Abstract
The evolutionary transition from invertebrates to vertebrates involved extensive gene duplication, but understanding precisely how such duplications contributed to this transition requires more detailed knowledge of specific cases of genes and gene families. Myogenic differentiation (MyoD) has long been recognized as a master developmental control gene and member of the MyoD family of bHLH transcription factors (myogenic regulatory factors [MRFs]) that drive myogenesis across the bilaterians. Phylogenetic reconstructions within this gene family are complicated by multiple instances of gene duplication and loss in several lineages. Following two rounds of whole-genome duplication (2R WGD) at the origin of the vertebrates, the ancestral function of MRFs is thought to have become partitioned among the daughter genes, so that MyoD and Myf5 act early in myogenic determination, whereas Myog and Myf6 are expressed later, in differentiating myoblasts. Comparing chordate MRFs, we find an independent expansion of MRFs in the invertebrate chordate amphioxus, with evidence for a parallel instance of subfunctionalization relative to that of vertebrates. Conserved synteny between chordate MRF loci supports the 2R WGD events as a major force in shaping the evolution of vertebrate MRFs. We also resolve vertebrate MRF complements and organization, finding a new type of vertebrate MRF gene in the process, which allowed us to infer an ancestral two-gene state in the vertebrates corresponding to the early- and late-acting types of MRFs. This necessitates a revision of previous conclusions about the simple one-to-four origin of vertebrate MRFs.
Keywords: myogenesis, chordate evolution, gene duplication
Introduction
Gene duplication is widespread, but various evolutionary routes are possible for the postduplicate daughter genes. Studying which of these evolutionary routes is taken in specific cases is required to better understand the frequency, significance, and precise nature of these modes of evolution. In addition, duplication can have functional consequences and has, for example, been implicated in the evolution of vertebrate novelties (e.g., neural crest cells; Blomme et al. 2006; MacKintosh and Ferrier 2018) and the high species diversity among teleost fish (Hoegg et al. 2004).
Myogenic differentiation (MyoD) was first discovered as a cell-fate determinant that could convert undifferentiated cells into myoblasts (Davis et al. 1987). It has since become apparent that it is an archetypal developmental regulator of myogenesis. MyoD and its orthologs (e.g., fruitfly Nautilus, sea urchin SUM1, tunicate Ci-MRF) constitute a family of bHLH transcription factors involved in myogenesis across the bilaterians, exemplifying the ancient homology and high level of conservation of significant components of the developmental toolkit. This level of conservation allows for comparative studies of MyoD orthologs across the bilaterians. Throughout the evolution of bilaterians, duplication has shaped this gene family, both on the small scale via segmental duplications (e.g., sea urchin, see below) and the large, as in the two rounds of whole-genome duplication (2R WGD) at the origin of the vertebrates or 3R WGD at the origin of teleost fish.
Following WGD and rediploidization, redundant duplicate genes can be either nonfunctionalized, subfunctionalized, or neofunctionalized (reviewed by Wagner 1998; Shimeld 1999; Lynch and Conery 2000; Prince and Pickett 2002; Innan and Kondrashov 2010; MacKintosh and Ferrier 2018). Gene paralogs that arise from WGD are often referred to as ohnologs (Wolfe 2000). For housekeeping genes, one of the daughter genes tends to be nonfunctionalized (Nakatani et al. 2007), whereas developmental genes with higher regulatory complexity, especially those involved in signaling and transcription regulation like the myogenic regulatory factors (MRFs) are often retained and undergo subfunctionalization. The Duplication–Degeneration–Complementation (DDC) hypothesis explains how mutations in the regulatory regions of duplicated genes eliminates components of their expression so that both genes are expressed as a subset of the ancestral gene’s expression, and both are then required to retain the total ancestral functionality (Force et al. 1999). Complex regulatory elements provide greater opportunity for partitioning of the ancestral gene’s functions among its daughters (Blomme et al. 2006; Jimenez-Delgado et al. 2009). Neofunctionalization can arise when a novel beneficial mutation occurs in one of the daughter genes or its regulatory regions, though this seems to be far rarer than subfunctionalization (MacCarthy and Bergman 2007). Subfunctionalization consistent with the DDC hypothesis, as well as via alternative routes involving dosage sensitivity, is widely observed among genes retained in duplicate following WGDs (Lynch and Conery 2000; Zhang 2003; Kleinjan et al. 2008; Innan and Kondrashov 2010; Braasch et al. 2016). To detect subfunctionalization, preduplicate genomes are essential for comparison.
Amphioxus provide important points of comparison for the assessment of the effect of WGDs since they diverged from the rest of the chordates before the 2R WGD events at the origin of vertebrates, and can be used to infer the ancestral vertebrate preduplicate state more directly than other more distantly related invertebrates. Large-scale comparisons of gene linkage revealed the general one-to-four pattern of synteny between amphioxus and human genomes, thus supporting the role of 2R WGD in the evolution of vertebrates (Putnam et al. 2008). This has resulted in many gene families showing one to four (or fewer due to losses) paralogous relationships (Meyer and Schartl 1999), as has previously been assumed for the vertebrate MRFs. Furthermore, whole-genome-scale comparisons between the expression domains of single amphioxus genes and their vertebrate ohnologs revealed that vertebrate genes retained in duplicate had subfunctionalized and their combined distinct expression domains broadly equated to the single amphioxus genes’ expression, whereas there was no obvious difference in expression between the genes that had reverted to single copy in vertebrates (Marlétaz et al. 2018). Use of a chordate comparison allowed gene expression in more similar homologous structures to be compared between invertebrate chordates and vertebrates, and directly addressed the 2R WGD that occurred on the vertebrate stem. From this recent example, and others, there is clear evidence of subfunctionalization via DDC, but the extent to which this model applies to the MRF family is uncertain.
In vertebrates, there are four types of MRF generally recognized, which fall into two overarching functional groups: MyoD and Myf5 are involved in muscle cell determination, an “early” function, whereas Myog and Myf6 are principally involved in differentiation, a “late” function that is activated by the “early” MRFs (Megeney and Rudnicki 1995; reviewed by Perry and Rudnicki 2000; Moncaut et al. 2013; Buckingham and Rigby 2014; Zammit 2017), possibly as a result of postduplication subfunctionalization. Although each of the four MRFs does have a distinct role in myogenesis, there is some amount of functional redundancy between family members. For instance, knockouts of MyoD or Myf5 result in fairly normal mice, whereas double-knockout of MyoD and Myf5 are lethal and prevent any skeletal muscle forming in the embryo (Rudnicki et al. 1993). In the differentiation phase, Myog is required to activate downstream muscle genes, since Myog knockouts specify the muscle lineage but fail to form muscle fibers, and this cannot be rescued by Myf6 (Buckingham and Rigby 2014). Myf6 seems to act both “early” and “late” in mammals, but its function is redundant to MyoD and Myf5 in the “early” phase, and Myog in the “late,” since Myf6 knockouts have a normal phenotype, though Myf6–MyoD double mutants show a similar phenotype to Myog mutants (Rawls et al. 1995). Although Myf6 can activate the same genes as Myog, Myf6 has been implicated in downregulation of Myog in fully differentiated myoblasts, a distinct role from any of the other MRFs, and one which defines its function as “late” (Buckingham 2017).
In zebrafish, a 3R teleost, there are the same MRFs as in tetrapods, but their expression patterns are somewhat different. As in mice, zebrafish double mutants of Myf5 and MyoD have no muscle, although either single mutant is normal, but for Myf6 and Myog, both are only expressed “late,” downstream of MyoD and Myf5, and expression of either of them is only affected by MyoD/Myf5 double mutants, not single ones (Schnapp et al. 2009). In teleosts with the typical five-gene arrangement (MyoD1, MyoD2, Myog, Myf5, and Myf6), the two paralogs of MyoD have overlapping but distinct expression profiles, consistent with subfunctionalization (Tan and Du 2002). In the 4R salmon, the three reported MyoD paralogs showed two distinct expression patterns (Bower and Johnston 2010), which is consistent with a partitioning of MyoD expression following 3R and 4R WGDs. In Xenopus laevis, the 3R frog, although there are two Myog genes with some common targets, only one activates the expression of the predominant isoform of the myosin heavy chain gene in adult muscle (Charbonnier et al. 2002). These examples show that MRFs have different, though overlapping functions following the various duplication events in different vertebrate lineages. Across all the vertebrates, we see the conservation of this general “early”–“late” distinction, where MyoD and Myf5 act in determination, whereas Myog and Myf6 control differentiation.
In invertebrate bilaterians, MRFs also regulate myogenesis, though most invertebrates have only one MRF gene (e.g., Drosophila Nautilus [Nau], Misquitta and Paterson 1999 and Caenorhabditis elegans hlh-1, Chen et al. 1994). In the echinoderms, the sea urchin has undergone an independent duplication of its MRFs, such that only MyoD2 is required for muscle development, whereas MyoD1 appears to have been co-opted to the skeletogenic lineage (Andrikou et al. 2013). The invertebrate chordate tunicates Ciona spp. have one MRF which activates expression of muscle genes in the tailbud (Izzi et al. 2013), whereas the earliest-branching chordate lineage, the cephalochordates (amphioxus or lancelets), have been reported to have varying numbers of MRFs, from one to three and with confusing nomenclature reflecting ambiguous orthology relationships among these amphioxus MRFs (supplementary information 1, Supplementary Material online; Schubert et al. 2003; Urano et al. 2003; Yuan et al. 2003; Somorjai et al. 2008; Bertrand et al. 2011; Tan et al. 2014). The amphioxus genes for which there is in situ hybridization data show expression in the myotomal component of the somites through development, and distinct patterns for the different genes (Schubert et al. 2003; Urano et al. 2003; Bertrand et al. 2011). However, because these amphioxus data are incomplete and somewhat confusing, the complement of MRFs for the chordates remains unclear.
Aside from the independent duplications in the urchin and amphioxus, invertebrates tend to have one MRF gene that is thought to be pro-orthologous to the four vertebrate MRF ohnolog clades. This might suggest a relationship marked by the vertebrate 2R WGD, where the four vertebrate MRFs are assumed to have arisen from a single invertebrate ancestral gene and duplicated twice in the 2R event, whereas invertebrate lineages typically retained a descendant of the single ancestral gene. This typical view is complicated by the fact that there are multiple amphioxus MRFs, which potentially diverged before the vertebrate expansion (Araki et al. 1996; Yuan et al. 2003). Further complicating the one-to-four assumption, the four vertebrate MRFs lie in only three distinct genomic loci; Myf5 and Myf6 reside in the same locus with extensively overlapping shared regulatory elements (Carvajal et al. 2008), whereas MyoD and Myog each exist as single loci on distinct chromosomes. A straightforward 2R relationship, however, would be expected to generate four paralogous loci in distinct regions of the genome, each containing one MRF.
It appears that both gene duplications and losses have obscured the expected pattern resulting from 2R WGD, though the evolution of the chordate MRFs requires more detailed phylogenetic and synteny analyses to determine the nature of their origins around, and their roles in, the invertebrate-to-vertebrate transition. Here, we show that the ancestral reconstruction for the vertebrate condition for MRFs was misled by an incomplete coverage of the vertebrate MRF gene complement and that the four-gene state of nonteleost vertebrates did not arise from a single ancestral gene undergoing two rounds of duplication. Instead, the pre-2R vertebrate ancestor had two clustered MRFs that, after the 2R events, resulted in a larger number of MRFs that then experienced secondary losses resulting in just the four ohnologs in humans, for example. Independently from this vertebrate MRF expansion, we show that amphioxus tandemly duplicated the MRF gene to produce a five-gene cluster, which has in-turn experienced subfunctionalization of the constituent genes, such that distinct amphioxus MRFs have differing expression patterns during myogenesis in the somites.
Results
Finding MRF Genes: New Genes and Clusters across the Bilaterians
A thorough gene search revealed several sequences previously unannotated or missing from the literature, especially in a few species with interesting duplication histories (supplementary table 1, Supplementary Material online). Although the echinoderm duplication is well-characterized for the urchin Strongylocentrotus purpuratus, we also found orthologs of MyoD2 in Lytechinus variegatus, Acanthaster planci, and Patiria miniata, and confirmed the clustered arrangement for the first three of these species (the low-quality assembly around this locus in the P. miniata genome prevented confirmation of these genes’ arrangement). The clustered arrangement of these genes in two species of urchin and one sea star suggests they originated via an echinoderm-specific tandem duplication.
In the salmon genome, which we included to assess the effect of a 4R WGD, previous reports describe single Myf5 and Myf6 genes, located on separate chromosomes (Bower and Johnston 2010). However, we found that there are in fact two Myf5–Myf6 clusters at two distinct chromosomal locations. Furthermore, we identified a fourth MyoD ortholog, which brings the total number of MRFs up to nine, when previously only six had been identified (Macqueen and Johnston 2006; Bower and Johnston 2010) (supplementary table 1, Supplementary Material online). This arrangement appears to be consistent with a duplication of the typical teleost arrangement (2× [MyoD1, MyoD2, Myog, Myf5, and Myf6]) followed by loss of one of the Myog paralogs.
Searches through representative vertebrate genomes revealed the existence of a novel MRF, which we here name Myf7, in the genomes of the coelacanth, spotted gar, and sterlet (supplementary information 2, Supplementary Material online; though the sterlet genome assembly available at the time was not good enough for its MRFs to be included in our subsequent analyses). This gene is located upstream of MyoD (10 kb in gar, 32 kb in coelacanth), with a gene related to ferritin in-between (zgc:172145; GO:0008199).
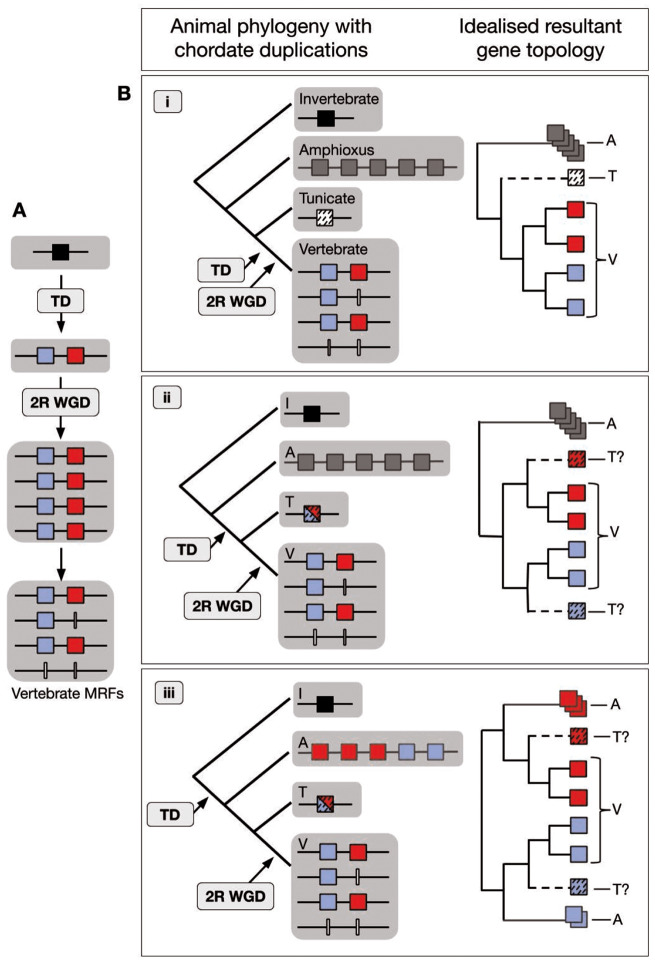
Building on the various numbers of MRFs reported for amphioxus (see Introduction; supplementary information 1, Supplementary Material online), we found a five-gene cluster of MRFs in the amphioxus species Branchiostoma floridae, B. belcheri, and B. lanceolatum (supplementary table 1, Supplementary Material online). We also found incomplete sequences of all five MRFs in the genome of Asymmetron lucayanum, a sister lineage to the Branchiostoma genus, but the quality of the assembly prevented confirmation of the cluster in this species and the sequences were not complete enough to include in our phylogenies (supplementary table 1, Supplementary Material online). Nevertheless, the presence of five MRFs in the four amphioxus species surveyed suggests that the expanded cluster may be typical for lancelets as a whole. In addition, these genes share a similar exon structure to other MRFs in both invertebrates and vertebrates. Taken together, the similar exon structure and clustered arrangement of the amphioxus MRFs led us to conclude that the genes originated via tandem duplications of a prototypical chordate MRF to produce a five-gene cluster in the ancestral cephalochordate. This expansion raises multiple possibilities for the relationships among the chordate MRFs, and the timing of the various duplications relative to the divergence of the different chordate groups. The three principal possibilities are illustrated in figure 1 and discussed below. Each of these three scenarios could generate a different phylogenetic topology. Therefore, from the phylogeny, the history of the genes could potentially be determined.
Fig. 1.
Schematic of chordate MRF evolution based on two-gene ancestral state hypothesis. (A) The tandem duplication (TD) predates the 2R WGD at the base of the vertebrates to generate the current vertebrate MRF gene complement. One ancestral MRF (black square) is duplicated in tandem to generate the two vertebrate MRF groups, (blue: Myog/Myf6 type; red: MyoD/Myf5 type), which via 2R WGD and subsequent gene losses (white lines), result in the vertebrate arrangement. (B) Three potential scenarios of the timing of the TD relative to the divergence of the three chordate phyla and the resultant topology of the MRF phylogeny. (i) Both amphioxus (A) and tunicates (T) diverge before the TD. (ii) The TD is after the divergence of A, so the A MRFs are an outgroup to the vertebrate (V) and T MRFs, but the TD predates the divergence of T from V so the single T MRF (red or blue, dashed) evolved from either the Myog/Myf6 or MyoD/Myf5 precursor, depending on which was retained. (iii) The TD predates the divergence of both the A and the T lineages. The T gene clusters either with the Myog/Myf6 or MyoD/Myf5 vertebrate groups depending on which precursor was retained. The five A MRFs similarly cluster with “either” or “both” vertebrate groups in any arrangement (shown here as three with red and two with blue), depending on which of the two post-TD genes were retained and/or duplicated.
Multiple Instances of Duplication and Loss and a Two-Gene State for the Vertebrate Ancestor
The phylogeny of MRFs from representative bilaterian taxa is consistent with several instances of duplication and subsequent gene loss (fig. 2). Our phylogenetic analyses are consistent with a single ancestral bilaterian MRF, since all the protostomes surveyed have one MRF orthologous to fruit fly Nautilus (fig. 3 and supplementary information 3a–c, Supplementary Material online). However, we infer that the two-gene state in echinoderms, where MyoD1 and MyoD2 are clustered in tandem is ancestral to the ambulacrarians. The hemichordates included in this study have an ortholog of MyoD1, but have likely lost their MyoD2 ortholog, as evidenced by the closer relationship between the hemichordate MyoD genes and the echinoderm MyoD1 than with the echinoderm MyoD2 (fig. 3). Furthermore, although this two-gene state for the ambulacrarians might indicate a possibility for a two-gene ancestral state for the deuterostomes as a whole, we consider this less likely than a single-gene ancestral state. This is due to the topology separating the ambulacrarian genes (ambulacrarian clade support: 49/0.84/26) from the chordate genes (17/0.44/35) and the amphioxus genes from those of the Olfactores (56/0.92/51). Had the duplication occurred earlier, we would expect a topology where the duplication node separates two clades with invertebrate deuterostome and vertebrate genes distributed in both, and the single protostome MRF as a sister outgroup. Though our node support values are low, our phylogeny is more consistent with a single-gene ancestral state that independently expanded by duplications in each of the ambulacrarian, amphioxus, and Olfactores clades (fig. 2). In the echinoderms, evidence of subfunctionalization, or potentially neofunctionalization, following duplication has been documented, as of the two MRF paralogs in the urchin, MyoD1 is co-opted to the skeletogenic lineage, whereas MyoD2 has retained the ancestral myogenic function (Andrikou et al. 2013). The hemichordates have retained just the MyoD1 paralog since the hypothesized ancestral ambulacrarian duplication, though it is not known if this gene has retained a myogenic function. Andrikou et al. (2013) report a third MRF in the urchin S. purpuratus, however, this gene showed no clear relationship to the other MRFs in our early phylogenetic analyses, so this gene was excluded from the final trees.
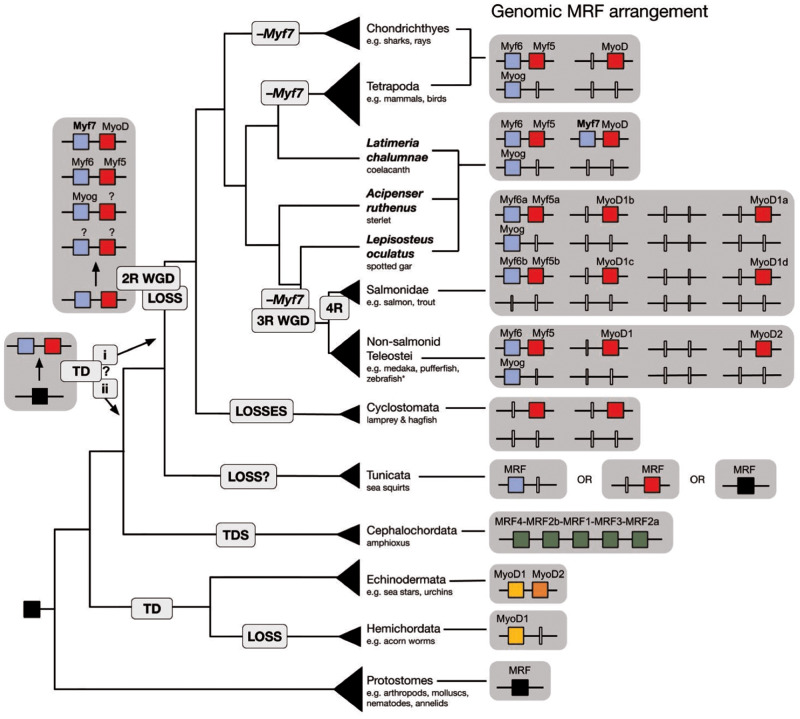
Fig. 2.
Schematic of bilaterian MRF evolution. Genomic MRF arrangement (right) for taxonomic groups (species cladogram, left) included in this study highlighting the vertebrate species that have retained Myf7 (coelacanth, sterlet, and spotted gar, bold text), as well as duplications and losses in several lineages. Retained MRFs are represented by colored boxes, whereas genes inferred to have been lost are represented by white vertical lines. Genes clustered in the genome are joined by black lines. The uncertainty as to the timing of the vertebrate early–late tandem duplication in the vertebrate (i) or Olfactores (ii) ancestor is denoted by the “?.” *Zebrafish has lost its MyoD2 paralog but is otherwise like the rest of nonsalmonid teleosts. TD, tandem duplication; 2R WGD, two rounds of whole-genome duplication; 3R WGD, teleost-specific third round of whole-genome duplication; 4R, salmonid-specific fourth round of whole-genome duplication; –Myf7, inferred loss of Myf7.
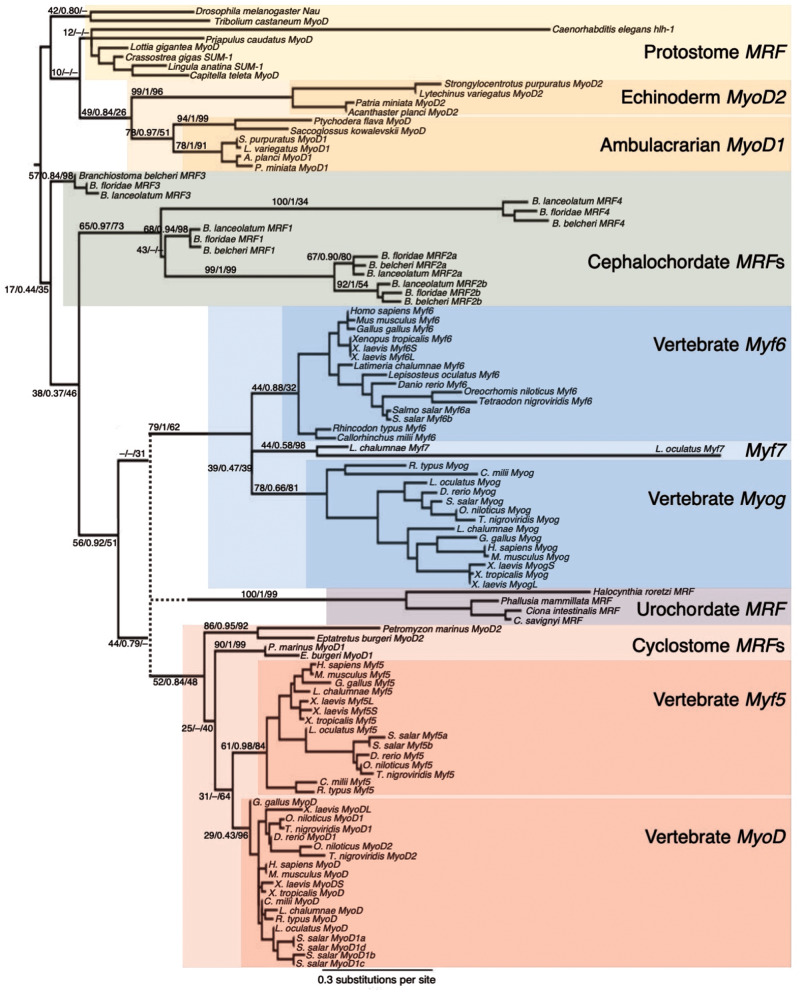
Fig. 3.
Maximum likelihood phylogeny of bilaterian MRFs. Major branches defining certain gene types, (e.g., the urochordate MRFs) are labeled with support values from three phylogenetic methods: ML (% 1,000 bootstraps)/BI (posterior probability)/NJ (% 1,000 bootstraps); values are represented by dashes when a certain topology was not present in that phylogeny. The tree is rooted on the branch separating the chordates from other bilaterians. The dotted line and corresponding support values represent the uncertainty of the placement of the urochordate sequences with support in ML and BI (44/0.79/–) and NJ (–/–/31). Colored boxes indicate gene types. The scale bar represents substitutions per site for the consensus ML tree (though not for the ambiguous urochordate branch; its node was manually drawn).
Within the chordates, the phylogeny supports the independence of the amphioxus expansion as first proposed by Araki et al. (1996), since all five amphioxus MRFs branch within the chordate clade, albeit with low support (chordate clade support: 17/0.44/35), but outside the Olfactores clade (Olfactores [vertebrates and urochordates] clade support: 56/0.92/51) (fig. 3). Thus, we infer that this five-gene amphioxus cluster diverged from a one-gene ancestral state, that is, before the duplication that gave rise to the two vertebrate MRF types (fig. 2). Two of the amphioxus MRFs share a first exon sequence (supplementary information 4, Supplementary Material online) and this exon is more similar within species than within genes (fig. 4B). Although a phylogeny of the second and third exons results in a topology where each gene forms a distinct clade (fig. 4C), the phylogeny of the first exon sequence has each species’ MRF2a and MRF2b clustering together within a larger overall MRF2a/MRF2b clade (fig. 4B).
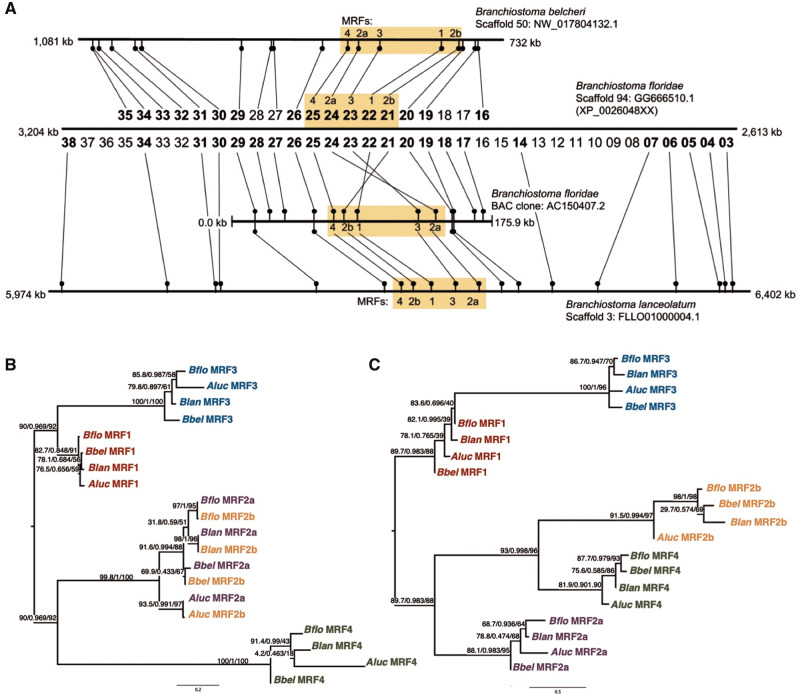
Fig. 4.
Physical and phylogenetic relationships of the five amphioxus MRFs. (A) Genomic arrangement of five-gene MRF cluster in three Branchiostoma spp. genomes. The MRF scaffolds from the B. belcheri, B. floridae, and B. lanceolatum genome with orientation of the MRF clusters compared with the orientation of flanking genes. Genes are from predicted B. floridae models with corresponding protein accession numbers in the format XP_00206048–.1 (e.g., MRF4 is XP_0020604825.1). Bold numbers are genes with orthologs confirmed on the other scaffolds using TBlastN. (B) Phylogeny of MRF exon one. IQ-TREE ML phylogenies with ultrafast bootstrap support, SH-aLRT support, and approximate Bayes posterior probability values. (C) Phylogeny of MRF exons two plus three. As in (B), separate exon alignments were made from sections of the full sequence of all five amphioxus MRFs (supplementary information 4a, Supplementary Material online).
The placement of the urochordate MRF gene could not be reliably determined by the three phylogenetic methods, probably because of the lack of conservation outside of the bHLH domain and its long, divergent sequence, especially that of the N-terminal domain (fig. 3, Ratcliffe et al. 2019). Even editing the alignment to reduce the inclusion of divergent urochordate sequence did not significantly improve resolution. Nevertheless, all three phylogenetic methods placed the urochordate sequences as sister to one of the vertebrate clades, and there is moderately high support for the branch that groups the Olfactores sequences together (support: 56/0.92/51) (fig. 3). From this, we tentatively conclude that the urochordate MRF represents an ortholog of one of the ancestral vertebrates’ two pre-2R genes, though to which one it is more similar could not be determined (supplementary information 3d and e, Supplementary Material online; clustered with MyoD/Myf5: BI: 0.79 and ML [with cyclostomes]: 44; with Myog/Myf6: ML [without cyclostomes]: 52 and NJ: 31), and this topology, albeit uncertain, supports the scenario where the urochordate sequence diverged from the ancestral two-gene state. However, the low node support values and differing topologies between the phylogenetic methods mean that we cannot confidently exclude the possibility that the urochordate MRF is instead directly descended from the ancestral single-copy state of the chordate ancestor.
In the vertebrates, we infer an ancestral two-gene state, originating from a tandem duplication of the ancestral chordate MRF. It was previously thought that the four vertebrate MRFs arose from a single gene, duplicated twice via 2R WGD, but this hypothesis would require a highly specific translocation to produce the Myf5–Myf6 cluster. This now seems unlikely because of our discovery of Myf7 (fig. 2). This new MRF is adjacent to MyoD in a two-gene cluster in the coelacanth, spotted gar, and sterlet, which share an ancestor at the origin of all bony fish (Osteichthyes) (fig. 2). Therefore, in three independent cases: at the base of the tetrapods, within the ray-finned fish (between the origins of the Neopterygii and Teleostei), and within the cartilaginous fish (Chondrichthyes), we infer losses of the ortholog of the Myf7 gene (fig. 2). Myf7 is located in the Myog/Myf6 clade in the phylogeny, which means the MyoD–Myf7 cluster with one gene of each type mirrors the Myf5–Myf6 cluster throughout the vertebrates. Based on the topology of the phylogeny (fig. 3), and the grouping of the urochordate MRFs within the vertebrate clade(s) (figs. 1B.ii and 2), we now tentatively infer that a tandem duplication at the base of the Olfactores generated two types of MRF, “early” like MyoD and Myf5, and “late” like Myog and Myf6. After the divergence of the urochordates, 2R WGD then duplicated this cluster twice, followed by several losses to generate the vertebrate MRF gene complement: the MyoD–Myf7 cluster, the Myf5–Myf6 cluster, and Myog. The fact that each of these clusters has one gene of each type of MRF, “early” and “late,” makes our “two-gene state followed by 2R” hypothesis the most straightforward explanation for the origin of the vertebrate MRF condition and suggests that the role of gene loss in the evolution of this gene family had previously been underestimated.
In addition to the urochordate MRF proving difficult to place in the phylogenetic tree, the cyclostome sequences also have a poorly resolved location in the phylogeny. Nevertheless, we can reliably infer that the two genes in the lamprey and hagfish genomes, named MyoD1 and MyoD2 (not corresponding to teleost or echinoderm paralogs), belong to the vertebrate MyoD/Myf5 clade (fig. 3). However, whether these genes represent divergent orthologs of MyoD and Myf5 themselves, or orthologs of the MyoD/Myf5-type genes lost from other vertebrates is not clear.
We also observe the effect of lineage-specific duplications on the MRF family, particularly following the 3R teleost-specific and the 4R salmonid-specific WGDs (fig. 2). Following the 3R teleost-specific WGD, the teleosts retained only one of the MRFs in duplicate (paralogs MyoD1 and MyoD2), whereas the other genes returned to the single-copy state. This is an instance of secondary gene loss (nonfunctionalization), which is even more pronounced in the zebrafish that have lost the ortholog of MyoD2 as well. In the salmon, we find nine MRFs: two orthologs each of Myf5 and Myf6, one of Myog, and four of MyoD. This appears to be a direct duplication of the typical teleost arrangement consistent with the 4R salmon-specific WGD, followed by the loss of a second Myog ortholog. However, interestingly the four salmon MyoD orthologs do not separate along the MyoD1/MyoD2 divide in the phylogeny (fig. 3), and previously, the three known MyoD genes were named MyoD1a, MyoD1b, and MyoD1c and were hypothesized to have originated independently of the 4R WGD. This intriguing salmon pattern is clarified by an examination of gene synteny and conservation across the vertebrate MRF loci (see below).
Conserved Synteny and Ghost Loci
Across the vertebrate phylogeny, we see evidence of gene losses, including the inferred loss of all MRFs from the fourth 2R WGD-generated locus, leaving a ghost MRF locus that is widespread across vertebrates. Investigation of the gene neighborhoods of the three retained MRF loci reveals a pattern of 4-fold synteny in the vertebrates, linking the three MRF-bearing chromosomes and another chromosome in certain species, an MRF ghost locus (supplementary information 5a–d, Supplementary Material online). In the human genome, the MRFs are located on chromosomes 1 (Myog), 11 (MyoD), and 12 (Myf5 and Myf6), whereas paralogs of MRF gene neighbors in gene families with four or fewer genes are also found on chromosome 19 (supplementary table 2 and information 5a–d, Supplementary Material online). Other studies highlighting 4-fold paralogy of the human genome also group chromosomes 1, 11, 12, and 19, (Dehal and Boore 2005; Putnam et al. 2008; Srivastava et al. 2008; Craxton 2010; Bertrand and Escriva 2011). From this network of paralogous genes linking these human chromosomes, we found the orthologs of these genes in other species, which revealed a pattern of conserved 4-fold synteny across the subset of vertebrate genomes analyzed (though the ghost locus was not intact in the chicken genome and it is uncertain if there is one in the lamprey given the state of the assembly) (supplementary table 2a–c, Supplementary Material online). There is also evidence for instances of rearrangement, including chromosomal fusion and fission in the mouse–human comparison, consistent with high levels of rearrangement in the mouse genome (Bourque et al. 2004).
Despite several instances of rearrangement, we were still able to find evidence of four conserved neighborhoods linked by ohnologous gene families, supporting a 2R history of the vertebrate MRF loci. Furthermore, 8-fold synteny was revealed in the zebrafish genome, as a result of the 3R WGD, which builds on Macqueen and Johnston’s (2008) identification of the MyoD2 ghost locus (supplementary table 2a–c, Supplementary Material online). In the case of the salmon MyoD gene phylogeny mentioned above, we find evidence that suggests a history consistent with direct duplication via 4R: the four salmon MyoD loci reside on two pairs of homologous chromosomes (Lien et al. 2016), and two of the loci appear to have homology to the MyoD1 locus in 3R teleosts (MyoD1b and MyoD1c), whereas the other two match more closely to the MyoD2 locus (MyoD1a and MyoD1d), including the ghost MyoD2 locus in zebrafish identified by Macqueen and Johnston (2008) (supplementary table 2d, Supplementary Material online). This then indicates that the four MyoD genes in salmon secondarily evolved to converge on a distinct MyoD sequence without particular similarity to the teleost MyoD1 versus MyoD2 genes, as evidenced by the phylogeny.
Conserved synteny can also be traced back to the base of the chordates, where we find significant conservation of orthologs shared between the amphioxus MRF locus and two of the four 2R ohnologous chromosomes in human. There was a strong signal between the B. lanceolatum MRF neighborhood and human chromosomes 11 and 12 (binomial test, 11: P < 2 × 10−3, 12: P < 1 × 10−2; Barnard’s exact test, 11: P < 1 × 10−10, 12: P < 2 × 10−8; supplementary information 6c–d, Supplementary Material online), whereas there was no significant association observed with chromosome 1, and only a marginally significant association with 19 in one of the tests (Barnard’s exact test, P < 0.05). Nevertheless, our findings are consistent with previous comparisons of human and amphioxus orthology. Putnam et al. (2008) found significant conserved synteny between several B. floridae scaffolds (which we found are homologous to the larger MRF scaffold in B. lanceolatum) and the ancestral linkage group #14, which corresponds to sections of human chromosomes 11 and 19. The homology between the reconstructed ancestral linkage groups and the human genome was updated by Srivastava et al. (2008), who link the same regions of chromosomes 11 and 19 found by Putnam et al. (2008), as well as regions of chromosomes 1 and 12 in an ancestral linkage group renamed as #2 (supplementary information 6e, Supplementary Material online). Many of the human orthologs of B. lanceolatum MRF neighbors that we assigned are located in regions of the human chromosomes corresponding to this ancestral linkage group, suggesting that this linkage group contains the ancestral chordate MRF locus. This further supports the 2R history of the vertebrate MRF arrangement and illustrates the one-to-four relationship of the MRF loci in amphioxus and human (supplementary information 6, Supplementary Material online).
Subfunctionalization of Amphioxus MRFs
The distinct roles of the two daughter types of MRF in vertebrates, as well as the functional distinction between the two MyoD genes in the urchin, have been well documented (see Introduction). Similarly, early studies characterized distinct roles for the few then-identified MRFs in amphioxus (see Introduction). Now, with the complete set of amphioxus genes, we generally see that the amphioxus MRFs are expressed in the mesodermal cells surrounding the blastopore in the late gastrula stage, and later in the somites continuing through the early larval stage (fig. 5 and details supplementary information 7, Supplementary Material online), consistent with previous descriptions of expression (Schubert et al. 2003; Urano et al. 2003), though there are clear differences in expression of these five genes indicating that they have undergone subfunctionalization. There are also several differences in expression between the two amphioxus species we studied, though some may be due to different developmental rates.
Fig. 5.
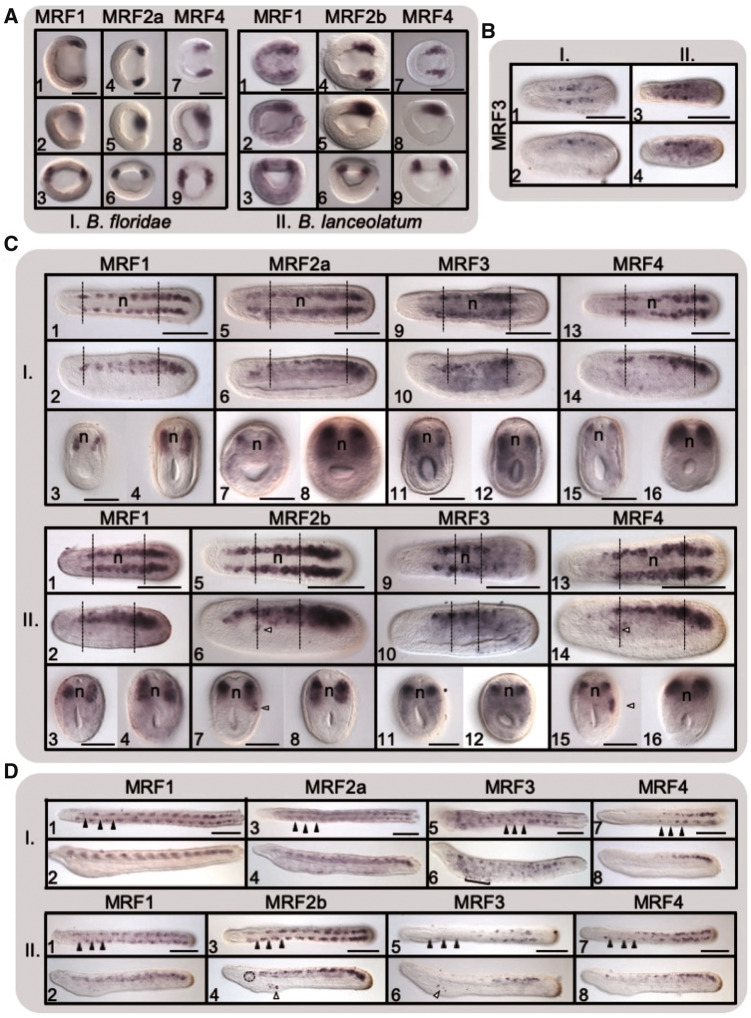
Expression time-course of amphioxus MRFs. WMISH of (A) late gastrula, (B) early neurula (up to 8 somites), (C) mid-late neurula (10–12 somites), and (D) early larva (12–15 somites) stages of (I) Branchiostoma floridae and (II) B. lanceolatum embryos. For both species at all stages, the top rows show dorsal views and the second rows show lateral views (anterior is to the left, scale bars represent 100 µm). For (A.I.) and (A.II.), the bottom row is the posterior view from the blastopore (dorsal to top). For (C.I.) and (C.II.), the bottom rows are cross-sections through the dotted lines in the anterior (left) and posterior (right) somites (scale bars represent 50 µm). Full arrowheads in (D.I.) and (D.II.) represent somite boundaries. Nonmyotomal expression is denoted in the neurulae with the open arrows (C.II.6 and II.7: expression of MRF2b in the somatic [parietal] layer left of the third somite and C.II.14 and II.15: expression of MRF4 in the somatic [parietal] layer of mesoderm of the third anterior left somite) and in the larvae with the square bracket (D.I.6: pharyngeal expression of MRF3), open arrowheads (D.II.4: expression of MRF2b in pharyngeal gill slits and D.II.6: expression of MRF3 in the mouth rudiment) and the circle (D.II.4: expression of MRF2b in the wall of the preoral pit).
The amphioxus MRFs are expressed in distinct regions of the somites at different stages of development. In B. floridae, MRF1, MRF2a, and MRF4 are first detected in the gastrula in two regions at the lip of the blastopore that are consistent with locations for presomitic mesoderm (fig. 5A;MRF2b expression shown in supplementary information 7, Supplementary Material online). MRF3, however, is first detected in the early neurula, expressed only in the central somites (fig. 5B.I). In the late neurula stage, MRF1 and MRF2a are expressed in all the somites, with a stronger expression signal extending further ventrally in the more posterior somites (fig. 5C.I). At this stage, MRF3 expression pattern expands from just the central somites to all the somites except the most anterior pair and those in the tailbud (fig. 5C.I). In the late neurula, MRF4 expression is similar to that of MRF1 and MRF2a, but it is not detected in the central somites, leaving a gap between the anterior and posterior regions of expression (fig. 5C.I.13 and I.14). Within the somites, MRF1, MRF2a, and MRF4 expression extends ventrally in more posterior somites but is restricted dorsally in anterior somites (fig. 5C.I), whereas MRF3 expression extends ventrally in all the somites in which it is expressed (fig. 5C.I.12). The expression patterns of MRF1, MRF2a, and MRF3 continue as before along the anterior–posterior array of somites into the late larval stage (fig. 5D.I), but MRF4 is detected solely in the posterior somites (fig. 5D.I.7 and I.8), and by the late larval stage, only MRF1 is still detected (supplementary information 7, Supplementary Material online).
The MRF expression pattern is generally similar in B. lanceolatum, however it is MRF2b that is detected throughout most of the anterior–posterior extent of the somites just as MRF2a is in B. floridae (B. lanceolatum MRF2a expression in supplementary information 7, Supplementary Material online). MRF1, MRF2b, and MRF4 follow the pattern of B. floridae MRF1, MRF2a, and MRF4, respectively (fig. 5A–D.II). MRF4 remains expressed in the central somites in the late neurula stage, that is, there is no gap (fig. 5C.II.13 and II.14), but in the larva, we see the same restriction of MRF4 to the more posterior somites as in B. floridae (fig. 5D.II.7 and II.8). Within the more posterior somites, expression is expanded further ventrally than in the anterior somites for MRF1, MRF2b, and MRF4, as in B. floridae (fig. 5C.II). MRF3 is again first detected in the early neurula and is only seen in the central somites (fig. 5B.II), but it remains in a more restricted anterior–posterior array of somites than the more extensive expression seen in B. floridae (fig. 5C–D.II). Overall, we observe some differences in the expression of the MRFs, but for the most part there are similar expression patterns for each gene in the two species, therefore, we see a similar pattern of subfunctionalization of the five amphioxus MRFs.
Some of the MRFs possibly also exhibited nonmyotomal expression in the presumptive precursors to the orobranchial musculature. This was not detected in all the larvae and differs between the species; therefore, these patterns are reported with caution. MRF3 was detected in the prospective gill slits of one-third of B. floridae larvae (fig. 5D.I.6), and in the mouth rudiment of half of the B. lanceolatum larvae (fig. 5D.II.6). Branchiostoma lanceolatum MRF2b was detected in the first primary gill slit (fig. 5D.II.4) in all larvae.
Discussion
Careful manual curation of available genomic sequence data combined with newly available whole-genome sequences has enabled us to produce a picture of MRF evolution at the invertebrate–vertebrate transition that is significantly different from the previous consensus. Automatic or pipeline annotations frequently make errors in predicting gene orthologies, which can drastically mislead evolutionary reconstructions and biological inferences. This is particularly evident in the case of Myf7 (supplementary information 2, Supplementary Material online), a vertebrate MRF described here for the first time. The way these Myf7 genes were previously annotated gave the impression that the Myf5 gene had translocated from adjacent to Myf6, to be adjacent to MyoD. It is now clear that Myf7 represents a novel MRF, linked to MyoD in the genomes of the coelacanth, sterlet, and gar. Based on our phylogeny, Myf7 is more similar to the Myog and Myf6 clade than the MyoD and Myf5 clade, so that the MyoD–Myf7 cluster mirrors the Myf5–Myf6 cluster conserved across the vertebrates. The presence of two parallel clusters suggests the origin of the vertebrate MRFs from a single two-gene cluster that predates the 2R WGDs at the base of the vertebrates.
It has long been recognized that the vertebrate MRFs fall into two distinct functional types, which also relates to their paired grouping in phylogenetic trees. MyoD and Myf5 are most closely related to each other in phylogenies and these are the MRFs expressed first during myogenesis, during the determination phase. Myf6 and Myog are then expressed in the next phase of myogenesis controlling the differentiation into muscle fiber cells, and Myf6 and Myog are in-turn most closely related to each other. The grouping of Myf7 with the Myog/Myf6 group suggests that Myf7 might also have a “late” rather than “early” role in myogenesis, though its function has yet to be addressed. Our finding that the pre-2R vertebrate ancestor had two rather than one MRF correlates precisely with this expression and functional data and we hypothesize that the vertebrate ancestor had a two-gene cluster with a MyoD/Myf5 precursor that controlled myogenic determination and a Myf6/Myog precursor that directed myogenic differentiation. This suggests that there was a partitioning of the ancestral MRFs role following the duplication that generated the two MRF types, leading to the early- and late expression of the subfunctionalized daughter genes.
The duplication that gave rise to the two functional types of vertebrate MRF, “early” and “late,” predates the 2R WGDs. These large duplication events have been implicated in many vertebrate advancements that also occurred at this time (Holland et al. 1994; Shimeld and Holland 2000; Blomme et al. 2006; MacKintosh and Ferrier 2018). Before that, however, is the duplication followed by subfunctionalization that resulted in the two functional types of MRF. This ancestral vertebrate MRF duplication may be linked to increasing complexity not only in the myogenic regulatory network but also in possible vertebrate-specific muscle innovations like myoblast fusion to produce multinucleate muscle fibers compared with the ancestral chordate, which may have had mononucleate muscle fibers or lamellae as in amphioxus (Peachey 1961; Flood 1967). Furthermore, the four vertebrate MRFs act in distinct but overlapping patterns, inferred to be specific subsets of the expression of an ancestral gene, but they have also subfunctionalized in sequence. The helix-III domain confers target specificity and differs between the two gene types, “early” and “late,” and is linked to the differentiation of their roles in regulating myogenesis (Bergstrom and Tapscott 2001). The pre-2R tandem duplication may have had important functional consequences for myogenesis, as has been also inferred for the 2R WGDs and development as a whole.
Among the vertebrates, further WGDs have shaped the MRFs. Although the instances of non- and subfunctionalization of the MRFs following the teleost 3R are well characterized (e.g., the subfunctionalization of MyoD1 and MyoD2; the losses of paralogs of Myog, Myf5, and Myf6 and MyoD2 in zebrafish), only using the new assembly of the salmon genome from 2016 (Lien et al. 2016) could we identify a complement of nine salmon MRFs, thus extending beyond the six previously described genes. The synteny analyses of Lien et al. (2016) revealed the distribution of paralogous segments of the salmon chromosomes that originated in the 4R salmon-specific-WGD, which in-turn informed our synteny analysis, especially when characterizing the four MyoD loci. This new extended complement and classification of the salmon MRFs has implications for the expression studies of these genes, as the high-sequence similarity between MyoD1a and the novel MyoD1d suggest that previous assays on MyoD1a would also detect MyoD1d and the biphasic expression observed by Bower and Johnston (2010) could reflect two different or overlapping expression domains of MyoD1a and MyoD1d, respectively.
The new assembly of the B. lanceolatum genome (Marlétaz et al. 2018) was also instrumental in our analyses, as it allowed for a more thorough comparison of a larger contiguous amphioxus locus than were available from the B. floridae or B. belcheri genome assemblies. This MRF scaffold also had many more genes annotated to it than the previous assembly, which allowed us to use it as a starting point for the reciprocal BlastP searches to detect synteny with vertebrate loci. In the comparison between the human and amphioxus loci, we only found a strong signal of orthology for human chromosomes 11 and 12 to the amphioxus MRF locus, but we did not find strong signal associating the amphioxus MRF neighborhood to human chromosomes 1 or 19. These chromosomes (11 and 12) also have a larger component of ancestral linkage group #2 (Srivastava et al. 2008) (supplementary information 6, Supplementary Material online), which is only homologous to small regions on human 19 and 1. Although human chromosome 19 has the ghost locus (supplementary table 2 and information 5, Supplementary Material online), and therefore, we know has lost an MRF and potentially several neighbors, it still retains more orthologs to the B. lanceolatum MRF neighborhood than expected by chance, as detected by one of our tests (supplementary information 6, Supplementary Material online). Chromosome 1, despite having the third largest number of 1:1 orthologs with the B. lanceolatum MRF scaffold, did not show statistical significance, perhaps because it is the largest, most gene-rich chromosome, and/or has undergone more rearrangements, as this chromosome has homology to more linkage groups than any of the others considered, all of which may have diluted the signal of the 2R homology.
Although there is only one MRF locus in amphioxus, compared with three (and a ghost) in human and other tetrapods, amphioxus underwent an independent expansion of their MRFs at this locus. Because the five-gene cluster was found in three Branchiostoma species, and the presence of the five genes was confirmed in Asymmetron lucayanum, we can infer that this expansion occurred in their ancestor and, due to the clustered arrangement, the expansion likely arose via a series of tandem duplications. Interestingly, the protein sequence of the first exon of MRF2a and MRF2b is identical, hence their naming, whereas the sequences for the second and third exons are not more closely related than to other genes, rather MRF4 is grouped with MRF2b, before MRF2a. The phylogeny of the first exon amino acid sequences groups the MRF2a and 2b proteins together with a topology that matches the species tree, whereas the phylogeny for the second and third exons separates the two genes into distinct clades (fig. 4B and C). Perhaps this is a result of gene conversion between the first exons of the two MRF2 genes, or it reflects a more recent duplication and more constraint on the sequence of the first exon. However, both of these hypotheses are somewhat compromised by the locations of the genes in the cluster, with MRF2a and 2b being separated by the MRF3 and 1 genes.
There is some discrepancy between genome assemblies with regards to the arrangement of the amphioxus MRFs (fig. 4A). In the B. floridae and B. belcheri genome assemblies, the order is MRF4–MRF2a–MRF3–MRF1–MRF2b, whereas in the B. floridae BAC clone and the newer B. lanceolatum assembly, the order is MRF4–MRF2b–MRF1–MRF3–MRF2a, though the neighboring genes in the B. floridae genome are also present around the MRF cluster in the same arrangement in B. lanceolatum, such that it is only four of the five MRFs that are differently arranged (fig. 4A). Long-range PCR experiments have confirmed the arrangement of the BAC clone (Coll-Lladó C, unpublished data), but have not resolved the gene arrangement in either B. floridae or B. lanceolatum genomic DNA. Whether this discrepancy is due to an assembly error in one or other amphioxus genome, perhaps conflated by the near-identical nucleotide sequences for the first exon of MRF2a and MRF2b, which sit at either side of the four genes that are “flipped” (fig. 4A), remains to be resolved.
The independence of the amphioxus MRF expansion was first hypothesized by Araki et al. (1996), based on their phylogeny of vertebrate MRFs and the two amphioxus MRF genes known at that time in B. floridae. They also were the first to present the question as to whether there was an ancestral one- or two-gene state. Critically, they did not include the occurrence of the 2R WGD on the vertebrate stem in their analysis and inferred instead that the vertebrate arrangement arose via a tandem gene duplication to generate a two-gene cluster (either before or after the divergence of amphioxus from the vertebrates), which then itself duplicated in vertebrate evolution, followed by the separation of the MyoD and Myog genes onto distinct chromosomes. This was echoed by Yuan et al. (2003) and Schubert et al. (2003). We have updated their possible scenarios to include the five-gene cluster in amphioxus, the one-gene state of the urochordates, and the impact of 2R on the vertebrate arrangement. This gave three possible scenarios (fig. 1B). The combination of our phylogeny and synteny analysis allowed us to confirm the independence of the amphioxus expansion, and that the vertebrate arrangement arose from a two-gene state.
The many instances of subfunctionalization, or reductions of the expression of retained semiohnologs, observed between amphioxus and the vertebrates (Marlétaz et al. 2018) could help explain the increased complexity of vertebrates, specifically via their gene regulatory landscapes, and the MRFs contribute to this model. However, the MRFs are a particular case where there has been an independent expansion and subfunctionalization in the preduplicate lineage as well. It has even been suggested that complexity evolved convergently among the chordates since the single Ciona MRF gene has two splice variants that are differentially expressed in an early-versus-late pattern which mirrors that of the vertebrates (Meedel et al. 1997). It is unclear whether the Ciona MRF splice variants are also expressed in different parts of the embryo, and whether it does indeed have an autoregulatory role in the switch between the short and long variants (Meedel et al. 2002). Still, coupled with the independent amphioxus expansion, this shows that myogenesis in the preduplicate invertebrate chordates is more complex than previously thought.
In the invertebrate chordate amphioxus, we find overlapping but distinct expression patterns of the five MRFs, as well as a few differences in expression in the two species assessed. The pattern of MRF1 expression was the same for both species as has been previously reported for these species (Somorjai et al. 2008). In addition, MRF3 is first expressed at a later stage relative to the other MRFs in both species, and MRF4 is restricted to the posterior of the larvae of both species. Between the species, MRF2a and MRF2b likely show differences in their expression patterns, however, this data must be interpreted with caution due to the similarity of the two sequences. Amid similar patterns of expression between the species, there are a few key differences. Notwithstanding the different rates of development between these two species, MRF4 expression differed between the species throughout the neurula stage, as it was present in central somites in B. lanceolatum but not B. floridae. MRF3 differed between the species in the region of the somite in which it was expressed in the late neurula stage. The different potential nonmyotomal expression of the MRFs is not conclusive at this point in time, but may demonstrate differences between the species in MRF expression in the orobranchial musculature. In total, with the conclusion that the amphioxus MRF expansion is ancestral to the cephalochordates, our results suggest that subfunctionalization of these five genes is also ancestral, and any differences between species are more recent, as the pattern of gene expression is generally consistent across the two species throughout the stages of development assessed here.
This expansion is consistent with the DDC hypothesis and also indicates a more complex regulatory network for myogenesis in the amphioxus than previously thought, as the number of MRFs has previously been underestimated. Although distinct expression patterns for MRF1 and MRF2a were characterized previously (Schubert et al. 2003; Urano et al. 2003; Bertrand et al. 2011), now the full extent of the subfunctionalization of the amphioxus MRFs has been revealed. Furthermore, we found instances of possible nonsomitic expression of some of the MRFs which may represent an example of neofunctionalization, or perhaps the conserved use of the myogenic toolkit in nonmyomeric structures, akin to the formation of head musculature in vertebrates (Buckingham and Rigby 2014). The nonsomitic expression observed for B. floridae MRF3 and B. lanceolatum MRF3 and MRF4 is consistent with a role in the developing striated muscle fibers in the orobranchial region (Yasui et al. 2014), which has not been reported in previous analyses of amphioxus MRF expression because these specific MRFs have not been assessed until now. The five-gene cluster of amphioxus illustrates multiple fates of duplicated genes in parallel with the already understood duplication and subfunctionalization in the vertebrates.
In other gene families, there are examples of independent duplications in the various chordate lineages. For instance, the urochordates have one Pax3/7 gene, the pro-ortholog of vertebrate Pax3 and Pax7, but amphioxus have a cluster of two genes, Pax3/7a and Pax3/7b (Barton-Owen et al. 2018), indicating that the ancestral Pax3/7 gene underwent a tandem duplication in the amphioxus ancestor after divergence from the lineage leading to the Olfactores. Pax3 and Pax7 are upstream regulators of the MRFs in vertebrate myogenesis (Moncaut et al. 2013; Buckingham and Rigby 2014; Buckingham 2017), which suggests there may be multiple levels of increased complexity driving amphioxus myogenesis that have previously been underestimated. Independent expansions were also observed for the amphioxus opsin genes (Holland et al. 2008), amphioxus globin genes (Storz et al. 2011), and Ciona T-box genes (Dehal et al. 2002), among many others (Minguillón et al. 2002). These examples show how the one-to-four expectation can be obscured not only by losses but also by independent duplications, as we also see for the MRF family. Though the amphioxus body plan has remained similar to the presumed ancestral chordate and its genome is considered to exhibit remarkable stasis compared with other chordate groups (Paps et al. 2012), we find higher than expected levels of complexity in the amphioxus MRFs.
MyoD and its relatives play an important and highly conserved role in regulating muscle development across bilaterian animals. Through multiple instances of gene duplication in several animal lineages, these genes are linked to increased complexity in the myogenic regulatory network. This is particularly notable in the vertebrates, where the four vertebrate MRFs were thought to display the typical “one-to-four” ohnologous relationship arising from the 2R WGDs. Instead, we find evidence for a tandem duplication that predates 2R and created the two distinct clades of vertebrate MRFs. In parallel, amphioxus, an invertebrate chordate, has undergone an independent expansion of its MRFs into a cluster of five linked and highly similar genes with distinct but overlapping expression patterns in myogenesis. These two events illustrate subfunctionalization of developmental genes following duplication where daughter genes are expressed as subsets of the ancestral gene, as has widely been assumed for most genes retained following duplication. Use of the early-branching chordate amphioxus sheds light on the invertebrate-to-vertebrate transition and demonstrates the role of duplication and subfunctionalization in the increasing complexity of vertebrates. More studies using these early-branching chordate lineages can further elucidate the fate of duplicated genes and their impact on complexity, especially those with important developmental roles and key morphological consequences, like the MRF family and other developmental toolkit genes.
Materials and Methods
Bioinformatics
Phylogeny
Amphioxus sequences were predicted using TBlastN (Altschul et al. 1990) searches of the genome assemblies of B. floridae, B. belcheri, and B. lanceolatum. The exon boundaries were confirmed by comparison of cDNA against genomic sequences. The genomic organization of the MRFs was determined by TBlastN against genomic assemblies, as well as long-range PCR of the intergenic region between MRF2b and MRF4 of the B. floridae BAC clone (F = AGAGGAGGTCAGTAGAGGGGTACAGGTC and R = CTGGGACGACATGATGACAGCCG.).
Aside from the amphioxus sequences, MRF protein sequences from other species were taken from Ensembl (Zerbino et al. 2018), NCBI protein database, or Uniprot (The UniProt Consortium 2019), as well as other databases including Xenbase (Karimi et al. 2018), Echinobase (Cameron et al. 2009; Kudtarkar and Cameron 2017), BeetleBase (Wang et al. 2007), SalmoBase (Samy et al. 2017), FlyBase (Thurmond et al. 2019), WormBase (Harris et al. 2010), and ANISEED (Brozovic et al. 2018). For genomes where the MRFs have not been annotated, a TBlastN search of the transcriptome or whole-genome assembly was used to find MRFs, using the closest relative’s sequences available. Some genes were manually curated, based on the known structure of verified or predicted gene models and transcriptome data, as well as the known structure of MRFs. For instance, there were no annotation records of the S. purpuratus MyoD2 orthologs in the genomes of P. miniata or L. variegatus, but these genes were predicted using alignments to sequences from Acanthaster planci (XP_022107336.1) and S. purpuratus (XP_011672159.1), respectively. Accession numbers and state of assembly/prediction for all sequences are listed in supplementary table 1, Supplementary Material online.
Alignment of the full coding sequence of the 106 MRFs from the 36 species included in the phylogeny was made in Jalview (Waterhouse et al. 2009) using MUSCLE (Edgar 2004) with default settings, and manually curated. A trimmed alignment, spanning human MyoD sequence from positions 78–186 contained the bHLH domain as well as conserved domains flanking it, and 19 residues from the highly conserved helix-III domain (Bergstrom and Tapscott 2001), when removing much of the long nonconserved sequence in between, was made for use in the ML and Bayesian tree inference. Both alignments were submitted to ProtTest (Abascal et al. 2005) to determine the best substitution model, the closest equivalent of which was used for each tree method. The full coding sequence alignment was used for the NJ method, but with a pairwise cut-off of 95%. NJ trees were run on MEGAX (Kumar et al. 2016) (1,000 bootstrap replicates, JTT substitution model, Gamma distribution of rates among sites, shape parameter = 0.77, homogeneous pattern among lineages, pairwise deletion cut-off 95%). ML trees were made using the trimmed alignment and were run with IQ-TREE (Nguyen et al. 2015) on the CIPRES science gateway (Miller et al. 2010), and the model was selected automatically with ModelFinder (Kalyaanamoorthy et al. 2017) (JTT+I+G4) with 1,000 bootstrap replicates (Hoang et al. 2018). Bayesian inference trees were also made using the trimmed alignment and were run using MrBayes 3.2.6 (Ronquist and Huelsenbeck 2003) on CIPRES as well (Miller et al. 2010) (500,000,000 generations, 500,000 print frequency, 50,000 sample frequency, 4 chains, rates invgamma, mixed aamodel, stoprule = yes, when 0.01). Appropriate burn-in was determined in Tracer v.1.7.7 (Rambaut et al. 2018) (5,000,000 generations), and the maximum clade credibility tree made using TreeAnnotator (http://beast.community/treeannotator, last accessed August 9, 2018) (MCC, median node heights). The final tree figure is the ML topology, with shared branch support values listed for major branches (ML/BI/NJ), and missing branch support values represented as dashes.
Synteny
A candidate chromosome for the human ghost locus was identified using the human genome paralogon database (McLysaght et al. 2002). This was then confirmed with the since-updated human genome (GrCh38.p12) on Genomicus v94.01 (Nguyen et al. 2018). This was used to generate a list of gene families shared across at least two of the three MRF loci, which was then pared down to those with four or fewer paralogs, in order to include only those with a probable 2R origin. From there, many paralogs of these genes were found on the inferred ghost locus, chromosome 19. This list was also applied to a subset of the vertebrates to compare across species, namely Mus musculus, Gallus gallus, Lepisosteus oculatus, and Danio rerio, and identify the 4- and 8-fold paralogy of the MRF and ghost loci in these species.
The synteny of the fish MyoD1 and MyoD2 loci was also examined by focusing on nearby neighbors of the respective loci, with both 3R ohnologs shared between the two loci and genes characteristic of each locus. Salmon orthologs of these genes were found using reciprocal BlastP (Altschul et al. 1990) searches against salmon proteins on NCBI using the zebrafish (or if there was not a zebrafish gene, the medaka) protein sequences as the first query. The locations were found using TBlastN searches of the salmon proteins against the salmon genome. To find all salmon-specific paralogs of the genes, the top two hits, rather than the top one hit, were used because of the 4R salmonid duplication.
Human–amphioxus synteny was assessed using reciprocal BLAST searches. The protein sequences of the gene models annotated to the MRF-bearing scaffold, scaffold 3 (FLLO01000004.1) in B. lanceolatum (http://amphiencode.github.io/Data/), as this scaffold is longer and better annotated than the other MRF-bearing scaffolds in B. floridae (scaffold 94) or B. belcheri (scaffold 50), was compiled. These proteins were queried against the NCBI database of all nonredundant reference human genome proteins using BlastP (Altschul et al. 1990). For each of the lancelet scaffold 3 proteins, the top hit human protein was then recorded and queried against a database of all B. lanceolatum proteins, and the top B. lanceolatum hit for each human protein was recorded. The two sets of results were compared, and reciprocal best hits were considered orthologs. The locations of these human proteins were downloaded using BIOMART (Smedley et al. 2015).
Counts of orthologs of lancelet MRF neighborhood genes located on the different human chromosomes were tested for significance using a cumulative probability binomial test (probabilities with replacement) and a one-tailed Barnard’s exact test (probabilities without replacement) (Barnard 1945). The total number of direct human orthologs of amphioxus genes on the MRF-bearing scaffold 3 found was 135, the total number of human protein-coding genes (PCG) is 20,313 (NCBI). For each chromosome, the ratio of the number of orthologs found on that chromosome (a) relative to the total number of orthologs (b = 135) was compared with the number of PCGs on that chromosome (c) relative to the total number of PCGs in the genome (d = 20,313), and P values were calculated using the Excel formula: 1-Binom.Dist(a, b,(c/d),TRUE), where TRUE determines the cumulative probability (left-tailed test), that is, the probability of there being at most that many successes (a) given that number of trials (b) and the probability of success (c/d); and the inverse (1-Binom.Dist()) gives the P value, or the probability that there could be more successes relative to the number of trials and the probability of success.
For the Barnard’s exact test, a 2×2 contingency table for each of the chromosomes with the number of orthologs on that chromosome, the number of orthologs on the other chromosomes, the number of nonorthologous PCGs on that chromosome and the number of nonorthologous PCGs on other chromosomes was used. The Barnard’s exact test was implemented in R (code downloaded from https://www.r-statistics.com/2010/02/barnards-exact-test-a-powerful-alternative-for-fishers-exact-test-implemented-in-r/, last accessed February 6, 2019).
The orthology of the MRF-bearing scaffolds in the three Branchiostoma spp. was assessed using reciprocal BlastP searches with the same approach as above. Due to the longer size of the B. lanceolatum scaffold (FLLO01000004.1: 8.7 Mb), we identified several scaffolds in each of the other species that had significantly more than expected orthologs to B. lanceolatum scaffold 3. These scaffolds in B. floridae, were namely 256 (0.2 Mb), 243 (3.1 Mb), 190 (2.3 Mb), 41 (4.4 Mb), 172 (1.6 Mb), 138 (2.6 Mb), and 94 (MRF, 3.5 Mb), which we then compared with the ancestral linkage groups associated with B. floridae scaffolds by Srivastava et al. (2008). The microsynteny of the amphioxus MRF loci was also assessed with the aim to determine the directionality of the MRF arrangement by comparing the immediate neighbors of the MRFs in B. floridae to the unannotated scaffolds in B. lanceolatum (FLLO01000004.1) and B. belcheri (NW_017804132.1), as well as the BAC clone from B. floridae (AC150407.2). We used the protein sequences of the predicted B. floridae genes neighboring the MRFs as queries in TBlastN searches against each of the different genomic sequences, with a score cut-off of 200.
Expression
Amphioxus WMISH
We used the amphioxus protocol for whole-mount in situ hybridization (WMISH) described by Holland et al. (1994). All Florida amphioxus (B. floridae) used in this thesis were collected from Tampa Bay (FL) by Dr Tom Butts and Dr Peter Osborne in July–August 2006. Spawning and embryo collection were performed as described in Holland and Yu (2004) for in situ hybridization. Embryos at different developmental stages (ranging from 7-h postfertilization to 3-day larvae) were fixed either for 1 h at room temperature or overnight at 4 °C with 4% (m/v) paraformaldehyde in MOPS buffer (0.5 M NaCl, 2 mM MgSO4, 1 mM EGTA, 0.1 M morpholinopropanesulfonic acid buffer, pH 7.5). After fixation, embryos were washed several times in 70% ethanol and stored at −20 °C.
Embryos, larvae, juveniles, and adults of European amphioxus (B. lanceolatum) were collected at the Laboratoire Aragó in Banyuls-sur-mèr (France) from May 31 to June 5, 2010. Spawning of ripe amphioxus was induced by heat stimulation (Fuentes et al. 2007) and different embryonic developmental stages (gastrula, early neurula, midneurula, late neurula, and early larval stage) were collected at regular intervals and fixed in 4% (m/v) paraformaldehyde in MOPS buffer for 1 h at room temperature or at 4 °C overnight. After fixation, embryos for WMISH were washed three times in 70% ethanol and stored in 70% ethanol at −20 °C. Larvae (first gill slit stage) and juvenile amphioxus from previous spawnings were kindly provided by Dr Héctor Escrivà and Dr Stéphanie Bertrand. Both developmental stages were fixed and stored in 70% ethanol or PBT following the same procedure described for the embryonic stages. Late developmental stages (late neurula and early larval stage) were kindly donated by Dr Ildikó Somorjai to complete the amphioxus developmental series.
Note added in proof.—During the final revision stages of this manuscript a new assembly of the Branchiostoma floridae genome was published (Simakov et al. 2020). The organisation of the B. floridae MRF gene cluster in this new assembly is the same as that in the earlier assembly, thus requiring no change to our figure 4. Also, our preliminary synteny analyses, using this new B. floridae assembly in place of the B. lanceolatum assembly, require no changes to our conclusions. The B. lanceolatum scaffold used in the synteny analyses described here was 8.74Mb in length with approximately 595 automatically annotated genes, and so the new B. floridae chromosome-scale assembly, with the MRFs on a 17.1Mb chromosome, provides little in the way of extra resolution.
Supplementary Material
Acknowledgments
The authors thank Prof Ian A. Johnston for comments on the article. We also thank Dr Hector Escriva and Dr Stephanie Bertrand for hosting us for the collection of B. lanceolatum material, and to Dr Ildiko Somorjai for advice on amphioxus in situ hybridizations. Madeleine Aase-Remedios and Dr Clara Coll-Lladó were supported by funding from the University of St Andrews, School of Biology and additional support from the CORBEL grant European Research Infrastructure cluster project.
References
- Abascal F, Zardoya R, Posada D.. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21(9):2104–2105. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Andrikou C, Iovene E, Rizzo F, Oliveri P, Arnone MI.. 2013. Myogenesis in the sea urchin embryo: the molecular fingerprint of the myoblast precursors. Evodevo 4(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki I, Terazawa K, Satoh N.. 1996. Duplication of an amphioxus myogenic bHLH gene is independent of vertebrate myogenic bHLH gene duplication. Gene 171(2):231–236. [DOI] [PubMed] [Google Scholar]
- Barnard GA. 1945. A new test for 2 × 2 tables. Nature 156(3954):177–177. [Google Scholar]
- Barton-Owen TB, Ferrier DEK, Somorjai I.. 2018. Pax3/7 duplicated and diverged independently in amphioxus, the basal chordate lineage. Sci Rep. 8(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom DA, Tapscott SJ.. 2001. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Mol Cell Biol. 21(7):2404–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S, Camasses A, Somorjai I, Belgacem MR, Chabrol O, Escande M-L, Pontarotti P, Escriva H.. 2011. Amphioxus FGF signaling predicts the acquisition of vertebrate morphological traits. Proc Natl Acad Sci U S A. 108(22):9160–9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S, Escriva H.. 2011. Evolutionary crossroads in developmental biology: amphioxus. Development 138(22):4819–4830. [DOI] [PubMed] [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y.. 2006. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 7(5):R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Pevzner PA, Tesler G.. 2004. Reconstructing the genomic architecture of ancestral mammals: lessons from human, mouse, and rat genomes. Genome Res. 14(4):507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower NI, Johnston IA.. 2010. Paralogs of Atlantic salmon myoblast determination factor genes are distinctly regulated in proliferating and differentiating myogenic cells. Am J Physiol Integr Comp Physiol. 298(6):R1615–R1626. [DOI] [PubMed] [Google Scholar]
- Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, et al. 2016. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 48(4):427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brozovic M, Dantec C, Dardaillon J, Dauga D, Faure E, Gineste M, Louis A, Naville M, Nitta KR, Piette J, et al. 2018. ANISEED 2017: extending the integrated ascidian database to the exploration and evolutionary comparison of genome-scale datasets. Nucleic Acids Res. 46(D1):D718–D725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M. 2017. Gene regulatory networks and cell lineages that underlie the formation of skeletal muscle. Proc Natl Acad Sci U S A. 114(23):5830–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Rigby P.. 2014. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 28(3):225–238. [DOI] [PubMed] [Google Scholar]
- Cameron RA, Samanta M, Yuan A, He D, Davidson E. 2009. SpBase: the sea urchin genome database and web site. Nucleic Acids Res. 37(Suppl 1):D750–D754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal JJ, Keith A, Rigby P.. 2008. Global transcriptional regulation of the locus encoding the skeletal muscle determination genes Mrf4 and Myf5 Genes Dev. 22(2):265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnier F, Gaspera B, Della Armand A-S, Van der Laarse WJ, Launay T, Becker C, Gallien C-L, Chanoine C.. 2002. Two myogenin-related genes are differentially expressed in Xenopus laevis myogenesis and differ in their ability to transactivate muscle structural genes. J Biol Chem. 277(2):1139–1147. [DOI] [PubMed] [Google Scholar]
- Chen L, Krause M, Sepanski M, Fire A.. 1994. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development 120(6):1631–1641. [DOI] [PubMed] [Google Scholar]
- Craxton M. 2010. A manual collection of Syt, Esyt, Rph3a, Rph3al, Doc2, and Dblc2 genes from 46 metazoan genomes – an open access resource for neuroscience and evolutionary biology. BMC Genomics 11(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB.. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51(6):987–1000. [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL.. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3(10):e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, Tomaso A, De DB, Gregorio A, Di GM, Goodstein DM, et al. 2002. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298(5601):2157–2167. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood PR. 1967. Structure of the segmental trunk muscle in amphioxus – with notes on the course and “endings” of the so-called ventral root fibres. Z Zellforsch. 84(3):389–416. [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan Y, Postlethwait J.. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151(4):1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes M, Benito E, Bertrand S, Paris M, Mignardot A, Godoy L, Jimenez-Delgado S, Oliveri D, Candiani S, Hirsinger E, et al. 2007. Insights into spawning behavior and development of the European amphioxus (Branchiostoma lanceolatum). J Exp Zool B Mol Dev Evol. 308B(4):484–493. [DOI] [PubMed] [Google Scholar]
- Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, et al. 2010. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 38(Suppl 1):D463–D467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A.. 2004. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 59(2):190–203. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Albalat R, Azumi K, Benito-Gutiérrez È, Blow MJ, Bronner-Fraser M, Brunet F, Butts T, Candiani S, Dishaw LJ, et al. 2008. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 18(7):1100–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland LZ, Yu J-K.. 2004. Cephalochordate (Amphioxus) embryos: procurement, culture, and basic methods In: Ettensohn CA, Wray GA, Wessel GM, editors. 1st ed Development of sea urchins, Ascidians, and other invertebrate Deuterostomes: experimental approaches. San Diego: Elsevier Academic Press; p. 195–215. [Google Scholar]
- Holland PWH, Garcia-Fernàndez J, Williams NA, Sidow A.. 1994. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994:125–133. [PubMed] [Google Scholar]
- Innan H, Kondrashov F.. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 11(2):97–108. [DOI] [PubMed] [Google Scholar]
- Izzi SA, Colantuono BJ, Sullivan K, Khare P, Meedel TH, Khare P, Colantuono BJ, Sullivan K, Izzi SA.. 2013. Functional studies of the Ciona intestinalis myogenic regulatory factor reveal conserved features of chordate myogenesis. Dev Biol. 376(2):213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Delgado S, Pascual-Anaya J, Garcia-Fernandez J.. 2009. Implications of duplicated cis-regulatory elements in the evolution of metazoans: the DDI model or how simplicity begets novelty. Brief Funct Genomics Proteomics. 8(4):266–275. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi K, Fortriede JD, Lotay VS, Burns KA, Wang DZ, Fisher ME, Pells TJ, James-Zorn C, Wang Y, Ponferrada VG, et al. 2018. Xenbase: a genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. 46(D1):D861–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, Bancewicz RM, Gautier P, Dahm R, Schonthaler HB, Damante G, Seawright A, Hever AM, Yeyati PL, van Heyningen V, et al. 2008. Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence. PLoS Genet. 4(2):e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudtarkar P, Cameron RA.. 2017. Echinobase: an expanding resource for echinoderm genomic information. Database 2017:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, Hvidsten TR, Leong JS, Minkley DR, Zimin A, et al. 2016. The Atlantic salmon genome provides insights into rediploidization. Nature 533(7602):200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2000. The evolutionary fate and consequences of duplicate genes. Science 290(5494):1151–1155. [DOI] [PubMed] [Google Scholar]
- MacCarthy T, Bergman A.. 2007. The limits of subfunctionalization. BMC Evol Biol. 7(1):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKintosh C, Ferrier DEK.. 2018. Recent advances in understanding the roles of whole genome duplications in evolution. F1000Res. 6:1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macqueen DJ, Johnston IA.. 2006. A novel salmonid myoD gene is distinctly regulated during development and probably arose by duplication after the genome tetraploidization. FEBS Lett. 580(21):4996–5002. [DOI] [PubMed] [Google Scholar]
- Macqueen DJ, Johnston IA.. 2008. An update on MyoD evolution in teleosts and a proposed consensus nomenclature to accommodate the tetraploidization of different vertebrate genomes. PLoS One 3(2):e1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlétaz F, Firbas PN, Maeso I, Tena JJ, Bogdanovic O, Perry M, Wyatt CDR, de la Calle-Mustienes E, Bertrand S, Burguera D, et al. 2018. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 564(7734):64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Hokamp K, Wolfe KH.. 2002. Extensive genomic duplication during early chordate evolution. Nat Genet. 31(2):200–204. [DOI] [PubMed] [Google Scholar]
- Meedel TH, Farmer SC, Lee JJ.. 1997. The single MyoD family gene of Ciona intestinalis encodes two differentially expressed proteins: implications for the evolution of chordate muscle gene regulation. Development 1721:1711–1721. [DOI] [PubMed] [Google Scholar]
- Meedel TH, Lee JJ, Whittaker JR.. 2002. Muscle development and lineage-specific expression of CiMDF, the MyoD-family gene of Ciona intestinalis. Dev Biol. 241(2):238–246. [DOI] [PubMed] [Google Scholar]
- Megeney LA, Rudnicki MA.. 1995. Determination versus differentiation and the MyoD family of transcription factors. Biochem Cell Biol. 73(9–10):723–732. [DOI] [PubMed] [Google Scholar]
- Meyer A, Schartl M.. 1999. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 11(6):699–704. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T.. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE). New Orleans (LA): IEEE. p. 1–8. [Google Scholar]
- Minguillón C, Ferrier DEK, Cebrián C, Garcia-Fernàndez J.. 2002. Gene duplications in the prototypical cephalochordate amphioxus. Gene 287(1–2):121–128. [DOI] [PubMed] [Google Scholar]
- Misquitta L, Paterson BM.. 1999. Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation. Proc Natl Acad Sci U S A. 96(4):1451–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncaut N, Rigby PWJ, Carvajal JJ.. 2013. Dial M(RF) for myogenesis. FEBS J. 280(17):3980–3990. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S.. 2007. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 17(9):1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NTT, Vincens P, Roest Crollius H, Louis A.. 2018. Genomicus 2018: karyotype evolutionary trees and on-the-fly synteny computing. Nucleic Acids Res. 46(D1):D816–D822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paps J, Holland PWH, Shimeld SM.. 2012. A genome-wide view of transcription factor gene diversity in chordate evolution: less gene loss in amphioxus? Brief Funct Genomics. 11(2):177–186. [DOI] [PubMed] [Google Scholar]
- Peachey LD. 1961. Structure of the longitudinal body muscles of amphioxus. J Cell Biol. 10(4):159–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RLS, Rudnicki MA.. 2000. Molecular mechanisms regulating myogenic determination and differentiation Front Biosci. 5(3):D750–D767. [DOI] [PubMed] [Google Scholar]
- Prince VE, Pickett FB.. 2002. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 3(11):827–837. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu J-K, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453(7198):1064–1071. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst Biol. 67(5):901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe LE, Asiedu EK, Pickett CJJ, Warburton MA, Izzi SA, Meedel TH.. 2019. The Ciona myogenic regulatory factor functions as a typical MRF but possesses a novel N-terminus that is essential for activity. Dev Biol. 448(2):210–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls A, Morris JH, Rudnicki M, Braun T, Arnold HH, Klein WH, Olson EN.. 1995. Myogenin’s functions do not overlap with those of MyoD or Myf-5 during mouse embryogenesis. Dev Biol. 172(1):37–50. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Arnold H-H, Jaenisch R.. 1993. Targeted inactivation of the muscle regulatory genes Myf-5 and MyoD: effect on muscle and skeletal development In: Wagner EF, Theuring F, editors. Transgenic animals as model systems for human diseases. Heidelberg (Berlin: ): Springer; p. 143–151. [Google Scholar]
- Samy JKA, Mulugeta TD, Nome T, Sandve SR, Grammes F, Kent MP, Lien S, Våge DI.. 2017. SalmoBase: an integrated molecular data resource for Salmonid species. BMC Genomics 18(1):482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp E, Pistocchi AS, Karampetsou E, Foglia E, Lamia CL, Cotelli F, Cossu G.. 2009. Induced early expression of mrf4 but not myog rescues myogenesis in the myod/myf5 double-morphant zebrafish embryo. J Cell Sci. 122(4):481–488. [DOI] [PubMed] [Google Scholar]
- Schubert M, Meulemans D, Bronner-Fraser M, Holland LZ, Holland ND.. 2003. Differential mesodermal expression of two amphioxus MyoD family members (AmphiMRF1 and AmphiMRF2). Gene Expr Patt. 3(2):199–202. [DOI] [PubMed] [Google Scholar]
- Shimeld SM. 1999. Gene function, gene networks and the fate of duplicated genes. Semin Cell Dev Biol. 10(5):549–553. [DOI] [PubMed] [Google Scholar]
- Shimeld SM, Holland P.. 2000. Vertebrate innovations. Proc Natl Acad Sci U S A. 97(9):4449–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simakov O Marlétaz F Yue J-X O’Connell B Jenkins J Brandt A Calef R Tung C-H Huang T-K Schmutz J, et al. . 2020. Deeply conserved synteny resolves early events in vertebrate evolution. Nat Ecol Evol. 4(6):820–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, Arnaiz O, Awedh MH, Baldock R, Barbiera G, et al. 2015. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 43(W1):W589–W598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somorjai IML, Bertrand S, Camasses A, Haguenauer A, Escriva H.. 2008. Evidence for stasis and not genetic piracy in developmental expression patterns of Branchiostoma lanceolatum and Branchiostoma floridae, two amphioxus species that have evolved independently over the course of 200 Myr. Dev Genes Evol. 218(11–12):703–713. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. 2008. The Trichoplax genome and the nature of placozoans. Nature 454(7207):955–960. [DOI] [PubMed] [Google Scholar]
- Storz JF, Opazo JC, Hoffmann FG.. 2011. Phylogenetic diversification of the globin gene superfamily in chordates. IUBMB Life 63(5):313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Du SJ.. 2002. Differential expression of two MyoD genes in fast and slow muscles of gilthead seabream (Sparus aurata). Dev Genes Evol. 212(5):207–217. [DOI] [PubMed] [Google Scholar]
- Tan X, Zhang PJ, Du SJ.. 2014. Evolutionary aspects of a new MyoD gene in amphioxus (Branchiostoma belcheri) and its promoter specificity in skeletal and cardiac muscles. Cell Mol Biol. 69(9):1210–1221. [Google Scholar]
- The UniProt Consortium. 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47(D1):D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, the FlyBase Consortium, et al. 2019. FlyBase 2.0: the next generation. Nucleic Acids Res. 47(D1):D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano A, Suzuki MM, Zhang P, Satoh N, Satoh G.. 2003. Expression of muscle-related genes and two MyoD genes during amphioxus notochord development. Evol Dev. 5(5):447–458. [DOI] [PubMed] [Google Scholar]
- Wagner A. 1998. The fate of duplicated genes: loss or new function? BioEssays 20(10):785–788. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang S, Li Y, Paradesi MSR, Brown SJ.. 2007. BeetleBase: the model organism database for Tribolium castaneum. Nucleic Acids Res. 35(Database):D476–D479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ.. 2009. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. 2000. Robustness – it’s not where you think it is. Nat Genet. 25(1):3–4. [DOI] [PubMed] [Google Scholar]
- Yasui K, Kaji T, Morov AR, Yonemura S.. 2014. Development of oral and branchial muscles in lancelet larvae of Branchiostoma japonicum. J Morphol. 275(4):465–477. [DOI] [PubMed] [Google Scholar]
- Yuan J, Zhang S, Liu Z, Luan Z, Hu G.. 2003. Cloning and phylogenetic analysis of an amphioxus myogenic bHLH gene AmphiMDF Biochem Biophys Res Commun. 301(4):960–967. [DOI] [PubMed] [Google Scholar]
- Zammit PS. 2017. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol. 72:19–32. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. 2018. Ensembl 2018. Nucleic Acids Res. 46(D1):D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol Evol. 18(6):292–298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.