Abstract
Many reagents have emerged to study the function of specific enzymes in vitro. On the other hand, target specific reagents are scarce or need improvement, allowing investigations of the function of individual enzymes in their native cellular context. Here we report the development of a target-selective fluorescent small-molecule activity-based DUB probe that is active in live cells and an in vivo animal model. The probe labels active ubiquitin carboxy-terminal hydrolase L1 (UCHL1), also known as neuron-specific protein PGP9.5 (PGP9.5) and Parkinson disease 5 (PARK5), a DUB active in neurons that constitutes 1 to 2% of the total brain protein. UCHL1 variants have been linked with neurodegenerative disorders Parkinson’s and Alzheimer’s diseases. In addition, high levels of UCHL1 also correlate often with cancer and especially metastasis. The function of UCHL1 activity or its role in cancer and neurodegenerative disease is poorly understood and few UCHL1-specific activity tools exist. We show that the reagents reported here are specific to UCHL1 over all other DUBs detectable by competitive activity-based protein profiling and by mass spectrometry. Our cell-penetrable probe, which contains a cyanimide reactive moiety, binds to the active-site cysteine residue of UCHL1 in an activity-dependent manner. Its use is demonstrated by the fluorescent labeling of active UCHL1 both in vitro and in live cells. We furthermore show that this probe can selectively and spatiotemporally report UCHL1 activity during the development of zebrafish embryos. Our results indicate that our probe has potential applications as a diagnostic tool for diseases with perturbed UCHL1 activity.
Introduction
The ubiquitin system relies to a great extent on cysteine catalysis. Ubiquitin is a small protein that consists of 76 amino acids that can modify target proteins through lysine residues, although it is also occasionally found to modify N-termini as well as cysteine and threonine residues.1−3 The addition of ubiquitin is catalyzed by E1 (2), E2 (∼40), and E3 (>600) enzymes in an ATP-dependent conjugation reaction by specific combinations of E1, E2, and E3 enzymes, and it is reversed by any of ∼100 deubiquitylating enzymes (DUBs) in humans.4,5 The ubiquitin carboxy-terminal hydrolase L1 (UCHL1) enzyme, also known as neuron-specific protein PGP9.5 (PGP9.5) and Parkinson’s disease 5 (PARK5), is a small protease that is thought to remove ubiquitin from small substrates, and it belongs to the small family of ubiquitin C-terminal hydrolases (UCHs).6
It is clear that UCHL1 can cleave ubiquitin, and that the mutation and reduced activity of this enzyme have been associated with neurodegenerative diseases, including Parkinson’s and Alzheimer’s diseases.7−12 High UCHL1 levels correlate with malignancy and metastasis in many cancers13,14 and have also been attributed to cellular stress, although the molecular mechanism of all of these processes is not very clear.
We earlier observed extreme levels of UCHL1 activity in lysates from prostate and lung cancer cells using a ubiquitin-derived activity-based probe that targets all cysteine DUBs.15 We reasoned that a good cell-permeable activity-based probe that targets UCHL1 specifically among other cysteine DUBs would be a highly valuable tool for understanding its normal function during embryogenesis and in adult tissues and how its dysfunction contributes to the malignant transformation and development of neurodegenerative diseases.
UCHL1, like many DUBs, is a cysteine protease, a class of enzymes considered extremely difficult to inhibit with small molecules as this class of enzymes is associated with nonspecific reactions with cysteine alkylating agents and with redox-cycling artifacts in assays.16 In addition, DUBs intrinsically bind ubiquitin through a protein–protein interaction, which is by definition difficult to interfere with using small molecules. Many DUBs, including UCHL1, are inactive without a substrate, and substrate binding aligns the catalytic triad for cleavage.17 Nevertheless, recently significant successes have been booked in the development of reversible and irreversible selective small-molecule inhibitors of the DUB USP7.18−23 We have recently reported the development of a selective covalent small-molecule inhibitor of the DUB ovarian tumor (OTU) protease OTUB2 using a covalent fragment approach and parallel X-ray crystallography.24 We reasoned that such covalent molecules are a good inroad for the further elaboration of specific activity-based probes (ABPs) also inspired by earlier work from the Tate laboratory that reported a small-molecule broadly acting DUB probe.25 We were pleased to find a good starting point in the patent literature26 that we used in our studies for the design of fluorescent ABPs. We report here the development of a fluorescent small-molecule ABP that can report UCHL1 activity in human cells and in zebrafish embryos.
Results and Discussion
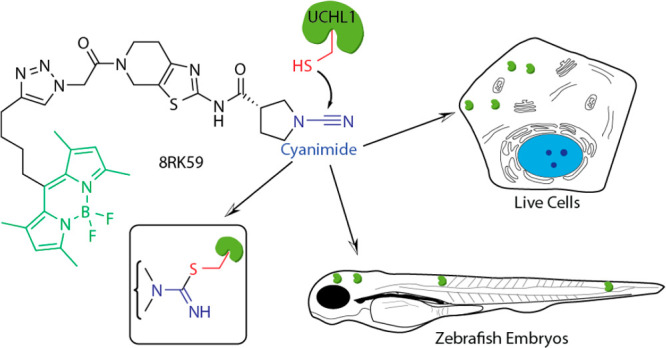
The development of a small-molecule-based DUB ABP starts with the identification of an appropriate DUB-selective small-molecule covalent binder. We reasoned that an ideal compound needed to meet two criteria: (1) it binds covalently to the active-site cysteine residue of a DUB and (2) it can easily be modified by chemical synthesis. Our attention was drawn to a collection of (S)-1-cyanopyrrolidine-3-carboxamide-based compounds reported to inhibit UCHL1 activity with submicromolar affinity.26 These compounds are equipped with a cyanimide moiety that is known to react with thiols to form an isothiourea covalent adduct (Figure 1A) and is thought to react reversibly.27 Despite the expected reversible nature, we decided to investigate this compound as a potential probe starting point.
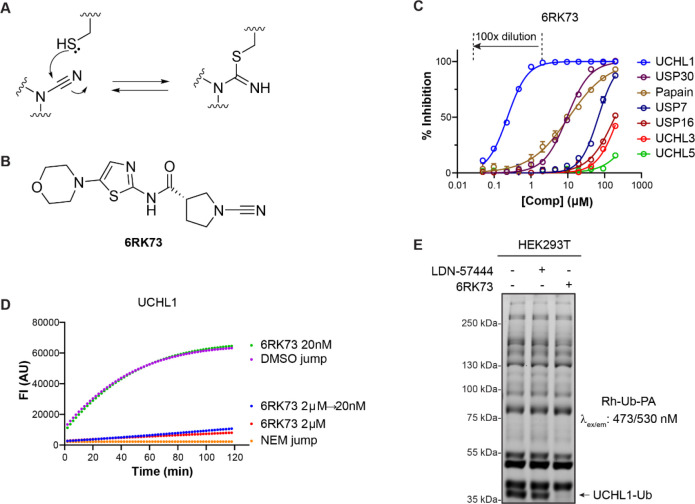
Figure 1.
Biochemical characterization of UCHL1 inhibitor 6RK73. (A) Reacting a thiol with a cyanimide results in the formation of an isothiourea adduct. (B) Structure of UCHL1 inhibitor 6RK73. (C) IC50 determination of 6RK73 for the indicated DUBs and papain. (D) Progress curves for UCHL1 proteolytic activity after jump dilution. (See also panel C.) DMSO and N-ethylmaleimide (NEM) are used as controls. (E) Fluorescence labeling of the remaining DUB activity in HEK293T cells upon treatment with UCHL1 inhibitors LDN-57444 and 6RK73.
Characterizing UCHL1 Cyanimide Inhibitors
In order to gain insight into the mode of action and DUB selectivity of these inhibitors, we synthesized and characterized one compound (compound 6RK73, Figure 1B) that in our hands inhibits UCHL1 with an IC50 of 0.23 μM after 30 min of incubation in a biochemical activity assay using a fluorogenic Ub-Rho-morpholine28 substrate (for preparation, see the Supporting Information) in the presence of 2 mM cysteine. Beneficially, 6RK73 proved to be almost unreactive toward the closest DUB family members, UCHL3 and UCHL5 (Figure 1C). The selectivity for UCHL1 was further confirmed by IC50 determination against a panel of other cysteine DUBs (including USP7, USP30, and USP16) and the non-DUB cysteine protease papain, showing a >50-fold difference in the IC50 value (Figure 1C and Supporting Information Table S1). We next performed a jump dilution experiment29 in which a 100× final assay concentration of UCHL1 was treated with 2 μM 6RK73 followed by 100× dilution into a substrate-containing buffer and direct fluorescence read-out (Figure 1C,D). Only after 50 min could a negligible increase in the fluorescence signal compared to the 2 μM inhibitor control be detected, which indicates that the inhibitor acts practically irreversibly. The formation of a covalent complex between UCHL1 and a single 6RK73 molecule was confirmed in an experiment where UCHL1 was incubated with 6RK73, and the reaction was followed by LC-MS analysis (Supporting Information Figure S1A). Next, we investigated whether the compound would inhibit UCHL1 in live cells. HEK293T cells were treated with 5 μM 6RK73 or commercially available active-site-directed reversible UCHL1 inhibitor LDN-5744430 for 24 h, followed by cell lysis and treatment with fluorescent broad-spectrum DUB probe rhodamine-ubiquitin-propargylamide (Rh-Ub-PA) to label all residual cysteine–DUB activity.31,32 The samples were denatured, resolved by SDS-PAGE, and scanned for rhodamine fluorescence (Figure 1E). Each band represents an active DUB that reacted with the probe, and the ability of a compound to inhibit a DUB is reflected by the disappearance of its corresponding band. Indeed, the band belonging to UCHL133 disappears upon treatment with 6RK73, whereas all other bands remain unchanged, indicating that 6RK73 selectively inhibits UCHL1 in the presence of other DUBs in cells. In comparison, UCHL1 is hardly inhibited by LDN-57444 in this experiment, despite their comparable IC50 values (0.88 μM for LDN-57444), which might be attributed to the fast reversible nature of this inhibitor.30
From Inhibitor to Probe
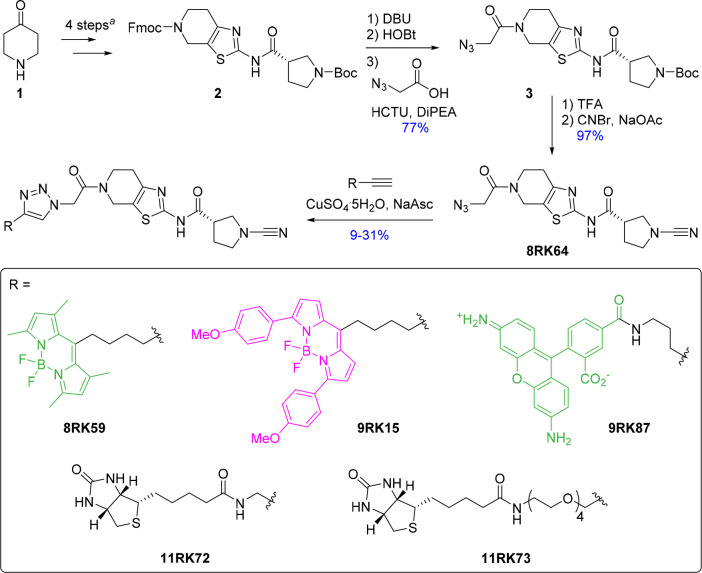
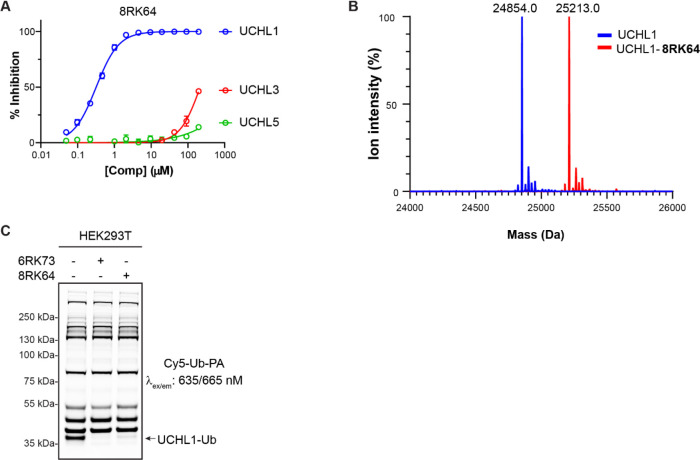
Given the high inhibitory potency and UCHL1 selectivity both in vitro and in cells and the fact that it forms a covalent bond, we envisioned that this type of cyanimide-containing molecule can serve as an ideal starting point for the construction of small-molecule-selective DUB ABPs. This would require the instalment of a reporter group (e.g., fluorescent label) onto the molecule. Compound 6RK73, however, does not provide an appropriate site for modification, so we searched for a structurally related compound with similar inhibitory characteristics from the same source.26 On the basis of this compound, we generated azide 8RK64 (the parent compound lacks the azide moiety) to which several reporter groups were coupled using the copper(I)-catalyzed azide alkyne cycloaddition (CuAAC) or “click reaction”. The compounds and their synthesis routes are shown in Scheme 1. Compound 2 was synthesized from 4-piperidinone (1) in four steps according to a reported procedure.26 The Fmoc-protected piperidine amine was liberated with DBU and coupled to 2-azidoacetic acid, resulting in compound 3. Next, the Boc protecting group was removed from the pyrrolidine amine, followed by a reaction with cyanogen bromide to install the cyanimide moiety, resulting in 8RK64. Treatment of UCHL1 with this compound followed by IC50 determination and LC–MS analysis gave results comparable to those for 6RK73 (Figure 2A,B, Supporting Information), which indicates that 8RK64 also functions as a UCHL1 covalent inhibitor. With IC50 values of 0.32 μM for UCHL1 and 216 μM and >1 mM for UCHL3 and UCHL5, respectively (Figure 2A, Supporting Information Table S1), this compound also retained its UCHL1 selectivity. In addition, 8RK64, like 6RK73, also inhibits UCHL1 activity in cells as shown in a DUB profiling experiment in HEK293T cells using a Cy5-Ub-PA probe (Figure 2C). Notably, 8RK64 could potentially be used as two-step ABP by taking advantage of its azide moiety.34
Scheme 1. Synthesis of Azide-Containing UCHL1 Inhibitor 8RK64 and Fluorescent and Biotinylated Probe Derivatives.
Synthesis steps are described in the literature.26
Figure 2.
Biochemical characterization of 8RK64. (A) IC50 determination of 8RK64 for UCHL1, UCHL3, and UCHL5. (B) Deconvoluted mass spectra of UCHL1 before (blue) and after (red) reaction with 8RK64. (C) Fluorescence labeling of the remaining DUB activity in HEK293T cells upon treatment with UCHL1 inhibitors 8RK64 and 6RK73.
Installation of a Dye Preserves Inhibitory Properties
Because it was unclear what the effect of coupling a bulky fluorescent group would have on the UCHL1 inhibition profiles and cell permeability, we decided to test three commonly used fluorophores. BodipyFL-alkyne, BodipyTMR-alkyne,35 and rhodamine110-alkyne (preparation procedures in the Supporting Information) were coupled to the azide of 8RK64 using copper(I)-mediated click chemistry, resulting in compounds 8RK59, 9RK15, and 9RK87 (Scheme 1). These “one-step” ABPs can potentially be used for the visualization of UCHL1 activity without the need for additional bio-orthogonal chemistry procedures. IC50 determination of these probes against UCHL1 revealed that the installment of the dyes affected the inhibitory potency only marginally (Figure 3A and Supporting Information Table S1). Rhodamine110 probe 9RK87 is almost as potent as its azide precursor 8RK64, with IC50 values of 0.44 μM and 0.32 μM, respectively. The installment of BodipyTMR (9RK15), on the other hand, resulted in a 10-fold potency decrease, although the data points could not be fitted properly to a dose–response function. The less bulky BodipyFL-ABP 8RK59, although not as potent as 8RK64, showed a very acceptable inhibition of UCHL1 with an IC50 close to 1 μM. The ability of 8RK59 to form a covalent complex with UCHL1 was confirmed in an LC–MS experiment as described above (Supporting Information).
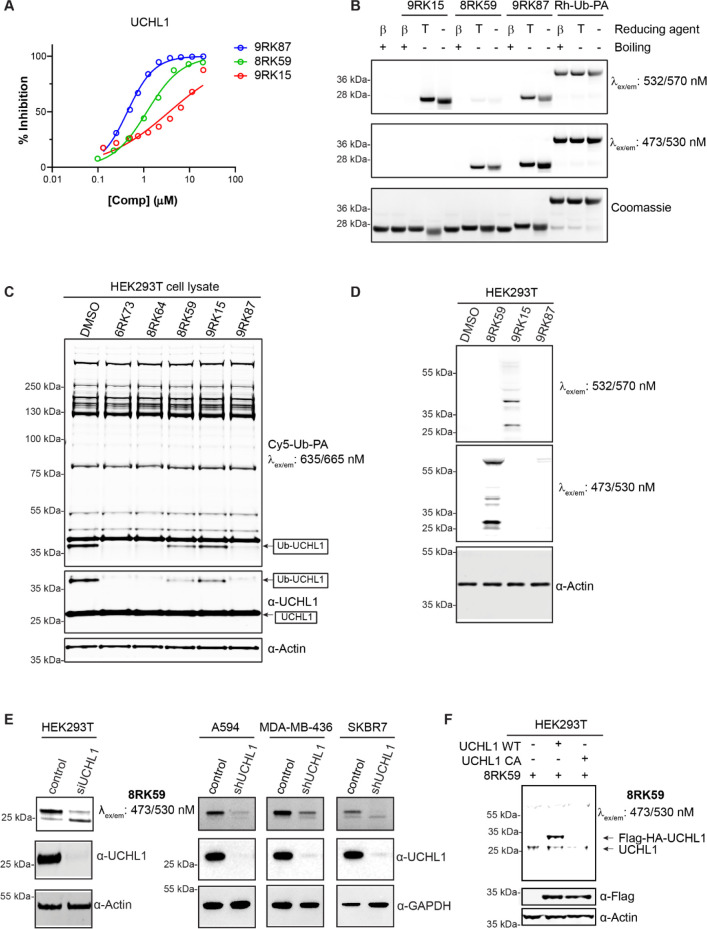
Figure 3.
Characterization of the fluorescent UCHL1 probes in vitro and in cells. (A) IC50 determination of 8RK59, 9RK15, and 9RK87 for UCHL1. (B) Labeling of purified recombinant human UCHL1 by the three probes and Rh-Ub-PA. β: β-Mercaptoethanol. T: TCEP. (C) Fluorescence labeling by Cy5-Ub-PA of remaining DUB activity in the HEK293T cell lysate upon treatment with UCHL1 inhibitors and probes. (D) Fluorescence scans showing the labeling pattern in HEK293T cells of the three probes. (E) Fluorescence labeling of UCHL1 activity in HEK293T, A549, MDA-MB-436, and SKBR7 cells with 8RK59. (F) 8RK59 labels overexpressed Flag-HA-UCHL1 wt but not the C90A active site mutant in HEK293T cells.
ABPs Can Exhibit UCHL1 Activity, and the Covalent Linkage Is Thermally Reversed
We next set out to investigate whether the probes can be used to label and visualize UCHL1 activity after SDS-PAGE and fluorescence gel scanning similar to that for the Rh-Ub-PA probe. To our surprise, for none of the three small-molecule probes could a clear band corresponding to probe-labeled UCHL1 be detected after incubation with purified recombinant human UCHL1. We reasoned that the isothiourea bond between UCHL1 and the probe, which is stable under the conditions used for inhibition and LC–MS experiments (vide supra), might be susceptible to the conditions used for protein denaturation (e.g., boiling in the presence of ∼300 mM β-mercaptoethanol). Indeed, when the same samples were resolved by SDS-PAGE under nondenaturing conditions (no boiling and the absence of β-mercaptoethanol), a clear band appeared that corresponds to probe-labeled UCHL1 for all three probes (Figure 3B). We also investigated if the ABP-UCHL1 bond would survive when β-mercaptoethanol is replaced with tris(2-carboxyethyl)phosphine) (TCEP), both of which are used to create a reducing environment. Figure 3B clearly shows that the ABP-UCHL1 bands remain intact in the presence of 50 mM TCEP and show a better-resolved profile (less smearing) compared to that of the nonreducing samples. The Rh-Ub-PA control samples show that nearly all UCHL1 is labeled and that the formed bond for this probe is stable under denaturing conditions, which corroborates earlier findings.31 The bands corresponding to Rh-Ub-PA and 9RK87 bound to UCHL1 (both bearing the same dye and present in equal amounts) are of similar intensity, which indicates that the small-molecule probes bind UCHL1 efficiently and that all UCHL1 is active upon probe engagement.
ABPs Bind to the Active-Site Cysteine Residue of UCHL1 and Exhibit UCHL1 Activity in Various Cell Lines
We next assessed the ability of the probes to bind and inhibit UCHL1 in a cell lysate by treating HEK293T cell extracts with the three fluorescent probes (at 5 μM) as well as their azide precursor 8RK64 and inhibitor 6RK73 for 1 h, followed by the labeling of all residual DUB activity with Cy5-Ub-PA. The Cy5-labeled Ub probe was used here to circumvent spectral interference with either of the other dyes used in the small-molecule probes. Fluorescent scanning of the gel after SDS-PAGE as well as Western blotting (WB) using anti-UCHL1 antibody clearly showed that rhodamine probe 9RK87 inhibits UCHL1 activity similarly to 8RK64 and 6RK73 (Figure 3C). Both Bodipy probes also potently inhibit UCHL1 in a cell lysate, although to a somewhat lesser extent, which could be expected on the basis of their IC50 values. All other bands are unchanged, which demonstrates that all compounds are able to bind UCHL1 selectively with respect to other DUBs in a cell lysate.
Encouraged by these results, we set out to assess the ability of the probes to penetrate the cell membrane and to label active UCHL1 in cells. HEK293T cells were treated with the probes (5 μM) for 24 h followed by cell lysis, SDS-PAGE (in the absence of β-mercaptoethanol and boiling), and fluorescence scanning at two wavelengths to detect all fluorescent dyes (Figure 3D). A clear band just above 25 kDa is observed for both Bodipy probes (8RK59 and 9RK15), which likely corresponds to ABP-labeled UCHL1 with an expected mass of ∼25.5 kDa. In addition to this band, a few extra bands are visible including one just below UCHL1 and one more pronounced band at around 55 kDa. Interestingly, hardly any band can be seen for the so-far most potent probe, 9RK87. We attributed this effect to the difference in cell permeability between Bodipy and rhodamine dyes, with the latter known to be less capable of crossing the cell membrane.36 Indeed, upon further investigation using microscopy in ABP-treated HeLa and HEK293T cells we confirmed that rhodamine probe 9RK87 is unable to enter these cells, whereas both Bodipy ABPs clearly are (Supporting Information Figure S2). For this reason and because BodipyFL-ABP proved to be a better inhibitor compared to its BodipyTMR analogue, we decided to continue with 8RK59 as the preferred probe for all further experiments.
The ability of 8RK59 to label UCHL1 activity in different cell lines was further explored in HEK293T cells and in three cancer cell lines known to express high levels of endogenous UCHL1: non-small-cell lung cancer (NSCLC) A549 cells, triple-negative breast cancer (TNBC) MDA-MB-436 cells, and SKBR7 cells.37 Cells infected with UCHL1 shRNA knockdown (shUCHL1) lentivirus or transfected with siUCHL1 as well as empty vector control or scrambled oligo (si control) were treated with each probe (5 μM) for 24 h, followed by cell lysis, SDS-PAGE (without boiling and β-mercaptoethanol), and fluorescence scanning (Figure 3E). A clear band appears in the fluorescence scan at the expected height (∼25.5 kDa) in all four cell lines, and this band is significantly decreased in the UCHL1 knockdown samples, indicating that this band indeed corresponds to ABP-labeled UCHL1.
To confirm that 8RK59 binds the active-site cysteine residue in UCHL1, we overexpressed Flag-HA-tagged UCHL1 and its C90A catalytic inactive mutant in HEK293T cells and incubated these cells with 5 μM 8RK59 for 24 h. Fluorescence scanning and anti-FLAG Western blotting show that 8RK59 binds only to wild-type UCHL1 but not to catalytically inactive UCHL1, indicating that the probe binding site is the active-site cysteine (Figure 3F).
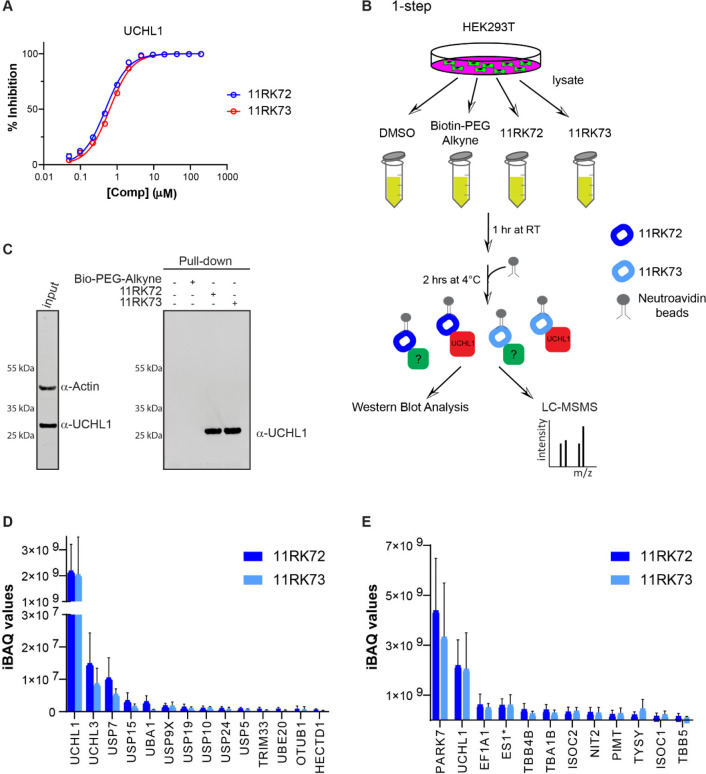
Determination of DUB Selectivity and Potential Off-Targets of the ABP
As mentioned before, besides the band corresponding to ABP-labeled UCHL1, a few other bands appeared on the gel (Figure 3D), but on the basis of the DUB profiling results (Figure 3C), these bands can most likely not be attributed to other DUBs. In order to gain more insight into potential off-targets, we performed pull-down experiments coupled to mass spectrometry to identify the proteins binding to our probe. We started with a two-step ABP approach in which HEK293T cells were incubated with azide-containing compound 8RK64 or DMSO control, followed by a postlysis click reaction with biotin-alkyne38 and subsequent pull-down with neutravidin-coated beads (Supporting Information Figure S3A,B). Samples were run (1 cm) on an SDS-PAGE gel, lanes were cut into two pieces, and the proteins were subjected to trypsin digestion and analyzed with LC–MSMS. As expected, the most enriched protein (with respect to the DMSO control) identified from this experiment was UCHL1 (Supporting Information Figure S3C). Only one other protein was also enriched, a protein deglycase named DJ-1 (PARK7) with a molecular weight of 20 kDa, which most likely corresponds to the band just below UCHL1 in Figure 3D. This enzyme also harbors an active-site cysteine residue which could bind to our probe. Indeed, the incubation of UCHL1 and PARK7 knockdown cells with 8RK59, followed by anti-UCHL1 and anti-PARK7 Western blotting, revealed that PARK7 also reacts with 8RK59 and that the gel band just below UCHL1 corresponds to PARK7 (Supporting Information Figure S3D).
In addition to UCHL1 and PARK7, a few other bands can be seen on the gel (Figure 3D), yet we identified only these two enzymes in the two-step ABP approach. We therefore performed a one-step pull-down experiment where we used two biotinylated versions of 8RK64: compound 11RK72 where biotin is directly linked to the inhibitor and compound 11RK73 with a PEG spacer in between. Both compounds show high inhibitory potential toward UCHL1 (Figure 4A) and form a covalent bond with UCHL1 (Supporting Information Figure S1B,C). The HEK293T cell lysate was incubated with both biotin-ABPs, biotin-alkyne, and DMSO controls followed by pull down with neutravidin-coated beads and subjected to full proteome LC–MSMS analysis (Figure 4B, Supporting Information Figure S3E). Efficient UCHL1 pull-down was confirmed for both biotinylated probes, and no UCHL1 could be detected in the DMSO and biotin-alkyne-treated control samples by Western blotting using anti-UCHL1 antibody (Figure 4C). From the LC–MSMS data, the intensity based absolute quantification (iBAQ) values were calculated using Maxquant software in the pull-down samples and compared to control samples. The list of identified proteins was ranked with respect to the iBAQ value to identify the highest enriched proteins (Supporting Information). An inspection of the list of all enzymes related to Ub (DUBs, E1, E2 conjugating enzymes, and E3 ligases) further substantiates the specificity of the probes for UCHL1 within the Ub system as shown in Figure 4D. Only a few of these enzymes were identified in the pull-down experiment, albeit with a substantial lower iBAQ value compared to that of UCHL1. Importantly, no UCHL1 could be identified by LC–MSMS in the control samples, which corroborates the results from the WB analysis in Figure 4C. The iBAQ values of the top-12 highest-ranking proteins are shown in Figure 4E. In line with the results obtained with the two-step approach, the highest-ranking proteins are UCHL1 and PARK7. PARK7 has a higher iBAQ value here, which contradicts our previous results from the in-cell labeling and two-step pull-down experiments and might be attributed to the use of a different (biotinylated) version of the ABP or the use of intact cells (in the two-step approach) versus the cell lysate (in the one-step approach). The next-highest-ranking group of proteins, albeit at much lower levels, includes amidase NIT2, also harboring an active-site cysteine residue, isochorismatase domain-containing proteins 1 and 2 (ISOC1 and ISOC2), and ES1, which has been annotated as glutamine amidotransferase-like class-1 domain-containing proteins 3A and 3B (GATD3A and GATD3B, respectively). Native protein abundances can influence the pull-down efficiencies. In order to gain insight into the protein abundances for each of the identified targets, we searched the PRIDE repository for a recent data set of a total cellular proteome quantitation MS analysis in HEK293T cells and used the one published by Joshi et al. (PRIDE project PXD015828).39 The iBAQ values corresponding to our identified targets are shown in Supporting Information Figure S4. The pull-down efficiency between UCHL1 and PARK7 is hardly affected by their relative protein abundances because these are very similar. On the other hand, some of the other targets, such as elongation factor 1α (EF1A1) and both tubulin chains (TBB4B and TBA1B), have very high WT abundance levels, which likely explains their abundance in our ABP pull-down experiment. Overall, the shorter (11RK72) and longer (11RK73) biotin probes give similar results, so the distance between the probe and biotin does not seem to influence the binding or the pull-down efficiency.
Figure 4.
Proteomics experiments with biotinylated ABP analogs to identify ABP targets. (A) IC50 determination of 11RK72 and 11RK73 for UCHL1. (B) Schematic representation of pull-down experiment to identify ABP binding proteins. (C) Confirmation of UCHL1 pull-down with biotinylated ABP analogs by Western blot analysis. WB was performed using UCHL1 and actin antibodies. Actin was used as a loading control and incubated together with UCHL1 antibody in the input sample. (D) iBAQ values of enzymes related to the Ub system identified in the pull-down LC–MSMS experiment averaged over three replicates. (E) iBAQ values of the top-12 highest-ranking proteins from the pull-down LC–MSMS experiment averaged over three replicates. *ES1 has been annotated as glutamine amidotransferase-like class 1 domain-containing protein 3A or 3B (GATD3A or GATD3B). Reference intensities (iBAQ values) of corresponding proteins in wild-type HEK293T cells39 are shown in Supporting Information Figure S4.
Upon comparison of the pull-down data (Figure 4) with the fluorescent probe labeling (Figure 3), we were unable to assign all bands to proteins. A majority of the most abundant proteins in the pull-down experiment have a molecular weight of between 20 and 35 kDa. In particular, the pronounced band at around 55 kDa in Figure 3D remains elusive. In a final attempt to assign this band, we resolved the pull-down protein sample from the one-step labeling experiment by SDS-PAGE. All proteins were visualized by silver staining, after which the bands were excised and analyzed by LC–MSMS (Supporting Information Figure S3F). Again, UCHL1 and PARK7 were clearly the main proteins identified from the bands at ∼25 kDa. The proteins corresponding to the other bands were less clear, but the main candidates were GAPDH at ∼40 kDa and elongation factor 1α, tubulin, or glutathione reductase (GSR) at ∼60 kDa. Whether these proteins actually bind to the probe or these results are due to their high expression levels remains elusive. On the basis of the result that we identified UCHL1 as the major probe target in three individual experiments and that we found PARK7 to be the only major off-target, we reasoned that 8RK59 could well be used for in-cell and in-vivo labeling of UCHL1 activity.
Probing UCHL1 Activity in Cells with 8RK59
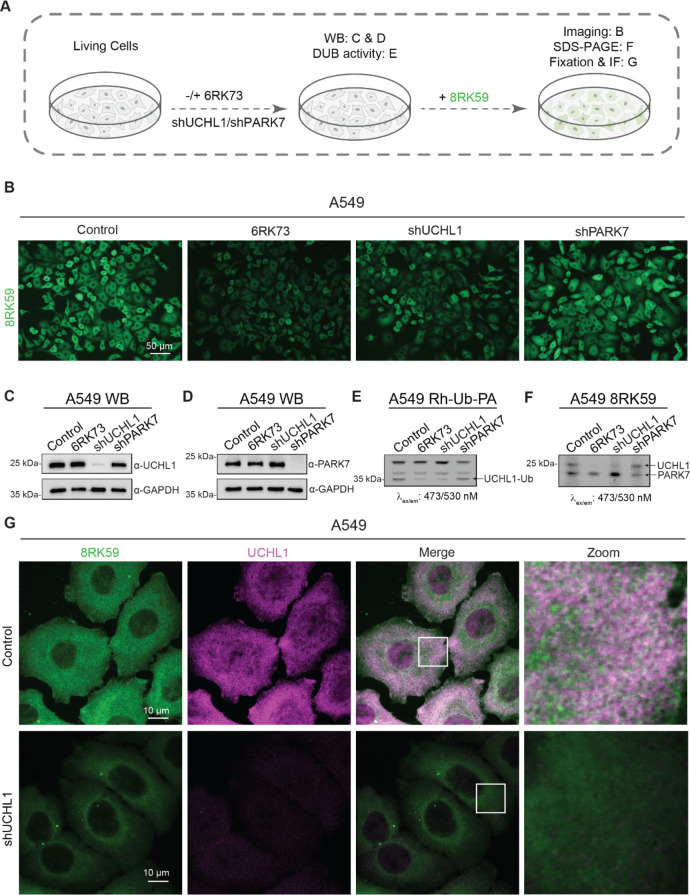
To assess the application and selectivity of 8RK59 in live cells, we first generated A549 cell lines that were selectively depleted of UCHL1 or PARK7 by shRNA-mediated lentiviral transduction. We also used 6RK73 to pharmacologically inhibit UCHL1 activity before adding the 8RK59 probe. The specificity of 8RK59 in live cells and cell lysate was analyzed by the fluorescence imaging of live cells, fluorescence scanning of cell lysates in which proteins were separated by SDS-PAGE gel, and immunofluorescence (IF) staining of fixed cells (Figure 5A). Our results demonstrated that 8RK59 could penetrate and label living cells and that the 8RK59 signal was significantly decreased in 6RK73 and shUCHL1 groups compared to that in control and shPARK7 groups (Figure 5B). The WB results showed an efficient shRNA-mediated depletion of UCHL1 and PARK7 (Figure 5C,D).
Figure 5.
Probing UCHL1 activity in cells with 8RK59. (A) Schematic overview of labeling UCHL1 activity with 8RK59 in cells with/without the depletion of UCHL1 or PARK7 and subsequent assays that were performed to characterize the staining specificity. (B) Live-cell fluorescence imaging of the 8RK59-labeled control (PLKO), 6RK73 (PLKO pretreated with 6RK73), and shUCHL1 and shPARK7 A549 cells. (C) Western blotting (WB) of UCHL1 in the control, 6RK73, and shUCHL1 and shPARK7 A549 cells. WB for GAPDH was included as a loading control. (D) WB of PARK7 in the control, 6RK73, and shUCHL1 and shPARK7 A549 cells. WB for GAPDH was included as a loading control. (E) DUB activity assay of the control, 6RK73, and shUCHL1 and shPARK7 A549 cells with Rh-Ub-PA. The UCHL1-Ub band is indicated with an arrow. (F) Fluorescence scanned SDS-PAGE gel of the 8RK59-labeled control, 6RK73, and shUCHL1 and shPARK7 A549 cells. UCHL1 and PARK7 bands are indicated with arrows. (G) Immunofluorescence (IF) staining of UCHL1 in an 8RK59-labeled control and shUCHL1 A549 cells. A 10 μm scale bar is included. Squares indicate areas that were used for close-up images. All cells from B to G were treated with 5 μM 8RK59 overnight, and 6RK73 group cells were pretreated with 5 μM 6RK73 for 4 h.
Next, we performed a DUB activity assay with the Rh-Ub-PA probe on cell lysates corresponding to the same four conditions that were used in Figure 5C,D. We observed that the UCHL1 activity in 6RK73 and shUCHL1 groups was significantly reduced compared to that in the control and shPARK7 groups (Figure 5E). The Coomassie staining of the DUB activity SDS-PAGE gel was used as a loading control (Supporting Information Figure S5A). The 8RK59 signal in the cell lysate was analyzed by fluorescence scanning of the SDS-PAGE gel of four group cells, and the results displayed that the probe indeed labeled UCHL1 and PARK7 in the cell lysate. However, only 6RK73 and shUCHL1 groups but not the shPARK7 group showed an obvious decrease in the 8RK59 signal compared with the control group (Figure 5F). The Coomassie staining of the 8RK59 SDS-PAGE gel was used as a loading control (Supporting Information Figure S5B). Similar experiments were performed in another cell line MDA-MB-436, where the shUCHL1 group showed a significant decrease in the 8RK59 signal compared with that of the PLKO group both in live cells (Supporting Information Figure S4C) and the cell lysate (Supporting Information Figure S5D). WB of UCHL1 has been used to confirm sufficient UCHL1 knockdown in MDA-MB-436 (Supporting Information Figure S5D). Taken together, these results indicate that 8RK59 selectively detects UCHL1 activity in live A549 and MDA-MB-436 cells.
To further study the difference between UCHL1 expression and activity inside the cells, we performed immunofluorescence (IF) staining of UCHL1 on a fixed control and shUCHL1 A549 cells, which were labeled with the 8RK59 probe overnight (Figure 5G). We observed that the 8RK59 signal in the shUCHL1 group showed a significant decrease compared with the control group, which is consistent with the results in Figure 5B of live-cell fluorescence images of the 8RK59-labeled control and shUCHL1 A549 cells. Besides, we found that the UCHL1 activity pattern is not fully overlapping with the UCHL1 expression pattern. The latter is consistent with the notion that UCHL1 activity is regulated by protein interaction partners or post-translational modifications that have been described previously.40 This finding illustrates the added value of examining UCHL1 activity above the UCHL1 protein level. It also suggests that this reagent can be used in drug screens in intact cells to identify modulators of UCHL1 activity.
Probing UCHL1 Activity in Zebrafish Embryos with 8RK59
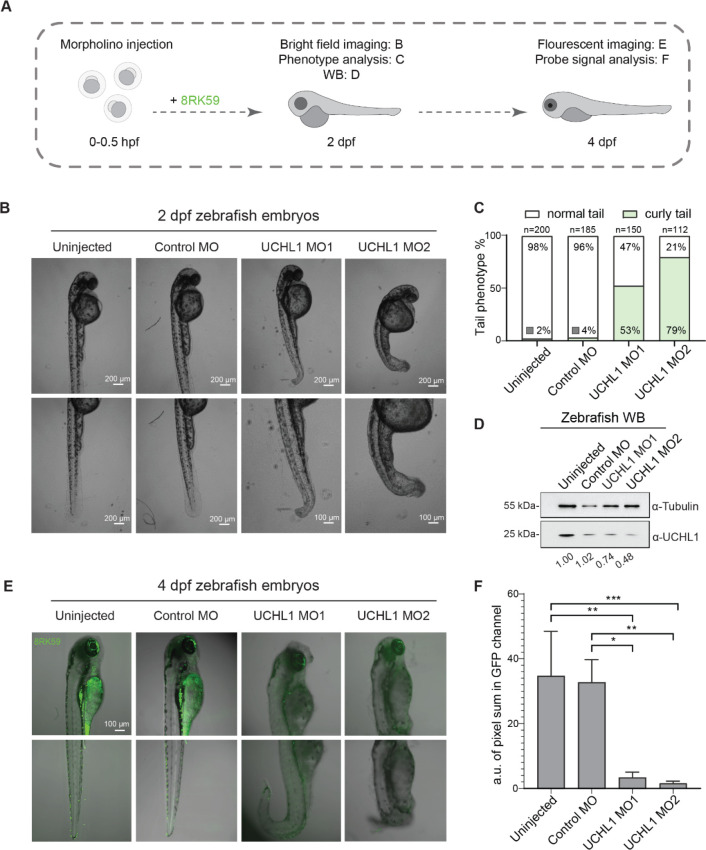
To investigate the application and specificity of 8RK59 in tracking UCHL1 activity in an in vivo animal model, we chose the zebrafish (Danio rerio) due to its high genetic homology to humans and the transparency of their embryos.41 We first generated UCHL1 knockdown zebrafish embryos by separately injecting two independent morpholinos (i.e., MO1 and MO2) that target UCHL1 at the single-cell stage of the embryos. After injection, we labeled the embryos with the 8RK59 probe and performed bright field (BF) microscopy imaging, phenotype analysis, and WB for UCHL1 protein levels 2 days postfertilization (dpf) embryos, followed by fluorescent microscopy imaging and signal analysis for 4 dpf embryos (Figure 6A). Two days after injection, two independent MOs that target UCHL1 showed a similar phenotype of curly tail with 53% of 150 embryos in the UCHL1MO1 group and 79% of 112 embryos in UCHL1MO2 group, while the group without injection (Uninjected) and the standard control MO (control MO) injection group did not show obvious curly tail zebrafish embryos (Figure 6B,C). The curly tail phenotype in UCHL1MO groups became more obvious after 6 dpf, but this also led to a higher mortality rate of the embryos (Supporting Information Figure S6A). WB analysis in 2 dpf zebrafish embryos with/without UCHL1MO injection showed an obvious UCHL1 decrease in UCHL1MOs groups compared with uninjected and control MO groups (Figure 6D). After the validation of UCHL1 depletion in zebrafish embryos, we took the fluorescent images of the 8RK59-labeled 4 dpf zebrafish embryos with/without UCHL1MO injection. The results showed a significantly reduced 8RK59 signal in two UCHL1MO injection groups compared with uninjected and control MO groups (Figure 6E,F).
Figure 6.
Probing UCHL1 activity with 8RK59 in morpholino (MO)-mediated UCHL1 knockdown zebrafish embryos. (A) Schematic overview of labeling UCHL1 activity with 8RK59 in zebrafish embryos with/without UCHL1MO injection. (B) Bright field images of 2 dpf zebrafish embryos without injection (uninjected) after injection with standard control morpholino (control MO) or with two independent UCHL1MOs (UCHL1MO1 or UCHL1MO2). (C) Quantification of the curly tail phenotype in 2 dpf zebrafish embryos with/without UCHL1MO injection. (D) WB of UCHL1 in 2 dpf zebrafish embryos with/without UCHL1MO injections. WB for tubulin was included as a loading control. The expression levels of UCHL1 normalized to tubulin are indicated below. (E) Probing UCHL1 activity with 5 μM 8RK59 in 4 dpf zebrafish embryos with/without UCHL1MO injections. (F) Statistical analysis of 8RK59 signal in 4 dpf zebrafish embryos with/without UCHL1MO injections. The intensity of 8RK59 was measured in three zebrafish embryos of each group by calculating the pixel sum in the GFP channel of both the head and tail areas. *, P < 0.05, **, P < 0.01, ***, P < 0.001, and two-way ANOVA.
Since we identified PARK7 as off-target in the pull-down experiments (Figure 4), we also generated MO-mediated PARK7 depleted zebrafish embryos and performed 8RK59-labeled experiments in those zebrafish embryos (Supporting Information Figure S6). We found that PARK7MOs zebrafish embryos did not display high mortality like UCHL1MOs zebrafish embryos at 6 dpf and revealed no very obvious phenotypic changes (Supporting Information Figure S6A). However, when we zoomed in and analyzed each zebrafish embryo at 2 dpf, we noticed that the PARK7MO1 group has around an 18% abnormal enlarged heart phenotype compared with other control groups (Supporting Information Figure S6B,C). The similar phenotype has been reported in a previous publication on PARK7–/– mice with increased heart/body weight ratios.42 The WB results showed efficient PARK7 knockdown in two PARK7MO groups compared with uninjected and control MO groups (Supporting Information Figure S6D). Importantly, no significant decrease in the 8RK59 signal was detected in the PARK7 knockdown zebrafish embryos based on the fluorescence imaging and signal analysis results of 4 dpf embryos (Supporting Information Figure S6E,F). Taken together, the results indicate that 8RK59 selectively detects UCHL1 in zebrafish embryos. When the selectivity of 8RK59 in cell lysates versus live cell measurements is compared, we observed a higher selectivity toward UCHL1 activity in live cells, which might be attributed to a possible change in protein activity during cell lysis.
Monitoring Spatiotemporal UCHL1 Activity during the Development of Zebrafish Embryos with 8RK59
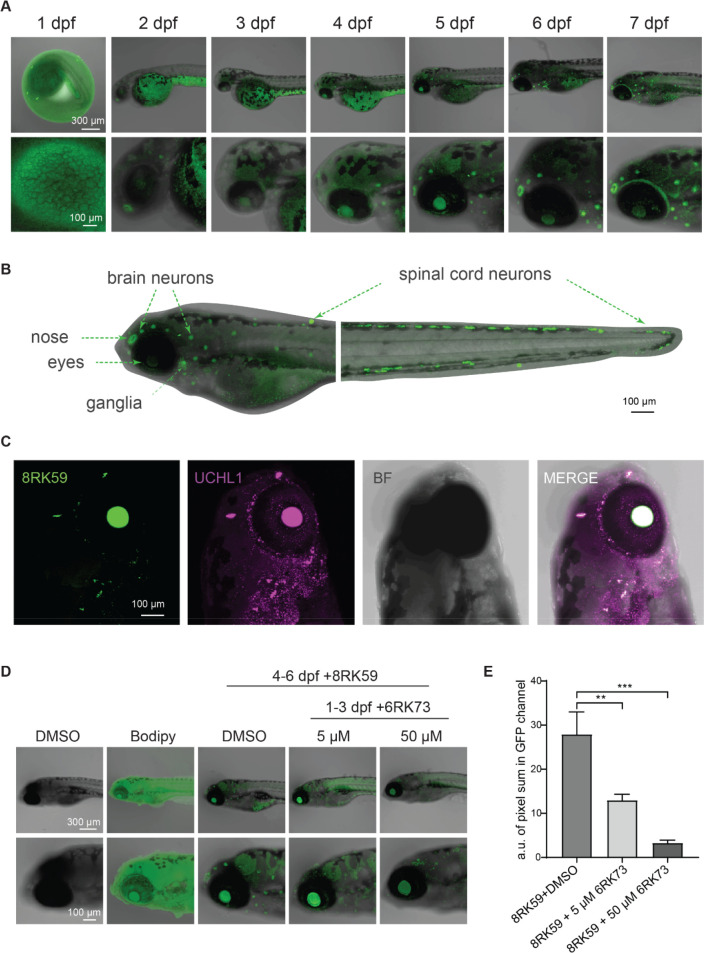
In order to investigate the application of 8RK59 in monitoring UCHL1 activity during the development of zebrafish, we treated zebrafish embryos with 8RK59 and recorded the temporal and spatial distribution of UCHL1 activity during the development of embryos from 1 to 7 dpf. Our results showed that the 8RK59 probe labeled nose, eyes, and brain neurons of the 2 to 3 dpf zebrafish embryos, and a large number of neurons were highlighted from 4 to 7 dpf embryos (Figure 7A). Importantly, all of these organs are enriched in neuronal cells and highly express UCHL1 mRNA in a previous study.43 Moreover, when we compare the 8RK59 signal of the whole zebrafish embryos with the published UCHL1-GFP transgenic zebrafish embryos,43 a similar signal distribution is observed between UCHL1-GFP transgenic and 8RK59-labeled zebrafish embryos, in particular, in the ganglia, eyes, nose, brain neurons, and spinal cord neurons that are enriched with sensory neurons (Figure 7B).
Figure 7.
Monitoring spatiotemporal UCHL1 activity during the development of zebrafish embryos with 8RK59. (A) Tracking the localization of active UCHL1 with 5 μM 8RK59 of zebrafish embryos from 1 to 7 dpf. (B) Landscape of UCHL1 activity in a 6 dpf zebrafish embryo. UCHL1 activity-enriched areas (ganglia, eyes, nose, brain neurons, and spinal cord neurons) are indicated with arrows. (C) IF staining of UCHL1 in a 5 μM 8RK59-labeled 6 dpf zebrafish embryo. A representative image of UCHL1-enriched areas in the brain of 6 dpf zebrafish embryo is shown. BF, brightfield image. (D) Monitoring UCHL1 activity staining intensity of 8RK59-labeled 6 dpf zebrafish embryos pretreated with/without UCHL1 activity inhibitor 6RK73 for 1–3 dpf. DMSO and BodipyFL dye were used as controls. (E) Statistical analysis of the 8RK59 signal in 6 dpf zebrafish embryos pretreated with/without UCHL1 activity inhibitor 6RK73. The intensity of 8RK59 was measured in three zebrafish embryos of each group by calculating the pixel sum in the GFP channel. **, P < 0.01, ***, P < 0.001, and two-way ANOVA.
To further study the difference between the UCHL1 expression and activity in zebrafish embryos, we fixed the 8RK59-labeled embryos and performed IF staining with the UCHL1 antibody. The results showed that both the 8RK59 probe and UCHL1 antibody labeled the ganglia, eyes, nose and brain neurons of zebrafish embryo brain (Figure 7C) and spinal cord neurons in the tail (Supporting Information Figure S7A). This supports the notion that in zebrafish embryos UCHL1 expression is a main determinant of UCHL1 activity. However, there are still some areas with UCHL1 expression but with no/low UCHL1 activity, which may be useful for understanding pools of active/inactive UCHL1 protein.
To assess whether 8RK59 could detect the UCHL1 activity changes in zebrafish embryos, we pretreated the zebrafish embryos with UCHL1 activity inhibitor 6RK73 from 1 to 3 dpf and then labeled the embryos with 8RK59 from 4 to 6 dpf. We found that 6RK73-pretreated zebrafish embryos resulted in a significantly lower 8RK59 signal labeling in a 6RK73-dose dependent manner (Figure 7D,E). In addition, the 6RK73 cotreated zebrafish embryos showed a decreased 8RK59 signal in the 6 dpf zebrafish embryos (Supporting Information Figure S7B). The fluorescence scanned SDS-PAGE gel of 6RK73 cotreated zebrafish lysate displayed significantly reduced UCHL1 activity compared with DMSO-cotreated zebrafish lysate (Supporting Information Figure S7C). These in vivo animal experiments indicate that 8RK59 can specifically visualize and track spatiotemporal UCHL1 activity during the development of zebrafish embryos. Compared with traditional IF and ISH methods used to study UCHL1 protein and mRNA level in fixed animals, this probe provides a new tool for researchers to study the function of active UCHL1 in its native cellular environment in live cells and animals. This opens a new window to investigate UCHL1 (dys)function in pathophysiological processes, including embryogenesis and maintaining tissue homeostasis, but also in cancer, tissue fibrosis, and neurodegenerative diseases (Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis). Moreover, this probe may have potential applications in enabling better diagnosis and treatment of diseases with perturbed UCHL1 activity.
Conclusions
One of the key challenges in DUB research is the creation of activity-based probes that target a single DUB type and at the same time are able to cross the cell membrane in order to study these enzymes inside living cells or even living organisms.44 It has recently been shown by us and others that Ub-based tools (such as ABPs) can be made subtype specific by engineering the amino acid sequence in Ub;32,45,46 however, these ABPs are not cell-permeable, although the use of cell-penetrating peptides has recently been applied to deliver Ub ABPs into cells.47 ABPs based on small-molecule inhibitors, on the other hand, are often cell-permeable and can be tuned chemically to become selective,48,49 although such ABPs for DUBs have been lacking so far. Here we provide evidence of the first fluorescent small-molecule target-specific DUB ABP (8RK59) that hits UCHL1 activity in vitro, in cells, and in vivo. We based our design on a cyanimide-containing inhibitor and show, in contrast to what has been reported in the literature,27 that cyanimides can act as (near to) irreversible binders. Whether the irreversible bond formation results from the chemical nature of the cyanimide used here or from its binding mode within the UCHL1 active site and whether this property can be extended to other DUBs remains to be investigated. The installment of a fluorescent group onto a small-molecule inhibitor can have a detrimental effect on its inhibitory properties. Our data show that the installation of a rhodamine fluorophore hardly affected and a BodipyFL fluorophore only marginally affected the inhibitory potency toward UCHL1, whereas our Ub-ABP experiments confirmed the preservation of their selectivity for UCHL1 among other cysteine DUBs. From these two probes, rhodamine-tagged 9RK87 showed better in vitro characteristics (e.g., a lower IC50 value and more potency in the cell lysate) but unfortunately proved to be unable to cross the cell membrane. As such, this probe could be preferred for in vitro experiments and might be optimized for in-cell use by chemically improving the cell-penetrating properties of rhodamine.28
Small-molecule inhibitors or probes almost inevitably result in nonspecific interactors, and this is not different for our compounds. We have considerably invested in the identification of potential off-targets of our probes by means of a proteomics approach. The data generated in this effort not only are useful for our own study but also provide valuable information for others working on this type of cyanimide-containing compound. The proteomics data is in line with the Ub-probe experiments, confirming that these compounds are UCHL1-specific within the Ub system. We indeed found a few potential off-targets in HEK293T cell lysates, the main ones being the protein and nucleotide deglycase PARK7. These cyanimide compounds may therefore provide a good starting point for small-molecule probes targeting PARK7, which, in spite of its important enzymatic function in protein and DNA repair in virtually any cell, has not yet been developed. On the basis of our data, we expect that the potency and selectivity of the probe can be further improved by means of chemical alterations of the inhibitor. A better knowledge of the structural determinants of the interactions between the probe and UCHL1 will be of great value for this. Unfortunately, despite several crystallization attempts we were unable to obtain appropriately diffracting crystals. During the preparation of our manuscript, Flaherty and co-workers50 reported on a related (S)-1-cyanopyrrolidine-2-carboxamide-based UCHL1 inhibitor, and they applied NMR and molecular modeling to gain insight into the interactions between inhibitor and UCHL1, which could provide useful information to further optimize our probes. In addition, they modified their inhibitor with an alkyne moiety, which, unlike our molecules, resulted in a decrease in potency toward UCHL1 and selectivity with respect to UCHL3. This two-step probe was then used to identify off-targets in KMS11 cells, but remarkably none of their identified proteins show overlap with our list.
In conclusion, we have developed a fluorescent small-molecule activity-based probe that labels UCHL1 activity in vitro, in live cells, and in an in vivo animal model. It is the first example of a one-step DUB-selective, cell-permeable ABP and therefore serves as a unique addition to the Ub toolbox, concomitantly addressing two of the outstanding challenges within this field. Our results show that the probe works in several different cell lines, and we therefore foresee a potential wide application of the probe in studying spatiotemporal UCHL1 activity in future studies of embryonic development and diseases such as Parkinson’s, Alzheimer’s, and cancer. In fact, we recently showed that 6RK73 decreases UCHL1 activity and thereby inhibits TGFβ-induced breast cancer metastasis.51 In a recent study, while our manuscript was under review, Tate and colleagues reported on the identification of a similar UCHL1 activity-based probe as investigated by us that inhibited TGF-β-induced primary human lung fibroblast conversion to myofibroblasts.52 We are convinced that the strategy reported here for small-molecule cyanimide-based probes can be expanded to other cysteine proteases and specifically DUBs. With the increasing importance of the Ub system as a source of practical drug targets, we believe that these ABP tools will fill an unmet need in allowing us to study active DUBs in their native environment in live cells or animals and as such will aid in the development of future therapeutics that target diseases associated with ubiquitination.
Methods
IC50 Determination
The in vitro enzyme inhibition assays were performed in non-binding-surface, flat-bottom, low-flange, black 384-well plates (Corning) at room temperature in a buffer containing 50 mM Tris·HCl, 100 mM NaCl at pH 7.6, 2.0 mM cysteine, 1 mg/mL 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonic acid (CHAPS), and 0.5 mg/mL γ-globulins from bovine blood (BGG) in triplicate. Each well had a final volume of 20.4 μL. All dispensing steps involving buffered solutions were performed on a Biotek MultiFlowFX dispenser. The compounds were dissolved in DMSO as 10, 1, and 0.1 mM stock solutions, and appropriate volumes were transferred from these stocks to the empty plate using a Labcyte Echo550 acoustic dispenser and accompanying dose–response software to obtain a 12-point serial dilution (3 replicates) of 0.05 to 200 μM. A DMSO backfill was performed to obtain equal volumes of DMSO (400 μL) in each well. N-Ethylmaleimide (NEM, 10 mM) was used a positive control (100% inhibition), and DMSO was used as a negative control (0% inhibition). Buffer (10 μL) was added, and the plate was vigorously shaken for 20 s. Next, 5 μL of a 4× final concentration enzyme stock was added, followed by incubation for 30 min. The substrate (5 μL of Ub-Rho-morpholine at a final concentration 400 nM or Cbz-PheArg-AMC at a final concentration of 10 μM in the case of Papain) and the increase in fluorescence intensity over time were recorded using a BMG Labtech CLARIOstar or PHERAstar plate reader (excitation 487 nm, emission 535 nm). The initial enzyme velocities were calculated from the slopes, normalized to the positive and negative controls, and plotted against the inhibitor concentrations (in M) using the built-in equation “[inhibitor] vs response – variable slope (four parameters), least-squares fit” with constraints “Bottom = 0” and “Top = 100” in GraphPad Prism 7 software to obtain the IC50 values.
Jump Dilution Assay
All assays were performed in triplicate. The assay was performed in a buffer containing 50 mM Tris·HCl, 100 mM NaCl at pH 7.6, 2.0 mM cysteine, 1 mg/mL 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonic acid (CHAPS), and 0.5 mg/mL γ-globulins from bovine blood (BGG). The final concentrations used were 1 nM UCHL1, 400 nM Ub-Rho-morpholine, and 2 μM or 20 nM or a jump dilution of 2 μM to 20 nM inhibitor. Samples of 20 μL containing 200 nM UCHL1 and 4 μM inhibitor (2% DMSO), 2% DMSO, or 20 mM N-ethylmaleimide (NEM) were incubated for 30 min at room temperature. Each sample (4 μL) was then diluted into a 400 μL solution containing 400 nM Ub-Rho-morpholine. After a brief mixing, 20 μL of each of these solutions was quickly transferred to a non-binding-surface, flat-bottom, low-flange, black 384-well plate (Corning), and the increase in fluorescence over time was recorded using a BMG Labtech PHERAstar plate reader (excitation 485 nm, emission 520 nm). As a control, samples were taken in which 40 μL of a 4 μM and 40 nM inhibitor solution in buffer (2% DMSO) was added to 35 μL of a 2.3 nM UCHL1 solution. After 30 min of incubation, 5 μL of a 6.4 μM Ub-Rho-morpholine solution was added, after which 20 μL of each solution was transferred to the same 384-well plate mentioned above, and the increase in fluorescence intensity was measured concomitantly. Fluorescent intensities were plotted against time using GraphPad Prism 7.
Covalent Complex Formation Mass Spectrometry Analysis
Samples of 1.4 μM UCHL1 in 70 μL buffer containing 50 mM Tris·HCl, 100 mM NaCl at pH 7.6, 2.0 mM cysteine, and 1 mg/mL 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonic acid (CHAPS) were prepared. These samples were treated with 1 μL of DMSO or 1 μL of a 10 mM inhibitor/probe stock solution in DMSO (140 μM final concentration) and incubated for 30 min at room temperature. Samples were then diluted 3-fold with water and analyzed by mass spectrometry by injecting 1 μL into a Waters XEVO-G2 XS Q-TOF mass spectrometer equipped with an electrospray ion source in positive mode (capillary voltage 1.2 kV, desolvation gas flow 900 L/h, T = 60 °C) with a resolution of R = 26 000. Samples were run using two mobile phases: (A) 0.1% formic acid in water and (B) 0.1% formic acid in CH3CN on a Waters Acquity UPLC protein BEH C4 column [300 Å, 1.7 μm (2.1 × 50 mm2), flow rate = 0.5 mL/min, run time = 14.00 min, column T = 60 °C, and mass detection 200–2500 Da]. Gradient: 2–100% B. Data processing was performed using Waters MassLynx mass spectrometry software 4.1, and ion peaks were deconvoluted using the built-in MaxEnt1 function.
Probe Labeling of Purified Recombinant UCHL1
The assay was performed in a buffer containing 50 mM Tris·HCl, 100 mM NaCl at pH 7.6, 2.0 mM cysteine, and 1 mg/mL 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonic acid (CHAPS). A stock solution containing 8 μM UCHL1 and stock solutions containing 20 μM 8RK59, 9RK15, 9RK87, and Rho-Ub-PA in buffer were prepared. The UCHL1 stock solution (50 μL) was mixed with 50 μL of all probe solutions followed by incubation for 60 min at 37 °C. Three aliquots of 10 μL of each sample were taken and treated with (1) 5 μL of loading buffer with β-mercaptoethanol, followed by 5 min of heating at 95 °C; (2) 5 μL of loading buffer with 50 mM TCEP; and (3) 5 μL of loading buffer. Samples were resolved by SDS-PAGE using a 4–12% Bis-Tris gel (Invitrogen, NuPAGE) with MES SDS running buffer (Novex, NuPAGE) for 45 min at 190 V. Gels were scanned for fluorescence on a GE Typhoon FLA 9500 using a green channel (λex/em 473/530 nm) and a red channel (λex/em 532/570 nm), followed by staining with InstantBlue Coomassie protein stain (Expedeon), after which the gel was scanned on a GE Amersham Imager 600.
Cell Lines and Cell Culture
HEK293T, HeLa, A549, and MDA-MB-436 cells were originally obtained from the American Type Culture Collection (ATCC), and SKBR7 cells were obtained from Dr. J. Martens (Erasmus University Medical Center, Rotterdam, The Netherlands). Cells were cultured in Dulbecco’s modified Eagles’ medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL penicillin–streptomycin (15140122, Gibco). Stable shUCHL1 A549, shPARK7, and shUCHL1MDA-MB-436 cell lines were generated by lentiviral infection, and the cell lines were continuously cultured under puromycin selection. Four UCHL1 and PARK7 shRNAs were identified and tested, and the most effective shUCHL1 (TRCN0000007273, Sigma) and shPARK7 (TRCN0000004920, Sigma) for lentiviral infections were used for experiments. All cell lines were regularly tested for the absence of mycoplasma and were authenticated.
Transfection
For shRNA expression, lentiviruses were produced by transfecting shRNA-targeting plasmids together with helper plasmids pCMV–VSVG, pMDLg–RRE (gag–pol), and pRSV–REV into HEK293T cells. Cell supernatants were collected 48 h after transfection and were used to infect cells to generate stable shRNA-mediated UCHL1/PARK7 knockdown cell lines.
For siRNA transfection, siRNAs targeting UCHL1 (set of 4: siGENOME, MQ-004309-00-0002 2 nmol) and PARK7 (set of 4: siGENOME, MQ-005984-00-0002 2 nmol) were obtained from Dharmacon. Knockdown of UCHL1 and PARK7 in HEK293T cells was performed as follows: for the six-well plate format, 200 μL of siRNA (500 nM stock) was incubated with 4 μL of Dharmafectin reagent 1 (Dharmacon) diluted in 200 μL of medium without supplements by shaking for 20 min at room temperature. The transfection mixture was added to cells and cultured at 37 °C and 5% CO2. Forty-eight hours after transfection, 8RK59 was added to the cells and incubated for 24 h. Cells were harvested and analyzed as described under the section “DUB activity profiling and competition with Ub-PA DUB probes”.
For the expression of UCHL1 in HEK293T cells, the Flag-HA-UCHL1 construct was obtained from Addgene (22563). Catalytically inactive mutant (C90A) UCHL1 was generated using site-directed mutagenesis. Wild-type and C90A mutant UCHL1 were transfected into HEK293T cells using the PEI transfection reagent. Twenty-four hours after transfection, 8RK59 was added to the cells and incubated for 24 h. Cells were harvested and analyzed as described under the section “DUB activity profiling and competition with Ub-PA DUB probes”.
Western Blotting
Cells were lysed in HR lysis buffer (50 mM Tris, 5 mM MgCl2, 250 mM sucrose, and 2 mM DTT at pH 7.4) with protease inhibitor cocktail for 10 min on ice. The lysates were sonicated using 10 cycles of 30 s pulse on, 30 s pulse off. Twenty Zebrafish embryos in each group were lysed in Laemmli buffer and boiled for 5 min. The lysates were centrifuged at maximun speed for 20 min at 4 °C. Thereafter, protein concentrations were measured using the DC protein assay (500-0111, Bio-Rad), and equal amounts of proteins were used for each condition that was analyzed by WB with following antibodies: UCHL1 (ab27053, Abcam) for cells, UCHL1 (HPA005993, ATLAS) for zebrafish embryos, PARK7 (ab76008, Abcam) for zebrafish embryos, Tubulin (2148, Cell Signaling) for cells and zebrafish embryos, GAPDH (MAB374, Millipore) for cells, and Actin (A5441, Sigma-Aldrich) for cells.
Immunofluorescence Staining
Cells were fixed for 20 min in 4% paraformaldehyde and then permeabilized in 0.1% Triton-X for 10 min. Nonspecific binding was blocked with blocking buffer (1% BSA in 0.1% PBS-Tween) for 30 min. Primary antibody UCHL1 (ab27053, Abcam) was diluted in blocking buffer and added to the cell for 1 h. After three washings with PBS, secondary antibody donkey anti-rabbit IgG Alexa fluorescence 555 (Invitrogen, no. A31572) was added and incubated for 30 min. After three washings with PBS, samples were mounted with VECTASHIELD antifade mounting medium with DAPI (H-1200, Vector Laboratories). Fluorescence images were acquired with the TCS SP8 confocal microscope (Leica).
Zebrafish embryos were fixed with 4% paraformaldehyde for 2 h at room temperature. Samples were dehydrated with 33, 66, and 100% methanol in PBS, followed by a rehydration step. Thereafter, the embryos were successively treated with 10 μg/mL proteinase K for 60 min at 37 °C, permeabilized with 0.25% Triton in PBS for 30 min on ice, and blocked with 10% FBS in PBS for 1 h at room temperature. Embryos were incubated with primary antibody (ab27053, Abcam) for at least 12 h at 4 °C. After washing with 0.1% Triton in PBS three times for 10 min, the samples were incubated with fluorescein-conjugated secondary antibody donkey anti-rabbit IgG Alexa fluorescence 555 (Invitrogen no. A31572) for 2 h at room temperature. After being washed with PBS (0.1% Triton), samples were analyzed using an SP5 STED confocal microscope (Leica, Rijswijk, The Netherlands).
DUB Activity Profiling and Competition with Ub-PA DUB Probes
HEK293T cells were treated with a 5 μM final concentration of the indicated compounds for 24 h. Cells were lysed in HR lysis buffer supplemented with protease inhibitor cocktail (11836145001, Roche). Samples were kept on ice and lysed by sonication (10 cycles of 30 s on and 30 s off). The protein extract (25 μg) was labeled with either 1 μM Rh-Ub-PA probe or 0.5 μM Cy5-Ub-PA probe for 30 min at 37 °C. For the cell lysate incubation, HEK293T cells were lysated as described above. HEK293T cell lysates were preincubated with a 5 μM final concentration of compounds for 1 h, followed by incubation with a 0.5 μM Cy5-Ub-PA probe for 30 min at 37 °C. Labeling reactions were terminated with sample buffer and heating to 100 °C for 10 min. Samples were size-separated in SDS-PAGE gels. In-gel fluorescence signals were scanned by employing the Typhoon FLA 9500 molecular imager (GE Healthcare). Images were analyzed using ImageJ software.
Probe Labeling of Endogenous UCHL1 in Living Cells
Cell lines were transfected with shRNAs, siRNAs, or UCHL1 constructs as described above. A final concentration of probes (5 μM) was added to the cell a day before harvesting. Fluorescent images were acquired with a DMi8 inverted fluorescence microscope (Leica). Cells were harvested in HR buffer as described above. The NuPAGE LDS sample buffer containing 50 mM TCEP was added to cell lysates. Samples were resolved by SDS-PAGE using a 4–12% bis–tris gel (Invitrogen, NuPAGE) with MES SDS running buffer (Novex, NuPAGE) for 45 min at 190 V. Gels were scanned for fluorescence on a GE Typhoon FLA 9500 using a green channel (λex/em 473/530 nm) and a red channel (λex/em 532/570 nm), followed by transferring proteins to nitrocellulose membrane (Amersham) and Western blot analysis.
Proteomics
For one-step approach, 4 × 106 HEK293T cells were seeded into 10 cm dishes for each treatment. Forty-eight hours later, HEK293T cells were harvested in lysis buffer containing 50 mM HEPES at pH 7.3, 150 mM NaCl, and a 1% NP-40 and 1× protease inhibitor cocktail and incubated for 30 min on ice. Cell lysates were centrifuged at maximum speed for 20 min. The lysates were incubated with a 5 μM final concentration of biotin-PEG4-alkyne, 11RK72, or 11RK73 or the same volume of DMSO for 1 h at room temperature. A 30 μL neutravidin beads slurry (50%) was added to each sample. The samples were then incubated for 2 h at 4 °C. Beads were washed six times in wash buffer containing 50 mM HEPES at pH 7.3, 150 mM NaCl, and 1% NP-40. After the washing buffer was completely removed, the NuPAGE LDS sample buffer (containing 7.5% β-mercaptoethanol) was added to the beads, followed by 15 min of incubation at 95 °C.
For the two-step approach, 4 × 106 HEK293T cells were seeded into 10 cm dishes for each treatment. Twenty-four hours later, a 5 μM final concentration of 8RK64 or the same volume of DMSO was added to the cells. After 24 h of incubation, HEK293T cells were harvested in lysis buffer containing 50 mM HEPES at pH 7.3, 150 mM NaCl, and a 1% NP-40 and 1× protease inhibitor cocktail and incubated for 30 min on ice. Cell lysates were centrifuged at maximum speed for 20 min. A 1× volume of click cocktail [100 mM CuSO4·5H2O, 1 M sodium ascorbate, 100 mM TBTA (tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine) ligand, 0.1 M HEPES at pH 7.3, and 5 μM biotin-alkyne] was added to a 2× volume of cell lysates and incubated for 45 min. A (30 μL) neutravidin bead slurry (50%) was added to each sample. The samples were then incubated for 2 h at 4 °C. Beads were washed six times in wash buffer containing 50 mM HEPES at pH 7.3, 150 mM NaCl, and 1% NP-40. After the washing buffer was completely removed, SDS sample buffer (containing 7.5% β-mercaptoethanol) was added to the beads, followed by 15 min of incubation at 95 °C. For MS analysis, proteins were run for 1 to 2 cm on 4–12% PAGE (NuPAGE Bis-Tris Precast Gel, Life Technologies) and stained with silver (SilverQuest Silver Stain, Life Technologies). The lane was cut into four equal parts, and gel slices were subjected to reduction with dithiothreitol, alkylation with iodoacetamide, and in-gel trypsin digestion using a Proteineer DP digestion robot (Bruker).
Tryptic peptides were extracted from the gel slices, lyophilized, dissolved in 95/3/0.1 v/v/v water/acetonitrile/formic acid, and subsequently analyzed by online C18 nanoHPLC MS/MS with a system consisting of an Easy nLC 1000 gradient HPLC system (Thermo, Bremen, Germany) and a LUMOS mass spectrometer (Thermo). Fractions were injected onto a homemade precolumn (100 μm × 15 mm; Reprosil-Pur C18-AQ 3 μm, Dr. Maisch, Ammerbuch, Germany) and eluted via a homemade analytical nano-HPLC column (15 cm × 50 μm; Reprosil-Pur C18-AQ 3 μm). The gradient was run from 0 to 50% solvent B (20/80/0.1 water/acetonitrile/formic acid v/v/v) in 20 min. The nano-HPLC column was drawn to a tip of ∼5 μm and acted as the electrospray needle of the MS source. The LUMOS mass spectrometer was operated with data-dependent MS/MS (top-10 mode) with collision energy at 32 V and recording of the MS2 spectrum in the orbitrap. In the master scan (MS1), the resolution was 120 000 and the scan range was 400–1500 at an AGC target of 400 000 at a maximum fill time of 50 ms. Dynamic exclusion occurred after n = 1 with an exclusion duration of 10 s. Charge states 2–5 were included. For MS2, precursors were isolated with the quadrupole with an isolation width of 1.2 Da. The HCD collision energy was set to 32 V. The first mass was set to 110 Da. The MS2 scan resolution was 30 000 with an AGC target of 50 000 at a maximum fill time of 60 ms.
Protein identification and label-free quantification were performed using Maxquant version 1.6.7.0, with all default parameters, using the Uniprot Homo sapiens minimal database (20 205 entries). In addition, iBAQ53 was ticked in the global parameters tab. These iBAQ values were averaged over the three replicates, and the values were used to produce the relevant bar graphs. Proteins were filtered (iBAQ value >0 in all probe-treated samples and unique peptides >3) and ranked for iBAQ values, and the top 12 (iBAQ > 108) were selected for display. In addition, all Ub system-related enzymes (DUBs, E1, E2, and E3) from the list were selected, and corresponding iBAQ values were plotted. The reference intensities of proteins in wild type HEK293T cells were taken from the Maxquant protein groups output file from Joshi et al.39 (from the PRIDE data archive with entry number PDX015828).
Probe Labeling of Endogenous UCHL1 in Zebrafish Embryos
Transgenic zebrafish lines Tg (kdrl: mTurquois) were raised, staged, and maintained according to standard procedures in compliance with the local Institutional Committee for Animal Welfare of the Leiden University. Zebrafish embryos were treated with 5 μM 8RK59 or a gradient 6RK73 concentration in the egg water. Fluorescent image acquisition was performed with a Leica SP5 STED confocal microscope (Leica, Rijswijk, Netherlands). The quantification of the 8RK59 signal was analyzed with Leica microscope software platform LAS X. Thirty zebrafish were treated in each group, and three representative images were taken and analyzed. Statistical analysis was performed using Graphpad Prism 8 software. Numerical data from triplicates are presented as the mean ± SD. Two-way analysis of variance (ANOVA) has been used to analyze multiple subjects.
Morpholino Injections in Zebrafish Embryos
Five morpholinos (Genetools, USA) were designed and used, consisting of standard control MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′; UCHL1 ATG MO1, 5′-TATTTCCATCGGTTTCCACTCCATG-3′; UCHL1 splice MO2 (target exon 4), 5′-GTTCCTTAAACATATCCACTTACCA-3′; PARK7 splice MO1 (target exon 2), 5′-TATGTAAAGTCAGACCTGTTTGTG-3′; and PARK7 splice MO2 (target exon 3), 5′-AAAACAGATTTGTACCTCAGAAAGG-3′. The single-cell stage of zebrafish embryos was injected with 2 ng of morpholinos into the yolk area. Approximately 200 embryos were injected for each group within 30 min. Bright-field images of 2 dpf zebrafish embryos were acquired with a DMi8 inverted fluorescence microscope (Leica). Fluorescent images of 4 dpf zebrafish embryos were acquired with a Leica SP5 confocal microscope (Leica). Three representative images were taken and analyzed. Statistical analysis was performed using Graphpad Prism 8 software. Numerical data from triplicates are presented as the mean ± SD. Two-way analysis of variance (ANOVA) has been used to analyze multiple subjects. Bright-field images of 6 dpf zebrafish embryos were acquired with a M50 stereo zoom microscope (Leica). Representative images are shown.
Acknowledgments
We thank Yves Leestemaker for assistance with the DUB activity-based profiling assays and Bjorn van Doodewaerd for LC–MS and HPLC purification assistance. P.t.D. and H.O. are supported by the Oncode Institute. S.L. is supported by the China Scholarship Council. This work was supported by the Dutch Organization for Scientific Research (NWO VICI grant 724.013.002 to H.O.), the Investment Grant NWO Medium Grant (91116004, partially financed by ZonMw to P.A.v.V.), and the Cancer Genomics Centre Netherlands (CGC.NL to P.t.D.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c07726.
Detailed synthesis procedures and compound characterization (including Ub–Rho–morpholine and rhodamine110-alkyne), NMR and LC-MS spectral data, proteomics data, IC50 data, and figures. The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with data set identifier PXD021557 (PDF)
Protein lists of one-step pull-down with 11RK72 and 11RK73 11RK72 and 11RK73 in bold (XLSX)
Protein lists from PRIDE project PDX015828 (XLSX)
Protein lists of two-step pull-down with 8RK64 8RK64 in bold (XLSX)
Author Contributions
§ R.K., S.L., and A.S. contributed equally to this work.
Zebrafish were raised, staged, and maintained according to standard procedures in compliance with the local Institutional Committee for Animal Welfare of the Leiden University.
The authors declare the following competing financial interest(s): H.O. was shareholder of the reagent company UbiQ.
Dedication
∥ In memory of Huib Ovaa, who passed away too young on May 19, 2020.
Supplementary Material
References
- Komander D.; Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Cadwell K.; Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 2005, 309 (5731), 127–130. 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- Pao K. C.; Wood N. T.; Knebel A.; Rafie K.; Stanley M.; Mabbitt P. D.; Sundaramoorthy R.; Hofmann K.; van Aalten D. M. F.; Virdee S. Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature 2018, 556 (7701), 381–385. 10.1038/s41586-018-0026-1. [DOI] [PubMed] [Google Scholar]
- Mevissen T. E. T.; Komander D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 2017, 86, 159–192. 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- Behrends C.; Harper J. W. Constructing and decoding unconventional ubiquitin chains. Nat. Struct. Mol. Biol. 2011, 18 (5), 520–528. 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- Wilkinson K. D.; Lee K.; Deshpande S.; Duerksenhughes P.; Boss J. M.; Pohl J. The neuron-specific protein PGP-9.5 is a ubiquitin carboxyl-terminal hydrolase. Science 1989, 246 (4930), 670–673. 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- Leroy E.; Boyer R.; Auburger G.; Leube B.; Ulm G.; Mezey E.; Harta G.; Brownstein M. J.; Jonnalagada S.; Chernova T.; Dehejia A.; Lavedan C.; Gasser T.; Steinbach P. J.; Wilkinson K. D.; Polymeropoulos M. H. The ubiquitin pathway in Parkinson’s disease. Nature 1998, 395 (6701), 451–452. 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- Maraganore D. M.; Farrer M. J.; Hardy J. A.; Lincoln S. J.; McDonnell S. K.; Rocca W. A. Case-control study of the ubiquitin carboxy-terminal hydrolase L1 gene in Parkinson’s disease. Neurology 1999, 53 (8), 1858–1860. 10.1212/WNL.53.8.1858. [DOI] [PubMed] [Google Scholar]
- Lunati A.; Lesage S.; Brice A. The genetic landscape of Parkinson’s disease. Rev. Neurol. 2018, 174 (9), 628–643. 10.1016/j.neurol.2018.08.004. [DOI] [PubMed] [Google Scholar]
- Bilguvar K.; Tyagi N. K.; Ozkara C.; Tuysuz B.; Bakircioglu M.; Choi M.; Delil S.; Caglayan A. O.; Baranoski J. F.; Erturk O.; Yalcinkaya C.; Karacorlu M.; Dincer A.; Johnson M. H.; Mane S.; Chandra S. S.; Louvi A.; Boggon T. J.; Lifton R. P.; Horwich A. L.; Gunel M. Recessive loss of function of the neuronal ubiquitin hydrolase UCHL1 leads to early-onset progressive neurodegeneration. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (9), 3489–3494. 10.1073/pnas.1222732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Cai F.; Zhang S.; Zhang S.; Song W. Overexpression of ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) delays Alzheimer’s progression in vivo. Sci. Rep. 2015, 4, 7298. 10.1038/srep07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop P.; Rocca D.; Henley J. M. Ubiquitin C-terminal hydrolase L1 (UCH-L1): structure, distribution and roles in brain function and dysfunction. Biochem. J. 2016, 473 (16), 2453–2462. 10.1042/BCJ20160082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S.; Foreman O.; Perkins S. L.; Witzig T. E.; Miles R. R.; van Deursen J.; Galardy P. J. The de-ubiquitinase UCH-L1 is an oncogene that drives the development of lymphoma in vivo by deregulating PHLPP1 and Akt signaling. Leukemia 2010, 24 (9), 1641–1655. 10.1038/leu.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst-Kennedy J.; Chin L. S.; Li L. Ubiquitin C-terminal hydrolase l1 in tumorigenesis. Biochem. Res. Int. 2012, 2012, 123706. 10.1155/2012/123706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong A.; Merkx R.; Berlin I.; Rodenko B.; Wijdeven R. H.; El Atmioui D.; Yalcin Z.; Robson C. N.; Neefjes J. J.; Ovaa H. Ubiquitin-based probes prepared by total synthesis to profile the activity of deubiquitinating enzymes. ChemBioChem 2012, 13 (15), 2251–2258. 10.1002/cbic.201200497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrigan J. A.; Jacq X.; Martin N. M.; Jackson S. P. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discovery 2018, 17 (1), 57–78. 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C.; Hoang Q. Q.; Kreinbring C. A.; Luchansky S. J.; Meray R. K.; Ray S. S.; Lansbury P. T.; Ringe D.; Petsko G. A. Structural basis for conformational plasticity of the Parkinson’s disease-associated ubiquitin hydrolase UCH-L1. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (12), 4675–4680. 10.1073/pnas.0510403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavory G.; O’Dowd C. R.; Helm M. D.; Flasz J.; Arkoudis E.; Dossang A.; Hughes C.; Cassidy E.; McClelland K.; Odrzywol E.; Page N.; Barker O.; Miel H.; Harrison T. Discovery and characterization of highly potent and selective allosteric USP7 inhibitors. Nat. Chem. Biol. 2018, 14 (2), 118–125. 10.1038/nchembio.2528. [DOI] [PubMed] [Google Scholar]
- O’Dowd C. R.; Helm M. D.; Rountree J. S. S.; Flasz J. T.; Arkoudis E.; Miel H.; Hewitt P. R.; Jordan L.; Barker O.; Hughes C.; Rozycka E.; Cassidy E.; McClelland K.; Odrzywol E.; Page N.; Feutren-Burton S.; Dvorkin S.; Gavory G.; Harrison T. Identification and Structure-Guided Development of Pyrimidinone Based USP7 Inhibitors. ACS Med. Chem. Lett. 2018, 9 (3), 238–243. 10.1021/acsmedchemlett.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull A. P.; Ioannidis S.; Krajewski W. W.; Pinto-Fernandez A.; Heride C.; Martin A. C. L.; Tonkin L. M.; Townsend E. C.; Buker S. M.; Lancia D. R.; Caravella J. A.; Toms A. V.; Charlton T. M.; Lahdenranta J.; Wilker E.; Follows B. C.; Evans N. J.; Stead L.; Alli C.; Zarayskiy V. V.; Talbot A. C.; Buckmelter A. J.; Wang M.; McKinnon C. L.; Saab F.; McGouran J. F.; Century H.; Gersch M.; Pittman M. S.; Marshall C. G.; Raynham T. M.; Simcox M.; Stewart L. M. D.; McLoughlin S. B.; Escobedo J. A.; Bair K. W.; Dinsmore C. J.; Hammonds T. R.; Kim S.; Urbe S.; Clague M. J.; Kessler B. M.; Komander D. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 2017, 550 (7677), 481–486. 10.1038/nature24451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberto I.; Liu X.; Seo H. S.; Schauer N. J.; Iacob R. E.; Hu W.; Das D.; Mikhailova T.; Weisberg E. L.; Engen J. R.; Anderson K. C.; Chauhan D.; Dhe-Paganon S.; Buhrlage S. J. Structure-Guided Development of a Potent and Selective Non-covalent Active-Site Inhibitor of USP7. Cell Chem. Biol. 2017, 24 (12), 1490–1500. 10.1016/j.chembiol.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kategaya L.; Di Lello P.; Rouge L.; Pastor R.; Clark K. R.; Drummond J.; Kleinheinz T.; Lin E.; Upton J. P.; Prakash S.; Heideker J.; McCleland M.; Ritorto M. S.; Alessi D. R.; Trost M.; Bainbridge T. W.; Kwok M. C. M.; Ma T. P.; Stiffler Z.; Brasher B.; Tang Y.; Jaishankar P.; Hearn B. R.; Renslo A. R.; Arkin M. R.; Cohen F.; Yu K.; Peale F.; Gnad F.; Chang M. T.; Klijn C.; Blackwood E.; Martin S. E.; Forrest W. F.; Ernst J. A.; Ndubaku C.; Wang X.; Beresini M. H.; Tsui V.; Schwerdtfeger C.; Blake R. A.; Murray J.; Maurer T.; Wertz I. E. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 2017, 550 (7677), 534–538. 10.1038/nature24006. [DOI] [PubMed] [Google Scholar]
- Pozhidaeva A.; Valles G.; Wang F.; Wu J.; Sterner D. E.; Nguyen P.; Weinstock J.; Kumar K. G. S.; Kanyo J.; Wright D.; Bezsonova I. USP7-Specific Inhibitors Target and Modify the Enzyme’s Active Site via Distinct Chemical Mechanisms. Cell Chem. Biol. 2017, 24 (12), 1501–1512. 10.1016/j.chembiol.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Resnick E.; Bradley A.; Gan J.; Douangamath A.; Krojer T.; Sethi R.; Geurink P. P.; Aimon A.; Amitai G.; Bellini D.; Bennett J.; Fairhead M.; Fedorov O.; Gabizon R.; Gan J.; Guo J.; Plotnikov A.; Reznik N.; Ruda G. F.; Diaz-Saez L.; Straub V. M.; Szommer T.; Velupillai S.; Zaidman D.; Zhang Y.; Coker A. R.; Dowson C. G.; Barr H. M.; Wang C.; Huber K. V. M.; Brennan P. E.; Ovaa H.; von Delft F.; London N. Rapid Covalent-Probe Discovery by Electrophile-Fragment Screening. J. Am. Chem. Soc. 2019, 141 (22), 8951–8968. 10.1021/jacs.9b02822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. A.; McLellan L.; Stockley M.; Gibson K. R.; Whitlock G. A.; Knights C.; Harrigan J. A.; Jacq X.; Tate E. W. Quantitative Chemical Proteomic Profiling of Ubiquitin Specific Proteases in Intact Cancer Cells. ACS Chem. Biol. 2016, 11 (12), 3268–3272. 10.1021/acschembio.6b00766. [DOI] [PubMed] [Google Scholar]
- Jones A.; Kemp M. I.; Stockley M. L.; Gibson K. R.; Whitlock G. A.; Madin A.. Novel Compounds. Internation patent WO 2016046530 A1, 2016.
- Falgueyret J. P.; Oballa R. M.; Okamoto O.; Wesolowski G.; Aubin Y.; Rydzewski R. M.; Prasit P.; Riendeau D.; Rodan S. B.; Percival M. D. Novel, nonpeptidic cyanamides as potent and reversible inhibitors of human cathepsins K and L. J. Med. Chem. 2001, 44 (1), 94–104. 10.1021/jm0003440. [DOI] [PubMed] [Google Scholar]
- Ub-Rho-morpholine is an improved version of the classically used Ub-Rho110Gly and releases the brighter Rh110-morpholinecarbonyl species upon enzymatic cleavage. See also; Lavis L. D.; Chao T. Y.; Raines R. T. Fluorogenic label for biomolecular imaging. ACS Chem. Biol. 2006, 1 (4), 252–260. 10.1021/cb600132m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland R. A.; Basavapathruni A.; Moyer M.; Scott M. P. Impact of enzyme concentration and residence time on apparent activity recovery in jump dilution analysis. Anal. Biochem. 2011, 416 (2), 206–210. 10.1016/j.ab.2011.05.029. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Lashuel H. A.; Choi S.; Xing X.; Case A.; Ni J.; Yeh L. A.; Cuny G. D.; Stein R. L.; Lansbury P. T. Jr. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem. Biol. 2003, 10 (9), 837–846. 10.1016/j.chembiol.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Ekkebus R.; van Kasteren S. I.; Kulathu Y.; Scholten A.; Berlin I.; Geurink P. P.; de Jong A.; Goerdayal S.; Neefjes J.; Heck A. J.; Komander D.; Ovaa H. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J. Am. Chem. Soc. 2013, 135 (8), 2867–2870. 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjonaj L.; Sapmaz A.; Gonzalez-Prieto R.; Vertegaal A. C. O.; Flierman D.; Ovaa H. USP7: combining tools towards selectivity. Chem. Commun. 2019, 55 (35), 5075–5078. 10.1039/C9CC00969H. [DOI] [PubMed] [Google Scholar]
- Altun M.; Kramer H. B.; Willems L. I.; McDermott J. L.; Leach C. A.; Goldenberg S. J.; Kumar K. G. S.; Konietzny R.; Fischer R.; Kogan E.; Mackeen M. M.; McGouran J.; Khoronenkova S. V.; Parsons J. L.; Dianov G. L.; Nicholson B.; Kessler B. M. Activity-Based Chemical Proteomics Accelerates Inhibitor Development for Deubiquitylating Enzymes. Chem. Biol. 2011, 18 (11), 1401–1412. 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Cravatt B. F.; Wright A. T.; Kozarich J. W. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 2008, 77, 383–414. 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Verdoes M.; Hillaert U.; Florea B. I.; Sae-Heng M.; Risseeuw M. D. P.; Filippov D. V.; van der Marel G. A.; Overkleeft H. S. Acetylene functionalized BODIPY dyes and their application in the synthesis of activity based proteasome probes. Bioorg. Med. Chem. Lett. 2007, 17 (22), 6169–6171. 10.1016/j.bmcl.2007.09.025. [DOI] [PubMed] [Google Scholar]
- Butkevich A. N.; Mitronova G. Y.; Sidenstein S. C.; Klocke J. L.; Kamin D.; Meineke D. N. H.; D’Este E.; Kraemer P. T.; Danzl J. G.; Belov V. N.; Hell S. W. Fluorescent Rhodamines and Fluorogenic Carbopyronines for Super-Resolution STED Microscopy in Living Cells. Angew. Chem., Int. Ed. 2016, 55 (10), 3290–3294. 10.1002/anie.201511018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul P. J.; Åkesson L.; Wiking M.; Mahdessian D.; Geladaki A.; Ait Blal H.; Alm T.; Asplund A.; Björk L.; Breckels L. M.; Bäckström A.; Danielsson F.; Fagerberg L.; Fall J.; Gatto L.; Gnann C.; Hober S.; Hjelmare M.; Johansson F.; Lee S.; Lindskog C.; Mulder J.; Mulvey C. M.; Nilsson P.; Oksvold P.; Rockberg J.; Schutten R.; Schwenk J. M.; Sivertsson Å.; Sjöstedt E.; Skogs M.; Stadler C.; Sullivan D. P.; Tegel H.; Winsnes C.; Zhang C.; Zwahlen M.; Mardinoglu A.; Pontén F.; von Feilitzen K.; Lilley K. S.; Uhlén M.; Lundberg E. A subcellular map of the human proteome. Science 2017, 356, eaal3321 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- Meier J. L.; Mercer A. C.; Rivera H.; Burkart M. D. Synthesis and evaluation of bioorthogonal pantetheine analogues for in vivo protein modification. J. Am. Chem. Soc. 2006, 128 (37), 12174–12184. 10.1021/ja063217n. [DOI] [PubMed] [Google Scholar]
- Joshi A.; Dai L.; Liu Y.; Lee J.; Ghahhari N. M.; Segala G.; Beebe K.; Jenkins L. M.; Lyons G. C.; Bernasconi L.; Tsai F. T. F.; Agard D. A.; Neckers L.; Picard D. The mitochondrial HSP90 paralog TRAP1 forms an OXPHOS-regulated tetramer and is involved in mitochondrial metabolic homeostasis. BMC Biol. 2020, 18 (1), 10. 10.1186/s12915-020-0740-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler B. M.; Edelmann M. J. PTMs in conversation: activity and function of deubiquitinating enzymes regulated via post-translational modifications. Cell Biochem. Biophys. 2011, 60 (1–2), 21–38. 10.1007/s12013-011-9176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J.; Liu S.; Cui C.; Ten Dijke P. Invasive Behavior of Human Breast Cancer Cells in Embryonic Zebrafish. J. Visualized Exp. 2017, (122), 55459. 10.3791/55459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billia F.; Hauck L.; Grothe D.; Konecny F.; Rao V.; Kim R. H.; Mak T. W. Parkinson-susceptibility gene DJ-1/PARK7 protects the murine heart from oxidative damage in vivo. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (15), 6085–6090. 10.1073/pnas.1303444110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son O. L.; Kim H. T.; Ji M. H.; Yoo K. W.; Rhee M.; Kim C. H. Cloning and expression analysis of a Parkinson’s disease gene, uch-L1, and its promoter in zebrafish. Biochem. Biophys. Res. Commun. 2003, 312 (3), 601–607. 10.1016/j.bbrc.2003.10.163. [DOI] [PubMed] [Google Scholar]
- Hewings D. S.; Flygare J. A.; Bogyo M.; Wertz I. E. Activity-based probes for the ubiquitin conjugation-deconjugation machinery: new chemistries, new tools, and new insights. FEBS J. 2017, 284 (10), 1555–1576. 10.1111/febs.14039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A.; Avvakumov G.; Tong J. F.; Fan Y. H.; Zhao Y. L.; Alberts P.; Persaud A.; Walker J. R.; Neculai A. M.; Neculai D.; Vorobyov A.; Garg P.; Beatty L.; Chan P. K.; Juang Y. C.; Landry M. C.; Yeh C.; Zeqiraj E.; Karamboulas K.; Allali-Hassani A.; Vedadi M.; Tyers M.; Moffat J.; Sicheri F.; Pelletier L.; Durocher D.; Raught B.; Rotin D.; Yang J. H.; Moran M. F.; Dhe-Paganon S.; Sidhu S. S. A Strategy for Modulation of Enzymes in the Ubiquitin System. Science 2013, 339 (6119), 590–595. 10.1126/science.1230161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjonaj L.; Sapmaz A.; Flierman D.; Janssen G. M. C.; van Veelen P. A.; Ovaa H. Development of a DUB-selective fluorogenic substrate. Chem. Sci. 2019, 10 (44), 10290–10296. 10.1039/C9SC02226K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui W. J.; Ott C. A.; Yang K.; Chung J. S.; Shen S. Q.; Zhuang Z. H. Cell-Permeable Activity-Based Ubiquitin Probes Enable Intracellular Profiling of Human Deubiquitinases. J. Am. Chem. Soc. 2018, 140 (39), 12424–12433. 10.1021/jacs.8b05147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanman L. E.; Bogyo M. Activity-Based Profiling of Proteases. Annu. Rev. Biochem. 2014, 83, 249–273. 10.1146/annurev-biochem-060713-035352. [DOI] [PubMed] [Google Scholar]
- Verdoes M.; Verhelst S. H. L. Detection of protease activity in cells and animals. Biochim. Biophys. Acta, Proteins Proteomics 2016, 1864 (1), 130–142. 10.1016/j.bbapap.2015.04.029. [DOI] [PubMed] [Google Scholar]
- Krabill A. D.; Chen H.; Hussain S.; Feng C.; Abdullah A.; Das C.; Aryal U. K.; Post C. B.; Wendt M. K.; Galardy P. J.; Flaherty D. P. Biochemical and cellular characterization of a cyanopyrrolidine covalent Ubiquitin C-terminal hydrolase L1 inhibitor. ChemBioChem 2020, 21 (5), 712–722. 10.1002/cbic.201900434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; González-Prieto R.; Zhang M.; Geurink P. P.; Kooij R.; Iyengar P. V.; van Dinther M.; Bos E.; Zhang X.; Le Dévédec S. E.; van de Water B.; Koning R. I.; Zhu H. J.; Mesker W. E.; Vertegaal A. C. O.; Ovaa H.; Zhang L.; Martens J. W. M.; Ten Dijke P. Deubiquitinase Activity Profiling Identifies UCHL1 as a Candidate Oncoprotein That Promotes TGFβ-Induced Breast Cancer Metastasis. Clin. Cancer Res. 2020, 26 (6), 1460–1473. 10.1158/1078-0432.CCR-19-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyain N.; Godinat A.; Lanyon-Hogg T.; Lachiondo-Ortega S.; Will E. J.; Soudy C.; Mondal M.; Mason K.; Elkhalifa S.; Smith L.; Harrigan J. A.; Tate E. W. Discovery of a potent and selective covalent inhibitor and activity-based probe for the deubiquitylating enzyme UCHL1, with anti-fibrotic activity. J. Am. Chem. Soc. 2020, 142, 12020–12026. 10.1021/jacs.0c04527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B.; Busse D.; Li N.; Dittmar G.; Schuchhardt J.; Wolf J.; Chen W.; Selbach M. Global quantification of mammalian gene expression control. Nature 2011, 473 (7347), 337–342. 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.