Vibrio natriegens is a naturally occurring marine bacterium that is emerging as a microbiological model system. Here, we describe Aquatic Killer 99 (AQKL99), a novel phage that infects Vibrio natriegens 14048. The genome of the phage is 58,464 bp long, has a GC content of 45.9%, and contains 51 protein-coding genes.
ABSTRACT
Vibrio natriegens is a naturally occurring marine bacterium that is emerging as a microbiological model system. Here, we describe Aquatic Killer 99 (AQKL99), a novel phage that infects Vibrio natriegens 14048. The genome of the phage is 58,464 bp long, has a GC content of 45.9%, and contains 51 protein-coding genes.
ANNOUNCEMENT
Vibrio spp. are Gram-negative proteobacteria that inhabit saline and estuarine environments; however, several species are human pathogens (1). The characterization of phages infecting Vibrio spp. is of broad interest for the fields of ecology, evolution, human health, and biotechnology (2). Vibrio natriegens is emerging as a microbiological model system due to its rapid doubling time (<10 min) under standard laboratory conditions and the availability of tools for genetic manipulation (3, 4). Here, we describe the isolation and genome annotation of Aquatic Killer 99 (AQKL99), a novel phage that infects Vibrio natriegens 14048.
In June 2019, 3 liters of surface seawater from Bayboro Harbor (Tampa Bay, FL, USA) was prefiltered through a 0.8-μm filter, and viruses were concentrated using iron chloride flocculation (5). A double-layer plaque assay on Vibrio natriegens 14048 on tryptic soy agar with 10 g per L sodium chloride at 25°C led to the isolation of a novel phage, which was subsequently plaque purified three times (6). Phage lysates were adsorbed to a copper grid, negatively stained with 2% (wt/vol) uranyl acetate (7), and viewed on a Hitachi 7100 transmission electron microscope equipped with a Gatan Orius high-resolution digital camera. The morphology of phage AQKL99 is consistent with that of members of the family Siphoviridae, with an average capsid diameter of 74 ± 18 nm and a long, flexible tail averaging 131 ± 38 nm, based on pixel measurements relative to the scale bar of 40 phage particles (Fig. 1). Phage AQKL99 was unable to produce plaques on 53 Vibrio colonies isolated from the same seawater sample on thiosulfate-citrate-bile salts-sucrose (TCBS) agar, suggesting a narrow host range. However, neither the identity nor the uniqueness of these Vibrio isolates was determined.
FIG 1.
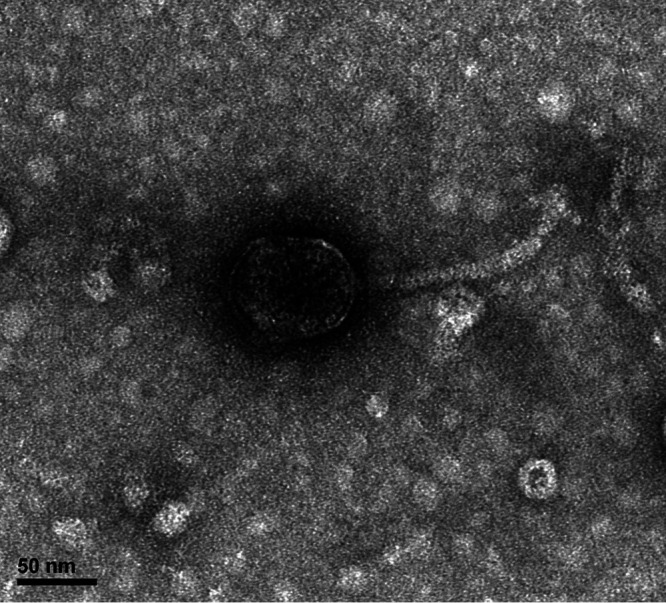
Transmission electron micrograph of Vibrio phage vB_VnaS-AQKL99.
Phage DNA was isolated from a 0.2-μm-filtered overnight lysate with the Qiagen MinElute virus spin kit and was sent to the Microbial Genome Sequencing (MiGS) Center for library preparation with the Illumina Nextera kit and sequencing on the NextSeq 550 platform. Sequences were processed using FastQC v0.11.5 (8) with default parameters, assembled using SPAdes v3.11.1 (9), and evaluated using QUAST v5.0.1 (10) to reveal a single contig with ∼2,900× coverage. This contig, which was presumed to represent the complete linear genome of phage AQKL99, is 58,464 bp long with a GC content of 45.9%. As determined with Geneious Prime v2020.1.2 (11), phage AQKL99 shares 75% genome-wide pairwise identity with Vibrio phage VhaS-tm (GenBank accession number KX198614), which was isolated from oyster tissue and can prevent Vibrio harveyi from causing disease in greenlip abalone (Haliotis laevigata) (12).
Open reading frames (ORFs) were predicted through Geneious Prime v2020.1.2 using default parameters, which resulted in the identification and annotation of 51 protein-coding genes, mostly transcribed in the reverse direction (11). Amino acid sequence similarity searches were performed against the nonredundant database using BLASTp with default parameters and an E value cutoff value of <0.001 (13). Approximately 75% of the genes are of unknown function; however, the phage AQKL99 genome encodes several putative structural proteins (major capsid protein, minor head protein, portal protein, tail assembly protein, tail length tape measure protein, and large and small terminase subunits), DNA replication proteins (DNA helicase, DNA polymerase, DNA replicative clamp, and primase), and other notable proteins (autolysin N-acetylmuramoyl-l-alanine amidase, AAA family ATPase, and multimodular transpeptidase-transglycosylase). Interestingly, the phage AQKL99 genome encodes three proteins putatively involved in deazaguanine DNA modification (QueC, QueD, and QueD), which is a common nucleotide substitution used to protect phage DNA from host restriction enzymes (14).
Data availability.
The genome sequence and associated metadata for Vibrio phage vB_VnaS-AQKL99 are available in GenBank under the accession number MT795651.1. Raw reads are available in the SRA database under the accession number SRX9058260.
ACKNOWLEDGMENTS
This work was supported by funding to M.B. from the National Science Foundation (award OCE-1722761). N.Y. was part of the 2020 National Summer Undergraduate Research Project (NSURP). K.M. was supported by a National Science Foundation Graduate Research Fellowship (award 3900101301).
REFERENCES
- 1.Turner JW, Good B, Cole D, Lipp EK. 2009. Plankton composition and environmental factors contribute to Vibrio seasonality. ISME J 3:1082–1092. doi: 10.1038/ismej.2009.50. [DOI] [PubMed] [Google Scholar]
- 2.Comeau A, Chan A, Suttle C. 2006. Genetic richness of vibriophages isolated in a coastal environment. Environ Microbiol 8:1164–1176. doi: 10.1111/j.1462-2920.2006.01006.x. [DOI] [PubMed] [Google Scholar]
- 3.Weinstock MT, Hesek ED, Wilson CM, Gibson DG. 2016. Vibrio natriegens as a fast-growing host for molecular biology. Nat Methods 13:849–851. doi: 10.1038/nmeth.3970. [DOI] [PubMed] [Google Scholar]
- 4.Dalia TN, Hayes CA, Stolyar S, Marx CJ, McKinlay JB, Dalia AB. 2017. Multiplex genome editing by natural transformation (MuGENT) for synthetic biology in Vibrio natriegens. ACS Synth Biol 6:1650–1655. doi: 10.1021/acssynbio.7b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.John S, Mendez C, Deng L, Poulos B, Kauffman A, Kern S, Brum J, Polz M, Boyle E, Sullivan M. 2011. A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ Microbiol Rep 3:195–202. doi: 10.1111/j.1758-2229.2010.00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. 2009. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol Biol 501:69–76. doi: 10.1007/978-1-60327-164-6_7. [DOI] [PubMed] [Google Scholar]
- 7.Ackermann H-W. 2009. Basic phage electron microscopy. Methods Mol Biol 501:113–126. doi: 10.1007/978-1-60327-164-6_12. [DOI] [PubMed] [Google Scholar]
- 8.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Institute, Cambridge, United Kingdom: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. [Google Scholar]
- 9.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Barton M, Elliott L, Li X, Abraham S, O'Dea M, Munro J. 2017. Bacteriophage therapy for the control of Vibrio harveyi in greenlip abalone (Haliotis laevigata). Aquaculture 473:251–258. doi: 10.1016/j.aquaculture.2017.01.003. [DOI] [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Hutinet G, Kot W, Cui L, Hillebrand R, Balamkundu S, Gnanakalai S, Neelakandan R, Carstens AB, Fa Lui C, Tremblay D, Jacobs-Sera D, Sassanfar M, Lee Y-J, Weigele P, Moineau S, Hatfull GF, Dedon PC, Hansen LH, de Crécy-Lagard V. 2019. 7-Deazaguanine modifications protect phage DNA from host restriction systems. Nat Commun 10:5442. doi: 10.1038/s41467-019-13384-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence and associated metadata for Vibrio phage vB_VnaS-AQKL99 are available in GenBank under the accession number MT795651.1. Raw reads are available in the SRA database under the accession number SRX9058260.


