Abstract
The capecitabine and oxaliplatin (CapeOX) regimen is a commonly used adjuvant chemotherapeutic regimen for gastric cancer (GC). However, some patients exhibit a poor chemotherapy response due to genetic differences among individuals. Therefore, finding an effective sensitization strategy for CapeOX is important in the treatment of GC. The present study aimed to investigate the predictive biomarkers of the CapeOX chemotherapeutic outcomes for patients with GC. A total of 30 differentially expressed genes (DEGs) were identified using the gene expression profiles from The Cancer Genome Atlas capecitabine and oxaliplatin treatment GC cases and seven key DEGs [uroplakin-1b (UPK1B), fatty acid-binding protein, heart (FABP3), cystatin-M, caspase-5 (CASP5), corticosteroid 11-β-dehydrogenase isozyme 2, cytochrome P450 4X1 (CYP4X1) and epidermal growth factor receptor kinase substrate 8-like protein 3] were associated with survival. Gene validation was performed in clinical samples divided into recurrence and nonrecurrence groups. Patients with high or low expression of UPK1B, FABP3, CASP5 and CYP4X1 had markedly different overall survival rates. A model was established and the area under the curve of the receiver operating characteristic reached 0.875 (0.793–0.957), indicating that the model had good sensitivity and specificity.
Keywords: gastric cancer, chemotherapy, capecitabine and oxaliplatin, capecitabine, oxaliplatin, biomarkers
Introduction
There were 1,000,000 new cases of gastric cancer (GC) and an estimated 783,000 deaths worldwide in 2018 (1). Surgery has always been the main method of treating advanced GC. However, the effect of surgery alone is unsatisfactory and adjuvant chemotherapy is indispensable for the comprehensive treatment of GC (2).
Chemotherapeutic regimens based on 5-fluorouracil (5-Fu) and platinum are widely used in the adjuvant treatment of GC (3). Among these regimens, the capecitabine and oxaliplatin (CapeOX) regimen is one of the most commonly used regimens for adjuvant chemotherapy following GC surgery (3). Capecitabine is an oral preparation of antimetabolites that act specifically on tumor cells. After oral administration, capecitabine is rapidly absorbed through the intestinal mucosa and converted into 5-Fu through a three-step enzyme chain reaction in vivo (4). Capecitabine blocks the intracellular thymidine synthase from deoxyribonucleotide uridine and interferes with DNA synthesis (4). Oxaliplatin has stronger cytotoxic effect and fewer side effects compared with cisplatin and can bind to DNA after entering the nucleus (5). This phenomenon results in the formation of a variety of intra- and interstrand crosslinking structures, thereby leading to DNA damage and initiating apoptosis through a variety of damage mechanisms (5).
Although the effect of the CapeOX regimen on GC is clear, the clinical response of patients with GC differs and some patients have a poor response to chemotherapy (6). Sensitization strategies for effective GC CapeOX regimens remain to be found. Studies on genome-wide research have revealed the diversity and the complex genetic alterations of patients with GC, which can explain the differing responses of different patients during chemotherapy (7,8). Currently, few biomarkers for the prediction of CapeOX chemotherapy have been validated for clinical use. Therefore, establishing a multibiomarker prediction tool is required.
In the present study, the genes associated with capecitabine and oxaliplatin drug response were explored using The Cancer Genome Atlas (TCGA) database and further validated in clinical samples. A model was established to analyze the probability of the treatment resistance of patients with GC and predict their outcomes. The present study may help physicians to administer the appropriate chemotherapy regimens and provide a theoretical basis for further intervention and the reversal of drug resistance in GC treatment.
Materials and methods
TCGA database and CapeOX-related gene analysis
Datasets were downloaded from TCGA database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). The data on patient survival, history of medication and mRNA expression profiles were downloaded from the TCGA-Stomach Adenocarcinoma data (STAD) repository (https://portal.gdc.cancer.gov/). Subsequently, two expression profiles were established for further analysis. One profile contained 29 cases previously treated with capecitabine and the other profile included 17 cases treated with oxaliplatin. The genes that met the cut-off criteria (fold-change >2 and adjusted P<0.1) were considered differentially expressed genes (DEGs). The integrative analysis of the two sets of DEGs was performed using cross analysis. Clustering analyses were performed to show the expression patterns of the DEGs. Online biological tools were used to investigate the functions and pathways of the candidate DEGs. Gene Ontology (GO) enrichment analysis (http://www.geneontology.org/) was used to explore the biological functions associated with the DEGs.
Identification of survival-related DEGs
The TCGA-STAD dataset included 279 patients with GC and newly updated survival information and gene expression arrays. The patients were assigned to high- or low-gene expression groups in accordance with the median of the RNA expression level data.
Patient information and tissue samples
A total of 74 GC tissue samples were obtained from patients in the Affiliated Suzhou Hospital of Nanjing Medical University. The inclusion criteria for patients were: i) At least 18 years of age and signed written informed consent and ii) radical gastrectomy for GC and adjuvant CapeOX chemotherapy. Patients who received any chemoradiotherapy or immunotherapy before surgery were excluded. The patients included 45 males and 29 females with a median age of 63 years. All fresh tissues were collected between December 2017 and May 2019. The tissue samples were immediately frozen and stored at −80°C for gene expression analysis. All patients were diagnosed consistently by clinicians and pathologists and followed up from the time of surgery to death or last contact. The follow-up was mainly conducted over the phone. The protocols of the study were approved by The Ethics Committee of the Affiliated Suzhou Hospital of Nanjing Medical University (Suzhou, China). Written informed consent was obtained from each patient.
Reverse transcription-quantitative (RT-q) PCR
Frozen tissues were homogenized and TRIzol reagent (Takara Bio, Inc.) was used to exact total tissue RNA following the manufacturer's instruction at 4°C and the total extraction time was ~1 h. Individual RNA was subjected to RT-qPCR by using the titanium one-step RT-PCR detection kit (Takara Bio, Inc.) and the HT7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. Tissue (~10 mg) was used for RNA extraction in each reaction. The reaction volume was 20 µg. The PCR thermocycling conditions were as follows: 95°C pre-denaturation for 3 min; then 35 cycles of denaturation at 95°C for 30 sec followed by annealing and extension at 55°C for 45 sec. Each experiment was repeated 3 times. The 2−ΔΔCq method was used for gene quantification (9). The sequences of the primers were: Uroplakin-1b (UPK1B), forward: 5′-GTGGGCCTCGATTCTACAGG-3′ and reverse: 5′-GATGTCACGTATCCAGCAAATCT-3′; fatty acid-binding protein, heart (FABP3), forward: 5′-GGCACCTGGAAGCTAGTGG-3′ and reverse: 5′-CTGCCTGGTAGCAAAACCC-3′; cystatin-M (CST6), forward: 5′-TACTTCCTGACGATGGAGATGG-3′ and reverse: 5′-GAGTTCTGCCAGGGAACCAC-3′; caspase-5 (CASP5), forward: 5′-TCACCTGCCTGCAAGGAATG-3′ and reverse: 5′-TCTTTTCGTCAACCACAGTGTAG-3′; corticosteroid 11-β-dehydrogenase isozyme 2 (HSD11B2), forward: 5′-ATTAGCCGCGTGCTAGAGTTC-3′ and reverse: 5′-CCGCATCAGCAACTACTTCAT-3′; cytochrome P450 4X1 (CYP4X1), forward: 5′-TGAGCAGAACAGATCCCAAGT-3′ and reverse: 5′-CAGAATGAGCCATCACCTCAAT-3′; epidermal growth factor receptor kinase substrate 8-like protein 3 (EPS8L3), forward: 5′-AGCCATTTACTTGCACCGGAA-3′, and reverse: 5′-CTCCCCTGCTTGCATGTCAT-3′; GADPH, forward: 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse: 5′-GAAGATGGTGATGGGATTTC-3′. GADPH served as an endogenous control to normalize gene expression. The comparative cycle threshold method was used to calculate the relative abundance of RNA compared with the expression of GADPH, hence the fold difference relative to GADPH was determined.
DEG validation and predictive model construction
All patients were divided into the recurrence and the non--recurrence groups through follow-up. A t-test was applied to validate the DEGs between the recurrence and the nonrecurrence groups. The genes that met the cut-off criteria (fold-change >2 and adjusted P<0.05) were validated DEGs. A predictive regression model was constructed using the four validated DEGs (UPK1B, FABP3, CASP5 and CYP4X1). Subsequently, a nomogram was constructed to visualize the results. For each patient, the nomogram helped calculate the total score and then predict the probability of the result.
Statistical analysis
For TCGA database analysis, the Deseq2 method was applied to standardize the data and eliminate the influence of the platform effect using R language version 3.6.3 (R Core Team, Vienna, Austria) (10). A unpaired t-test was applied to identify the DEGs between complete response (CR) and non-CR GC tissues. For GO enrichment analysis, P<0.1 was set as the threshold. For identification of survival-related DEGs, univariate Cox regression was performed. Statistical significance was assessed using the Tarone-Ware test and P<0.05 was set as the threshold. For the survival analysis of a single gene, Kaplan-Meier survival analysis was performed for patients with high/low differential gene expression. A Multivariate Cox regression model was used to evaluate the weight of the effect of genes on survival.
Receiver operating characteristic (ROC) curve analysis was performed to evaluate the sensitivity and the specificity of the DEGs and the model for GC CapeOX treatment response prediction was evaluated using the SPSS software version 20.0 (IBM Corp.). An area under the curve (AUC) value was calculated and used to designate the ROC effect. The nomogram was built with R language version 3.6.3 (10). In the nomogram, the regression coefficients of all independent variables were used to determine the proportion of scores. P<0.05 was considered to indicate a statistically significant difference.
Results
DEG identification and data integration
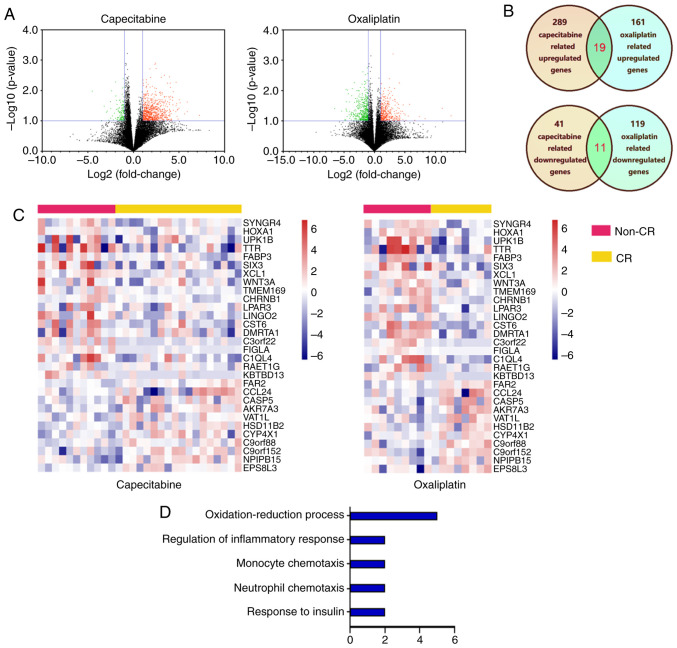
The datasets of GC were downloaded from TCGA database. Among the cases, 17 oxaliplatin (including eight CR and nine non-CR) and 29 capecitabine (including 18 CR and 11 non-CR) treatment cases were obtained. The gene expression profiles from the capecitabine treatment cases identified 330 DEGs (289 upregulated genes and 41 downregulated genes) between non-CR and CR cases (Fig. 1A). The gene expression profiles from the oxaliplatin treatment cases identified 280 DEGs (161 upregulated genes and 119 downregulated genes) between non-CR and CR cases. Through cross analysis, 30 consistent DEGs were identified from the aforementioned profiles (Fig. 1B) and the hierarchical cluster heatmap of the integrated data demonstrated that these genes can distinguish between patients with CR and non-CR significantly (P<0.05) (Fig. 1C). GO functional analysis was performed to characterize the function of the DEGs and identify the key candidate pathways. The GO analysis demonstrated that the DEGs were enriched in processes such as ‘oxidation-reduction process’, ‘regulation of inflammatory response’, ‘monocyte chemotaxis’, ‘neutrophil chemotaxis’ and ‘response to insulin’ (Fig. 1D). The results of GO enrichment indicated that the DEGs in the network serve an important role in GC treatment with capecitabine and oxaliplatin.
Figure 1.
DEGs identification and data integration. (A) Volcano plot of genome-wide gene expression profiles in non-CR cancer and CR cases tissues from capecitabine treatment cases and oxaliplatin treatment cases from The Cancer Genome Atlas database. Red plots represent upregulated DEGs in non-CR cases with P<0.1 and log2 (fold-change) >1. Black plots represent normally expressed mRNAs. Green plots represent downregulated expressed mRNAs in non-CR cases with P<0.1 and log2 (fold-change) >1. (B) Consistently expressed DEGs identified from the above profiles though cross analysis, dark yellow represented capecitabine related genes, light green represented oxaliplatin related genes and green represented consistently expressed DEGs. (C) Heatmap analysis of differential expression profiles between non-CR cases and CR cases. (D) DEG functional and signaling pathway enrichment were conducted using online websites for Gene Oncology, P<0.05 was set as the threshold. DEGs, differentially expressed genes; CR, complete response.
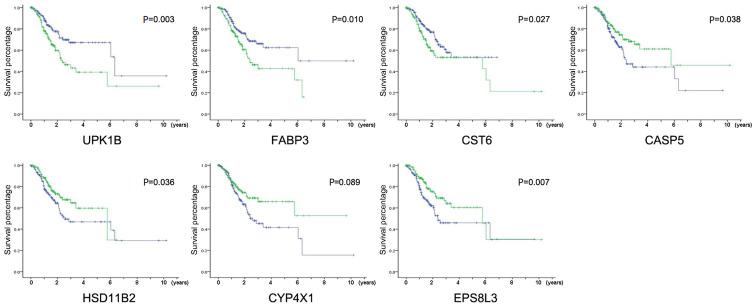
Identification of key DEGs associated with GC survival
The TCGA-STAD dataset contained the transcriptome sequencing data and the latest follow-up clinical data of 279 cases. The Cox univariate regression analysis of the 30 DEGs was performed using these data. As shown in Table I, seven genes were independent risk factors for patient prognosis. The high expression of UPK1B (P=0.002), FABP3 (P=0.006) and CST6 (P=0.041) and the low expression of CASP5 (P=0.022), HSD11B2 (P=0.045), CYP4X1 (P=0.026) and EPS8L3 (P=0.013) were significantly associated with poor prognosis. The survival curves of all the above genes also showed significant differences between the high expression and low expression groups (P<0.05; Fig. 2). These results suggested that the expression of these seven genes can reflect the short-term response of patients to chemotherapeutic drugs and may be associated with the long-term survival of patients.
Table I.
Seven genes are significant independent risk factors for the prognosis of patients with CR and non-CR gastric cancer.
| Gene | Regulation | Hazard ratio | P-value |
|---|---|---|---|
| SYNGR4 | Up | −0.103 | 0.633 |
| HOXA1 | Up | −0.277 | 0.208 |
| UPK1B | Up | 0.681 | 0.002b |
| TTR | Up | 0.128 | 0.552 |
| FABP3 | Up | 0.609 | 0.006b |
| SIX3 | Up | 0.116 | 0.593 |
| XCL1 | Up | −0.010 | 0.962 |
| WNT3A | Up | −0.144 | 0.511 |
| TMEM169 | Up | 0.332 | 0.126 |
| CHRNB1 | Up | 0.037 | 0.866 |
| LPAR3 | Up | 0.241 | 0.266 |
| LINGO2 | Up | 0.125 | 0.563 |
| CST6 | Up | 0.446 | 0.041a |
| DMRTA1 | Up | 0.072 | 0.741 |
| C3orf22 | Up | −0.093 | 0.668 |
| FIGLA | Up | 0.276 | 0.204 |
| C1QL4 | Up | 0.144 | 0.506 |
| RAET1G | Up | 0.248 | 0.252 |
| KBTBD13 | Up | 0.136 | 0.555 |
| FAR2 | Down | −0.254 | 0.248 |
| CCL24 | Down | −0.113 | 0.601 |
| CASP5 | Down | −0.504 | 0.022a |
| AKR7A3 | Down | −0.278 | 0.207 |
| VAT1L | Down | 0.036 | 0.868 |
| HSD11B2 | Down | −0.442 | 0.045a |
| CYP4X1 | Down | −0.490 | 0.026a |
| C2orf88 | Down | −0.044 | 0.838 |
| C9orf152 | Down | −0.116 | 0.594 |
| NPIPB15 | Down | −0.304 | 0.164 |
| EPS8L3 | Down | −0.545 | 0.013a |
P<0.05
P<0.001. UPK1B, uroplakin-1b; FABP3, fatty acid-binding protein, heart; CST6, cystatin-M; CASP5, caspase-5; HSD11B2, corticosteroid 11-β-dehydrogenase isozyme 2; CYP4X1, cytochrome P450 4X1; EPS8L3, epidermal growth factor receptor kinase substrate 8-like protein 3; CR, complete response.
Figure 2.
Identification of key DEGs associated with GC survival. Kaplan-Meier survival curves for patients of GC with high and low indicated gene expression in TCGA-STAD dataset, P-values were calculated using the Tarone-Ware test, green line represented high expression group and blue line represented low expression group. DEGs, differentially expressed genes; GC, gastric cancer; TCGA-STAD, The Cancer Genome Atlas Stomach Adenocarcinoma data; UPK1B, uroplakin-1b; FABP3, fatty acid-binding protein, heart; CST6, cystatin-M; CASP5, caspase-5; HSD11B2, corticosteroid 11-β-dehydrogenase isozyme 2; CYP4X1, cytochrome P450 4X1; EPS8L3, epidermal growth factor receptor kinase substrate 8-like protein 3.
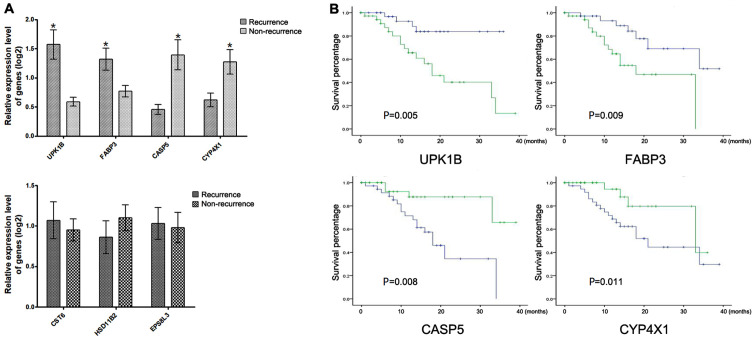
Validation of the DEGs in clinical tissue samples
The mRNA expression of the seven genes in 74 clinical GC tissues with adjuvant CapeOX treatment was detected using RT-qPCR to confirm the reliability of the identified DEGs. Through follow-up, 31 patients with recurrence and 43 patients without recurrence were found. Results demonstrated a significant upregulation of UPK1B and FABP3 and a significant downregulation of CASP5 and CYP4X1 in the recurrence group compared with those in the non-recurrence group (Fig. 3A). The Kaplan-Meier survival analysis demonstrated that patients with high or low expression of UPK1B, FABP3, CASP5 and CYP4X1 had significantly different overall survival times (Fig. 3B). Each gene expression was divided into high or low expression by using the median as the cut-off.
Figure 3.
Validation of the DEGs in clinical tissue samples. (A) The DEGs with and without significantly differential expression in patients with recurrence compared with non-recurrence patients, P-values were calculated using the t-test. (B) Kaplan-Meier survival curves for patients of GC with high/low indicated gene expression in clinical GC tissues, P-values were calculated using the Tarone-Ware test. *P<0.05, green line represented high expression group and blue line represented low expression group. DEGs, differentially expressed genes; GC, gastric cancer; UPK1B, uroplakin-1b; FABP3, fatty acid-binding protein, heart; CASP5, caspase-5; CYP4X1, cytochrome P450 4X1.
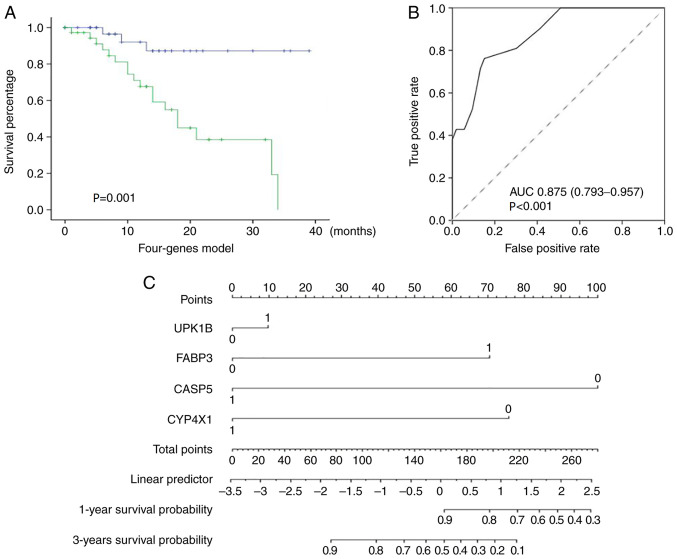
Role of UPK1B, FABP3, CASP5 and CYP4X1 in the prediction of the GC CapeOX treatment outcomes
The combination of different markers as prognostic indices appeared encouraging. Therefore, the prediction value of the combination of the validated genes was further evaluated. A multivariate Cox regression model was implemented using the four genes UPK1B, FABP3, CASP5 and CYP4X1. The patients were classified into high- or low-risk group by using the regression model. The estimated risk was calculated using the following formula: Risk = (1.209 × the status of UPK1B) + (1.601 × the status of FABP3) - (1.517 × the status of CASP5) - (0.786 × the status of CYP4X1). The gene status was marked 1 if the expression was high. Otherwise, the gene status was marked 0. The Kaplan-Meier survival analysis demonstrated that patients in the high-risk group exhibited significantly shorter survival rates compared with those in the low-risk group (P<0.001; Fig. 4A). ROC curve analysis was also performed. The AUC of ROC reached 0.875 (95% CI, 0.793–0.957; P<0.001), indicating that the model had good sensitivity and specificity (Fig. 4B). A new nomogram was constructed to facilitate clinical use in predicting 1-year and 3-year survival rates (Fig. 4C). The comprehensive analysis indicated that the combination of the four genes could predict the outcome of GC CapeOX treatment.
Figure 4.
Prediction value of the model for GC adjuvant CapeOX chemotherapy outcomes. (A) Kaplan-Meier survival curves for patients in the high-risk and low-risk groups using the Cox regression model, P-values were calculated using the Tarone-Ware test, green line represented high-risk group and blue line represented low-risk group. (B) Receiver operating characteristic analysis of the model. (C) A nomogram was established to predict one-year and three-year survival rates following adjuvant CapeOX chemotherapy. GC, gastric cancer; CapeOX, capecitabine and oxaliplatin; AUC, area under the curve.
Discussion
Chemotherapy resistance, whether primary or acquired, is common (6). The differences in molecular expression between tumors have resulted in different responses of patients with cancer to chemotherapeutic drugs (4–5). The accurate selection of chemotherapeutic drugs, which maximize the effectiveness of chemotherapy and reduce the toxic and side effects, is one of the most critical issues in the clinical application of chemotherapy (4–5).
In GC, platinum- and 5-Fu-based CapeOX therapy is one of the common postoperative adjuvant chemotherapy regimens for GC (3). Literature has reported that various genes serve an important role in the occurrence of resistance to capecitabine and oxaliplatin (8,11–14). Low levels of tubulin β-3 chain and AMBP in serum can predict an improved response and survival rates for patients with advanced GC receiving capecitabine (11,12). When the proline rich 13 (PRR13) gene is silenced, tumor suppressor region 1 (TSP1) expression is increased and oxaliplatin resistance is significantly decreased in GC cells (13). Consistently, the therapeutic resistance to oxaliplatin of GC is significantly weakened when gap junction α-1 protein is transfected (14). In addition, a variety of noncoding RNAs are associated with capecitabine and oxaliplatin resistance in GC cells (8,15).
To predict the sensitivity of chemotherapy, a multiple genes panel can obtain higher sensitivity and specificity compared with a single gene. However, in order to facilitate a practical application, the number of genes in the majority of panels is >100. For example, a tumor microenvironment risk score prediction panel could predict whether patients with colon cancer benefit from adjuvant chemotherapy (16). Thus far, no study has reported a model of multiple gene combinations to predict the response of GC to the CapeOX chemotherapy regimens, to the best of our knowledge. Therefore, the introduction of new biomarkers could expand the clinical options currently available to physicians.
In the present study, four genes associated with CapeOX chemotherapy outcomes were finally identified through multilevel screening and a prediction model was established. The high expression of UPK1B and FABP3 and the low expression of CASP5 and CYP4X1 in adjuvant CapeOX-treated patients with GC were associated with poor outcomes.
Studies have shown that UPK1B is closely associated with the occurrence and development of malignant tumors (17–20). The differential expression of UPK1B in multiple tumors, including GC, has been demonstrated and can predict the OS rates of patients (17,18). However, the molecular mechanism underlying UPK1B function in tumors has yet to be fully elucidated. UPK1B-knockdown significantly reduces the expression of key genes in the Wnt/β-catenin signaling pathway in cancer cells (19). The Wnt pathway is the key pathway for the development of oxaliplatin resistance in tumors (20). Thus, UPK1B may regulate the drug sensitivity of oxaliplatin through the Wnt signaling pathway.
FABP3 belongs to the FABP family of proteins, which control the metabolism and transport of long-chain fatty acids (21–26). Studies have shown that fatty acid metabolism is involved in tumorigenesis and tumor progression in numerous types of cancer by affecting tumor proliferation and tumor migration (21,22). Recent studies have demonstrated FABP3 abnormal expression in various types of cancer, including breast cancer and brain tumors (23,24). Elevated FABP3 expression is associated with tumor progression, invasiveness and poor prognosis in GC and non-small cell lung cancer (25,26). The findings of the present study suggested that fatty acid metabolism may also serve an important role in chemotherapy drugs.
CASP5 is an inflammatory caspase that serves a role in the immune system (27–29). Inflammation and cancer development are closely associated and a distinct inflammatory environment often exists within tumors (27). Multiple CASPs are abnormally expressed in GC and associated with the survival rates of GC (28). CASP5 gene overexpression can significantly promote the angiogenesis ability of vascular endothelial cells by promoting the VEGF signaling pathway (29). The present study indicated that CASP5 could improve the OS rates of patients with GC following CapeOX chemotherapy, suggesting that the inflammation level may also serve an important role in the response of tumors to chemotherapy drugs.
The CYP4X1 gene is located in the cytochrome P450 ABXZ gene cluster and its function is largely unknown (30). Cytochrome P450 is a superfamily of heme-thiolate proteins involved in the oxidative metabolism of endogenous compounds, including fatty acids and arachidonic acid derivatives (31). Arachidonic acid and its derivatives can be found widely in human tissues and serve an important role as signaling mediators in the nerve, immune and cardiovascular systems (31). CYP4X1 has also been reported to be abnormally expressed in oral squamous cell cancer or breast cancer (32,33). A previous study reported that CYP4X1 is involved in important physiological processes in the metabolism of arachidonic acid derivatives (34). As such, CYP4X1 has been suggested as a potential drug target for cancer therapy (34). However, the mechanism of interaction between CYP4X1 and chemotherapy drugs remains to be elucidated.
The present study suggested that a variety of different action pathways and different types of genes may serve a role in the chemosensitivity of capecitabine and oxaliplatin. The present study had limitations and further verification of the results in a prospective and multicenter study is needed. Evidently, validating the roles of UPK1B, FABP3, CASP5 and CYP4X1 in GC and the CapeOX chemotherapy process through further in vitro and in vivo experiments is necessary. The current established model requires more samples for the validation of its reliability.
Acknowledgements
Not applicable.
Funding
This study was funded by The Natural Science Foundation of Jiangsu (grant no. BK20180215).
Availability of data and materials
All data generated or analyzed during this study are included in this published article. The datasets generated and/or analyzed during the current study are available in The Cancer Genome Atlas repository (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and the Gene Ontology repository (http://www.geneontology.org/).
Authors' contributions
XG and YZ designed the study. ZY, RS, YJ and MG performed the experiments. YJ, WX and MG analyzed the data. YZ wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Specimen collection was approved by The Ethics Committee of the Affiliated Suzhou Hospital of Nanjing Medical University (Suzhou, China) (approval no. 20180215). Written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Cainap C, Vlad C, Seicean A, Balacescu O, Seicean R, Constantin AM, Balacescu L, Crisan O, Marta MM, Cainap S. Gastric cancer: Adjuvant chemotherapy versus chemoradiation. A clinical point of view. J BUON. 2019;24:2209–2219. [PubMed] [Google Scholar]
- 3.Xue K, Ying X, Bu Z, Wu A, Li Z, Tang L, Zhang L, Zhang Y, Li Z, Ji J. Oxaliplatin plus S-1 or capecitabine as neoadjuvant or adjuvant chemotherapy for locally advanced gastric cancer with D2 lymphadenectomy: 5-year follow-up results of a phase II–III randomized trial. Chin J Cancer Res. 2018;30:516–525. doi: 10.21147/j.issn.1000-9604.2018.05.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu Z, Zhang X, Liu W, Liu T, Hu B, Li W, Fan Q, Xu J, Xu N, Bai Y, et al. A multicenter, randomized trial comparing efficacy and safety of paclitaxel/capecitabine and cisplatin/capecitabine in advanced gastric cancer. Gastric Cancer. 2018;21:782–791. doi: 10.1007/s10120-018-0809-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmad S. Platinum-DNA interactions and subsequent cellular processes controlling sensitivity to anticancer platinum complexes. Chem Biodivers. 2010;7:543–566. doi: 10.1002/cbdv.200800340. [DOI] [PubMed] [Google Scholar]
- 6.Peng J, Tan C, Zeng X, Liu S. Cost-effectiveness analysis of capecitabine monotherapy versus capecitabine plus oxaliplatin in elderly patients with advanced gastric cancer. PLoS One. 2018;13:e0199553. doi: 10.1371/journal.pone.0199553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim ST, Ahn S, Lee J, Lee SJ, Park SH, Park YS, Lim HY, Kang WK, Kim KM, Park JO. Value of FGFR2 expression for advanced gastric cancer patients receiving pazopanib plus CapeOX (capecitabine and oxaliplatin) J Cancer Res Clin Oncol. 2016;142:1231–1237. doi: 10.1007/s00432-016-2143-2. [DOI] [PubMed] [Google Scholar]
- 8.Fan B, Shen C, Wu M, Zhao J, Guo Q, Luo Y. MiR-17-92 cluster is connected with disease progression and oxaliplatin/capecitabine chemotherapy efficacy in advanced gastric cancer patients: A preliminary study. Medicine (Baltimore) 2018;97:e12007. doi: 10.1097/MD.0000000000012007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 10.Integrated Development for R. RStudio, Inc.; Boston, MA: RStudio Team (2015). RStudio. [Google Scholar]
- 11.Yu J, Gao J, Lu Z, Li Y, Shen L. Serum levels of TUBB3 correlate with clinical outcome in Chinese patients with advanced gastric cancer receiving first-line paclitaxel plus capecitabine. Med Oncol. 2012;29:3029–3034. doi: 10.1007/s12032-012-0292-y. [DOI] [PubMed] [Google Scholar]
- 12.Huang H, Han Y, Gao J, Feng J, Zhu L, Qu L, Shen L, Shou C. High level of serum AMBP is associated with poor response to paclitaxel-capecitabine chemotherapy in advanced gastric cancer patients. Med Oncol. 2013;30:748. doi: 10.1007/s12032-013-0748-8. [DOI] [PubMed] [Google Scholar]
- 13.Bi J, Bai Z, Ma X, Song J, Guo Y, Zhao J, Yi X, Han S, Zhang Z. Txr1: An important factor in oxaliplatin resistance in gastric cancer. Med Oncol. 2014;31:807. doi: 10.1007/s12032-013-0807-1. [DOI] [PubMed] [Google Scholar]
- 14.Liu D, Zhou H, Wu J, Liu W, Li Y, Shi G, Yue X, Sun X, Zhao Y, Hu X, et al. Infection by Cx43 adenovirus increased chemotherapy sensitivity in human gastric cancer BGC-823 cells: Not involving in induction of cell apoptosis. Gene. 2015;574:217–224. doi: 10.1016/j.gene.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Danza K, Silvestris N, Simone G, Signorile M, Saragoni L, Brunetti O, Monti M, Mazzotta A, De Summa S, Mangia A, Tommasi S. Role of miR-27a, miR-181a and miR-20b in gastric cancer hypoxia-induced chemoresistance. Cancer Biol Ther. 2016;17:400–406. doi: 10.1080/15384047.2016.1139244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou R, Zeng D, Zhang J, Sun H, Wu J, Li N, Liang L, Shi M, Bin J, Liao Y, et al. A robust panel based on tumour microenvironment genes for prognostic prediction and tailoring therapies in stage I–III colon cancer. EBioMedicine. 2019;42:420–430. doi: 10.1016/j.ebiom.2019.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pichler R, Fritz J, Tulchiner G, Klinglmair G, Soleiman A, Horninger W, Klocker H, Heidegger I. Increased accuracy of a novel mRNA-based urine test for bladder cancer surveillance. BJU Int. 2018;121:29–37. doi: 10.1111/bju.14019. [DOI] [PubMed] [Google Scholar]
- 18.Dai J, Li ZX, Zhang Y, Ma JL, Zhou T, You WC, Li WQ, Pan KF. Whole genome messenger RNA profiling identifies a novel signature to predict gastric cancer survival. Clin Transl Gastroenterol. 2019;10:e00004. doi: 10.14309/ctg.0000000000000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang FH, Ma XJ, Xu D, Luo J. UPK1B promotes the invasion and metastasis of bladder cancer via regulating the Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci. 2018;22:5471–5480. doi: 10.26355/eurrev_201809_15807. [DOI] [PubMed] [Google Scholar]
- 20.Kukcinaviciute E, Jonusiene V, Sasnauskiene A, Dabkeviciene D, Eidenaite E, Laurinavicius A. Significance of Notch and Wnt signaling for chemoresistance of colorectal cancer cells HCT116. J Cell Biochem. 2018;119:5913–5920. doi: 10.1002/jcb.26783. [DOI] [PubMed] [Google Scholar]
- 21.Levi L, Wang Z, Doud MK, Hazen SL, Noy N. Saturated fatty acids regulate retinoic acid signalling and suppress tumorigenesis by targeting fatty acid-binding protein 5. Nat Commun. 2015;6:8794. doi: 10.1038/ncomms9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yarla NS, Bishayee A, Sethi G, Reddanna P, Kalle AM, Dhananjaya BL, Dowluru KS, Chintala R, Duddukuri GR. Targeting arachidonic acid pathway by natural products for cancer prevention and therapy. Semin Cancer Biol. 2016;40-41:48–81. doi: 10.1016/j.semcancer.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Hyvonen M, Enback J, Huhtala T, Lammi J, Sihto H, Weisell J, Joensuu H, Rosenthal-Aizman K, El-Andaloussi S, Langel U, et al. Novel target for peptide-based imaging and treatment of brain tumors. Mol Cancer Ther. 2014;13:996–1007. doi: 10.1158/1535-7163.MCT-13-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopes-Coelho F, Andre S, Felix A, Serpa J. Breast cancer metabolic cross-talk: Fibroblasts are hubs and breast cancer cells are gatherers of lipids. Mol Cell Endocrinol. 2018;462:93–106. doi: 10.1016/j.mce.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 25.Hashimoto T, Kusakabe T, Sugino T, Fukuda T, Watanabe K, Sato Y, Nashimoto A, Honma K, Kimura H, Fujii H, Suzuki T. Expression of heart-type fatty acid-binding protein in human gastric carcinoma and its association with tumor aggressiveness, metastasis and poor prognosis. Pathobiology. 2004;71:267–273. doi: 10.1159/000080061. [DOI] [PubMed] [Google Scholar]
- 26.Tang Z, Shen Q, Xie H, Zhou X, Li J, Feng J, Liu H, Wang W, Zhang S, Ni S. Elevated expression of FABP3 and FABP4 cooperatively correlates with poor prognosis in non-small cell lung cancer (NSCLC) Oncotarget. 2016;7:46253–46262. doi: 10.18632/oncotarget.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deshmukh SK, Srivastava SK, Poosarla T, Dyess DL, Holliday NP, Singh AP, Singh S. Inflammation, immunosuppressive microenvironment and breast cancer: Opportunities for cancer prevention and therapy. Ann Transl Med. 2019;7:593. doi: 10.21037/atm.2019.09.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Ni F, Yu F, Cui Z, Zhu X, Chen J. Prognostic significance of mRNA expression of CASPs in gastric cancer. Oncol Lett. 2019;18:4535–4554. doi: 10.3892/ol.2019.10816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Li Y, Cai L, Bai B, Wang Y. Effects of CASP5 gene overexpression on angiogenesis of HMEC-1 cells. Int J Clin Exp Pathol. 2015;8:15794–15800. [PMC free article] [PubMed] [Google Scholar]
- 30.Carver KA, Lourim D, Tryba AK, Harder DR. Rhythmic expression of cytochrome P450 epoxygenases CYP4×1 and CYP2c11 in the rat brain and vasculature. Am J Physiol Cell Physiol. 2014;307:C989–C998. doi: 10.1152/ajpcell.00401.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manikandan P, Nagini S. Cytochrome P450 structure, function and clinical significance: A review. Curr Drug Targets. 2018;19:38–54. doi: 10.2174/1389450118666170125144557. [DOI] [PubMed] [Google Scholar]
- 32.Russo A, Biselli-Chicote PM, Kawasaki-Oyama RS, Castanhole-Nunes MMU, Maniglia JV, de Santi Neto D, Pavarino ÉC, Goloni-Bertollo EM. Differential expression of prostaglandin i2 synthase associated with arachidonic acid pathway in the oral squamous cell carcinoma. J Oncol. 2018;2018:6301980. doi: 10.1155/2018/6301980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray GI, Patimalla S, Stewart KN, Miller ID, Heys SD. Profiling the expression of cytochrome P450 in breast cancer. Histopathology. 2010;57:202–211. doi: 10.1111/j.1365-2559.2010.03606.x. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S. Computational identification and binding analysis of orphan human cytochrome P450 4X1 enzyme with substrates. BMC Res Notes. 2015;8:9. doi: 10.1186/s13104-015-0976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. The datasets generated and/or analyzed during the current study are available in The Cancer Genome Atlas repository (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and the Gene Ontology repository (http://www.geneontology.org/).