Abstract
The world is experiencing the worst global health crisis in recent decades since December/2019 due to a new pandemic coronavirus. The COVID-19 disease, caused by SARS-CoV-2, has resulted in more than 30 million cases and 950 thousand deaths worldwide as of September 21, 2020. Determining the extent of the virus on public surfaces is critical for understanding the potential risk of infection in these areas. In this study, we investigated the presence of SARS-CoV-2 RNA on public surfaces in a densely populated urban area in Brazil. Forty-nine of 933 samples tested positive (5.25%) for SARS-CoV-2 RNA, including samples collected from distinct material surfaces, including metal and concrete, and distinct places, mainly around hospital care units and public squares. Our data indicated the contamination of public surfaces by SARS-CoV-2, suggesting the circulation of infected patients and the risk of infection for the population. Constant monitoring of the virus in urban areas is required as a strategy to fight the pandemic and prevent further infections.
Keywords: COVID-19, Coronavirus, Genome detection, Environment surveillance
Graphical abstract
1. Introduction
Since late December 2019, the world is experiencing the worst global health crisis in recent decades due to the ongoing transmission of a novel coronavirus. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing coronavirus disease 2019 (COVID-19) spread globally and affected several sectors, including those related to medicine, economics, and politics, among others (Cutler, 2020; Gorbalenya et al., 2020; Lai et al., 2020; Zhu et al., 2020). COVID-19 has already affected over 200 countries, resulting in more than 30.6 million cases and 950 thousand deaths worldwide as of September 21, 2020 (WHO, 2020). In Brazil, the first official case was registered on February 26, 2020, and more than 4,490,000 confirmed cases and 135,000 deaths had been registered across the country as of September 21, 2020 (WHO, 2020).
Belo Horizonte is the capital of Minas Gerais State, one of Brazil's most populous metropolitan regions (6 million inhabitants). The city recorded 39,379 confirmed cases and 1168 deaths due to COVID-19 as of September 21 (Secretaria Municipal de Saúde, 2020). The areas within Belo Horizonte where most of the deaths and confirmed cases have occurred correspond precisely to areas with public squares, bus stations/terminals, and hospital areas, i.e., where a large flow and concentration of people is commonly observed (Secretaria Municipal de Saúde, 2020).
Recent studies have identified the presence of SARS-CoV-2 RNA on different surfaces and environments inside hospitals, revealing the dynamics of viral dissemination within these places (Guo et al., 2020; Liu et al., 2020; Wang et al., 2020). SARS-CoV-2 remains viable on different types of surfaces, such as metal and plastic, for up to 72 h, depending on the type of surface material, and can remain infectious in aerosols for at least 3 h (Van Doremalen et al., 2020). Assessing the presence of the virus in the environment, objects, and surfaces in public areas is fundamental for understanding the risk of infection in the population. In addition, any information gained can be used by health managers to control population movements in these areas, as well as implement environmental disinfection measures.
2. Material and methods
2.1. Sample collection
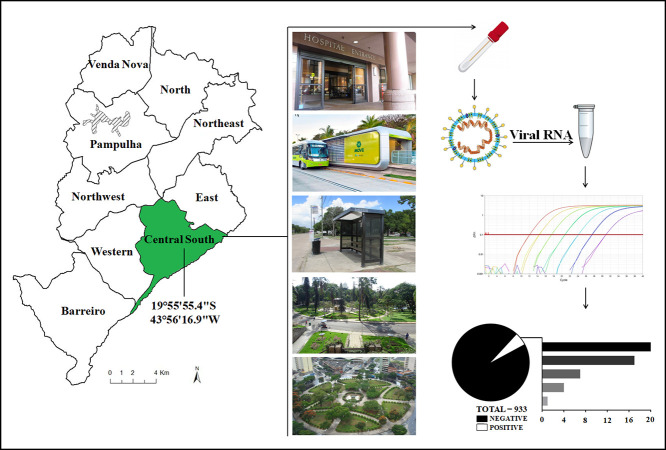
We investigated the presence of SARS-CoV-2 RNA in a downtown area of Belo Horizonte between April and June 2020, in a total of nine days. Belo Horizonte's climate is classified as tropical with a dry season, with moderately hot and humid summers and dry and pleasant winters. The temperature is mild throughout the year, with averages ranging from 19 °C to 24 °C, with the annual compensated average of 22 °C and 1430 mm is the average annual rainfall (INMET, 2020). All samples were collected between 14:00 pm and 17:00 pm. Temperature was between 20 °C to 25 °C (average temperature 22 °C) and relative humidity varying between 36% to 83% (average humidity 54%). Environmental data was retrieved from Time and Date AS website (https://www.timeanddate.com/weather/brazil/belo-horizonte/). This region of the city (central-southeast: 19°55′55.4″S 43°56′16.9″W) has the highest concentration of hospitals and health units and one of the highest number of notified COVID-19 cases. Importantly, this part of the city also has a large number of people accessing public transportation and transportation facilities daily. A total of 933 samples were collected from eight different categories of places (Supplementary Table 1), including: a) 38 health care units (hospitals, medical centers, and emergency care units); b) 17 public squares; c) two public parks; d) one public market; e) six bus terminals; f) one shopping mall; g) 10 education centers (universities and schools); 8) 21 other public places, including banks, government departments, among others (Supplementary Table 1). Samples were collected from different sources including entrance doors, handrails, benches and tables, bus stops, and ground, and from distinct materials, including concrete, metal, rock, brickwork, and others. For the collection of environmental samples, swabs with sterile phosphate-buffered saline were vigorously rubbed on surfaces (10 cm2) of the aforementioned local and objects. The swabs were then transferred to tubes containing transport solution (1 mL of guanidine isothiocyanate buffer, 4 M) and taken directly to the laboratory for testing.
2.2. RNA extraction and RT-qPCR
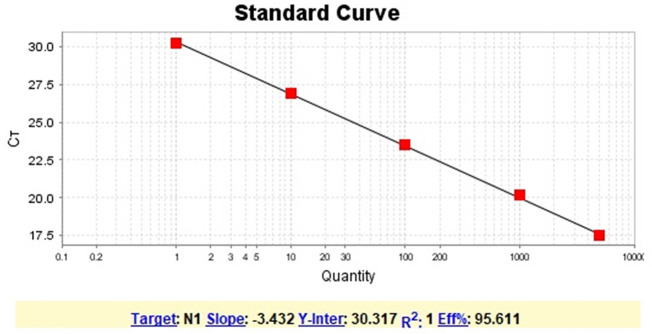
For each sample, 70 μL of transport solution containing a sample was submitted to nucleic acid extraction using the QIAmp Viral RNA Mini Kit (QIAGEN, Maryland, USA). Total RNA (5 μL) was used as a template for one-step qPCR (Promega, Wisconsin, USA) (in a final volume of 20 μL per reaction, GoTaq1-sept qPCR system, Promega), using primers and probes specific for the N1 and N2 regions of the SARS-CoV-2 genome (CDC, USA 2020). RNA extraction was performed in batches of 13 samples plus one negative control. Samples were considered positive when they presented amplification for N1 an N2 targets region, considering the threshold for cycle quantification value (Cq) of 40 (CDC, USA 2020). Since between the range of 37–40 Cq indicate minimal quantities of DNA, Cq ≥ 40 were considered negative. The results of RT-qPCR runs were manually inspected for the correction of baseline and threshold parameters whenever necessary due to heterogeneity in the amount of input RNA among different samples (Bustin et al., 2009). Negative (extraction control and non-template control) and positive controls (RNA extracted from inactivated SARS-CoV-2, kindly provided by Dr. Danielle Durigon and Dr. Edison Durigon, USP, Brazil) were used. To confirm the results, all positive samples were submitted to a second round of RNA extraction and RT-qPCR. Quantification of viral RNA in the environmental samples was based on a standard curve generated from serial dilutions (1:10) of SARS-CoV-2 RNA and converted to genomic units per ten square centimeters of surface (the area we made the sampling for this study). Quantification was based only on N1 target gene given the high efficiency of our standard curve (Supplementary Fig. 1). N2 target gene quantification was not performed due to a low efficiency achieved for the standard curve for this target. Nevertheless, the Cq values for N2 target for all positive samples are included in supplementary Table 2. SARS-CoV-2 RNA control was previously quantified as described elsewhere (de Almeida et al., 2020).
Supplementary Fig. 1.
Standard curve (N1 target gene) for SARS-CoV-2 RNA quantification. The curve was generated from serial dilutions (1:10) of SARS-CoV-2 RNA.
3. Results
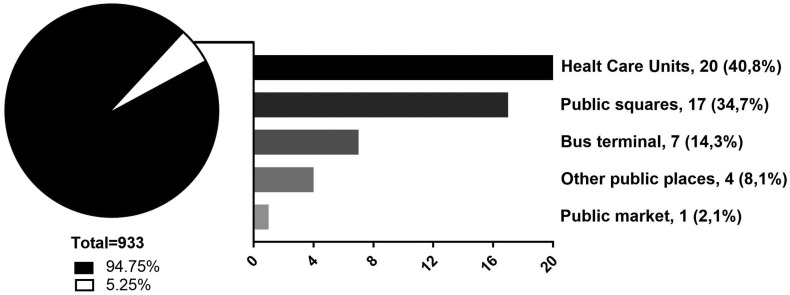
A total of 49 samples (5.25%) were positive for the presence of SARS-CoV-2 RNA (Table 1 and Fig. 1). SARS-CoV-2 RNA was detected in 20 samples collected around health care units, corresponding to 40.8% of the total positive samples, with Ct values ranging from 23.3 to 37.7 (N1) and 22.2 to 39.4 (N2). These samples are distributed in 12 different health care units and were detected mainly at bus' stops near the entrance of the hospitals and emergency care units (Supplementary Table 1). Seventeen samples were positive for SARS-CoV-2 RNA at public squares distributed across seven different places, corresponding to 34.7% of the positive samples found in this study. Most of the positive samples were detected in benches, with Ct values ranging from 32 to 37.5 (N1) and 34.4 to 39 (N2). We also detected SARS-CoV-2 in different bus terminals in the city, being seven positive samples (14.3%) with Ct values ranging from 29 to 34.7 (N1) and 30.5 to 38.5 (N2). All of these samples were collected at entrance handrails of the bus terminals (Table 1). Finally, we detected the presence of SARS-CoV-2 RNA at very low concentration in one sample collected in the wall of the major public market of the city (N1 Ct = 36.9; N2 Ct = 39.6), and four samples collected at bus stops [floor (1) and benches (3)] in front of public banks (2), sport club (1), and a government department (1), with Ct values ranging from 34.7 to 38.1 (N1) and 37.8 to 39.5 (N2) (Table 1 and Supplementary Table 1). After RNA quantification, we observed that most positive samples had very low viral amount (≤1 genomic unit/cm2), and the higher viral load were detected in samples collected from health care units (Table 1).
Table 1.
Positive samples for SARS-CoV-2 RNA in surfaces at different locations in Belo Horizonte, Brazil.
| Category | Sample ID | Surface (material) | Ct value (N1) | Concentration (N1) (genomic units/10 cm2) |
|---|---|---|---|---|
| Health care units | HCU 29C | Wall (brickwork) | 25.9 | 67.9 |
| HCU 2 B | Floor (concrete) | 34.1 | ≤1 | |
| HCU 3 N | Floor (concrete) | 37.2 | ≤1 | |
| HCU 8 A | Floor (concrete) | 28.5 | 11.7 | |
| HCU 12 E | Floor (concrete) | 37.7 | ≤1 | |
| HCU 13 J | Wall (concrete) | 34.5 | ≤1 | |
| HCU 29 A | Sidewalk (concrete) | 34.2 | ≤1 | |
| HCU 29 M | Sidewalk (concrete) | 36.8 | ≤1 | |
| HCU 30C | Floor (concrete) | 36.2 | ≤1 | |
| HCU 3 D | Bench (metal) | 34.1 | ≤1 | |
| HCU 3 I | Bench (metal) | 34 | ≤1 | |
| HCU 1 D | Bench (metal) | 31.6 | ≤1 | |
| HCU 9H | Bench (metal) | 36.7 | ≤1 | |
| HCU 11C | Handrail (metal) | 36.4 | ≤1 | |
| HCU 9 A | Bench (metal) | 33.1 | ≤1 | |
| HCU 13 E | Bench (metal) | 36.9 | ≤1 | |
| HCU 21 D | Pillar (rock) | 36 | ≤1 | |
| HCU 29 F | Bench (plastic) | 23.7 | 299.8 | |
| HCU 29H | Bench (plastic) | 23.3 | 392.8 | |
| HCU 12 D | (glass) | 36 | ≤1 | |
| Public squares | PS 18 R | Wall (brickwork) | 37.4 | ≤1 |
| PS 1 E | Table (concrete) | 33.1 | ≤1 | |
| PS 3C | Bench (concrete) | 35.8 | ≤1 | |
| PS 6 G | Bench (concrete) | 37.5 | ≤1 | |
| PS 18 X | Sidewalk (concrete) | 35.4 | ≤1 | |
| PS 15 F | Bench (wood) | 34.8 | ≤1 | |
| PS 3 K | Bench (metal) | 35.7 | ≤1 | |
| PS 17 J | Handrail (metal) | 36.5 | ≤1 | |
| PS 18 I | Handrail (metal) | 35.1 | ≤1 | |
| PS 2 G | Bench (rock) | 35.6 | ≤1 | |
| PS 2H | Bench (rock) | 34 | ≤1 | |
| PS 2 I | Bench (rock) | 33.9 | ≤1 | |
| PS 9C | Bench (rock) | 35.5 | ≤1 | |
| PS 17 A | Bench (rock) | 32.3 | ≤1 | |
| PS 17 D | Bench (rock) | 32 | ≤1 | |
| PS 17 E | Bench (rock) | 34 | ≤1 | |
| PS 18 M | Bench (rock) | 36.9 | ≤1 | |
| Bus terminals | BT 1 A | Handrail (metal) | 32.8 | ≤1 |
| BT 1 B | Handrail (metal) | 34.5 | ≤1 | |
| BT 1C | Handrail (metal) | 29 | 8.4 | |
| BT 1 Q | Handrail (metal) | 32.1 | ≤1 | |
| BT 1 S | Handrail (metal) | 32.8 | ≤1 | |
| BT 1 W | Handrail (metal) | 29 | 8.4 | |
| BT 2 D | Handrail (metal) | 34.7 | ≤1 | |
| Other public places | OPP 11C | Floor (concrete) | 36.7 | ≤1 |
| OPP 5 A | Bench (metal) | 36.2 | ≤1 | |
| OPP 10 A | Bench (metal) | 38.1 | ≤1 | |
| OPP 11 A | Bench (metal) | 34.7 | ≤1 | |
| Public market | PM 1 G | Wall (concrete) | 36.9 | ≤1 |
For PCR quantification, we consider 1 SARS-CoV-2 plaque forming unit as 1 SARS-CoV-2 genomic unit.
Fig. 1.
Distribution of positive samples for SARS-CoV-2 RNA. 933 samples were collected at different locations in Belo Horizonte, Brazil, with 49 being positive for viral genome detection, distributed among five different categories of locations. Raw numbers and percentages are indicated.
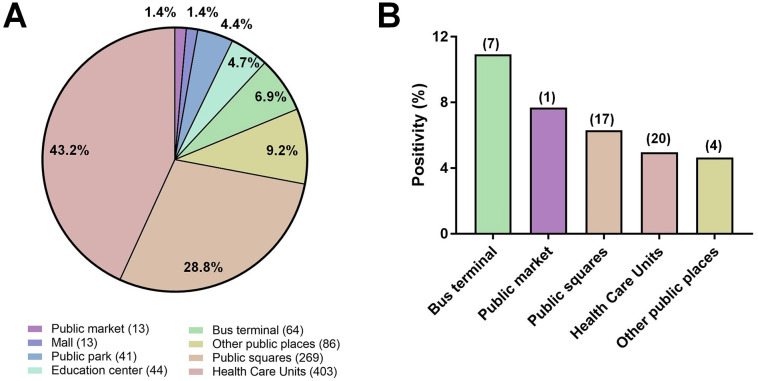
It is important to mention that samples were not collected evenly among the different categories of places. Most of the samples were collected from health care units (403 samples, 43.2%) and public squares (269 samples, 28.8%) (Supplementary Fig. 2), and it can be the reason why over 3/4 of positive samples were detected from samples collected at these locations. Considering the proportion of positivity in the different places, bus terminals exhibit the higher positivity rate among the evaluated places, followed by public market, public squares, and health care units. The Belo Horizonte City Hall was informed about the contaminated areas, and, after disinfection (laundry detergent followed by 1% sodium hypochlorite), viral RNA could no longer be detected, except for one sample recollected at the entrance of a hospital. We did not identify any positive samples in public parks, education centers, and a mall that were included in this study.
Supplementary Fig. 2.
Environmental samples collected from different locations. (A) Distribution of samples collected in different categories of places. Raw numbers of samples are indicated in front of the categories names; (B) SARS-CoV-2 RNA positivity (%) in different locations. Raw numbers of samples are indicated above columns.
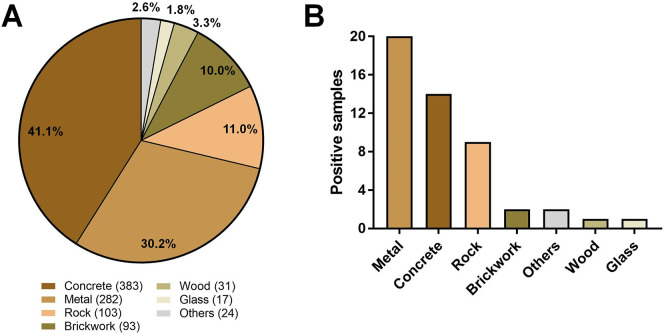
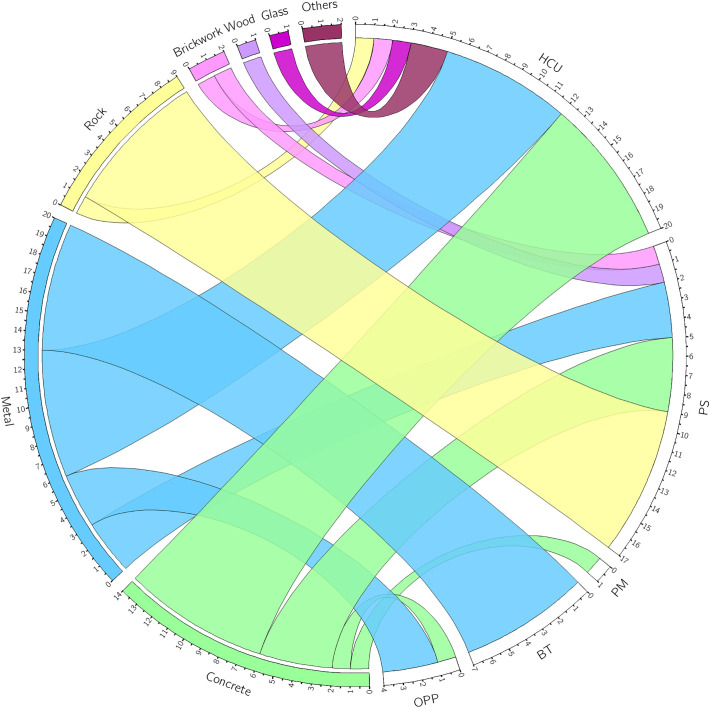
Samples were collected from different materials, the majority being from concrete (383 samples, 41.1%) and metal (282 samples, 30.2%), and also rock, brickwork, wood, glass, and a minority of other materials, including plastic and pottery (10 samples each) and asphalt (4 samples) (Fig. 2A). From the 49 positive samples, 20 were identified in metal surfaces, especially from benches of bus stops and handrails of hospitals entrances and bus terminals (Fig. 2B and Supplementary Table 1). Fourteen positive samples were recovered from concrete surfaces, mainly sidewalks near health care units and public squares (Figs. 2B and 3). Nine positive samples were found in rock surfaces, all being found in public squares but one sampled from a frontal pillar of a hospital (Fig. 3). SARS-CoV-2 RNA was also identified in two samples collected from brickworks (one from health care unit and other from a public square), and in two plastic surfaces from benches of an emergency care unit (Fig. 3 and Supplementary Table 1). Finally, SARS-CoV-2 RNA was detected in one sample from a wood bench located in a public square, and one sample from a glass surface in a bus stop in front of a hospital (Figs. 2B and 3).
Fig. 2.
Environmental samples collected from different materials. (A) Distribution of samples collected in different surfaces. Raw numbers of samples are indicated in front of the materials names; (B) Distribution of positive samples among different kinds of materials.
Fig. 3.
Association between locations and materials of positive samples for SARS-CoV-2 RNA. Circos plot associating locations and surface materials of positive samples. HCU: health care units; PS: public squares; PM: public market; BT: bus terminals; OPP: other public places.
4. Discussion
Although we sought to detect the presence of viral RNA, not infectious particles, it is possible that infectious particles were present in these environments, and care must be taken to avoid further contamination and the eventual collapse of the local health system. The infectious dose of SARS-CoV-2 virions to start a productive infection in humans is still unclear, and also if infectious particles can be recovered from surfaces with low viral load. Previous studies have reported the isolation of SARS-CoV-2 from nasopharyngeal and oropharyngeal samples from distinct patients in a nursing facility (USA) with high Ct value (≤34) (Arons et al., 2020). Furthermore, other studies have reported the recovery of the virus from different surfaces, including metal, cardboard, and plastic (Van Doremalen et al., 2020). Different studies have reported the detection of SARS-CoV-2 genome in different environmental surfaces, especially inside hospital facilities, suggesting that environmental contamination by the virus is possible (Carraturo et al., 2020; Chia et al., 2020; Jiang et al., 2020; Wu et al., 2020; Ye et al., 2020). Chin and coworkers observed that SARS-CoV-2 remains viable in smooth surfaces from distinct materials, including metal and plastic, up to 4 days at 22 °C and humidity around 65% (Chin et al., 2020). Therefore, it is possible that infectious particles could also be recovered from the surfaces found positive for the virus RNA in this study, especially considering that around 40% of the positive samples identified in this study had Ct values below 34. Although the real infectious potential of viruses detected in this study cannot be established, we believe the detection of SARS-CoV-2 by molecular assays indicates the potential risk of infection, and care must be taken to avoid further increases in the number of COVID-19 cases. It is important to mention that most of the positive samples in our study had very low amount of RNA (≤1 genomic unit/cm2) and such amount of virus might not be able to trigger COVID-19 in patients. Nevertheless, the viral RNA in different surfaces was detected in our assays, despite the low concentration, and care must be taken to avoid possible infection.
Although Belo Horizonte has relatively few cases when compared with other cities in Brazil, such as São Paulo and Rio de Janeiro, our data reinforce that the virus is circulating in the city and can be found on surfaces such as benches, tables, handrails, and floors, and in places with a large flow of people, such as public squares and hospital entrances. The detection of the viral RNA at these sites indicates that adequate cleaning of public environments and reinforcement of educational campaigns for hygienic and social distancing practices should be undertaken. Given the short sampling period in this study, our data do not support a correlation between different environmental parameters, such as local temperature or humidity, and the detection of the virus. Nevertheless, other studies addressed this matter and suggested that SARS-CoV-2 remains viable for up to 7 days at 22 °C, precisely the average temperature in the days we made the sampling for virus detection for this study (Chin et al., 2020). Furthermore, in our work, a mean of 54% of humidity was observed, and another human coronavirus, HCoV-229E, was more stable at 50% of humidity, a similar humidity value (Kampf et al., 2020). These data support the indication that viable viruses could be recovered from places where we detected the viral RNA. Further studies may contribute to elucidate better how frequent we can isolate SARS-CoV-2 from inanimate surfaces in the field.
It is important to notice that the transmission of the virus in buses seems to be considerably high, as demonstrated in China, where in a bus with 68 individuals, one person in a 100 min trip disseminated the virus to another 23 passengers (Shen et al., 2020). Although we did not evaluate the presence of the virus inside the buses of our city, the detection in the bus terminals and stations is an indicative that those places are an important risk factor for people to get infected, not only inside the buses. Our study highlights the need for the constant assessment of the presence of the virus, not only in hospital facilities, but also in places close to medical areas and with a large circulation of people. The presence of SARS-CoV-2 in these environments can result in an increase in the number of cases of the disease in the near future if control measures are not forcefully adopted.
The following are the supplementary data related to this article.
SARS-CoV-2 detection in public surfaces - samples, locations and details, complete list. Belo Horizonte, Brazil. Samples bold-highlighted: SARS-CoV-2 RNA detected.
Ct values (N2) of positive samples for SARS-CoV-2 RNA.
CRediT authorship contribution statement
Jônatas Santos Abrahão: Conceptualization, Supervision, Resources, Writing - original draft, Writing - review & editing. Lívia Sacchetto: Investigation, Writing - review & editing. Izabela Maurício Rezende: Investigation. Rodrigo Araújo Lima Rodrigues: Investigation, Visualization, Writing - original draft, Writing - review & editing. Ana Paula Correia Crispim: Investigation. César Moura: Investigation. Diogo Correia Mendonça: Investigation. Erik Reis: Investigation, Writing - review & editing. Fernanda Souza: Investigation. Gabriela Fernanda Garcia Oliveira: Investigation. Iago Domingos: Investigation. Paulo Victor de Miranda Boratto: Investigation. Pedro Henrique Bastos Silva: Investigation. Victoria Fulgêncio Queiroz: Investigation. Talita Bastos Machado: Investigation. Luis Adan Flores Andrade: Investigation. Karine Lima Lourenço: Investigation. Thaís Silva: Investigation. Graziele Pereira Oliveira: Investigation. Viviane de Souza Alves: Conceptualization, Writing - original draft. Pedro Augusto Alves: Investigation. Erna Geessien Kroon: Conceptualization, Writing - original draft. Giliane de Souza Trindade: Conceptualization, Writing - original draft. Betânia Paiva Drumond: Conceptualization, Supervision, Writing - original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank our colleagues from Belo Horizonte City Hall, in especial Coronel Genedempsey Bicalho Cruz (Superintendência de Limpeza Urbana – SLU/BH). In addition, we thank the Gabinete da Reitora da UFMG, the Pró Reitoria de Pesquisa da UFMG/Secretaria de Educação Superior/Ministério da Educação (number 23072.211119/2020-10), Finep/RTR/PRPq/Rede COVID-19 (number 0494/20-0120002600), Programa de Pós-graduação em Microbiologia da UFMG, CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and FAPEMIG (Fundação de Amparo à Pesquisa do estado de Minas Gerais) for their financial support. B.P.D, E.G.K., G.S.T. and J.S.A. are CNPq researchers. J.S.A. and P.A.A. are members of Rede Vírus MCTIC.
Editor: Jay Gan
References
- Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., Taylor J., Spicer K., Bardossy A.C., Oakley L.P., Tanwar S., Dyal J.W., Harney J., Chisty Z., Bell M., Methner M., Paul P., Carlson C.M., McLaughlin H.P., Thornburg N., Tong S., Tamin A., Tao Y., Uehara A., Harcourt J., Clark S., Brostrom-Smith C., Page L.C., Kay M., Lewis J., Montgomery P., Stone N.D., Clark T.A., Honein M.A., Duchin J.S., Jernigan J.A. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Carraturo F., Del Giudice C., Morelli M., Cerullo V., Libralato G., Galdiero E., Guida M. Persistence of SARS-CoV-2 in the environment and COVID-19 transmission risk from environmental matrices and surfaces. Environ. Pollut. 2020 doi: 10.1016/j.envpol.2020.115010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Division of Viral Diseases . 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. [Google Scholar]
- Chia P.Y., Coleman K.K., Tan Y.K., Ong S.W.X., Gum M., Lau S.K., Lim X.F., Lim A.S., Sutjipto S., Lee P.H., Son T.T., Young B.E., Milton D.K., Gray G.C., Schuster S., Barkham T., De P.P., Vasoo S., Chan M., Ang B.S.P., Tan B.H., Leo Y.S., Ng O.T., Wong M.S.Y., Marimuthu K., Lye D.C., Lim P.L., Lee C.C., Ling L.M., Lee L., Lee T.H., Wong C.S., Sadarangani S., Lin R.J., Ng D.H.L., Sadasiv M., Yeo T.W., Choy C.Y., Tan G.S.E., Dimatatac F., Santos I.F., Go C.J., Chan Y.K., Tay J.Y., Tan J.Y.L., Pandit N., Ho B.C.H., Mendis S., Chen Y.Y.C., Abdad M.Y., Moses D. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat. Commun. 2020 doi: 10.1038/s41467-020-16670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. The Lancet Microbe. 2020 doi: 10.1016/s2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler D. How will COVID-19 affect the health care economy? JAMA - J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.7308. [DOI] [PubMed] [Google Scholar]
- de Almeida P.R., Demoliner M., Antunes Eisen A.K., Heldt F.H., Hansen A.W., Schallenberger K., Fleck J.D., Spilki F.R. SARS-CoV2 quantification using RT-dPCR: a faster and safer alternative to assist viral genomic copies assessment using RT-qPCR. bioRxiv. 2020 doi: 10.1101/2020.05.01.072728. [DOI] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z.D., Wang Z.Y., Zhang S.F., Li X., Li L., Li C., Cui Y., Fu R. Bin, Dong Y.Z., Chi X.Y., Zhang M.Y., Liu K., Liu K., Cao C., Liu B., Zhang K., Gao Y.W., Lu B., Chen W. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg. Infect. Dis. 2020;26:1583–1591. doi: 10.3201/eid2607.200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INMET Normas Climatológicas do Brasil. 2020. https://portal.inmet.gov.br/ URL.
- Jiang F.C., Jiang X.L., Wang Z.G., Meng Z.H., Shao S.F., Anderson B.D., Ma M.J. Detection of severe acute respiratory syndrome coronavirus 2 RNA on surfaces in quarantine rooms. Emerg. Infect. Dis. 2020 doi: 10.3201/eid2609.201435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020 doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J., Ma S., Wang Y., Cai Z., Hu J., Wei N., Wu J., Du H., Chen T., Li R., Tan H., Kang L., Yao L., Huang M., Wang H., Wang G., Liu Z., Hu S. Factors associated with mental health outcomes among health care workers exposed to coronavirus disease 2019. JAMA Netw. Open. 2020 doi: 10.1001/jamanetworkopen.2020.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Yuan, Ning Z., Chen Y., Guo M., Liu Yingle, Gali N.K., Sun L., Duan Y., Cai J., Westerdahl D., Liu X., Xu K., Ho K. fai, Kan H., Fu Q., Lan K. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020 doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Secretaria Municipal de Saúde . 2020. Boletim epidemiológico e assistencial - COVID-19 N°108/2020. (Belo Horizonte) [Google Scholar]
- Shen Y., Li C., Dong H., Wang Z., Martinez L., Sun Z., Handel A., Chen Z., Chen E., Ebell M.H., Wang F., Yi B., Wang H., Wang X., Wang A., Chen B., Qi Y., Liang L., Li Y., Ling F., Chen J., Xu G. Community outbreak investigation of SARS-CoV-2 transmission among bus riders in eastern China. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., De Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Feng H., Zhang S., Ni Z., Ni L., Chen Y., Zhuo L., Zhong Z., Qu T. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Coronavirus disease (COVID) - Weekly Epidemiological and Operational updates September 2020–21 September 2020. [Google Scholar]
- Wu S., Wang Y., Jin X., Tian J., Liu J., Mao Y. Environmental contamination by SARS-CoV-2 in a designated hospital for coronavirus disease 2019. Am. J. Infect. Control. 2020 doi: 10.1016/j.ajic.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye G., Lin H., Chen S., Wang S., Zeng Z., Wang W., Zhang S., Rebmann T., Li Y., Pan Z., Yang Z., Wang Y., Wang F., Qian Z., Wang X. Environmental contamination of SARS-CoV-2 in healthcare premises. J. Inf. Secur. 2020 doi: 10.1016/j.jinf.2020.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SARS-CoV-2 detection in public surfaces - samples, locations and details, complete list. Belo Horizonte, Brazil. Samples bold-highlighted: SARS-CoV-2 RNA detected.
Ct values (N2) of positive samples for SARS-CoV-2 RNA.