, duration, and age. From this investigation, we observed a qualitatively similar pattern (supplementary sections 7.3 and 9, Supplementary Material online) of reduced power to infer population-size changes around 80 ka when basing the inference on single genomes. Thus, all human groups appeared to have suffered reduced Ne, of varying degrees, between ∼100 and ∼20 ka; declining to between 50% and 10% of an Ne of ∼30,000 at ∼300 ka. We note that Ne does not necessarily capture the census size and that population structure clearly can impact the estimates of Ne (Mazet et al. 2016). However, in terms of population genetics and understanding of past population histories, estimates of Ne are informative as they tell us about the rate of genetic drift, which in turn can be important for understanding the evolutionary history.
With the 25 complete genomes from Khoe-San individuals that represent one of two legs of the deepest population divergence in Homo sapiens, we have a unique opportunity to search for regions in the genome that display an unusual signal of high numbers of derived variants among all groups of humans. This pattern will be an indicator of distinct adaptation prior to the deepest population divergence, >300,000 years ago. We developed and investigated three Population Branch Statistic (PBS) - derived analyses (supplementary section 12, Supplementary Material online; Schlebusch et al. 2012) that target different parts of human evolutionary history (fig. 5A and supplementary section 12 and table S12.2, Supplementary Material online) and use the 3P-CLR (Racimo 2016) statistic to investigate adaptation in the lineage leading to Homo sapiens.
Fig. 5.
Signatures of adaptation in the genomes. (A) Schematic overview of the three different population branch statistic (PBS) based analyses. The different PBS-based statistics are designed to capture adaptation signals in different parts of the phylogeny. (B) Manhattan plot of the archaicPBS statistic across the genome (supplementary fig. S12.3, Supplementary Material online, displays the aPBS and the emhPBS results). The eight dashed red lines show all the top-five peaks among the three PBS statistics (they are highly correlated). The most likely candidate genes are written below the peaks with genes involved with brain functions, immune system, and other functions indicated in blue, green, and black, respectively. The dashed horizontal line shows the 99.9% percentile of the archaicPBS statistic for these data. (C) A close-up of the strongest peak for archaicPBS, which is located upstream of the gene LPHN3. (D) An example of a local selection signal in southern Khoe-San. |iHS| for southern Khoe-San is shown in green, |iHS| for northern Khoe-San in red, and XP-EHH in purple. The strong negative XP-EHH values suggest adaptation in southern Khoe-San.
Four of the top-ten 3P-CLR peaks and four of the eight top-five regions for the three PBS-statistics (because there is overlap among the PBS top lists, the three top-five lists sum up to eight genomic regions) can be linked to selection for brain development (supplementary section 12, Supplementary Material online). The region with the strongest signal common to all three PBS statistics implicates the LPHN3 (latrophilin 3) gene on chromosome 4 (fig. 5B and C), which has an important function in determining the connectivity rates between the principal neurons in the cortex, and the gene is associated with attention deficit–hyperactivity disorder (Lu et al. 2015). For several of these genes, there is also a strong effect on skull morphology, in addition to the brain-associated effect (see supplementary section 12, Supplementary Material online), a result that has been reported previously (Green et al. 2010; Schlebusch et al. 2012). Furthermore, the regions with strong signals of adaptation in the lineage leading to Homo sapiens are enriched for brain development genes in gene ontology (GO) analyses (Kofler and Schlotterer 2012) (supplementary tables S12.1 and S12.5, Supplementary Material online).
Immune response genes also overlap with signals of adaptation in the lineage leading to Homo sapiens. For instance, the third and fourth strongest 3P-CLR signals and two of the top-five regions for the PBS-statistics overlap with immune response genes (supplementary sections 12.1 and 12.2, Supplementary Material online). Additional strong signals are found for genes in sperm/flagellum motility (supplementary sections 12.1 and 12.2, Supplementary Material online); for example, the DNAL1 gene expressed in motile flagella is located in the region with the strongest 3P-CLR signal (supplementary section 12.1, Supplementary Material online) and the flagellum category is an enriched GO-term in two of the three PBS statistics (supplementary tables S12.3 and S12.5, Supplementary Material online).
We note that identifying targets of selection in early humans, several hundred thousands of years ago, is a difficult problem and that, similar to previous studies (Schlebusch et al. 2012; Racimo et al. 2014; Racimo 2016), our approach also results in a list of potential targets of selection, which need further investigation. However, although there is modest overlap with previous studies, the emerging trend of these investigations points to some similarity in gene functions (Green et al. 2010; Schlebusch et al. 2012; Racimo et al. 2014; Racimo 2016).
In addition to adaptation in the lineage leading to Homo sapiens, we searched for gene regions targeted by selection in specific groups, that is, local adaptation signals using haplotype-based methods for within population (iHS; Voight et al. 2006) and between population comparisons (XP-EHH; Sabeti et al. 2007) (see supplementary section 11, Supplementary Material online). Signals of local adaptation frequently overlapped with genes involved in immune response to infectious diseases in several of the analyses and on different levels of population groupings. For instance, within the northern Khoe-San the strongest signal overlapped with the MHC-region (supplementary table S11.1 and fig. S11.1, Supplementary Material online), and the two strongest signals in the southern Khoe-San were found close to the MHC region; near several genes coding for immunoglobins (supplementary table S11.3 and fig. S11.1, Supplementary Material online). When contrasting the northern and southern Khoe-San, two other regions within the MHC were identified as strong targets of adaptation (in the top-ten regions in XP-EHH analysis; supplementary table S11.2 and fig. S11.2, Supplementary Material online). GO-term analyses (Kofler and Schlotterer 2012) show enrichment for immune response genes among the adaptation signals in the southern Khoe-San as well as in other Africans (supplementary table S11.9, Supplementary Material online). Previous studies found the MHC region to be a common target of selection in various Khoe-San groups (Schlebusch et al. 2012; Owers et al. 2017; Sugden et al. 2018) as well as other populations (Pickrell et al. 2009). The greatest single iHS-value in the northern Khoe-San overlaps with the anthrax toxin receptor-like pseudogene 1 (ANTXRLP1, on chromosome 10), which is near the anthrax toxin receptor-like (ANTXRL) gene. Anthrax is endemic to Namibia, where many of the northern San groups live, and causes intense sporadic disease outbreaks affecting wild animals and humans (Turner et al. 2013). This signal has not been reported previously. In summary, immune system-related genes appear to be targets of adaptation irrespective of time and group, but with slightly different genes involved, which, sometimes, can be directly linked to local and endemic disease conditions.
Signals of local adaptation overlap with genes associated with diet, for instance the FRRS1 gene involved in dietary absorption of iron shows a strong signal in the northern Khoe-San (supplementary table S11.2 and supplementary section 11.5, Supplementary Material online), and the SLCO1B3 gene that mediates fat metabolism and uptake of xenobiotic compounds shows a strong adaptive signal in the southern Khoe-San (the genome-wide greatest single iHS-value; fig. 5D and supplementary table S11.3 and supplementary section 11.6, Supplementary Material online). Adaptation to increased metabolism of endo- and xenobiotics (Schuster et al. 2010) and fat storage (Sugden et al. 2018) have been reported previously for Khoe-San groups. The genome-wide greatest signal of group-specific adaptation (supplementary table S11.8, Supplementary Material online) overlaps with the MINPP1 gene-region, which codes for the only enzyme known to hydrolyze phytic acid in humans. Phytic acid is storing phosphorus in many plant tissues, particularly in bran, seeds, cereals, and grains. Phytic acid is not digested by humans, but it chelates minerals and vitamins and tends to decrease their uptake from food (supplementary section 11.8, Supplementary Material online). The sign of the signal indicates that this gene has been under much stronger selection in the non-Khoe-San group than in the Khoe-San group. This signal has not been reported previously and is an ideal candidate for future studies that focus on potential targets of selection, related to the change in food-producing lifeways.
Genes involved in skeletal muscle development show signals of adaptation, specifically among the Khoe-San populations (supplementary sections 11.5–11.7, Supplementary Material online). In southern Khoe-San, two strong selection signals (the second strongest XP-EHH signal and the widest XP-EHH signal) both implicate genes associated with muscle function (the DTNB gene and the NAA35 gene; supplementary table S11.4, Supplementary Material online), the SNTB1 gene was among the top-ten XP-EHH regions in northern Khoe-San (supplementary table S11.2, Supplementary Material online), and the strongest iHS signal in the Khoe-San group as whole overlaps with the PPP1R12B gene region that plays a regulatory role in muscle contraction (supplementary table S11.5, Supplementary Material online). Selection acting on genes related to muscle development and function has been reported previously for Khoe-San groups (Schlebusch et al. 2012) and other populations (Pickrell et al. 2009). Interestingly the DTNB gene specifically also appeared in the top 1% of selected genes in East Asians, the SNTB1 gene in the top 1% in Oceania (it was the top-11th iHS signal) and the PPP1R12B gene in the top 1% in Bantu-speaking groups (it was the top-14th iHS signal) (Pickrell et al. 2009).
Based on the complete genomes, we also examined the distribution of loss-of-function (LOF) variants in the Khoe-San and estimated levels of functional significance (supplementary section 5.9, Supplementary Material online). Biological functions associated with LOF variants in the Khoe-San included the detection of chemical stimuli (smell and taste), receptor activity, immune response, and keratin/intermediate filaments (supplementary table S5.7, Supplementary Material online). We found two examples of LOF variants which are close to completely lost in most non-African populations, but are found at moderate to high frequencies among the 25 Khoe-San individuals; CASP12 and FMO2. The functional form of the CASP12 gene was found at 48% among the 25 Khoe-San individuals, whereas the global average is around 5% and the loss of the Caspase-12 protein has been associated with an increased risk of sepsis as it is involved in the downregulation of inflammatory cytokines (Saleh et al. 2004, 2006). Although the nonfunctional form of FMO2 is close to fixation in most populations, the functional form was found among the Khoe-San at 60%. The gene product is an enzyme that metabolizes thiourea; however, in doing so produces toxic derivatives (Veeramah et al. 2008). Carriers of the functional allele may be at increased risk for pulmonary toxicity when exposed to thiourea, which is present in a wide range of industrial, household, and medical products. The high frequencies of these functional alleles in the Khoe-San may point to differing selective pressures experienced in the past by these populations.
Conclusion
The genetic diversity among the Khoe-San is the greatest among all human groups across the world, which, in part, is explained by relatively recent (pre-colonial) admixture. When the admixed DNA portion was excluded, the genetic diversity of the Khoe-San approached levels seen in other African populations. All human groups, including the Khoe-San, showed a reduction in Ne (between 1/3 and 1/10) between ∼100 and 20 ka (fig. 4). The early phase of the reduction coincides with the Out-of-Africa bottleneck for non-Africans. Sub-Saharan African populations would not have been impacted by this migration bottleneck, but they all (including the Khoe-San) show a reduction in Ne (fig. 4C). This observation suggests that an additional factor—beyond the migration out of Africa—impacted all humans at this time, perhaps the change in climate. For example, work on the Lake Malawi core indicates severe drought and low-lake stage occurring between ∼109 and 92 ka when the area is also shifting from leaf- to grass-dominated vegetation (Veeramah et al. 2008; Beuning et al. 2011; Scholz et al. 2011), which roughly aligns with a change from warm toward colder temperatures for Africa (fig. 4B; Caley et al. 2018). These events may have caused a reduction in the number of humans; potentially also driving them out of arid African regions, such as the Sahara, and into western Asia.
By revealing substantial and previously unknown genetic variation, we demonstrate that a sizable portion of human genetic variation, including common variants, remains undiscovered among populations often overlooked in medical genetics. We inferred adaptation signals in the genomes and found an overrepresentation of these signals overlapping immunity genes, irrespective of group or time period. This suggests that immunity genes have been under selection throughout human evolutionary history and across the globe.
Materials and Methods
A full description of materials and methods is included in the Supplementary Material online.
Supplementary Material
Acknowledgments
We are grateful to all subjects who participated in this research. The computations were performed at the Swedish National Infrastructure for Computing (SNIC-UPPMAX). We thank Johanna Lagensjö and the Uppsala SNP&Seq Platform for use of laboratory space and reagents. Sequencing was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure supported by the Swedish Research Council for Infrastructures and Science for Life Laboratory, Sweden. The SNP&SEQ Technology Platform is also supported by the Knut and Alice Wallenberg Foundation. We thank Joseph Lachance and Sarah Tishkoff for sharing the data published in Lachance et al. (2012) and Carolina Bernhardsson for help with the data upload. We thank the Working Group of Indigenous Minorities in Southern Africa (WIMSA) and the South African San Council for their support and facilitating fieldwork. The project was reviewed and approved by the University of Witwatersrand (South Africa) Human Research Ethics Committee (M180654), the Swedish Ethical Review Authority (Dnr 2019-05174), and the South African San Council. This work was supported by the Swedish Research Council (No. 621-2014-5211 to C.M.S. and No. 642-2013-8019 to M.J.], the Lars Hierta Foundation (to C.M.S.), the Nilsson-Ehle Endowments (to C.M.S.), the European Research Council (ERC—No. 759933 to C.M.S.), and the Knut and Alice Wallenberg Foundation (to M.J.).
The sequence data from the 25 individuals are deposited on the European Genome Phenome archive (https://www.ebi.ac.uk/ega/), accession number (EGAS00001004459), and are available for academic research use under controlled access policies.
References
- 1000 Genomes Project ConsortiumAuton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR.. 2015. A global reference for human genetic variation. Nature 526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuning KRM, Zimmerman KA, Ivory SJ, Cohen AS.. 2011. Vegetation response to glacial-interglacial climate variability near Lake Malawi in the southern African tropics. Palaeogeogr Palaeoclimatol Palaeoecol. 303(1–4):81–92. [Google Scholar]
- Breton G, Schlebusch CM, Lombard M, Sjodin P, Soodyall H, Jakobsson M.. 2014. Lactase persistence alleles reveal partial East African ancestry of southern African Khoe pastoralists. Curr Biol. 24(8):852–858. [DOI] [PubMed] [Google Scholar]
- Caley T, Extier T, Collins JA, Schefuß E, Dupont L, Malaizé B, Rossignol L, Souron A, McClymont EL, Jimenez-Espejo FJ, et al. 2018. A two-million-year-long hydroclimatic context for hominin evolution in southeastern Africa. Nature 560(7716):76–79. [DOI] [PubMed] [Google Scholar]
- Cann RL, Stoneking M, Wilson AC.. 1987. Mitochondrial DNA and human evolution. Nature 325(6099):31–36. [DOI] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. 2010. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327(5961):78–81. [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. 2010. A draft sequence of the Neandertal genome. Science 328(5979):710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A.. 2011. Bayesian inference of ancient human demography from individual genome sequences. Nat Genet. 43(10):1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldemann T. 2014. “Khoisan” linguistic classification today In: Güldemann T, Fehn A-M, editors. Beyond ‘Khoisan’: historical relations in the Kalahari Basin. Current Issues in Linguistic Theory 330. Amsterdam: John Benjamins; p. 1–41. [Google Scholar]
- Henn BM, Steele TE, Weaver TD.. 2018. Clarifying distinct models of modern human origins in Africa. Curr Opin Genet Dev. 53:148–156. [DOI] [PubMed] [Google Scholar]
- Kim HL, Ratan A, Perry GH, Montenegro A, Miller W, Schuster SC.. 2014. Khoisan hunter-gatherers have been the largest population throughout most of modern-human demographic history. Nat Commun. 5(1):5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Schlotterer C.. 2012. Gowinda: unbiased analysis of gene set enrichment for genome-wide association studies. Bioinformatics 28(15):2084–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance J, Vernot B, Elbers CC, Ferwerda B, Froment A, Bodo JM, Lema G, Fu W, Nyambo TB, Rebbeck TR, et al. 2012. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell 150(3):457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 475(7357):493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YC, Nazarko OV, Sando R 3rd, Salzman GS, Li NS, Sudhof TC, Arac D.. 2015. Structural basis of latrophilin-FLRT-UNC5 interaction in cell adhesion. Structure 23(9):1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholdt E, Lede V, Barbieri C, Mpoloka SW, Chen H, Slatkin M, Pakendorf B, Stoneking M.. 2014. Tracing pastoralist migrations to southern Africa with lactase persistence alleles. Curr Biol. 24(8):875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A, et al. 2016. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538(7624):201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazet O, Rodriguez W, Grusea S, Boitard S, Chikhi L.. 2016. On the importance of being structured: instantaneous coalescence rates and human evolution–lessons for ancestral population size inference? Heredity (Edinb) 116(4):362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338(6104):222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo T, Xu J, Vicente M, Malmström H, Soodyall H, Jakobsson M, Schlebusch CM. Forthcoming 2020. Y-chromosome variation in southern African Khoe-San populations based on whole genome sequences. Genome Biol Evol. doi:10.1093/gbe/evaa098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Akey JM, Jakobsson M, Pritchard JK, Tishkoff S, Willerslev E.. 2017. Tracing the peopling of the world through genomics. Nature 541(7637):302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owers KA, Sjodin P, Schlebusch CM, Skoglund P, Soodyall H, Jakobsson M.. 2017. Adaptation to infectious disease exposure in indigenous Southern African populations. Proc Biol Sci. 284:20170226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, et al. 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19(5):826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Patterson N, Barbieri C, Berthold F, Gerlach L, Guldemann T, Kure B, Mpoloka SW, Nakagawa H, Naumann C, et al. 2012. The genetic prehistory of southern Africa. Nat Commun. 3(1):1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Patterson N, Loh PR, Lipson M, Berger B, Stoneking M, Pakendorf B, Reich D.. 2014. Ancient west Eurasian ancestry in southern and eastern Africa. Proc Natl Acad Sci U S A. 111(7):2632–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505(7481):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F. 2016. Testing for ancient selection using cross-population allele frequency differentiation. Genetics 202(2):733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F, Kuhlwilm M, Slatkin M.. 2014. A test for ancient selective sweeps and an application to candidate sites in modern humans. Mol Biol Evol. 31(12):3344–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW Jr, Orlando L, Metspalu E, et al. 2014. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505(7481):87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL.. 2005. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci U S A. 102(44):15942–15947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R; The International HapMap Consortium, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449(7164):913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M, Mathison JC, Wolinski MK, Bensinger SJ, Fitzgerald P, Droin N, Ulevitch RJ, Green DR, Nicholson DW.. 2006. Enhanced bacterial clearance and sepsis resistance in caspase-12-deficient mice. Nature 440(7087):1064–1068. [DOI] [PubMed] [Google Scholar]
- Saleh M, Vaillancourt JP, Graham RK, Huyck M, Srinivasula SM, Alnemri ES, Steinberg MH, Nolan V, Baldwin CT, Hotchkiss RS, et al. 2004. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature 429(6987):75–79. [DOI] [PubMed] [Google Scholar]
- Scerri EML, Chikhi L, Thomas MG.. 2019. Beyond multiregional and simple out-of-Africa models of human evolution. Nat Ecol Evol. 3(10):1370–1372. [DOI] [PubMed] [Google Scholar]
- Scerri EML, Thomas MG, Manica A, Gunz P, Stock JT, Stringer C, Grove M, Groucutt HS, Timmermann A, Rightmire GP, et al. 2018. Did our species evolve in subdivided populations across Africa, and why does it matter? Trends Ecol Evol (Amst). 33(8):582–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Durbin R.. 2014. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 46(8):919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch C. 2010. Issues raised by use of ethnic-group names in genome study. Nature 464(7288):487; author reply 487. [DOI] [PubMed] [Google Scholar]
- Schlebusch CM, Jakobsson M.. 2018. Tales of human migration, admixture, and selection in Africa. Annu Rev Genomics Hum Genet . 19(1):405–428. [DOI] [PubMed] [Google Scholar]
- Schlebusch CM, Malmstrom H, Gunther T, Sjodin P, Coutinho A, Edlund H, Munters AR, Vicente M, Steyn M, Soodyall H, et al. 2017. Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science 358(6363):652–655. [DOI] [PubMed] [Google Scholar]
- Schlebusch CM, Skoglund P, Sjodin P, Gattepaille LM, Hernandez D, Jay F, Li S, De Jongh M, Singleton A, Blum MG, et al. 2012. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science 338(6105):374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz CA, Cohen AS, Johnson TC, King J, Talbot MR, Brown ET.. 2011. Scientific drilling in the Great Rift Valley: the 2005 Lake Malawi Scientific Drilling Project—an overview of the past 145,000 years of climate variability in Southern Hemisphere East Africa. Palaeogeogr Palaeoclimatol Palaeoecol. 303(1–4):3–19. [Google Scholar]
- Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, et al. 2010. Complete Khoisan and Bantu genomes from southern Africa. Nature 463(7283):943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Thompson JC, Prendergast ME, Mittnik A, Sirak K, Hajdinjak M, Salie T, Rohland N, Mallick S, Peltzer A, et al. 2017. Reconstructing prehistoric African population structure. Cell 171(1):59–71.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden LA, Atkinson EG, Fischer AP, Rong S, Henn BM, Ramachandran S.. 2018. Localization of adaptive variants in human genomes using averaged one-dependence estimation. Nat Commun. 9(1):703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner WC, Imologhome P, Havarua Z, Kaaya GP, Mfune JKE, Mpofu IDT, Getz WM.. 2013. Soil ingestion, nutrition and the seasonality of anthrax in herbivores of Etosha National Park. Ecosphere 4(1):art13. [Google Scholar]
- Veeramah KR, Thomas MG, Weale ME, Zeitlyn D, Tarekegn A, Bekele E, Mendell NR, Shephard EA, Bradman N, Phillips IR.. 2008. The potentially deleterious functional variant flavin-containing monooxygenase 2*1 is at high frequency throughout sub-Saharan Africa. Pharmacogenet Genomics. 18(10):877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramah KR, Wegmann D, Woerner A, Mendez FL, Watkins JC, Destro-Bisol G, Soodyall H, Louie L, Hammer MF.. 2012. An early divergence of KhoeSan ancestors from those of other modern humans is supported by an ABC-based analysis of autosomal resequencing data. Mol Biol Evol. 29(2):617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente M, Jakobsson M, Ebbesen P, Schlebusch CM.. 2019. Genetic affinities among Southern Africa hunter-gatherers and the impact of admixing farmer and herder populations. Mol Biol Evol. 36(9):1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK.. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4(3):e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.