Abstract
The current coronavirus (COVID-19) pandemic is exacerbated by the absence of effective therapeutic agents. Notably, patients with COVID-19 and comorbidities such as hypertension and cardiac diseases have a higher mortality rate. An efficient strategy in response to this issue is repurposing drugs with antiviral activity for therapeutic effect. Digoxin (DIG) and ouabain (OUA) are FDA drugs for heart diseases that have antiviral activity against several coronaviruses. Thus, we aimed to assess antiviral activity of DIG and OUA against SARS-CoV-2 infection. The half-maximal inhibitory concentrations (IC50) of DIG and OUA were determined at a nanomolar concentration. Progeny virus titers of single-dose treatment of DIG, OUA and remdesivir were approximately 103-, 104- and 103-fold lower (> 99% inhibition), respectively, than that of non-treated control or chloroquine at 48 h post-infection (hpi). Furthermore, therapeutic treatment with DIG and OUA inhibited over 99% of SARS-CoV-2 replication, leading to viral inhibition at the post entry stage of the viral life cycle. Collectively, these results suggest that DIG and OUA may be an alternative treatment for COVID-19, with potential additional therapeutic effects for patients with cardiovascular disease.
Subject terms: Cell biology, Drug discovery, Immunology, Microbiology, Diseases
Introduction
Human coronaviruses (HCoV) are enveloped, positive-sense single-stranded RNA viruses. Six coronaviruses, namely HCoV-229E, HCoV-OC43, HCoV-HKU1, HCoV-NL63, severe acute respiratory syndrome coronavirus (SARS-HCoV) and Middle East respiratory syndrome coronavirus (MERS-HCoV) are responsible for respiratory illnesses in human. Of these, SARS-CoV and MERS-CoV infection results in severe acute respiratory disease and have caused worldwide awareness of these illnesses1.
In December 2019, a novel coronavirus was identified in a group of patients with pneumonia in Wuhan, Hubei province, China2. Subsequently, the International Committee on Taxonomy of Viruses (ICTV) named this virus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the causative agent of coronavirus disease 19 (COVID-19)3. Owing to the substantial increase in the number of confirmed cases of COVID-19 through human-to-human transmission, a large epidemic occurred in Wuhan4. As SARS-CoV-2 spread to more than 216 countries, the World Health Organization (WHO) declared an ongoing pandemic on March 11 20205. As of 5 June 2020, more than 6 million confirmed cases and 387,298 deaths have been reported worldwide6.
The common symptoms of patients with COVID-19 are mild and most are asymptomatic7–10. However, some cases have severe pneumonia, acute cardiac injury, multi-organ failure and death4,10–12. Notably, patients with COVID-19 and comorbidities such as hypertension and cardiac diseases show a higher mortality rate than patients without cardiac diseases (51.2% vs. 5.1%)7,8,11,13,14.
No approved vaccine or specific antiviral agent is yet available for COVID-19. One of the fastest and most practical strategies to address this issue is to identify preexisting approved drugs with antiviral activity against SARS-CoV-2. Accordingly, numerous clinical trials and studies are underway to evaluate vaccines and therapeutics, with the most well-studied clinical trial-repurposed drugs being (hydroxy)chloroquine and remdesivir (REM), the latter of which received emergency use authorization by the U.S. Food and Drug Administration (FDA) on May 1, 202015,16. Moreover, the WHO announced a large global clinical trial, SOLIDARITY, to assess the therapeutic effects of four repurposed drugs on March 18, 2020.
In addition to antiviral drugs based on viral protease inhibitors and nucleoside analogs, the cardiac glycoside (CG)-based drugs digoxin (DIG) and ouabain (OUA) have been shown to exhibit antiviral activity through various mechanisms against several DNA and RNA viruses, such as cytomegalovirus, herpes simplex virus, MERS-CoV, human immunodeficiency virus, respiratory syncytial virus, chikungunya virus, and the recently identified SARS-CoV-217–24.
Notably, these agents have been used to treat various heart diseases and were mainly identified to bind to the transmembrane protein sodium/potassium ATPase (Na+/K+-ATPase) and inhibit ion-exchange, leading to increased intracellular Ca++ concentration and heart muscle contraction25–27. Therefore, in this study, we evaluated the antiviral activity of DIG and OUA based on viral growth kinetics and inhibition at different stages of viral infection, and compared it to that of vehicle (DMSO), chloroquine (CHQ) and REM to identify a suitable and potent antiviral agent to treat COVID-19 patients with cardiac diseases.
Results
Determination of half maximal inhibitory, cell cytotoxic concentration and selective index value of DIG and OUA
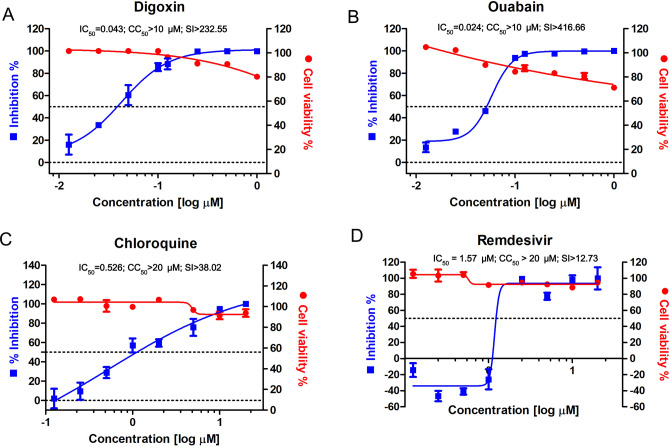
To assess the antiviral activity and cell cytotoxicity of DIG and OUA against SARS-CoV-2 infection, we investigated their half-maximal inhibitory concentration (IC50), cytotoxicity concentration 50% (CC50) and selective index (SI), and compared to those of CHQ. Vero cells were infected with BetaCoV/Korea/KCDC03/2020 at a multiplicity of infection (MOI) of 0.01 in the presence of OUA and DIG (0.0125, 0.025, 0.05, 0.1, 0.125, 0.25, 0.5 and 1 μM), CHQ (0.125, 0.25, 0.5, 1, 2, 5, 10 and 20 μM) and REM (0.125, 0.25, 0.5, 1, 2.5, 5, 10 and 20 μM) for 1 h and incubated with the respective drug for 24 h. The viral copy numbers in the cell culture supernatant were determined by amplifying the nucleocapsid (N) gene by quantitative real-time PCR (qRT-PCR), and cell viability was measured using the PrestoBlue Cell Viability reagent. The IC50 values of DIG (IC50 = 0.043 μM) and OUA (IC50 = 0.024 μM) were determined at a nanomolar concentration, and were over tenfold lower than those of CHQ (IC50 = 0.526 μM), and REM (IC50 = 1.57 μM) (Fig. 1A–D). In addition, the SI (CC50/IC50) values of DIG (CC50 > 10 μM, SI > 232.55) and OUA (CC50 > 10 μM, SI > 416.66) were over fivefold greater than those of either CHQ (CC50 > 20 μM, SI > 38.02) or REM (CC50 > 20 μM, SI > 12.73) (Fig. 1A–D).
Figure 1.
Half maximal inhibitory concentration (IC50), cytotoxic concentration (CC50) and selective index (SI) of digoxin, ouabain, chloroquine and remdesivir against SARS- CoV-2 infection. IC50 (left axis, blue square), CC50 (right axis, red circle), and SI of (A) digoxin (DIG), (B) ouabain (OUA), (C) chloroquine (CHQ) and (D) remdesivir (REM). Vero cells were infected with BetaCoV/Korea/KCDC03/2020 at an MOI of 0.01 in the presence of the indicated drug concentration for 1 h. Subsequently, the cells were washed and incubated in the presence of the indicated drug concentration for 24 h. IC50 and CC50 were determined from dose response curves based on treatment with eight concentrations. IC50 were determined by viral copy number based on standard curve, and CC50 was investigated by cell viability assay. Viral copy number in DMSO was set to 100%, and the remaining values as means ± SD (n = 3). Corresponding viral mRNA expression levels are shown in Supplementary Figure S1.
Antiviral activity of DIG and OUA based on SARS-CoV-2 growth kinetics
To evaluate antiviral activity of drugs based on SARS-CoV-2 growth kinetics, cells were treated with optimal concentrations of DIG (150 nM), OUA (100 nM), CHQ (10 μM) and REM (10 μM), which inhibited over 95% of viral N mRNA expression at 24 hpi (Supplementary Fig. S1A–E).
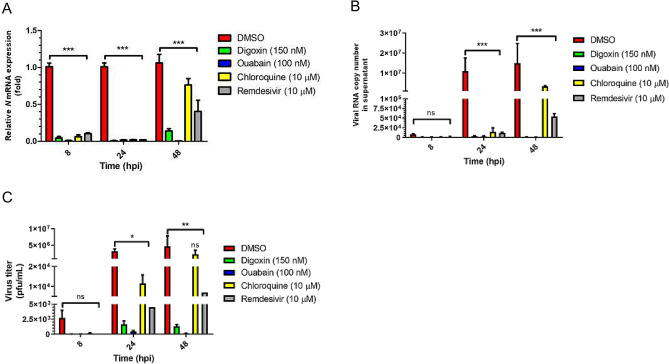
After treating the virus-infected cells with drugs, viral N mRNA expression, viral copy number, and progeny virus titer were measured at 8, 24 and 48 h (the duration of the complete virus replication was assessed via growth kinetics) (Fig. 2A–C). Viral N mRNA expression was almost fully suppressed (> 99%) by all drugs at 8 and 24 hpi. However, the expression of viral N mRNA was significantly restored in CHQ-treated cells at 48 hpi (Fig. 2A). Viral copy numbers were also markedly reduced at 48 h in the DIG-, OUA-, and REM-treated cell culture supernatants (Fig. 2B). Progeny virus titer in the culture medium of DIG, OUA and REM treatment, as measured using plaque assays, revealed a 103- and 104-fold reduction, respectively, and an inhibition ratio of > 99% for the drugs compared to those for the carrier (DMSO, 1.80 × 106 plaque forming units (pfu)/mL). Moreover, the virus titer was considerably reduced in the cells treated with DIG (1.83 × 103 pfu/mL), OUA (1.80 × 102 pfu/mL) and REM (7.0 × 103 pfu/mL), but not CHQ (2.45 × 106 pfu/mL) at 48 hpi (Fig. 2C, Table 1 and Supplementary Fig. S2).
Figure 2.
Antiviral activity of digoxin, ouabain, chloroquine and remdesivir on SARS-CoV-2 growth kinetics. Vero cells were infected with BetaCoV/Korea/KCDC03/2020 at an MOI of 0.01 in the presence of DIG (150 nM), OUA (100 nM), CHQ (10 μM) and REM (10 μM) for 1 h. Subsequently, the cells were washed and incubated in the presence of the indicated drug for 8, 24, and 48 hpi. At each time point, (A) viral mRNA expression, (B) viral copy number and (C) progeny titer were assessed using qRT-PCR and plaque assays. Viral mRNA expression was normalized to GAPDH expression. DMSO was set to 1, and the remaining values are represented as a relative value. Viral copy number was calculated using a standard curve. Values are presented as mean ± SD (n = 3). Statistically significantly differences between DMSO and drug treatment are represented as *P < 0.05, **P < 0.01 and ***P < 0.001 determined using the two-way ANOVA with Bonferroni post-tests (each column compared to control). ns not significant.
Table 1.
Progeny virus titer in cell supernatant.
| Time (hpi) | Drug | Pfu/mL |
|---|---|---|
| 8 | DMSO | 3.07 × 103 |
| Digoxin | 1.67 × 101 | |
| Ouabain | 1.67 × 101 | |
| Chloroquine | 1.60 × 102 | |
| Remdesivir | 0.66 × 101 | |
| 24 | DMSO | 3.90 × 106 |
| Digoxin | 1.43 × 103 | |
| Ouabain | 6.53 × 102 | |
| Chloroquine | 1.37 × 104 | |
| Remdesivir | 4.50 × 103 | |
| 48 | DMSO | 1.80 × 106 |
| Digoxin | 1.83 × 103 | |
| Ouabain | 1.80 × 102 | |
| Chloroquine | 2.45 × 106 | |
| Remdesivir | 7.00 × 103 |
Determination of the inhibition step in the SARS-CoV-2 life cycle by drug treatment
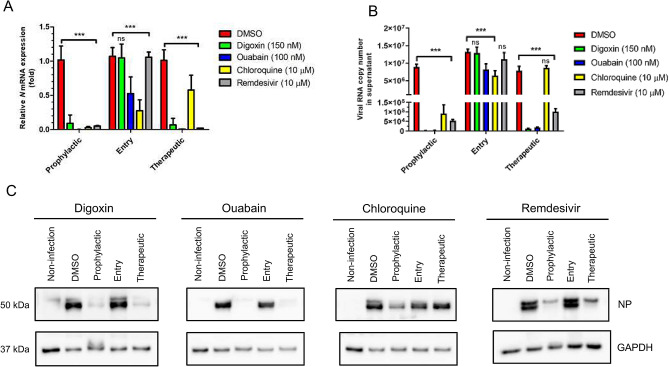
To determine which step of the virus life-cycle is inhibited by drug treatment, DIG, OUA, CHQ and REM were administered at the different time points of treatment: prophylactic (1 h prior to infection and maintenance for 24 h), entry (0 h of infection and maintenance for 2 h), and therapeutic (2 h following infection and maintenance for 24 h). All drugs demonstrated high efficacy upon prophylactic administration. Viral copy number, mRNA expression, and viral N protein expression were lower in prophylactic-treated cells than in non-treated cells by approximately 99% (Fig. 3). In entry-treated cells, CHQ and OUA treatment significantly inhibited viral RNA and protein levels to approximately 60% and 30% of those of DMSO, respectively, whereas DIG and REM treatment did not effectively inhibit virus propagation (Fig. 3A–C). In comparison, OUA, DIG, and REM, but not CHQ, treatment markedly reduced viral replication in the therapeutic-treated cells (Fig. 3A–C).
Figure 3.
Determination of the inhibition step of each drug in the SARS-CoV-2 life cycle. Vero cells were infected with BetaCoV/Korea/KCDC03/2020 at an MOI of 0.01 and treated with DIG (150 nM), OUA (100 nM), CHQ (10 μM) and REM (10 μM) in prophylactic, entry, and therapeutic conditions. Then, the cells were incubated with the indicated drug or without drug in fresh media for 24 h. Viral (A) mRNA expression, (B) copy number, and (C) N protein expression were investigated using qRT-PCR and western blotting (also see Supplementary Figure S3; full image of the blots). Viral mRNA expression was normalized to GAPDH levels and represented as relative values. Values are presented as mean ± SD (n = 3). Anti-GAPDH blots were used as loading controls. Viral NP protein and GAPDH protein were blotted in the same gel. Statistically significantly differences between DMSO and drug treatment are represented as **P < 0.01 and ***P < 0.001 determined using the two-way ANOVA with Bonferroni post-tests. ns not significant.
Discussion
SARS-CoV-2 infection causes not only multiple organ failure but also higher mortality rate in patients with underlying cardiac diseases4,28,29. Notably, the angiotensin-converting enzyme 2 (ACE2) receptor, which serves as a functional receptor for coronaviruses, is systemically distributed in multiple organs and is especially highly expressed in the heart and lungs29. Therefore, these organs may be directly attacked by SARS-CoV-2 or indirectly damaged by elevated levels of proinflammatory cytokines30–35.
The drugs DIG and OUA have been used to treat heart conditions of patients for over 10 decades, and thus their clinical dosage regimen, bioavailability, pharmacokinetic profile information, and safety are well known27. Hence, these drugs may exert multiple benefits in patients with COVID-19 in terms of antiviral and symptom management and safety. Although the IC50 in an in vitro study is a poor guide for clinically relevant concentrations of DIG and OUA, previous studies of cancer therapy with DIG using human cells reported IC50 ranges from 0.02 to 0.34 μM, and the correspond plasma concentrations were safe and in the acceptable range from 0.8 to 2.6 nM (1 ng/mL = 1.28 nmol/L) in patients with cardiac diseases27,39. Moreover, the serum levels of patients taking oral doses of 0.25 mg/day (3.4 to 5.1 μg/kg/day) DIG are in the range from 1 to 2.6 nM40,41. Therefore, the IC50 of DIG and OUA described in this study may be helpful for further pre-clinical and clinical studies.
In this study, single dose of DIG and OUA treatment consistently showed superior antiviral activity against human isolate BetaCoV/Korea/KCDC03/2020 infection, as evident from the evaluation of viral mRNA expression, copy number (released virions in cell supernatant), and progeny virus titer up to 48 hpi in vitro. The progeny virus titer at 48 hpi in the DIG and OUA treatment groups was comparable to that in REM and reduced more than 103–104-fold compared to either the non-treated vehicle (DMSO) or CHQ groups, indicating that DIG and OUA have effective antiviral activity with stability up to 48 hpi (this time point represents the peak viral titer on growth kinetics in Vero cells, corresponding to maximal SARS-CoV-2 replication)36. Moreover, DIG and OUA treatment significantly inhibited over 99% of viral mRNA expression, which is more effective than REM (> 60%) and CHQ (> 30%) at 48 hpi. These results suggest that DIG and OUA could be an alternative treatment against SARS-CoV-2 infection.
Notably, DIG and OUA significantly inhibited viral mRNA expression, copy number, and viral protein expression when administered at the post-entry stage, although DIG did not show effective antiviral activity at the host entry stage of the virus cycle. Clinically, these results are very important for therapeutics, as a large numbers of patients are asymptomatic at the initial stage of SARS-CoV-2 infection37,38. Moreover, the results indicate that the inhibition mechanism of SARS-CoV-2 by DIG may be similar to that of respiratory syncytial virus (RSV), wherein inhibition occurs at the step of viral RNA synthesis18. However, OUA may have another inhibition mechanism, as OUA treatment at the entry stage inhibited approximately 30% of viral mRNA and protein expression. This suggests that an OUA may have an alternative antiviral mechanism related to blocking Src-mediated endocytosis in the entry step of coronaviruses42. Interestingly, a recent study reported that digitoxin, a CG, suppresses proinflammatory cytokines in influenza A virus-infected cotton rat lung43, which suggests that DIG and OUA may have an additional therapeutic role against COVID-19 with hypercytokinemia.
Taken together, we demonstrated a more effective antiviral activity of DIG and OUA against SARS-CoV-2 infection in vitro than previously approved antiviral agents such as chloroquine and remdesivir. We propose that these agents may be used as therapeutic options for patients with COVID-19 and comorbid cardiac diseases.
Materials and methods
Virus, cells, compounds, and infection
BetaCoV/Korea/KCDC03/2020 (GISSAID accession ID: EPI_ISL_407193) was obtained from the National Culture Collection for Pathogens of Korea Center for Disease Control and prevention (KCDC)44. Vero cells were purchased from the American Type Culture Collection (ATCC CCL-81, Manassas, VA, USA) and cultured in Dulbecco’s minimal essential medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 2 (infection media) or 10% (growth media) v/v heat inactivated fetal bovine serum (FBS; Gibco) and 1% v/v penicillin/streptomycin (p/s; Gibco) in a humidified incubator of 5% CO2 atmosphere at 37 °C. OUA, CHQ and DIG were purchased from Sigma-Aldrich (St. Louis, MO, USA). REM was purchased from MedChemExpress (Monmouth Junction, NJ, USA). DIG tablets (inno.N, Seoul, Korea) were provided by Dr. Sang il Kim. OUA and DIG tablets were dissolved in H2O and CHQ, DIG and REM were dissolved in DMSO. Confluent Vero cells were infected at an MOI of 0.01 of BetaCoV/Korea/KCDC03/2020 in DMEM containing 2% FBS and 1% p/s for 1 h. Following incubation, cells were washed twice with phosphate buffered saline (PBS; Gibco) and incubated in the infection media. All experiments were performed in biosafety level 3 facilities according to KCDC guidelines.
Cloning and linearity determination of the RNA reference
The PCR products with SARS-CoV-2 N primers were cloned into pGEM-T Easy Vector (Promega, Madison, WI, USA) and in vitro transcribed using the RiboMAX Large Scale RNA Production System-T7 (Promega). The linearity of the RNA reference template was evaluated with a tenfold serial dilution of in vitro transcribed N RNA of SARS-CoV-2 (104–1010 copies). The copy number of RNA was calculated using: [X g/μL RNA/(transcript length in nucleotides – 340)] × 6.022 × 1023 = Y molecules/μL45. The linear range was determined using a standard curve generated with diluted reference RNAs and the best-fit line to the raw data was established by linear regression analysis with 95% confidence intervals using GraphPad Prism (version 5.01; La Jolla, CA, USA).
Quantitative real-time PCR (qRT-PCR)
For the quantification of viral copy number, total RNA was isolated from cell supernatants using the QIAamp viral RNA mini kit (Qiagen, Hilden, Germany) and cDNA was synthesized from 1 μg of total RNA using SuperScript IV (Invitrogen, Waltham, MA, USA) according to the manufacturer’s protocol. qRT-PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA) and an Applied Biosystems QuantStudio3 (Applied Biosystems) following the manufacturer’s protocol.
For measurement of viral mRNA, total RNA was isolated using TRIzol (Ambion, Leicestershire, UK) reagent. Subsequently, cDNA was synthesized from 1 μg of total RNA using a SuperScript IV (Invitrogen). qRT-PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems) and Applied Biosystems QuantStudio3 (Applied Biosystems) as follows: denaturation at 95 ℃ for 5 min, followed by 40 cycles of 95 ℃ for 30 s and 60 ℃ for 30 s. The sequences of the GAPDH primers used were: 5′-GAA CGG GAA GCT TGT CAT CAA TGG-3′ and 5′-TGT GGT CAT GAG TCC TTC CAC GAT-3′. The sequences of the N primers used were: 5′-GGG AGC CTT GAA TAC ACC AAA A-3′ and 5′-TGT AGC ACG ATT GCA GCA TTG-3′46.
Western blotting
Vero cells were lysed in Pierce RIPA buffer (Thermo Scientific, Waltham, MA, USA) and 20 μg of protein was separated using Bolt 4–12% Bis–Tris Plus (Invitrogen). Proteins were detected by probing the membranes with 1:1000 anti-SARS-CoV-2 nucleocapsid (Sino Bio., Beijing, China) and 1:2000 anti-GAPDH (Cell Signaling Technologies, Danvers, MA, USA) antibodies. Protein transfer was performed using iBlot 2 (Invitrogen) and iBlot 2 PVDF Regular Stacks (Invitrogen). Membranes were incubated with 1:2000 goat anti-rabbit antibody (Cell Signaling) conjugated with horseradish peroxidase for 1 h. Then, membranes were washed five times with tris-buffered saline with 5% Tween 20. Thereafter, the blots were detected using Super Signal (Thermo Scientific).
Plaque assay
Monolayers of Vero cells were prepared in 12-well plates. The cells were infected with tenfold serial dilutions of supernatant from treated cells and incubated at 37 °C for 1 h. The medium was removed and cells were washed with PBS. Each well was overlaid with MEM/agarose (Gibco) and maintained at room temperature until the overlay turned solid. The plates were incubated at 37 °C for 3 days. The cells were then fixed with 2% paraformaldehyde (Thermo Scientific) and stained with 1% crystal violet (Sigma-Aldrich) overnight.
IC50 and CC50 measurements
Confluent Vero cells in 12-well plates were pre-treated with eight concentrations of OUA and DIG (0.0125, 0.025, 0.05, 0.1, 0.125, 0.25, 0.5 and 1 μM), CHQ (0.125, 0.25, 0.5, 1, 2, 5, 10 and 20 μM) and REM (0.125, 0.25, 0.5, 1, 2.5, 5, 10 and 20 μM) for 1 h in DMEM containing 2% FBS and 1% p/s. After incubation, an MOI of 0.01 of BetaCoV/Korea/KCDC03/2020 was added to cells for 1 h, cells were washed twice with PBS, and a new medium was added with the indicated concentrations of the drugs for 24 h. Subsequently, cell viability was measured using the PrestoBlue Cell Viability reagent (Invitrogen), and viral RNA was isolated from cell supernatants and cDNA was synthesized. qRT-PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems) and Applied Biosystems QuantStudio3. The copy number was calculated based on the reference RNA template and the IC50 value was calculated using GraphPad Prism.
Drug treatment
Vero cells were pre-treated with DIG (150 nM), OUA (100 nM), CHQ (10 μM) and REM (10 μM) for 1 h, and then the virus was applied for 1 h to allow infection. The drug-virus mixture was removed and the cells were washed twice with PBS. Subsequently, the cells were incubated in the presence of fresh medium containing the optimal concentrations of the drugs, and the cells and supernatant were collected at 8, 24 and 48 hpi for quantification of viral mRNA, copy number, and progeny virus titer.
For the prophylactic condition, Vero cells were pre-treated with the drugs in infection medium for 1 h and the virus was applied for 2 h to allow infection. Subsequently, the cells were washed twice with PBS and incubated for 24 h in the presence of the drugs in fresh infection medium.
For the entry condition, the cells were treated with the drugs for the infection period (2 h), followed by removal of the drug-virus mixture and washing of the cells. Subsequently, the cells were incubated in infection medium without the drugs for 24 h.
For the therapeutic condition, following viral infection without the drugs for 2 h, the virus was discarded and the cells were washed. Then, drugs in fresh infection medium were added to the cells for 24 h.
Statistical analysis
The statistical significance of DMSO control and drug treatments were assessed by one-way ANOVA with Dunnett’s multiple comparison test. The statistical comparison of the viral copy number, mRNA expression, and progeny virus titer was performed using two-way ANOVA with Bonferroni post-tests. Data plotting and statistical analysis were performed using GraphPad Prism. A P value < 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
This research was supported by the Korea National Institute of Health fund (2019-NI-070-01).
Author contributions
J.C., J.H.C., and B.S.C. designed the experiments. J.C., and J.H.C. wrote the manuscript. J.C., Y.J.L., and J.H.K. conducted many of experiments (J.C. performed virus infection, qPCR, and western blotting; Y.J.L. conducted plaque assay; J.H.K. performed qRT-PCR and analysis data of virus copy number). S.I.K. provided digoxin tablet and S.I.K. and S.S.K. advised clinical and pharmacological aspect of the drugs. J.H.C. and B.S.C. supervised the experiments and revised the manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Byeong-Sun Choi, Email: byeongsun@Korea.kr.
Jang-Hoon Choi, Email: jhchoi@nih.go.kr.
Supplementary information
is available for this paper at 10.1038/s41598-020-72879-7.
References
- 1.Su S, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ICTV. Naming the 2019 Coronavirus. https://talk.ictvonline.org/ (2020).
- 4.Hui DS, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization (WHO). WHO Director-General's Opening Remarks at the Media Briefing on COVID-19. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-11-march-2020 (2020).
- 6.World Health Organization (WHO). Coronavirus Disease (COVID-19) Outbreak Situation. https://www.who.int/emergencies/diseases/novel-coronavirus-2019?gclid=Cj0KCQjwoPL2BRDxARIsAEMm9y_2o8RtKOqhgZuzXq1021Xw8yuLIV7iopd9crZU1zVmv73jUkTemN8aArjQEALw_wcB (2020).
- 7.Shi, S. et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. E1–E8 (2020). [DOI] [PMC free article] [PubMed]
- 8.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention (CDC). U. Symptoms of Coronavirus. https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html (2020).
- 11.Chen N, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Day M. Covid-19: Four fifths of cases are asymptomatic, China figures indicate. BMJ. 2020;369:m1375. doi: 10.1136/bmj.m1375. [DOI] [PubMed] [Google Scholar]
- 13.Madjid, M., Safavi-Naeini, P., Solomon, S. D. & Vardeny, O. Potential effects of coronaviruses on the cardiovascular system: A review. JAMA Cardiol. E1–E10 (2020). [DOI] [PubMed]
- 14.Yang X, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhama, K. et al. COVID-19, an emerging coronavirus infection: Advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum. Vaccin. Immunother. 1–7, 10.1080/21645515.2020.1735227 (2020). [DOI] [PMC free article] [PubMed]
- 16.Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A review. JAMA. 2020;323:1824–1836. doi: 10.1001/jama.2019.20153. [DOI] [PubMed] [Google Scholar]
- 17.Amarelle, L. & Lecuona, E. The antiviral effects of Na, K-ATPase inhibition: A minireview. Int. J. Mol. Sci.19, 10.3390/ijms19082154 (2018). [DOI] [PMC free article] [PubMed]
- 18.Norris MJ, et al. Targeting intracellular ion homeostasis for the control of respiratory syncytial virus. Am. J. Respir. Cell Mol. Biol. 2018;59:733–744. doi: 10.1165/rcmb.2017-0345OC. [DOI] [PubMed] [Google Scholar]
- 19.Dodson AW, Taylor TJ, Knipe DM, Coen DM. Inhibitors of the sodium potassium ATPase that impair herpes simplex virus replication identified via a chemical screening approach. Virology. 2007;366:340–348. doi: 10.1016/j.virol.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganesan VK, Duan B, Reid SP. Chikungunya virus: Pathophysiology, mechanism, and modeling. Viruses. 2017;9:368. doi: 10.3390/v9120368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapoor A, et al. Human cytomegalovirus inhibition by cardiac glycosides: Evidence for involvement of the HERG gene. Antimicrob. Agents Chemother. 2012;56:4891–4899. doi: 10.1128/AAC.00898-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laird GM, Eisele EE, Rabi SA, Nikolaeva D, Siliciano RF. A novel cell-based high-throughput screen for inhibitors of HIV-1 gene expression and budding identifies the cardiac glycosides. J. Antimicrob. Chemother. 2014;69:988–994. doi: 10.1093/jac/dkt471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C-W, Chang H-Y, Lee Y-Z, Hsu H-Y, Lee S-J. The cardenolide ouabain suppresses coronaviral replication via augmenting a Na+/K+-ATPase-dependent PI3K_PDK1 axis signaling. Toxicol. Appl. Pharmacol. 2018;356:90–97. doi: 10.1016/j.taap.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeon S, et al. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020 doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dvela M, Rosen H, Feldmann T, Nesher M, Lichtstein D. Diverse biological responses to different cardiotonic steroids. Pathophysiology. 2007;14:159–166. doi: 10.1016/j.pathophys.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Fürstenwerth H. Ouabain-the insulin of the heart. Int. J. Clin. Pract. 2010;64:1591. doi: 10.1111/j.1742-1241.2010.02395.x. [DOI] [PubMed] [Google Scholar]
- 27.Rahimtoola SH, Tak T. The use of digitalis in heart failure. Curr. Probl. Cardiol. 1996;21:781–853. doi: 10.1016/S0146-2806(96)80001-6. [DOI] [PubMed] [Google Scholar]
- 28.Kang, Y. J. Mortality rate of infection with COVID-19 in Korea from the perspective of underlying disease. Disaster Med. Public Health Prep. 1–3, 10.1017/dmp.2020.60 (2020). [DOI] [PMC free article] [PubMed]
- 29.Zou X, et al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akhmerov A, Marbán E. COVID-19 and the heart. Circ. Res. 2020;126:1443–1455. doi: 10.1161/CIRCRESAHA.120.317055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ong EZ, et al. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe. 2020;27:1–4. doi: 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin C, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi, Y. et al. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. MedRxiv, https://doi.org/10.1101/2020.03.12.20034736 (2020). [DOI] [PMC free article] [PubMed]
- 35.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Resp. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harcourt J, et al. Isolation and characterization of SARS-CoV-2 from the first US COVID-19 patient. BioRxiv. 2020 doi: 10.1101/2020.03.02.972935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Backer JA, Klinkenberg D, Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Eurosurveillance. 2020;25:2000062. doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng YY, Ma YT, Zhang JY, Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17:259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.López-Lázaro M, et al. Digitoxin inhibits the growth of cancer cell lines at concentrations commonly found in cardiac patients. J. Nat. Prod. 2005;68:1642–1645. doi: 10.1021/np050226l. [DOI] [PubMed] [Google Scholar]
- 40.Goldberger ZD, Goldberger AL. Therapeutic ranges of serum digoxin concentrations in patients with heart failure. Am. J. Cardiol. 2012;109:1818–1821. doi: 10.1016/j.amjcard.2012.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The Food and Drug Administration (FDA). Prescribing Information Lanoxin (Digoxin). https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/020405s013lbl.pdf (2011).
- 42.Burkard C, et al. ATP1A1-mediated Src signaling inhibits coronavirus entry into host cells. J. Virol. 2015;89:4434–4448. doi: 10.1128/JVI.03274-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollard, H. B., Pollard, B. S. & Pollard, J. R. Classical drug digitoxin inhibits influenza cytokine storm, with implications for COVID-19 therapy. bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 44.Kim J-M, et al. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res. Perspect. 2020;11:3. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J-H, et al. Clinical diagnosis of early dengue infection by novel one-step multiplex real-time RT-PCR targeting NS1 gene. J. Clin. Virol. 2015;65:11–19. doi: 10.1016/j.jcv.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 46.Centers for Disease Control and Prevention (CDC). Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Primer and Probe Information. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.