Abstract
In this study, the effect of sustainable probiotics on Campylobacter jejuni colonization and gut microbiome composition was evaluated using chicken as a model organism. Chickens were given Lactobacillus casei over-expressing myosin-cross-reactive antigen (LC+mcra). LC+mcra can generate bioactive compounds in larger quantity including conjugated linoleic acid. A total of 120 chickens were used in duplicate trials to investigate the effectiveness of LC+mcra in decreasing C. jejuni colonization by means of kanamycin resistant strain compared to the control group. We observed that LC+mcra can efficiently colonize various parts of the chicken gut and competitively reduce colonization of natural and challenged C. jejuni and natural Salmonella enterica. LC+mcra was found to reduce C. jejuni colonization in cecum, ileum and jejunum, by more than one log CFU/g when compared to the no-probiotic control group. Furthermore, 16S rRNA compositional analysis revealed lower abundance of Proteobacteria, higher abundance of Firmicutes, along with enriched bacterial genus diversity in gut of LC+mcra fed chicken. Decreased contamination of drinking water by C. jejuni and S. enterica was also observed, suggesting a potential function of reducing horizontal transfer of enteric bacteria in poultry. Outcomes of this study reveal high potential of LC+mcra as sustainable approach to decrease colonization of C. jejuni and S. enterica in poultry gut along with other beneficial attributes.
Subject terms: Microbiology, Antimicrobials, Applied microbiology, Pathogens
Introduction
Campylobacter jejuni causes foodborne enteric infection campylobacteriosis and is considered as one of the prominent zoonotic pathogens. During 2018, among over twenty-five thousand cases of foodborne infections reported by FoodNet, the incidence rate of Campylobacter infections was the highest, 19.5 in 1000 people1. Poultry is the major reservoir of C. jejuni and cross-contamination of chicken meat during processing causes the majority of sporadic occurrences of campylobacteriosis1, 2. The current trend of white meat consumption, particularly chicken meat, has been increased significantly in the US and around the world, which is believed to be the reason for the increasing numbers of campylobcateriosis3. C. jejuni is normally present in the gut microbiota of poultry and other birds, capable of growing optimally at 42 °C, which is the body temperature of chicken4. According to recent reports, the prevalence pattern of C. jejuni in pasture poultry and their sensitivity to various antibiotics indicate that animal farming without antibiotic and/or synthetic chemical growth promoters diminishes the prevalence of multiple antibiotic resistance C. jejuni in poultry, without altering colonization of other bacteria5,6. Another important zoonotic pathogen is Salmonella enterica, with a higher prevalence in organic farming systems than in conventional farming systems, but a lower incidence of multi-antibiotic resistance in their organic counterparts7, 8. Therefore, developing sustainable alternatives that can replace antibiotic growth promoters and synthetic chemicals is a priority in order to reduce multiple-drug resistant C. jejuni and S. enterica.
Probiotics can be considered as the prime candidates for the prevention and reduction of cross-contamination with foodborne bacterial pathogens in food products8–12, however, their effectiveness is heavily dependent on the variety and total amount of produced and secreted metabolites/byproducts11,13. Probiotics can break down undigested dietary components through colonizing in the gastrointestinal (GI) tract of the host and after reaching these undigested components in different portions of intestine, especially cecum, jejunum and ileum, the probiotics produce a remarkable quantity of metabolites or bacterial byproducts14,15. Probiotics and their metabolites also modulate the composition of gut microbiome by competitive colonizing or through the secretion of bioactive compounds in different gut intestinal portions16,17. For example, the increase of the beneficial microorganisms in gut microbial ecosystem ultimately decreases the detrimental microbiota, which can lead to a healthy gut state for the overall health of the host16,17.
The amount and types of bioactive metabolites produced by beneficial microbes are vastly influenced by undigested dietary components and/or prebiotics11,13. Any potential prebiotic intended for use has to comply with some regulations, like possessing the “Generally Recognized as Safe (GRAS)” status, being assessed for appropriate dose and potential side effects, must be free of contaminants and impurities and must not cause alterations to the intestinal microbiota that could negatively affect the host18. According to Wang, it is reported that prebiotics must not be digested in the upper portion of the gastrointestinal tract before reaching large intestine, where they are selectively fermented by beneficial and/or commensal intestinal microbes19. Such type of fermentation leads to the changes in metabolic processes of gut microbes and nutrients availability in gut ecosystem, thus possessing a beneficial effect on the host. But these beneficial effects vastly depend on the selective stimulation of intestinal probiotics with prebiotic19. With such regulations and conditions, it is difficult to find an ideal prebiotic. Instead of adding stimuli or prebiotic, designing a probiotic that can provide enhanced beneficial effects by itself is a better alternative approach20. Bacterial strains usually used for synbiotic include but not limited to Lactobacillus spp., Bifidobacteria spp. and several prebiotics mostly form natural food sources like cocoa, peanut21.
The secondary metabolites produced by beneficial microbes include lipids, particularly short chain and poly-unsaturated fatty acids22–25. Among the short chain and poly-unsaturated fatty acids, linoleic acid (LA) is considered as one of the most functional metabolites produced by different probiotic species of Bifidobacterium, Lactobacillus and Lactococcus26. Several research groups, including our own, have focused on enhancing the productivity of LA and conjugated LA (CLA) from probiotic sources, both in the human/animal gut environment and the industrial production11,13,27. Our previous study showed a relative high number of Lactobacillus and their metabolites exhibited antimicrobial activities against various enteric pathogens such as C. jejuni, S. enterica and enterohemorrhagic Escherichia coli28–31. Lactobacillus spp. also have the adherence and multiplying ability in the host32. They can produce acids, hydrogen peroxides and bacteriocins, but with no pathogenic traits and can persist as normal flora32. Among different species of Lactobacillus, L. casei has been reported as one of the crucial probiotic strain32. Considering this, our research team over-expressed the myosin-cross-reactive antigen (Mcra) (‘mcra’ gene obtained from L. rhamnosus, another probiotic strain)33 for linoleate isomerase over-production in Lactobacillus casei (LC). The mcra over-expressing probiotic, named as “LC+mcra”, showed enhanced activities, including anti-inflammatory and antimicrobial effects both in vitro and in vivo animal model (mice)13,31. It has also been reported that LC+mcra induced no detrimental effect on mice gut, rather it improved the murine gut health through the modulation gut microbiome13.
Here, we intended to evaluate the effectiveness of LC+mcra on the colonization of a marker-containing C. jejuni strain (CJRMKm) as well as the natural colonization of C. jejuni and S. enterica in different sections of chicken gut. We also aimed to compare the gut microbial composition of the control and LC+mcra fed group chickens through 16S rRNA compositional analysis. This study may help to evaluate the effect of LC+mcra in controlling the colonization of C. jejuni and S. enterica in chickens at the pre-harvest level, along with the effect of LC+mcra on modulation of gut microbiome.
Results
Efficient colonization capability of LC+mcra in chicken gut
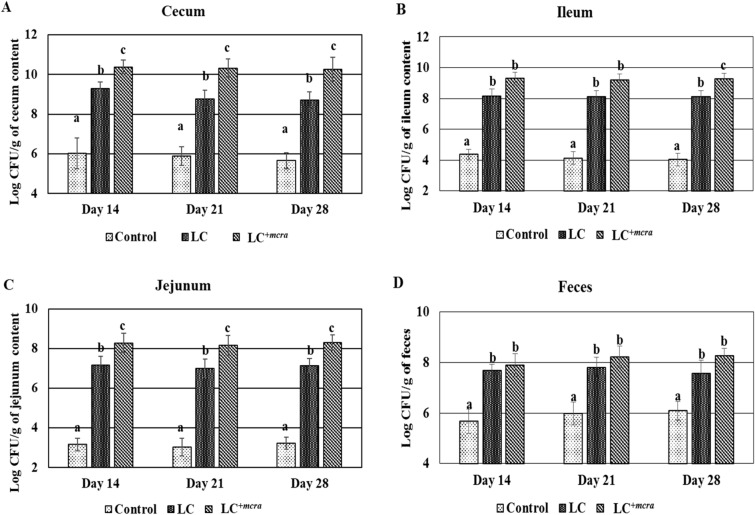
To compare the colonization capability of LC and LC+mcra in various sections of chicken gut, the colonization of Lactobacillus was enumerated and compared in our three groups of chicks from two separate trials (Fig. 1). The administration with LC+mcra showed higher and more stable cecal colonization level of Lactobacillus, above 10 log CFU/g throughout 4 weeks, which were substantially higher than wild-type LC fed group as well as natural colonization group (Fig. 1A). A similar pattern was found in LC+mcra fed chicken jejunum, which showed a significantly (p < 0.05) higher number of Lactobacillus at day 28 (Fig. 1B). Though this difference was only numerically higher in ileum when compared to wild type LC fed group (Fig. 1C). The number of Lactobacillus in the fecal shedding of probiotic-fed group were observed to rise when compared to the control group throughout the study period (Fig. 1D). Specifically, in the chicken group fed with LC+mcra, colonies of Lactobacillus in fecal shedding gradually increased, whereas fecal shedding of Lactobacillus in the wild-type LC fed group and control group (group fed with no probiotic) were observed to be noticeably lower than that in the LC+mcra fed group (Fig. 1D).
Figure 1.
Colonization of wild-type probiotic, LC and CLA over-producing probiotic, LC+mcra in different intestinal portions of chickens and their fecal shedding pattern compared to control in terms of Lactobacillus count. Colonization of LC/LC+mcra in cecum (A), ileum (B), jejunum (C) and fecal shedding of LC/ LC+mcra (D) in terms of Lactobacillus count. Results are mean ± SD and letters (a–c) are used to mark the significant differences at various time points compared with control and among the treatment groups at p < 0.05.
Control of colonization of C. jejuni strain in probiotic fed chickens
The chickens were orally administered with LC or LC+mcra in order to evaluate their roles in competitively reducing the colonization of the marker-containing strain CJRMKm. Even though, the colonization of CJRMkm was decreased notably by both LC and LC+mcra compared to the control group, in various parts of chicken gut, chickens that were fed with LC+mcra demonstrated a higher reduction in the colonization of CJRMKm than the LC fed groups. Chickens colonized with LC+mcra strain showed a notable reduction in CJRMKm colonization in different portions of intestine specifically ileum and jejunum by at least 1 log or more as compared to the chickens which were fed with wild type probiotic (LC) strain at day 14, 21 and 28 (Fig. 2). When comparing the colonization of CJRMKm in the cecum, we observed that chickens pre-administered with LC or LC+mcra showed lower CJRMKm colonization in cecum contents (Fig. 2A). Likewise, both probiotics (LC and LC+mcra) reduced the number of CJRMKm colonization in ileum contents compared to control group (Fig. 2B). The same pattern was found in jejunum contents compared to control group (Fig. 2C). The notable reduction on CJRMkm gastrointestinal colonization was also found in form of a lower number of CJRMKm fecal shedding when fed with LC or LC+mcra (Fig. 2D). Notable effectiveness of LC+mcra was observed in chickens after 1st week post-challenged with CJRMKm, at which there was an approximate 0.9 log CFU/g reduction in chicken feces compared to control group. In the following 2 weeks, CJRMKm fecal shedding of LC+mcra fed chickens continuously reduced by more than 1.0 log CFU/g and 1.10 log CFU/g at day 21 and 28, respectively, compared to the control group (Fig. 2D).
Figure 2.
Effect of wild-type probiotic, LC and CLA over-producing probiotic, LC+mcra on colonization and fecal shedding pattern of CJRMKm in different intestinal portions of chickens at days 14, 21 and 28 of infection compared to control. Colonization in cecum (A), ileum (B), jejunum (C) and fecal shedding of CJRMKm (D). Results are mean ± SD and letters (a–c) are used to mark the significant differences at various time points compared with control and among the treatment groups at p < 0.05.
Reduction on natural colonization of C. jejuni and S. enterica in probiotic fed chickens
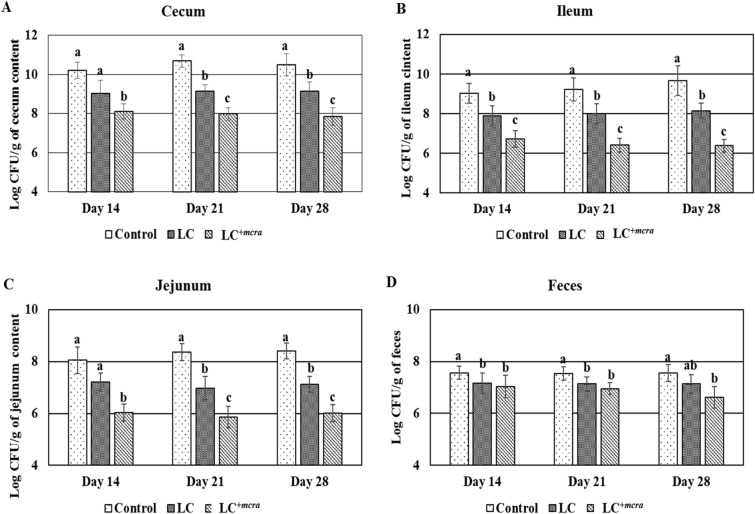
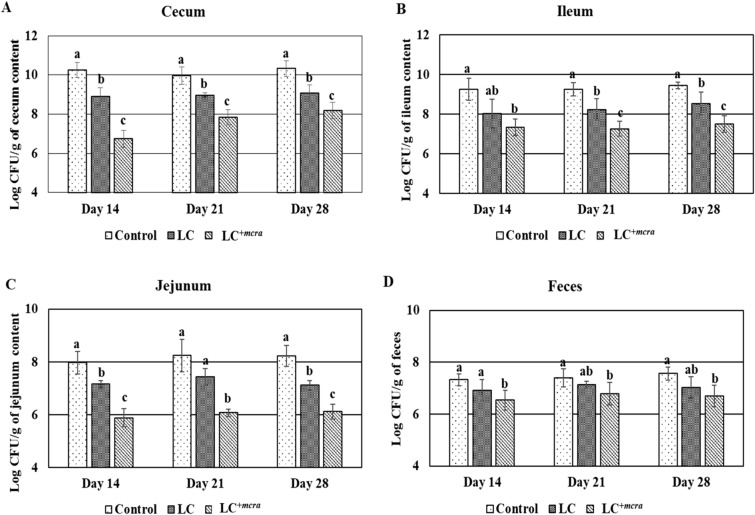
Chickens pre-treated with either LC or LC+mcra showed resistance against the natural colonization of C. jejuni and S. enterica in cecum, jejunum and ileum, but the chickens pre-treated with LC+mcra had a much effective influence on reduction of colonization of both bacterial pathogens (Figs. 3, 4). Specifically, chickens fed with LC+mcra showed remarkable reduction in the level of natural colonization of C. jejuni in cecum contents, ileum contents and in jejunum contents at days 14, 21 and 28, respectively when compared to the contents of the control groups (Fig. 3A–C). Meanwhile, a consequential decrease in fecal shedding of C. jejuni was also detected in LC+mcra fed chickens (Fig. 3D). For S. enterica, the natural colonization in cecum, ileum and jejunum contents at days 14, 21 and 28, respectively were remarkably lower when pre-treated with LC+mcra, compared to cecum contents, ileum contents and jejunum contents of the control group (Fig. 4A–C). The LC+mcra administration substantially reduced the S. enterica fecal shedding as well (Fig. 4D). A similar pattern of reduction was also found in the cecum, ileum, jejunum contents and fecal shedding of S. enterica when pre-treated with LC (Fig. 4A–D).
Figure 3.
Effect of wild-type probiotic, LC and CLA over producing probiotic, LC+mcra on colonization and fecal shedding pattern of C. jejuni in different intestinal portions of chickens at days 14, 21 and 28 compared to control. Colonization in cecum (A), ileum (B), jejunum (C) and fecal shedding of C. jejuni (D). Results are mean ± SD and letters (a–c) are used to mark the significant differences at various time points compared with control and among the treatment groups at p < 0.05.
Figure 4.
Effect of wild-type probiotic, LC and CLA over producing probiotic, LC+mcra on colonization and fecal shedding pattern of S. enterica in different intestinal portions of chickens at day 14, 21 and 28 compared to control. Colonization in cecum (A), ileum (B), jejunum (C) and fecal shedding of S. enterica (D). Results are mean ± SD and letters (a–c) are used to mark the significant differences at various time points compared with control and among the treatment groups at p < 0.05.
Reduced level of contamination in water provided to the probiotic fed chickens
At different time points (day 14, 21 and 28), water samples were collected to determine the cross-contamination with both S. enterica and C. jejuni. The contamination level of C. jejuni in water was reduced approximately by 1.0 log CFU/50 ml and more than 1.0 log CFU/50 ml in the groups of chickens fed with LC and LC+mcra, respectively, compared to the control group throughout the experimental period (Fig. 5A). A similar pattern of reduction in S. enterica contamination level was also found (Fig. 5B).
Figure 5.
Comparison of water contamination level among the probiotics fed (wild-type probiotic, LC and CLA over-producing probiotic, LC+mcra) and control groups of chickens. Contamination level of C. jejuni (A) and S. enterica (B) in water bottles. Results are mean ± SD and letters (a–b) are used to mark the significant differences at various time points compared with control and among the treatment groups at p < 0.05.
Gut microbiome alteration in the probiotic fed chickens
At the phylum level, we observed all three groups of chickens (control, LC fed and LC+mcra fed groups) were colonized with abundant number of Firmicutes. Over all 83.47%, 82.38% and 84.16% of Firmicutes were found in control, LC fed group and LC+mcra fed group, respectively (Fig. 6). Increased numbers of both Bacteroidetes and Tenericutes was also observed in the LC and LC+mcra fed groups compared to the control group. Higher number of Bacteroidetes was also observed in the gut of LC (21.72%) and LC+mcra (19.82%) fed chickens compared to the control group. The percentage of Tenericutes was approximately double for both LC and LC+mcra fed groups of chickens (Fig. 6). On the other hand, a decrease of the abundance of Proteobacteria in both LC (38.3%) and LC+mcra (39.8%) fed groups was observed. In fact, the lowest abundance of Proteobacteria was observed in the LC+mcra fed group (Fig. 6). In case of Verrucomicrobia, there was not much difference between control group and LC fed group, but there was a noteworthy reduction of Verrucomicrobia, approximately 80%, in the abundance in LC+mcra fed group. For Actinobacteria and other phyla, no remarkable differences were observed among the groups (Fig. 6).
Figure 6.
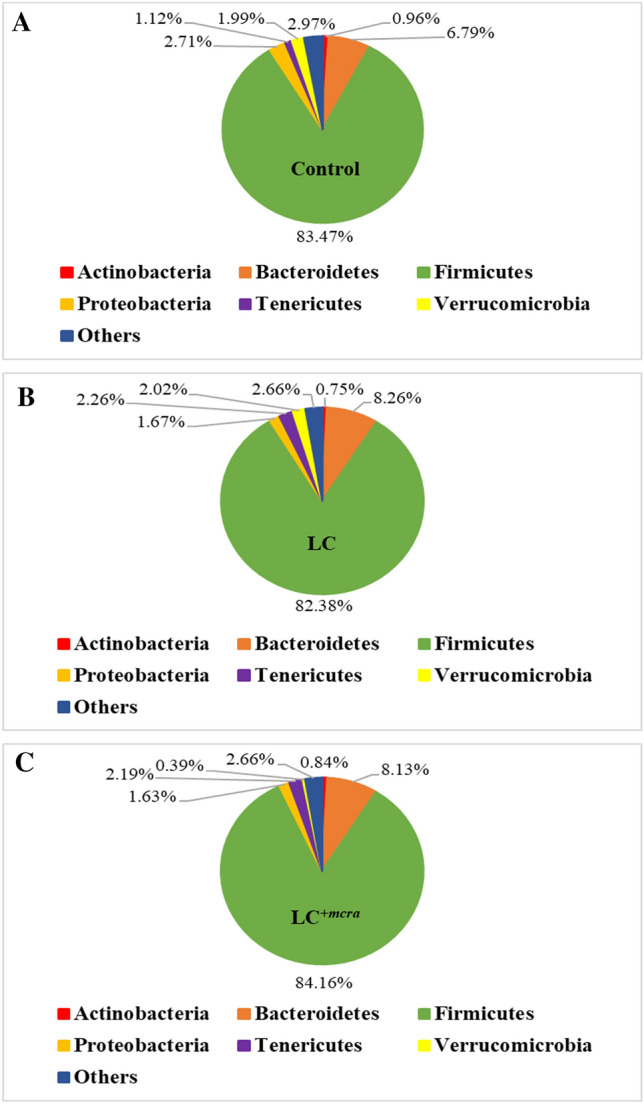
Relative abundance of sequences representing the chicken gut microbiota at phylum level. Comparison of bacterial population of chicken cecal contents collected from different groups at day 28, (A) control group, (B) wild-type probiotic, LC, fed group and (C) CLA over-producing probiotic, LC+mcra, fed group.
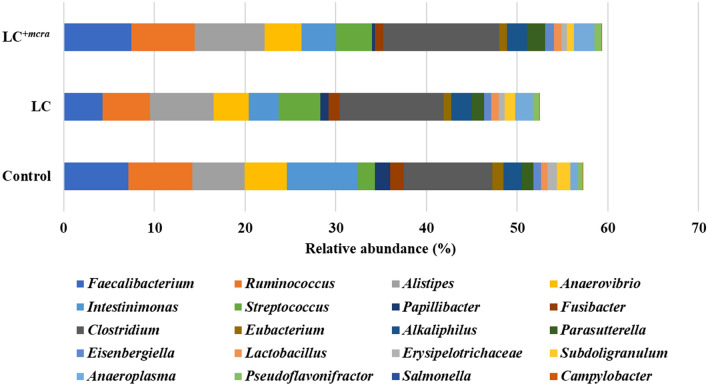
At the genus level, the distribution of bacteria in the three groups of chickens fed with LC, LC+mcra or control showed a distinct variation. For example, there was a decreased abundance in the LC or LC+mcra fed groups compared to the control for Ruminococcus, Anaerovibrio, Intestinimonas, Papillibacter, Fusibacter, Eubacterium, Erysipelotrichaceae, Subdoligranulum and most importantly for Salmonella and Campylobacter (Fig. 7). For Alistipes, Streptococcus, Clostridium, Alkaliphilus, Parasutterella, Eisenbergiella, Anaeroplasma and Pseudoflavonifractor, there was an increase of the abundance in either the LC or LC+mcra fed group compared to the control group (Fig. 7). There was also an increase of Lactobacillus in both groups fed with LC and LC+mcra, as expected. Increase in the genus level abundance of Anaeroplasma (2.31- and 2.71-fold increase in groups fed LC and LC+mcra, respectively) and Streptococcus (2.43- and 2.07-fold increase in groups fed LC and LC+mcra, respectively) was found to be the preeminent difference in groups fed LC and LC+mcra when compared to the control. Pseudoflavonifractor was also detected with an approximate 1.41 and 1.45-fold increase in LC and LC+mcra fed groups, respectively, as compared to the control group. Lactobacillus was found to have an increase of around 1.16- and 1.17-fold in groups fed LC and LC+mcra, respectively, as compared to the control group. On the other hand, the abundance of Papillibacter was decreased by 1.86- and 4.85-fold in LC and LC+mcra fed groups, respectively; along with a 1.54- and 1.71-fold decrease of Erysipelotrichaceae in LC and LC+mcra fed groups, respectively, as compared to the control group. Colonization of Salmonella was decreased by 2.13- and 3.54-fold in groups fed LC and LC+mcra, respectively, as compared to the control group. The decrease of Campylobacter colonization was approximately 3.16-fold in the LC+mcra fed group of chickens compared to the control group (Fig. 7).
Figure 7.
Comparison of bacterial composition at genus level of chicken cecal contents collected at day 28, among control group, wild-type probiotic, LC, fed group and CLA over-producing probiotic, LC+mcra, fed group.
We have also compared the diversity of bacterial species among the three groups using alpha indices (Shannon index, Simpson index and Margalef’s richness). There was no significant difference observed in bacterial diversity among the three (control, LC and LC+mcra fed) groups. For instance, the number of observed species in both LC and LC+mcra fed groups was only numerically higher than the control, with LC+mcra group having the highest, an average of 1592 species (Fig. 8A). There was also only numerical difference in Simpson index for control, LC and LC+mcra fed group (Fig. 8B). The average of Shannon index of LC+mcra fed group was numerically the highest followed by LC fed group and control group (Fig. 8C). The Margalef’s richness was also numerically the highest for LC+mcra fed group followed by LC fed group and control group (Fig. 8D).
Figure 8.
Comparison of chicken gut microbiome at species level. Chicken cecal contents were collected from control, wild-type probiotic, LC, fed group and CLA over-producing probiotic, LC+mcra, fed groups at day 28. Bacterial diversity at species level (A) Number of species, (B) Simpson index, (C) Shannon index and (D) Margalef’s richness was compared. Results are mean ± SD.
Discussion
Previously we showed the antagonistic effect of conjugated linoleic acid over-producing probiotic, LC+mcra, on enteric bacterial pathogens through in vitro studies28,30,31. Here, we verified the effectiveness of CLA over-producing LC+mcra against the colonization of the marker-containing CJRMKm strain, as well as on the natural colonization of C. jejuni and S. enterica in a chicken model. According to our in vitro study, the novel probiotic strain LC+mcra demonstrated at least 21 folds more CLA conversion rate in bacterial cell free cultural supernatant and it could also survive longer in comparison with the wild-type LC strain31. We also observed a remarkable decrease of C. jejuni and S. enterica growth in co-culture with LC+mcra and in the presence of the cell-free cultural supernatant collected from LC+mcra. LC+mcra could also alter the host cells-C. jejuni/S. enterica interactions, expression of virulence genes and other virulence properties30, 31.
Though, according to literature, efficiency of probiotics in reducing colonization of enteric pathogenic bacteria is yet inconclusive, several groups of research have proposed that the increased production of metabolites from probiotics (e.g., CLA) might improve the overall health benefits of probiotics on host, in addition to the exclusion of enteric pathogens through colonization competition in gut ecosystem11, 29–31,34,35. In this study, we used a CJRMKm strain containing kanamycin resistance gene which was inserted into cdt of C. jejuni chromosome as shown in a previous study demonstrating that deletion of cdt did not alter the colonization capability of C. jejuni in chicken gut36. The 8-day-old chicks challenged with CJRMKm showed a persistent resistance to CJRMKm colonization in their gut through the effect of probiotics. Specifically, probiotic treatments reduced the colonization of CJRMKm from chicken cecum, ileum and jejunum in a time-dependent manner at two, three and four weeks of age. The microbial balance of fecal shedding in the beneficial, commensal and detrimental microbes serves as the crucial indicator of gastrointestinal health37, correspondingly we observed the decreased colonization of CJRMKm in both fecal contents and various intestinal contents, as well as an increased colonization trend of probiotic strains. Probiotics themselves were able to limit gastrointestinal bacterial pathogens through competitive exclusion and colonization resistance9,38–41.
From the in vivo findings of this study, we observed that both probiotics (LC and LC+mcra) remarkably diminished C. jejuni and S. enterica colonization in gastrointestinal portions including cecum, jejunum and ileum, which supported our previous studies13. In addition, LC+mcra showed more aggressive reductions on the colonization of CJRMKm, C. jejuni and S. enterica as a result of the effects achieved from its increased amount of CLA conversion13,31. Previously, CLA has been reported and associated with antibacterial effect against different enteric pathogens including C. jejuni and S. enterica, though the distinctive mechanisms of interaction between CLA and bacterial cells are still under investigation30,31,42. Furthermore, our in vivo examination based on chicken model also confirmed the protective functions of LC+mcra against gastrointestinal bacterial pathogens, supporting the previous in vitro outcomes29–31. In mice model, CLA was observed to provide a protective mechanism through both stimulating beneficial bacterial colonization and maintaining the balance in microbial composition in gut ecosystem, which served as the first line defense against foodborne pathogenic bacterial colonization13.
The investigation of the chicken normal floral distribution was another goal of this study. Several research groups also reported that CLA-rich diets could stimulate the fatty acids metabolism and preserve homeostatic gut microbiota43,44. The balanced gut microbial ecosystem, along with the diversity of microbes, was modified by the introduction of probiotic capable of producing CLA. The researchers also suggested that higher abundances of beneficial bacteria such as Lactobacillus and Bifidobacterium could influence the gut homeostasis in a positive way that contributes to defending against enteric pathogens11. In this study, we found that the most abundant bacterial phyla in both probiotic fed and non-fed group of chickens were Firmicutes and Bacteroidetes. When the chickens were given probiotic either wild-type LC or bioactive LC+mcra strain, the abundance of the Firmicutes and Bacteroidetes were increased notably. Previously, Wang et al. also reported a similar finding45. When they treated the chickens with probiotics specifically Lactobacillus, they also observed that Firmicutes and Bacteroidetes were colonized notably higher in the cecum of the broiler. We also observed the relatively a smaller number of Proteobacteria in both groups of chickens treated with probiotics, specifically in the gut of the LC+mcra fed group. Proteobacteria compose with a large number of Gram-negative enteric bacteria including pathogens. By feeding probiotics, several research groups also observed reduced number of Proteobacteria in the animal models13,46,47. We also found that Clostridium, Faecalibacterium, Ruminococcus and Alistipes were the predominant genera in all three groups of chickens which is in agreement with the previous reports of Luo et al. and Shaufi et al.46,48. These Clostridium, Faecalibacterium, Ruminococcus and Alistipes genera are also found to involve in the production of different beneficial metabolites in poultry gut, including SCFA46. In our study, we observed that these Clostridium, Faecalibacterium, Ruminococcus and Alistipes genera were highly abundant in the probiotic fed group chickens specifically in the gut of LC+mcra fed chickens.
As two groups of chickens were given orally either LC or LC+mcra, higher abundance of the Lactobacillus species was observed in those groups. Previously Wang et al. also observed that probiotic feeding increased the abundance of the probiotics in poultry45. Furthermore, we observed that there was reduced number of Gram-negative bacteria including Fusibacter, S. enterica and C. jejuni in the gut of probiotic fed chickens. We also found that the decreasing pattern of the Gram-negative bacteria was much more noticeable in the LC+mcra fed group than the LC fed group which is agreed with the previous findings of Peng et al.13. The abundance and diversity of the available species were also compared in this study. Though there was no statistically significant difference, numerical differences among control, LC and LC+mcra fed groups of chickens were observed. Similar finding was also reported by Wang et al.45. The diversity of the species largely varies with the heredity, environment and diets, which might be responsible for the nonsignificant difference among the groups of chickens.
In addition to this, reduced numbers of C. jejuni and S. enterica were observed in the drinking water collected from the water containers in the probiotic-fed groups. This result is supported by our previous studies where we reported LC+mcra inhibited C. jejuni, S. enterica and E. coli13,28–31. The survival and persisting ability of C. jejuni in water has been reported, which means that reducing survivability in drinking water of chicken may limit both the horizontal transfer of this bacterial pathogen to other chicks and the colonization of C. jejuni in flocks49.
It has been reported that antibiotic growth promoter limits both the prevalence of C. jejuni at the pre-harvest level in conventional poultry farms as well as the poultry carcass cross-contamination with C. jejuni in conventionally processed chicken meat when compared to organic processors5,50. Previously, we observed that berry pomace extracts decreased the natural colonization of C. jejuni and colonization of marker-containing CJRMKm strain in chicken cecum remarkably with positive modulation of chicken gut microbiome51–53. Similarly, here we also observed noteworthy decrease in CJRMKm, C. jejuni and S. enterica in different portions of chicken gastrointestinal tract in the probiotics fed chicks (both wild-type and CLA over-producing LC) with the LC+mcra having a substantially better performance. In water samples of both probiotics fed groups, there was reduction in the cross-contamination with C. jejuni and S. enterica. Therefore, the combination of berry pomace extract and LC+mcra can be a potential approach for reducing the colonization of C. jejuni and S. enterica as well as improving the growth of chickens, but further investigation is needed.
Conclusively, this study indicated the effectiveness of CLA over-producing probiotic strain, LC+mcra, on the colonization of both marker-containing CJRMKm strain and naturally colonized C. jejuni and S. enterica in various sections of chicken intestine along with the modulation of gut microbiota towards healthier gut. Specifically, chickens fed with LC+mcra daily for 1 week showed a notable reduction on the colonization of CJRMKm. Natural colonization of chicken gut and fecal shedding with both C. jejuni and S. enterica were also observed to be notably reduced by probiotics feeding. Though the study was conducted in controlled laboratory environment, the outstanding roles of LC+mcra provides a promising option for reducing the colonization of the most predominant zoonotic pathogens, specifically S. enterica and C. jejuni in chicken gut, which may eventually lead to lower incidence of cross-contamination in poultry products. Thus, LC+mcra may have a beneficial effect on the quality of the poultry products and retain consumer satisfaction with regards to the safety of the poultry products and reducing the risk of zoonotic infections.
Methods
Bacterial strains and growth conditions
Lactobacillus casei (ATCC 334) (LC) and Campylobacter jejuni RM1221 (ATCC BAA-1062) were used in this study. To distinguish from other C. jejuni, a marker-containing Kanamycin cassette was inserted into the chromosomal DNA of C. jejuni and named as ‘CJRMKm’52,53. CJRMKm was grown in Karmali agar (Himedia, India) with selective supplements (Himedia, India) under microaerophilic conditions, in presence of 10% CO2, 5% O2 and 85% N2 at 37 °C. Over-production of conjugated linoleic acid in LC, was achieved by incorporating the myosin cross reactive antigen gene (mcra) in LC chromosome giving it the name of ‘LC+mcra’13,28–31. Both LC and LC+mcra were grown at 37 °C for 18 h in presence of 5% CO2 (Thermo Fisher Scientific Inc., USA) on MRS (de Man Rogosa Sharpe) agar (EMD Chemicals Inc., USA).
In vivo study design and chicken model
Experiments in the chicken model were carried out up to 4 weeks, in duplicate trials, using 934-1 Isolator Units (Dye Sheet Matter, USA), which were equipped with HEPA filter and air circulation system. Each animal trial consisted with 60 SPF chicks (one day-old), obtained from Charles River, USA. Institutional Animal Care and Use Committee (IACUC, protocol number R-16-33) recommended guidelines were followed for chicken husbandry. Commercially available crumbles without antibiotic supplementation (Purina Animal Nutrition, USA) were used as chicken feed. Chickens were divided into 3 groups (20 chickens per group) which corresponded to their respective 934-1 Isolator Units (Dye Sheet Matter, USA) (Tables 1, 2). For each treated/control group, chickens were further divided into 2 separate sub-groups (10 chickens in each case) and housed in individual 934-1 Isolator Unit (Dye Sheet Matter, USA). Water was given using Gravity-fed water containers.
Table 1.
Number of chickens allocated in the experimental groups with treatments.
| Treatments | Trial 1 | Trial 2 | Total |
|---|---|---|---|
| Phosphate buffered saline (A) | 20 | 20 | 40 |
| L. casei ATCC334 (B) | 20 | 20 | 40 |
| Mcra over-expressing L. casei (C) | 20 | 20 | 40 |
| Total | 60 | 60 | 120 |
Table 2.
Experimental procedures with timeline (per trial).
| Timeline | Number of chickens | |||||
|---|---|---|---|---|---|---|
| Daily probiotic colonization | Enteric bacterial challenge | Fecal sample collection | Euthanization | Cecum sample harvest | Live | |
| Day0 | – | – | 60 (1/chicken) | – | – | 60 |
| Day1–Day7 | 40 (B, C)a | – | – | – | – | 60 |
| Day8 | – | 60 (A, B, C)a | 60 (1/chicken) | – | – | 60 |
| Day14 | – | – | 60 (1/chicken) | 18 (6/group) | 18 (6/group) | 42 |
| Day21 | – | – | 42 (1/chicken) | 18 (6/group) | 18 (6/group) | 24 |
| Day28 | – | – | 24 (1/chicken) | 24 (8/group) | 24 (8/group) | 0 |
aA, corresponds to ‘control group’; B. corresponds to the ‘group fed with LC’ and C, corresponds to ‘group fed with LC+mcra’.
Feeding probiotics and C. jejuni challenge to chicken model
Both the probiotic strains LC and LC+mcra were grown at 37 °C overnight in shaking incubator (at 120 rpm) on MRS broth (EMD Chemicals Inc., USA) and suspensions were prepared using PBS. LC and LC+mcra solutions (100 μl) containing 1010 CFU/ml were fed to chicken groups as shown in Table 2. On day 8, all chickens were challenged with CJRMKm which was grown in Bolton broth (Himedia, India) in presence of 5% fetal bovine serum (FBS) (Corning, USA) with shaking (at 120 rpm) for 30 h under microaerophilic condition (10% CO2, 5% O2 and 85% N2). Suspension of CJRMKm (100 μl) containing 109 CFU/ml was fed to the 8-day-old chicks through oral gavage. Chickens were routinely checked for any physical injury after the gavage administration and then moved back to their respective cages.
Sample collection from chicken carcass and processing
Euthanization was performed on day 14 and day 21 for 6 chickens per group and on day 28 for the rest of the chickens, with the purpose of evaluating the degree of colonization of probiotics and CJRMKm in the chick cecum, ileum and jejunum. To determine the colonization pattern of probiotics, homogenized cecum, ileum or jejunum contents (~ 200 g) were diluted and then plated on MRS agar (EMD Chemicals Inc., USA) for enumeration. To test the CJRMKm colonization level, cecum, ileum, or jejunum contents (~ 200 g) were homogenized in 1 ml of phosphate buffer saline (PBS) and then plated on Karmali Agar containing selective supplements (Himedia, India) and Kanamycin (100 µg/ml) (Thermo Fisher Scientific, USA) following serial dilution techniques for enumeration. Colonization of C. jejuni (challenged and natural) and S. enterica (natural) was also determined following standard methods. For enumeration of C. jejuni and S. enterica, intestinal contents from cecum, ileum or jejunum (~ 200 g) were collected and homogenized in PBS solution (1 ml), diluted and then plated on Karmali Agar with selective supplement and Xylose Lysine Deoxycholate Agar (XLD Agar) (Himedia, India), respectively. Water samples were also collected from the drinking containers of cages. Water samples (50 ml) were collected in centrifuge tubes (VWR, USA) using a sterile 25 ml pipette (VWR, USA). Then the samples were centrifuged (3000 × g) for 20 min, bacterial pellets were re-suspended in PBS (1 ml) and plated on Karmali Agar with selective supplement and XLD Agar for enumeration.
16S rRNA compositional analysis
For comparison of the chicken gut microbiome, the cecum contents of six 28-day-old chickens from each group were randomly collected for 16S rRNA compositional analysis following the methods previously described by Peng et al.13. Briefly, DNA was extracted with QIAamp Fast DNA Stool Kit (QIAGEN, USA). The variable regions (V3 and V4) of 16S rRNA were targeted for phylogenic classifications. DNA libraries were prepared using Nextera DNA Library Preparation Kit and Nextera Index Kit (Illumina, USA) and pooled into equimolar concentration according to the protocol provided by the manufacturers. Sequencing (paired-end: 2 × 300 bp) was preformed using MiSeq v3 600-Cycle Kit (Illumina, USA) by Illumina MiSeq. Sequencing data were processed by MiSeq Reporter-BaseSpace for FASTQ workflow generation (Greengenes database). Demultiplexing was conducted using the perfect index recognition (mismatch = 0) and then by removing PhiX reads. A total 8,509,169 quality-filtered reads were used for analysis of the 16S rRNA composition. The data were analyzed to determine the differences of relative abundance at phylum, genus and species (number of species, Simpson index, Shannon index and Margalef’s richness) levels among three groups (control, LC or LC+mcra fed groups) following standard protocol.
Statistical analysis
For statistical analysis, cecum, ileum or jejunum from a bird was considered as an experimental unit. The data were ranked, and ANOVA was used to differentiate the level of colonization (CFU/g cecum, ileum or jejunum contents). For comparison of the mean ranks, Tukey’s test was used. Differences in the water samples were compared by performing ANOVA and Tukey’s test was used for comparison of mean ranks.
Ethical approval and informed consent
Institutional Animal Care and Use Committee (IACUC) approval was taken for the chicken trials (Protocol Number R-16-33). IACUC recommended guidelines were followed for chicken husbandry.
Acknowledgements
We thank Dr. Xiaoping Zhu, Chair, Department of Veterinary Medicine, UMD for providing support for this animal trail. We also thank Mr. Girmay Tekle Gebreluul, Ms. Johanna Lavigne and Mr. Yonas Araya, Department of Veterinary Medicine, UMD for their technical assistance during animal experiments. The authors would also like to thank USDA-NIFA for grant support (Exploratory Grant: 2018-67030-27426) for this study.
Author contributions
Z.T. designed and carried out the experiments, analyzed the data and wrote the manuscript. M.P. assisted in the experiments, performed 16S rRNA compositional analysis and revised the manuscript. Z.A.M. assisted in sample processing, microbiological experiments and reviewing the manuscript. A.A., J.B., P.D.B. and A.Y. assisted in experimental preparation, sample processing and microbiological experiments. D.B. supervised the research and finalized the manuscript. All authors approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Center for Disease Control and Prevention (CDC). Preliminary Incidence and Trends of Infections with Pathogens Transmitted Commonly Through Food—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2015–2018. Morb. Mortal. Wkly. Rep. (2019) [DOI] [PMC free article] [PubMed]
- 2.Facciolà A, Riso R, Avventuroso E, Visalli G, Delia SA, Laganà P. Campylobacter: from microbiology to prevention. J. Prev. Med. Hyg. 2017;58(2):E79–E92. [PMC free article] [PubMed] [Google Scholar]
- 3.Lin J. Novel approaches for Campylobacter control in poultry. Foodborne Pathog. Dis. 2009;6:755–765. doi: 10.1089/fpd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams LK, Fonseca BB, Humphrey TJ. In: Campylobacter jejuni in Poultry: A Commensal or a Pathogen? Fonseca BB, Fernandez H, Rossi HA, editors. Berlin: Springer; 2016. pp. 75–67. [Google Scholar]
- 5.Luangtongkum T, et al. Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl. Environ. Microbiol. 2006;72:3600–3607. doi: 10.1128/AEM.72.5.3600-3607.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salaheen S, et al. Bioactive extracts from berry byproducts on the pathogenicity of Salmonella typhimurium. Int. J. Food Microbiol. 2016;237:128–135. doi: 10.1016/j.ijfoodmicro.2016.08.027. [DOI] [PubMed] [Google Scholar]
- 7.Peng M, Salaheen S, Buchanan RL, Biswas D. Alterations of Salmonella enterica serovar typhimurium antibiotic resistance under environmental pressure. Appl. Environ. Microbiol. 2018;84(19):e01173–e10118. doi: 10.1128/AEM.01173-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng M, et al. Prevalence and antibiotic resistance pattern of Salmonella serovars in integrated crop-livestock farms and their products sold in local markets. Environ. Microbiol. 2016;18:1654–1665. doi: 10.1111/1462-2920.13265. [DOI] [PubMed] [Google Scholar]
- 9.Amalaradjou MAR, Bhunia AK. Modern approaches in probiotics research to control foodborne pathogens. Adv. Food Nutr. Res. 2012;67:185–239. doi: 10.1016/B978-0-12-394598-3.00005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes SR, Vargas AJ. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Explore (NY) 2016;12(6):463–466. doi: 10.1016/j.explore.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng M, Biswas D. Short chain and polyunsaturated fatty acids in host gut health and foodborne bacterial pathogen inhibition. Crit. Rev. Food Sci. Nutr. 2017;57:3987–4002. doi: 10.1080/10408398.2016.1203286. [DOI] [PubMed] [Google Scholar]
- 12.Nagarajan V, et al. Antimicrobial effect and probiotic potential of phage resistant Lactobacillus plantarum and its interactions with zoonotic bacterial pathogens. Foods. 2019;8(194):1–13. doi: 10.3390/foods8060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng M, et al. Prevention of enteric bacterial infections and modulation of cecal microbiota with conjugated linoleic acids producing Lactobacillus in mice. Gut Microbes. 2019;11(3):433–452. doi: 10.1080/19490976.2019.1638724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012;9(10):577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 15.Marcobal A, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7(10):1933–1943. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cammarota G, Ianiro G, Bibbo S, Gasbarrini A. Gut microbiota modulation: probiotics, antibiotics or fecal microbiota transplantation? Intern. Emerg. Med. 2014;9:365–373. doi: 10.1007/s11739-014-1069-4. [DOI] [PubMed] [Google Scholar]
- 17.Dahiya DK, et al. Gut microbiota modulation and its relationship with obesity using prebiotic fibers and probiotics: a review. Front. Microbiol. 2017;8:563. doi: 10.3389/fmicb.2017.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.FAO. Technical Meeting on Prebiotics: Food Quality and Standards Service (AGNS), Food and Agriculture Organization of the United Nations (FAO) 2017, 15–16 (2017)
- 19.Wang Y. Prebiotics: present and future in food science and technology. Food Res. Int. 2009;42:8–12. doi: 10.1016/j.foodres.2008.09.001. [DOI] [Google Scholar]
- 20.Peng M, Salaheen S, Biswas D. In: Encyclopedia of Agriculture and Food Systems. Alfen NKV, editor. Amsterdam: Elsevier Inc. Press; 2014. pp. 346–357. [Google Scholar]
- 21.Pandey KR, Naik SR, Vakil BV. Probiotics, prebiotics and synbiotics—a review. J. Food Sci. Technol. 2015;52(12):7577–7587. doi: 10.1007/s13197-015-1921-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2009;12(10):661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 23.O’Shea EF, Cotter PD, Stanton C, Ross RP, Hill C. Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: bacteriocins and conjugated linoleic acid. Int. J. Food Microbiol. 2012;152:189–205. doi: 10.1016/j.ijfoodmicro.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 24.Serini S, Piccioni E, Merendino N, Calviello G. Dietary polyunsaturated fatty acids as inducers of apoptosis: implications for cancer. Apoptosis. 2009;14(2):135–152. doi: 10.1007/s10495-008-0298-2. [DOI] [PubMed] [Google Scholar]
- 25.Bassaganya-Riera J, Viladomiu M, Pedragosa M. Probiotic bacteria produce conjugated linoleic acid locally in the gut that targets macrophage PPAR γ to suppress colitis. PLoS ONE. 2012;7(2):e31238. doi: 10.1371/journal.pone.0031238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rizos EC, Ntzani EE, Bika E, Kostapanos MS, Elisaf MS. Association between omega-3 fatty acid supplementation and risk of major cardiovascular disease events: a systematic review and meta-analysis. JAMA. 2012;308(10):1024–1033. doi: 10.1001/2012.jama.11374. [DOI] [PubMed] [Google Scholar]
- 27.Yadav H, Jain S, Sinha PR. Production of free fatty acids and conjugated linoleic acid in probiotic dahi containing Lactobacillus acidophilus and Lactobacillus casei during fermentation and storage. Int. Dairy J. 2007;17(8):1006–1010. doi: 10.1016/j.idairyj.2006.12.003. [DOI] [Google Scholar]
- 28.Peng M, Reichmann G, Biswas D. Lactobacillus casei and its byproducts alter the virulence factors of foodborne bacterial pathogens. J. Funct. Foods. 2015;15:418–428. doi: 10.1016/j.jff.2015.03.055. [DOI] [Google Scholar]
- 29.Tabashsum Z, Peng M, Kahan E, Rahaman SO, Biswas D. Effect of conjugated linoleic acid overproducing Lactobacillus with berry pomace phenolic extracts on Campylobacter jejuni pathogenesis. Food Funct. 2018;10:296–303. doi: 10.1039/C8FO01863D. [DOI] [PubMed] [Google Scholar]
- 30.Tabashsum Z, Peng M, Salaheen S, Comis C, Biswas D. Competitive elimination and virulence property alteration of Campylobacter jejuni by genetically engineered Lactobacillus casei. Food Control. 2018;85:283–291. doi: 10.1016/j.foodcont.2017.10.010. [DOI] [Google Scholar]
- 31.Peng M, Tabashsum Z, Patel P, Bernhardt C, Biswas D. Linoleic acids overproducing Lactobacillus casei limits growth, survival and virulence of Salmonella Typhimurium and enterohemorrhagic Escherichia coli. Front. Microbiol. 2018;9:2663. doi: 10.3389/fmicb.2018.02663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reid G. The scientific basis for probiotic strains of Lactobacillus. Appl. Environ. Microbiol. 1999;65(9):3763–3766. doi: 10.1128/AEM.65.9.3763-3766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Byeon JI, et al. Growth inhibition of foodborne and pathogenic bacteria by conjugated linoleic acid. J. Agric. Food Chem. 2009;57:3164–3172. doi: 10.1021/jf8031167. [DOI] [PubMed] [Google Scholar]
- 34.Garner CD, et al. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar Typhimurium murine model of infection. Infect. Immun. 2009;77:2691–2702. doi: 10.1128/IAI.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snel J, Born L, van der Meer R. Dietary fish oil impairs induction of gamma-interferon and delayed-type hypersensitivity during a systemic Salmonella enteritidis infection in rats. APMIS. 2010;118:578–584. doi: 10.1111/j.1600-0463.2010.02630.x. [DOI] [PubMed] [Google Scholar]
- 36.Biswas D, et al. Effect of cytolethal distending toxin of Campylobacter jejuni on adhesion and internalization in cultured cells and in colonization of the chicken gut. Avian Dis. 2006;50:586–593. doi: 10.1637/7514-020706R1.1. [DOI] [PubMed] [Google Scholar]
- 37.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenney PT, Pamer EG. From hype to hope: the gut microbiota in enteric infectious disease. Cell. 2015;163:1326–1332. doi: 10.1016/j.cell.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng M, Bitsko E, Biswas D. Functional properties of peanut fractions on the growth of probiotics and foodborne bacterial pathogens. J. Food Sci. 2015;80:M635–M641. doi: 10.1111/1750-3841.12785. [DOI] [PubMed] [Google Scholar]
- 41.Peng M, Aryal U, Cooper B, Biswas D. Metabolites produced during the growth of probiotics in cocoa supplementation and the limited role of cocoa in host-enteric bacterial pathogen interactions. Food Control. 2015;53:124–133. doi: 10.1016/j.foodcont.2015.01.014. [DOI] [Google Scholar]
- 42.Meraz-Torres LS, Hernandez-Sanchez H. Conjugated linoleic acid in dairy products: a review. Am. J. Food Technol. 2012;7:176–179. doi: 10.3923/ajft.2012.176.179. [DOI] [Google Scholar]
- 43.Marques TM, et al. Dietary trans-10, cis-12-conjugated linoleic acid alters fatty acid metabolism and microbiota composition in mice. Br. J. Nutr. 2012;113:1–11. doi: 10.1017/S0007114514004206. [DOI] [PubMed] [Google Scholar]
- 44.Kho ZY, Lal SK. The human gut microbiome—a potential controller of wellness and disease. Front. Microbiol. 2018;9:1–23. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, et al. Effect of probiotics on the meat flavour and gut microbiota of chicken. Sci. Rep. 2017;7(1):6400. doi: 10.1038/s41598-017-06677-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo YH, et al. Broilers fed dietary vitamins harbor higher diversity of cecal bacteria and higher ratio of Clostridium, Faecalibacterium, and Lactobacillus than broilers with no dietary vitamins revealed by 16S rRNA gene clone libraries. Poult. Sci. 2013;92(9):2358–2366. doi: 10.3382/ps.2012-02935. [DOI] [PubMed] [Google Scholar]
- 47.Oakley BB, et al. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014;360(2):100–112. doi: 10.1111/1574-6968.12608. [DOI] [PubMed] [Google Scholar]
- 48.Shaufi MA, Sieo CC, Chong CW, Gan HM, Ho YW. Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses. Gut Pathog. 2015;7:1–12. doi: 10.1186/s13099-015-0049-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newell DG, Fearnley C. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 2003;69(8):4343–4351. doi: 10.1128/AEM.69.8.4343-4351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salaheen S, Nguyen C, Hewes D, Biswas D. Cheap extraction of antibacterial compounds of berry pomace and their mode of action against the pathogen Campylobacter jejuni. Food Control. 2014;46:174–181. doi: 10.1016/j.foodcont.2014.05.026. [DOI] [Google Scholar]
- 51.Salaheen S, Almario JA, Biswas D. Inhibition of growth and alteration of host cell interactions of Pasteurella multocida with natural byproducts. Poult. Sci. 2014;93:1375–1382. doi: 10.3382/ps.2013-03828. [DOI] [PubMed] [Google Scholar]
- 52.Salaheen S, et al. Reduced Campylobacter jejuni colonization in poultry gut with bioactive phenolics. Food Control. 2018;84:1–7. doi: 10.1016/j.foodcont.2017.07.021. [DOI] [Google Scholar]
- 53.Salaheen S, Kim SW, Haley BJ, Van Kessel JAS, Biswas D. Alternative growth promoters modulate broiler gut microbiome and enhance body weight gain. Front. Microbiol. 2017;8:2088. doi: 10.3389/fmicb.2017.02088. [DOI] [PMC free article] [PubMed] [Google Scholar]