Abstract
Fusobacterium nucleatum (F. nucleatum) is frequently detected in primary colorectal cancer (CRC) and matching metastasis, and has been linked to a worse prognosis. We investigated the presence of F. nucleatum in gastric cancer (GC) and gastric preneoplastic conditions of the stomach, and its potential prognostic value in GC patients. Fusobacterium spp. and F. nucleatum were quantified in various specimens from gastrointestinal tract including paired CRC and GC tissues using probe-based qPCR. Fusobacterium spp. and F. nucleatum were more frequently found in tumorous tissue of CRC and GC compared to non-tumorous tissues. The frequency and bacterial load were higher in CRC compared to GC patients. F. nucleatum positivity showed no association to chronic gastritis or preneoplastic conditions such as intestinal metaplasia. F. nucleatum-positivity was associated with significantly worse overall survival in patients with Lauren’s diffuse type, but not with intestinal type GC. There was no association with gender, Helicobacter pylori-status, tumor stage or tumor localization. However, F. nucleatum was positively associated with patient’s age and a trend for a lower global long interspersed element-1 DNA methylation. In conclusion, our work provides novel evidence for clinical relevance of F. nucleatum in GC by showing an association between F. nucleatum positivity with worse prognosis of patients with Laurens’s diffuse type gastric cancer. Further studies are necessary to explore related mechanistic insights and potential therapeutic benefit of targeted antibiotic treatment in GC patients.
Subject terms: Cancer, Gastrointestinal cancer, Gastric cancer, Gastrointestinal cancer
Introduction
Microbiota of the gastrointestinal tract (GI) is increasingly appreciated in symbiotic relationship with host. GI-microbiota triggers an immune fine-tuning and may play a crucial role in induction of inflammation contributing to a multistep process of carcinogenesis, as proposed for colorectal cancer (CRC) and gastric cancer (GC)1–4.
Fusobacterium nucleatum (F. nucleatum), a gram-negative bacterium, is a common member of oral microbiota5,6 and has been linked to development of oral plaques and periodontitis7. Most intriguingly, it has been suggested to play a role in carcinogenesis as it has been detected in CRC tissues and even cultured from colon biopsies8. Recently, F. nucleatum has been also detected in several other tumours including oesophageal9 and pancreatic cancer tissue10.
Most extensive and compelling evidence for the potential role of F. nucleatum in carcinogenesis supported by the studies in CRC. F. nucleatum is found in tumorous tissues at higher bacterial load in comparison to adjacent non-tumorous mucosa8,11. Furthermore, it has been traced from primary tumours to liver metastases and was associated with a worse prognosis, suggesting its potential role not only in carcinogenesis but also possible therapeutic translational implications12. For instance, antibiotic therapy of mice with xenograft tumours positive for F. nucleatum led to a significant decrease in tumour growth in vivo experiments12. From molecular perspective, F. nucleatum has been linked to certain molecular alterations in CRC for instance with CpG island methylator phenotype (CIMP), TP53 wild-type, hMLH1 methylation, MSI and CHD7/8 mutation11,13. Moreover, F. nucleatum has been correlated with expression of proinflammatory genes, lower CD3+ T-cell density and increased TNF-α gene expression in CRC14–16.
The microbiome composition of the stomach is unique. Helicobacter pylori (H. pylori) is the predominant species and the key trigger for development of peptic ulcer disease and GC17,18. Despite years of research, the exact interaction of H. pylori with mucosa remains only partially understood. It is now clearly recognized that H. pylori is an infectious disease that causes chronic non-atrophic gastritis (CNAG) that can progress to preneoplastic conditions such as atrophic gastritis (AG), intestinal metaplasia (IM) and finally to dysplasia and cancer19. With new sequencing tools, it is increasingly appreciated that not H. pylori alone but rather the microbiome in whole complexity contributes to disease conditions. Several studies in detail reported about microbial alterations in stomach20,21. Fusobacterium spp. are frequently found in stomach mucosa20–23. According to few preliminary reports F. nucleatum have been found in tumorous GC tissues as well24,25, but there are still many unanswered questions. High-throughput techniques including 16 s RNA/DNA sequencing allow only a relative quantification of microbial community while polymerase chain reaction (PCR) based F. nucleatum analysis may provide an absolute quantification in relation to human cells. Next, whether F. nucleatum may be linked to preneoplastic conditions and contribute to carcinogenesis is still unknown. Most importantly, the clinical and prognostic relevance of F. nucleatum in GC has not been studied in detail.
In the present study, we performed in-depth characterization of Fusobacterium spp. and F. nucleatum in GC. To elaborate on its potential role in gastric carcinogenesis, we evaluated normal gastric mucosa (N), chronic gastritis samples with CNAG or with AG and IM, and correlated the positivity to clinicopathological characteristics and prognosis of GC patients.
Results
F. nucleatum in CRC
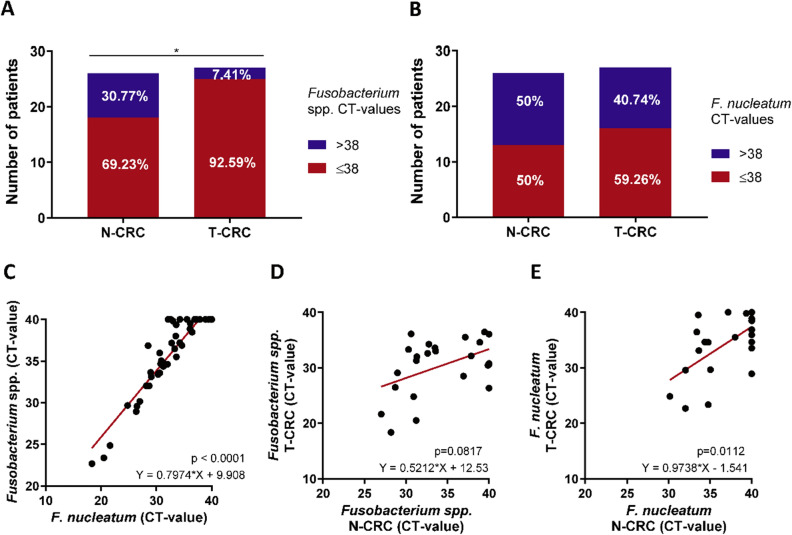
F. nucleatum have been previously evaluated in CRC tissues using PCR-based quantitative analysis. To confirm the analysis in our European cohort, we first validated the quantitative detection method and the reproducibility of Fusobacterium spp. and F. nucleatum analysis in a subset of samples from CRC patients. Based on our reproducibility results, the cycle threshold (Ct) values of ≤ 38 for both Fusobacterium spp. and F. nucleatum were classified as positive. In non-tumorous and tumorous CRC tissues we observed Fusobacterium spp. positivity in 69.23% (18/26) and 92.59% (25/27) (Fig. 1A, p = 0.0394), respectively. F. nucleatum positivity was present in 50% (13/26) N-CRC and 59.26% (16/27) T-CRC specimens (Fig. 1B). Overall, there was a significant correlation between F. nucleatum and Fusobacterium spp. (Fig. 1C, p < 0.0001). Analysis of the N-CRC and T-CRC samples (Fig. 1D,E) revealed only a trend for positive correlation for Fusobacterium spp. abundance in paired samples (p = 0.0817), while F. nucleatum load correlated significantly between N-CRC and T-CRC (p = 0.0112). Overall, we confirm that Fusobacterium spp. and F. nucleatum are more frequently detectable in T-CRC than in N-CRC and F. nucleatum load correlates significantly between tumorous and non-tumorous tissues.
Figure 1.
Abundance of Fusobacterium spp. and F. nucleatum in colorectal cancer patients. (A) Proportion of Fusobacterium spp. positivity in non-tumorous (N-CRC, n = 26) and tumorous colon tissues (T-CRC, n = 27). (B) Proportion of F. nucleatum in N-CRC (n = 26) and T-CRC (n = 27) tissues. (C) Correlation between Fusobacterium spp. and F. nucleatum in N- and T-CRC. (D) Correlation of Fusobacterium spp. abundance between N-CRC and T-CRC. (E) Correlation of F. nucleatum abundance between N-CRC and T-CRC. Data are presented as raw Ct-values; negative undetectable values were set to Ct of 40, CT-value > 38 were defined as negative and CT-value ≤ 38 were defined as positive. Fisher’s exact and Spearman’s tests were used for analyses.
F. nucleatum in GC
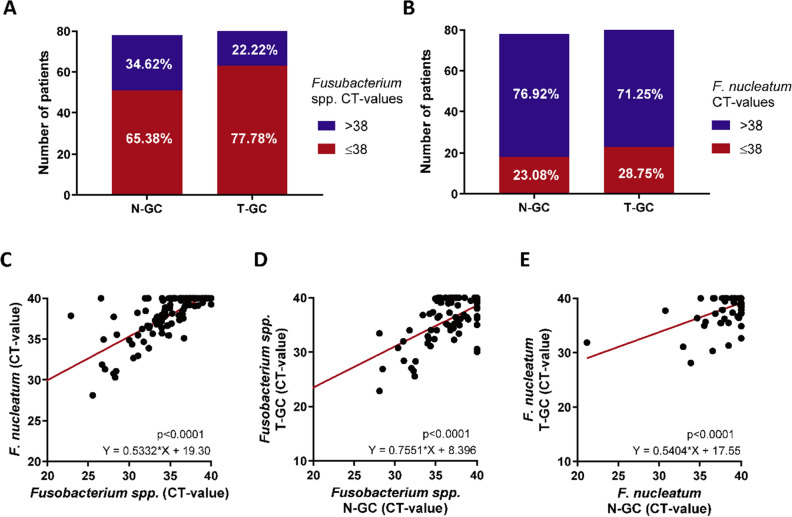
Next, we investigated F. nucleatum in tumour tissues of gastric cancer (T-GC) and its adjacent mucosa (N-GC). Based on our validation and reproducibility results, the Ct-value of ≤ 38 cycles were defined as positivity also in gastric mucosa. Fusobacterium spp. was detectable in 65.38% (51/78) of N-GC and 77.78% (63/81) of T-GC samples (Fig. 2A). F. nucleatum was positive in 23.08% (18/78) of N-GC and 28.75% (23/80) of T-GC samples (Fig. 2B). In similar fashion as in CRC, we observed a statistically significant correlation between abundance of Fusobacterium spp. and F. nucleatum (p < 0.0001) in mucosa of GC patients (Fig. 2C). Furthermore, the abundance of Fusobacterium spp. and F. nucleatum correlated significantly between N-GC and T-GC (each p < 0.0001, Fig. 2D,E).
Figure 2.
Abundance of Fusobacterium spp. and F. nucleatum in gastric cancer patients. (A) Proportion of Fusobacterium spp. positivity in non-tumorous (N-GC, n = 78) and tumorous gastric cancer tissues (T-GC, n = 81). (B) Proportion of F. nucleatum positivity in N-GC (n = 78) and T-GC (n = 80). (C) Correlation between Fusobacterium spp. and F. nucleatum in N-GC and T-GC. (D) Correlation of Fusobacterium spp. abundance between N-GC and T-GC. (E) Correlation of F. nucleatum abundance between N-GC and T-GC. Data are presented as raw Ct-values; negative undetectable values were set to Ct of 40, CT-value > 38 were defined as negative and CT-value ≤ 38 were defined as positive. Fisher’s exact and Spearman’s tests were used for analyses.
Differences in F. nucleatum abundance between CRC and GC
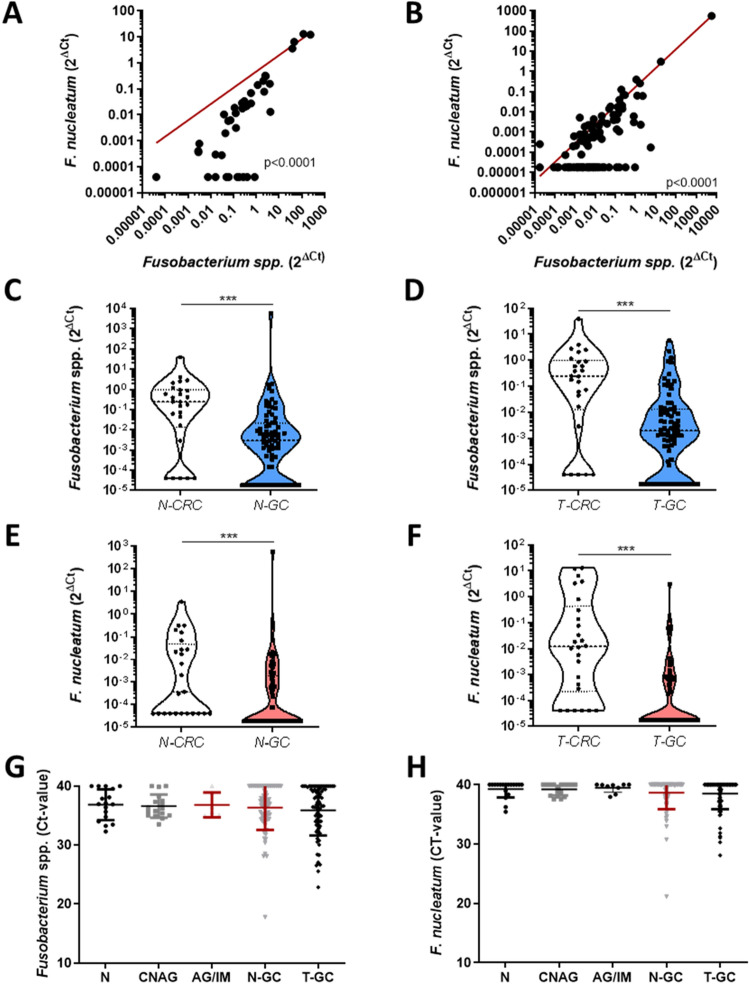
Following normalization to prostaglandin transporter (PGT), we observed a significant correlation between Fusobacterium spp. and F. nucleatum in CRC (Fig. 3A, p < 0,0001) and in GC (Fig. 3B, p < 0,0001). In comparison to non-normalized values presented in Figs. 1 and 2, the normalized abundance of Fusobacterium spp. and F. nucleatum was similar between N-CRC and T-CRC and between N-GC and T-GC, respectively (Supplementary Fig. S1). Next, we evaluated the differences in bacterial load of Fusobacterium spp. and F. nucleatum between CRC and GC. Despite the anatomical distance to oral cavity, abundance of Fusobacterium spp. in N-CRC and T-CRC was significantly higher than in N-GC and T-GC, respectively (Fig. 3C,D). In addition, F. nucleatum was higher in N-CRC and T-CRC compared to N-GC and T-GC, respectively (Fig. 3E,F).
Figure 3.
Difference in Fusobacterium spp. and F. nucleatum between colon, gastric mucosa and abundance in preneoplastic gastric mucosa. (A) Correlation between normalized Fusobacterium spp. and F. nucleatum in N-CRC and T-CRC specimens. (B) Correlation between normalized Fusobacterium spp. and F. nucleatum in N-GC and T-GC specimens. (C) Relative abundance of Fusobacterium spp. in N-CRC (n = 26) and N-GC (n = 78) (p < 0.0001). (D) Relative abundance of Fusobacterium spp. in T-CRC (n = 26) and T-GC (n = 79) (p < 0.0001). (E) Relative abundance of F. nucleatum in N-CRC (n = 25) and N-GC (n = 79) (p < 0.0001). (F) Relative abundance of F. nucleatum in T-CRC (n = 26) and T-GC (n = 80) (p < 0.0001). (G) Abundance of Fusobacterium spp. in N (n = 17), CNAG (n = 17), AG/IM (n = 6), N-GC (n = 78) and T-GC (n = 81) tissues in GC (p = 0.97). (H) Abundance of F. nucleatum in N (n = 18), CNAG (n = 17), AG/IM (n = 9), N-GC (n = 78) and T-GC (n = 80) in GC (p = 0.86). Relative abundance is presented as 2ΔCT values normalized to PGT. Undetectable values were set to the lowest measurable normalized value. Mann–Whitney-test was used for statistical analysis of two groups and Kruskal–Wallis test for more than two groups.
F. nucleatum in preneoplastic conditions in comparison to GC
To explore the potential involvement of Fusobacterium spp. and F. nucleatum we compared samples from patients with normal mucosa (N), CNAG, AG/IM, N-GC and T-GC. The analysis of Ct-values, revealed relatively similar pattern of Fusobacterium spp. and F. nucleatum abundance in normal and chronic gastritis with or without preneoplastic conditions in comparison to GC, suggesting that F. nucleatum may be probably involved in rather late stages of classical Correa’s cascade of gastric carcinogenesis (Fig. 3G,H). F. nucleatum was present in 16.7% (3/18) of N, 17.65% (3/17) CNAG mucosa and 0% (0/9) in AG/IM mucosa, which was not significantly different to N-GC and T-GC. Since none of the AG/IM mucosa samples were positive for F. nucleatum, we did not perform any correlation to OLGA/OLGIM. To evaluate potential association between F. nucleatum and H. pylori in non-neoplastic mucosa, we compared Fusobacterium spp. and F. nucleatum levels between subjects with and without active H. pylori infection irrespective of gastritis type or severity in total cohort of non-neoplastic mucosa. As shown in the Supplementary Fig. S2, we observed no difference in Fusobacterium spp. between H. pylori positive and negative gastric mucosa, while a slightly lower level of F. nucleatum was found in H. pylori positive mucosa (p = 0.046). However, only each three samples from each H. pylori positive and negative groups were below the defined Ct-value of 38 and further data are needed to elaborate on this topic.
F. nucleatum and GC subgroup analysis
To evaluate if F. nucleatum might be associated with specific GC characteristics, we divided the GC cohort in F. nucleatum positive and negative groups (Table 1). We compared the F. nucleatum-positive and -negative groups with regard to gender, tumour localization, UICC and TNM stages, grading, Lauren’s classification and H. pylori status but except for age there were no differences between the groups.
Table 1.
Clinicopathological characteristics of Gastric Cancer patients in relation to F. nucleatum positivity.
| F. nucleatum | |||||||
|---|---|---|---|---|---|---|---|
| All | Positive | Negative | p | ||||
| n = 81 | % | n = 23 | % | n = 57 | % | ||
| Age | 65.85 | ± 11.58 | 70.04 | ± 9.70 | 64.16 | ± 12.19 | 0.042* |
| Gender | 0.21 | ||||||
| Male | 47 | 58 | 11 | 48 | 36 | 63 | |
| Female | 34 | 42 | 12 | 52 | 21 | 37 | |
| Tumor localization | 0.37 | ||||||
| Cardia | 8 | 10 | 4 | 17 | 4 | 7 | |
| Corpus | 45 | 56 | 12 | 52 | 32 | 56 | |
| Antrum | 28 | 34 | 7 | 30 | 21 | 37 | |
| UICC | 0.75 | ||||||
| I | 16 | 20 | 4 | 17 | 11 | 19 | |
| II | 21 | 26 | 8 | 35 | 13 | 23 | |
| III | 36 | 44 | 9 | 39 | 27 | 47 | |
| IV | 8 | 10 | 2 | 9 | 6 | 11 | |
| T | 0.77 | ||||||
| 1 + 2 | 18 | 22 | 4 | 17 | 13 | 23 | |
| 3 | 36 | 45 | 10 | 43 | 26 | 46 | |
| 4 | 27 | 33 | 9 | 39 | 18 | 32 | |
| N | 0.26 | ||||||
| 0 | 29 | 36 | 9 | 39 | 19 | 33 | |
| 1 | 15 | 19 | 6 | 26 | 9 | 16 | |
| 2 | 13 | 16 | 1 | 4 | 12 | 21 | |
| 3 | 23 | 28 | 6 | 26 | 17 | 30 | |
| Unknown | 1 | 1 | 1 | 4 | 0 | 0 | |
| M | 1 | ||||||
| 0 | 72 | 89 | 20 | 87 | 51 | 89 | |
| 1 | 8 | 10 | 2 | 9 | 6 | 11 | |
| Unknown | 1 | 1 | 1 | 4 | 0 | 0 | |
| Grading | 0.37 | ||||||
| 1 | 3 | 4 | 0 | 0 | 3 | 5 | |
| 2 | 29 | 36 | 10 | 43 | 18 | 32 | |
| 3 | 49 | 60 | 13 | 57 | 36 | 63 | |
| Laurén-classification | 0.61 | ||||||
| Diffuse Type | 44 | 54 | 12 | 52 | 32 | 56 | |
| Intestinal Type | 26 | 32 | 6 | 26 | 19 | 33 | |
| Mixed Type | 7 | 9 | 3 | 13 | 4 | 7 | |
| Unknown | 4 | 5 | 2 | 9 | 2 | 4 | |
| H. pylori | 0.82 | ||||||
| Negative | 8 | 10 | 2 | 9 | 6 | 11 | |
| Positive | 17 | 21 | 5 | 22 | 12 | 21 | |
| Unknown | 56 | 69 | 16 | 70 | 39 | 68 | |
F. nucleatum positivity was defined by the cut-off of ≤ 38. *Unpaired t-test. ns: non-significant. F. nucleatum data were available only for 80 subjects. UICC: Union for International Cancer Control; T-primary tumor stage; N- lymphnode metastasis staging; M: metastasis staging.
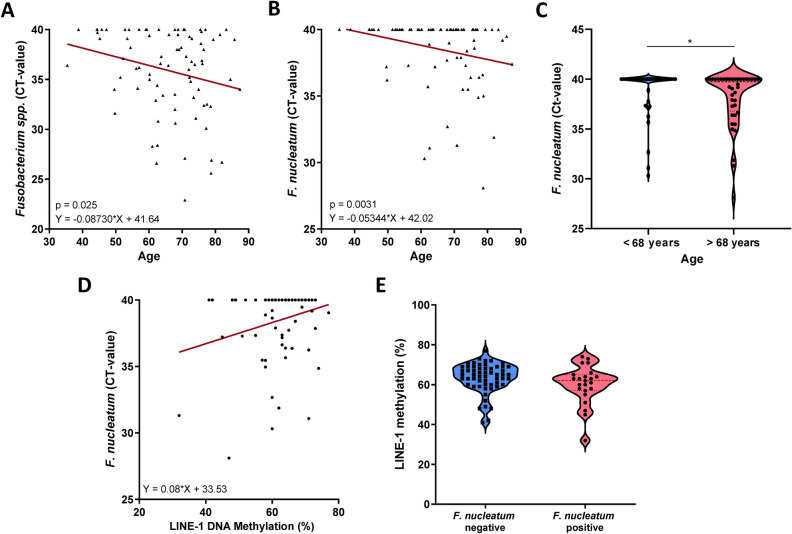
Fusobacterium spp. and F. nucleatum correlate with age
Correlation analysis revealed a positive correlation between patient’s age and Fusobacterium spp. and between patient’s age and F. nucleatum. Older patients had higher Fusobacterium spp. (p = 0.025, Fig. 4A) and higher F. nucleatum (p = 0.0031, Fig. 4B) abundance in T-GC. Furthermore, patients with F. nucleatum positive T-GC were overall older than patients with negative T-GCs. Based on the median age with cut-off of 68 years, older groups with GC had significantly higher F. nucleatum load compared to younger patients (Fig. 4C).
Figure 4.
Correlation between F. nucleatum, LINE-1- and patients age in tumorous tissue of GC patients. (A) Correlation between Fusobacterium spp. abundance in T-GC and patients age in GC patients (n = 81, p = 0.025). (B) Correlation between F. nucleatum abundance in T-GC and patients age in GC patients (n = 80, p = 0.0031). (C) Differences in F. nucleatum abundance based on the patients age defined as below or above median age (68 years). (D) Correlation between F. nucleatum and LINE-1 DNA methylation in T-CRC specimens (n = 80, p = 0.153). (E) LINE-1 DNA methylation differences between F. nucleatum-positive (n = 23) and –negative (n = 57) T-GC (p = 0.09). Mann–Whitney and Spearman’s tests were used for analysis.
Correlation with global and gene specific methylation changes
It has been recently suggested that F. nucleatum may be associated with distinct molecular alterations in cancer. We evaluated possible correlation between F. nucleatum and global DNA hypomethylation using surrogate long interspersed element-1 (LINE-1) methylation and miR-137 promoter methylation, which are frequently deregulated in GC and CRC. Overall, correlation analysis between LINE-1 and F. nucleatum revealed a non-significant trend for lower LINE-1 methylation in subjects with higher F. nucleatum load (p = 0.156, Fig. 4D). LINE-1 methylation in the F. nucleatum-positive group was slightly lower as in F. nucleatum-negative group although the difference did not reach statistical significance (60.1 ± 9.6 vs. 63.4 ± 7.4, p = 0.09) (Fig. 4E). For comparison, gene specific DNA methylation analysis of miR-137 and F. nucleatum revealed no difference between the groups (data not shown).
Survival analysis
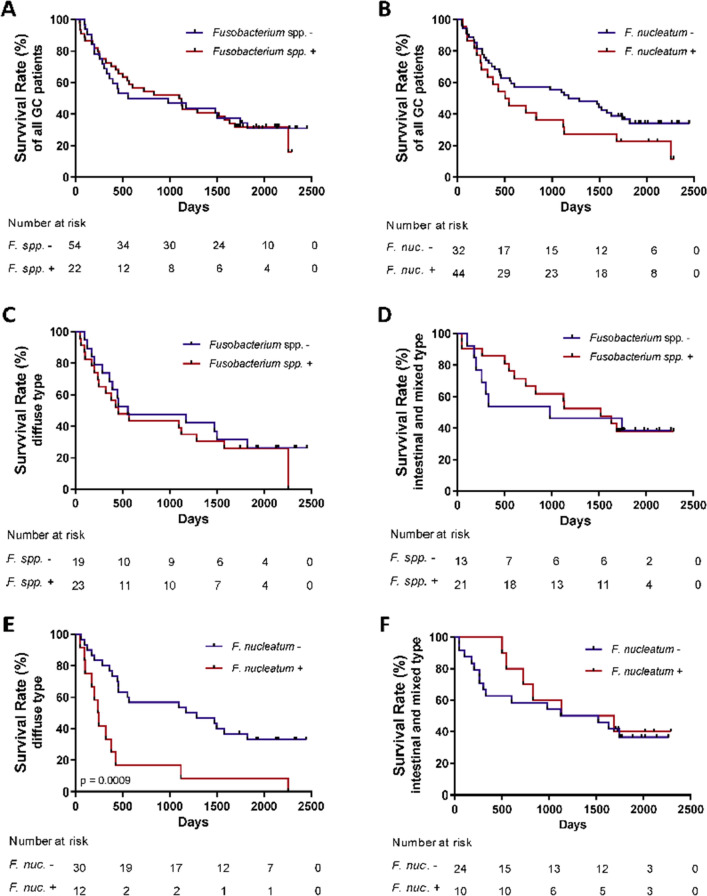
Survival data were obtained for GC subjects for a period of up to 2500 days. To avoid potential bias related to surgical complications, we excluded in total four patients from analysis due to the death within the first 30 days after receiving the diagnosis (1 with F. nucleatum positive and 3 with F. nucleatum negative T-GCs). Median survival of 76 patients was 981 days. Overall survival analysis revealed no difference between Fusobacterium spp. positive and negative gastric cancer patients (Fig. 5A, p = 0.997). In comparison (Fig. 5B), survival analysis based on the F. nucleatum positivity revealed a trend for a worse overall survival in the F. nucleatum positive group (524.5 days) in comparison to the F. nucleatum negative group (1287 days, p = 0.13). Survival analysis between different GC subgroups based on Lauren’s classification revealed no difference in regard to Fusobacterium spp. positivity both for diffuse and for intestinal or mixed-types of GC patients (Fig. 5C,D). Remarkably, survival of the patients with F. nucleatum-positive (n = 10) vs. –negative (n = 24) T-GC revealed no difference in the group with intestinal and mixed-type tumours (1406 vs 1323 days, p = 0.64, respectively) (Fig. 5E). However, patients with F. nucleatum-positive diffuse type of GC (n = 12) had significantly worse overall survival compared to F. nucleatum-negative (n = 30) GC (244.5 days vs. 1229.5, p = 0.009, respectively) (Fig. 5D). Comparison of clinicopathological characteristics of F. nucleatum positive and negative diffuse type of GC revealed only differences in age, but no other major differences, suggesting stage-independent effect of F. nucleatum positivity on the prognosis in diffuse type GC (Table 2).
Figure 5.
Overall survival rates of GC patients based on Fusobacterium spp. and F. nucleatum status. (A) Overall survival rates of GC patients with positive and negative Fusobacterium spp. status (p = 0.285). (B) Overall survival rates of GC patients with positive and negative F. nucleatum status (p = 0.129). (C) Overall survival rates of GC patients with Lauren’s diffuse type with positive and negative Fusobacterium spp. status (p = 0.536). (D) Overall survival rates of GC patients with Lauren’s intestinal and mixed types with positive and negative Fusobacterium spp. status (p = 0.798). (E) Overall survival rates of GC patients with Lauren’s diffuse type gastric cancer with positive and negative F. nucleatum status (p = 0.0009). (F) Overall survival rates of GC patients with Lauren’s intestinal and mixed types with positive and negative F. nucleatum status (p = 0.643). Log-rank (Mantel–Cox) test was used for survival data.
Table 2.
Comparison of the patients with F. nucleatum positive and negative Lauren’s diffuse subtype gastric cancer tumours.
| F. nucleatum positive | F. nucleatum negative | p | |||
|---|---|---|---|---|---|
| n = 12 | % | n = 30 | % | ||
| Age (years ± SD) | 70.75 | ± 10.25 | 61.07 | ± 11.89 | 0.0189* |
| Gender | |||||
| Male | 6 | 50.00 | 17 | 56.67 | ns |
| Female | 6 | 50.00 | 13 | 43.33 | |
| Tumor localization | |||||
| Cardia | 1 | 8.33 | 1 | 3.33 | ns |
| Corpus | 8 | 66.67 | 17 | 56.67 | |
| Antrum | 3 | 25.00 | 12 | 40.00 | |
| UICC | |||||
| I | 1 | 8.33 | 5 | 16.67 | ns |
| II | 4 | 33.33 | 6 | 20.00 | |
| III | 6 | 50.00 | 16 | 53.33 | |
| IV | 1 | 8.33 | 3 | 10.00 | |
| T | |||||
| 1 + 2 | 1 | 8.33 | 7 | 23.33 | ns |
| 3 | 5 | 41.67 | 15 | 50.00 | |
| 4 | 6 | 50.00 | 8 | 26.67 | |
| N | |||||
| 0 | 5 | 41.67 | 9 | 30.00 | ns |
| 1 | 4 | 33.33 | 5 | 16.67 | |
| 2 | 0 | 0.00 | 6 | 20.00 | |
| 3 | 3 | 25.00 | 10 | 33.33 | |
| M | |||||
| 0 | 10 | 83.33 | 27 | 90.00 | ns |
| 1 | 1 | 8.33 | 3 | 10.00 | |
| Unknown | 1 | 8.33 | 0 | 0.00 | |
| G | |||||
| 2 | 3 | 25.00 | 3 | 10.00 | ns |
| 3 | 9 | 75.00 | 27 | 90.00 | |
F. nucleatum positivity was defined by the cut-off of ≤ 38. *Unpaired t-test; ns—non-significant (p > 0.05).
Discussion
Increasing evidence suggests that F. nucleatum may be involved in tumour development and associated with worse prognosis in CRC and other cancers. However, only limited data is available on the role of F. nucleatum in GC and gastric preneoplastic conditions. Using a well-characterized cohort of GC patients, we showed that Fusobacterium spp. and F. nucleatum may be frequently found not only in N- and T-CRC, but also in N- and T-GC although less frequently and at lower abundance. F. nucleatum was furthermore detected in normal mucosa and chronic gastritis. Interestingly, F. nucleatum was found in N-CRC and T-CRC in higher abundance despite the anatomical distance compared to N-GC and T-GC, respectively. Overall survival analysis revealed a significantly worse prognosis of patients with F. nucleatum-positive T-GC only in Lauren’s diffuse type GC, but not in intestinal type GC.
Amounting evidence has been collected to confirm the presence of F. nucleatum in CRC. Our data are in the frame of existing reports showing the F. nucleatum positivity in up to 60% of CRC specimens8,11,26. Only few preliminary reports have been dealing with this topic in GC and no data to prognostic relevance of F. nucleatum in GC has been studied yet. Yamamura et al. studied 20 samples from various GI cancers and detected F. nucleatum in 2 out of 20 cases24. Hsieh et al. demonstrated an enrichment of F. nucleatum in GC and have suggested F. nucleatum as potential diagnostic biomarker for GC25. In our cohort, we observed F. nucleatum positivity in GC patients in up to 28.75%. Surprisingly, the absolute abundance of F. nucleatum in T-GC was not different to N-GC, which is different to CRC studies.
We next performed the comparison of F. nucleatum absolute load between tumorous and non-tumorous colon and gastric mucosa. Both Fusobacterium spp. and F. nucleatum were at higher abundance in N-CRC compared to N-GC, as well as in T-CRC compared to T-GC. It is remarkable as the anatomical distance and proximity to an oral cavity would probably rather suggest higher abundance of F. nucleatum in the stomach as in the colon. Two reports have recently published results elaborating on the potential mechanism of F. nucleatum transfer to the tumours. Abed et al.27 have recently shown that host polysaccharide Gal-GalNAc, which is overexpressed in CRC, recognizes fusobacterial Fap2, which may trigger binding of F. nucleatum to the tumours. In another report, the authors confirmed an increased Gal-GalNAc levels in various tumours including GC28. Although the level of Gal-GalNAc was high in both CRC and GC tissues, the level in non-tumorous CRC samples was much lower as in non-tumorous GC which may explain the load differences. Overall, our results may support the hypothesis of potential hematogenous route of F. nucleatum spreading.
Some time ago, several initial studies have reported the capability of F. nucleatum to form biofilms. For instance, Zilm et al. reported that F. nucleatum may form biofilms and optimize its adhesion characteristics29. This property of F. nucleatum was dependent on the host environment in response to alkaline pH30. In CRC using the 3-dimensional tumour spheroid model, Kasper et al. observed development of biofilm-like structure in the tumour spheroid microenvironment by F. nucleatum31. The pathogenicity of F. nucleatum in the stomach may however be different as its low pH creates a unique microenvironment and microbial interplay. Low abundance of F. nucleatum in stomach in comparison to colon allows us to speculate on protective properties of acidic milieu preventing F. nucleatum dissemination. From another side, we observe no clear pattern for an increased abundance of F. nucleatum in AG/IM tissues where higher pH due to mucosa atrophy is expected. Considering the increasing interest in biofilm formation in the colon, further studies will be also necessary to address this point in the stomach microenvironment.
To understand the functional role of F. nucleatum in GC, we next analysed F. nucleatum in non-/preneoplastic gastric mucosa under consideration if H. pylori status and performed survival analysis. Aviles-Jimenez et al. have recently linked certain alterations in stomach microbiota composition to Correa’s cascade stages from CNAG to IM to intestinal type gastric cancer32. In our specific quantitative analysis, we did not observe any difference in F. nucleatum in preneoplastic conditions as well as no clear signal was found for H. pylori status. Since the sample size was sufficient only for pilot analysis, further studies will be needed to take a closer look at the F. nucleatum abundance in preneoplastic conditions with its variables and influencing factors specifically.
F. nucleatum has been repeatedly associated with worse prognosis in patients with oesophageal cancer9, pancreatic cancer10 and colorectal cancer26,33, but the data on GC are not available, yet. Although the overall survival analysis revealed only a non-significant trend toward a worse prognosis, we further performed subgroup analysis based on the Lauren’s classification, which is one of the most simple and valuable classifications of GC that partially mirrors the molecular GC classification and is frequently underappreciated in scientific work related to GC34. While no pattern was observed for intestinal type, we observed significantly worse overall survival in diffuse type GC patients with F. nucleatum positive tumours. It has been reported that F. nucleatum may promote carcinogenesis in CRC via FadA adhesin, which binds to E-cadherin, activated β-catenin signalling and accordingly various inflammatory and oncogenic properties of the cells35. Since diffuse type of GC is strongly associated with E-cadherin deregulation one may speculate for potential molecular mimicry of F. nucleatum to diffuse type of GC and probably specific prognostic relevance.
In one of the pivotal reports, F. nucleatum was associated with CIMP positivity, hMLH1 methylation, MSI and CHD7/8 positivity11. We analysed correlation between F. nucleatum and LINE-1 as a global methylation marker and miR-137 methylation36. F. nucleatum positive GC tumours showed a trend to lower LINE-1 methylation with overall positive correlation, while no association was found for miR-137. Although this may suggest that indeed, F. nucleatum positivity could be associated with certain epigenetic alterations such as global DNA hypomethylation, from another side, the lower LINE-1 DNA methylation could also be related to the aging as F. nucleatum positivity correlated strongly also to older age.
Despite intriguing results, we would like to underline that this is one of the first analyses and multiple remaining questions need to be addressed in future work. First, the study aimed to evaluate specifically the translational role of F. nucleatum in GC, therefore the data acquired may allow only a partial view on the microbial changes. Microbiome-sequencing may provide in-depth view on microbial alterations in GC. Second, our work provides only some preliminary molecular analysis on correlation with LINE-1 methylation. Additional in vitro and in vivo studies should provide mechanistic insights and explanation. Third, in particular from the clinical point of view, the data to F. nucleatum may have substantial clinical consequences. It has been recently reported that antibiotic treatment of tumours harbouring F. nucleatum led to reduced tumour growth in mice12. Therefore, use of antibiotics (for example metronidazole) could be a possible therapeutic consequence in patients with diffuse type GC with F. nucleatum positivity. Furthermore, the impact of Fusobacterium on the treatment response especially in the era of immunotherapy may be quite intriguing. Recently, it has been reported that prudent diets rich in whole grains and dietary fibres were associated with lower risk of F. nucleatum positive CRC while diets that may promote intestinal inflammation were associated with increased risk of F. nucleatum positive tumours37,38. Diet has been shown to provide a great source of various microRNAs including xenomiRNAs39, therefore, taking into account an association between diet and F. nucleatum positivity one may speculate on the role of exogenous microRNA or even various drugs. Further studies will be needed to address the impact of proton-pump-inhibitors and antibiotics on positivity and variation of F. nucleatum in stomach and CRC.
In summary, the results of our work strongly support the potential involvement of F. nucleatum in gastric carcinogenesis. F. nucleatum is frequently found in normal, preneoplastic and neoplastic mucosa although substantially lower than in colon. Even though there were no specific clinicopathological differences related to F. nucleatum positive gastric cancer patients, F. nucleatum positivity was associated with significantly worse overall survival in diffuse Lauren’s type GC patients. Further studies are needed to evaluate possible therapeutic implications and molecular alterations responsible for this phenotype.
Materials and methods
Study design
Prospectively collected samples were evaluated in this study. Tissue samples were collected in the Departments of Gastroenterology and Surgery at the Hospital of Lithuanian University of Health Sciences (Kaunas, Lithuania) and in the Department of Gastroenterology, Hepatology and Infectious Diseases at the Otto-von-Guericke University Magdeburg (Germany) in the context of the ERA-Net PathoGenoMics project. The study was performed according to the principles of the Declaration of Helsinki. Kaunas Regional Bioethics Committee (No. BE-2-10) and Institutional Review Board of Otto-von-Guericke University Magdeburg (No. 80/2011) approved both studies. All patients participating in the study provided written informed consent.
Survival analysis
The Lithuanian Cancer Registry and the Hospital of Lithuanian University of Health Sciences collected survival data of the gastric cancer patients for up to 2500 days. The time of survival was measured as the time interval between the date of GC diagnosis and the date of death.
Samples collection
The collection and characterization of biological material was partly described in our previous studies36,40. Briefly, specimens from GC and CRC were prospectively collected during surgical interventions. Samples from controls (N) and patients with various stages of chronic gastritis were obtained during endoscopy. The samples were immediately snap-frozen in liquid nitrogen and placed in − 80 °C freezer. The updated Sydney classification was applied for histological characterization of gastritis41. The Lauren’s classification was used for histological assessment of GC tumours. H. pylori status was analysed either by H. pylori ELISA IgG test (Virion\Serion GmbH, Germany) for GC patients or using multistep approach via serology, microbiology and histology as previously reported42,43. We obtained 81-paired samples from patients with GC including tumour tissues (T-GC) and their corresponding adjacent non-tumorous gastric mucosa (N-GC). Histopathological assessment of GC tissues was performed by an experienced pathologist at the tertiary centre form Lithuania. For preliminary analysis we included samples from 18 patients with histologically confirmed normal gastric mucosa (N), 17 patients with CNAG and 9 patients with AG/IM. In addition, we included samples from 27 patients with colorectal cancer (T-CRC) and their corresponding adjacent non-tumorous colon mucosa (N-CRC). An overview for sample collection and methods are presented in Supplementary Table S1 and the clinical and demographic data in Table 1.
DNA isolation and quantitative real-time PCR
DNA was extracted from frozen tissue samples, pretreated with QIAzol Lysis reagent (Qiagen, Valencia, CA) and chloroform based on manufacturer’s recommendations as described previously36,40. Probe-based quantitative real-time PCR was performed using Bio-Rad CFX96 real-time PCR cycler (BioRad, CA). Following probe-based primer were used: Fusobacterium spp.44; F. nucleatum9; prostaglandin transporter (PGT), also known as solute carrier organic anion transporter family, member 2A1 (SLCO2A1), as endogenous control for normalization as previously described8. Primer and probe sequences are provided in Supplementary Table S2. Ct-values for Fusobacterium spp. and F. nucleatum were set to 40 if PCR analyses revealed a negative result. Normalization was performed using 2^deltaCt-method. The values of the samples with undetectable Fusobacterium spp. and F. nucleatum were set to the lowest measurable normalized values.
Methylation analysis
Purified genomic DNA from tissue samples was used for global long interspersed nucleotide element-1 (LINE-1) and miR-137 promoter methylation analyses. The procedure was in detail described in our previous reports36,40. Briefly, we applied Cells-to-CpG Bisulfite Conversion Kit (Life Technologies, Carlsbad, CA) for bisulphite modification, thereafter the standard PCR with biotin-labelled primers and eventually the pyrosequencing on PyroMark Q96 ID (Qiagen) using PyroMark Gold Q96 reagents (Qiagen). The mean methylation level of analysed CpG motifs was used for quantitative methylation analysis.
Statistical analysis
Statistical evaluation was conducted with GraphPad Prism 7.0 (San Diego, CA), statistical software. We applied χ2-test for qualitative analysis and for quantitative analysis we used either Wilcoxon test for paired samples or Mann–Whitney U test for unpaired samples. For comparison of more than two groups we used the Kruskal–Wallis test. Spearman’s test was applied for correlation analysis. Survival analyses were performed with the Mantel-Cox test. Two-sided p-values of < 0.05 were considered as statistically significant.
Ethical standards
The study was performed according to the principle of the Declaration of Helsinki. The study was approved by the Kaunas Regional Bioethics Committee No. BE2-10 and Institutional Review Board of Otto-von-Guericke University Magdeburg No. 80/2011. All patients provided written informed consent.
Supplementary information
Acknowledgements
The authors would like to thank Ursula Stolz for technical support.
Author contributions
E.T.B., C.T. and R.S. performed the experiments; J.K., J.S., P.M. provided clinical material; E.T.B. and A.L. analysis and interpretation of the data and drafting of the manuscript; A.L. study concept and design and guarantor of the study; all authors approved the final version of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was performed in the frame of the doctoral thesis and supported by institutional funds. Samples acquisition was supported by BMBF: Grant number: BMBF-0315905D to PM; Lithuanian Research Council of Lithuania: Grant number: APP-2/2016 to JK and JS. AL is supported by the funds of European Commission through the European Funds for regional development (EFRE) as well as by the regional Ministry of Economy, Science and Digitalization as part of the “Autonomie im Alter” research group for “LiLife”-Project.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
A.L. is a senior editorial board member of Scientific Reports. The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-73448-8.
References
- 1.Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science (80-) 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang L, Pei Z. Bacteria, inflammation, and colon cancer. World J. Gastroenterol. 2006;12:6741–6746. doi: 10.3748/wjg.v12.i42.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hope ME, Hold GL, Kain R, et al. Sporadic colorectal cancer - Role of the commensal microbiota. FEMS Microbiol. Lett. 2005;244:1–7. doi: 10.1016/j.femsle.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 4.Dickert NW, Kass NE. NIH Public Access. October 2008;141:520–529.
- 5.Zhou X, Liu X, Li J, et al. Real-time PCR quantification of six periodontal pathogens in saliva samples from healthy young adults. Clin. Oral Investig. 2014;19(4):937–946. doi: 10.1007/s00784-014-1316-0. [DOI] [PubMed] [Google Scholar]
- 6.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Signat B, Roques C, Poulet P, et al. Role of Fusobacterium nucleatum in periodontal health and disease. Curr. Issues Mol. Biol. 2011;13:25–36. [PubMed] [Google Scholar]
- 8.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamura K, Baba Y, Nakagawa S, et al. Human microbiome fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin. Cancer Res. 2016;22(22):5574–5581. doi: 10.1158/1078-0432.CCR-16-1786. [DOI] [PubMed] [Google Scholar]
- 10.Mitsuhashi K, Nosho K, Sukawa Y, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209–7220. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tahara T, Yamamoto E, Suzuki H, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science (80-) 2017;1448:1–10. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamada T, Zhang X, Mima K, et al. Fusobacterium nucleatum in colorectal cancer relates to immune response differentially by tumor microsatellite instability status. Cancer Immunol. Res. 2018;6(11):1327–1336. doi: 10.1158/2326-6066.CIR-18-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host. Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mima K, Sukawa Y, Nishihara R, et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 2015;1:653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dharmani P, Strauss J, Ambrose C, et al. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect. Immun. 2011;79:2597–2607. doi: 10.1128/IAI.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugano K, Tack J, Kuipers EJ, et al. Kyoto global consensus report on Helicobacter pylori gastritis. Gut. 2015;64:1353–1367. doi: 10.1136/gutjnl-2015-309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dzutsev A, Goldszmid RS, Viaud S, et al. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. Eur. J. Immunol. 2015;45:17–31. doi: 10.1002/eji.201444972. [DOI] [PubMed] [Google Scholar]
- 19.Correa P, Piazuelo MB. The gastric precancerous cascade. J. Dig. Dis. 2012;13:2–9. doi: 10.1111/j.1751-2980.2011.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulz C, Schütte K, Koch N, et al. The active bacterial assemblages of the upper GI tract in individuals with and without Helicobacter infection. Gut. 2018;67:216–225. doi: 10.1136/gutjnl-2016-312904. [DOI] [PubMed] [Google Scholar]
- 21.Vasapolli R, Schütte K, Schulz C, et al. Analysis of Transcriptionally Active Bacteria Throughout the Gastrointestinal Tract of Healthy Individuals. Gastroenterology. 2019;157(4):1081–1092. doi: 10.1053/j.gastro.2019.05.068. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2017;67(2):226–236. doi: 10.1136/gutjnl-2017-314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coker OO, Dai Z, Nie Y, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2017;67:1024–1032. doi: 10.1136/gutjnl-2017-314281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamura K, Baba Y, Miyake K, et al. Fusobacterium nucleatum in gastroenterological cancer: Evaluation of measurement methods using quantitative polymerase chain reaction and a literature review. Oncol. Lett. 2017;14:6373–6378. doi: 10.3892/ol.2017.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh Y-Y, Tung S-Y, Pan H-Y, et al. Increased abundance of clostridium and fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci. Rep. 2018;8:158. doi: 10.1038/s41598-017-18596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaoka Y, Suehiro Y, Hashimoto S, et al. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J. Gastroenterol. 2017;53(4):517–524. doi: 10.1007/s00535-017-1382-6. [DOI] [PubMed] [Google Scholar]
- 27.Abed J, Emgård JEM, Zamir G, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abed J, Maalouf N, Parhi L, et al. Tumor Targeting by Fusobacterium nucleatum: A Pilot Study and Future Perspectives. Front Cell Infect Microbiol. 2017;7:295. doi: 10.3389/fcimb.2017.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zilm PS, Rogers AH. Co-adhesion and biofilm formation by Fusobacterium nucleatum in response to growth pH. Anaerobe. 2007;13:146–152. doi: 10.1016/j.anaerobe.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Chew J, Zilm PS, Fuss JM, et al. A proteomic investigation of Fusobacterium nucleatum alkaline-induced biofilms. BMC Microbiol. 2012;12:189. doi: 10.1186/1471-2180-12-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasper SH, Morell-Perez C, Wyche TP, et al. Colorectal cancer-associated anaerobic bacteria proliferate in tumor spheroids and alter the microenvironment. Sci. Rep. 2020;10(1):1–13. doi: 10.1038/s41598-020-62139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, et al. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 2014;4:1–11. doi: 10.1038/srep04202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33(8):1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 34.Link A, Kupcinskas J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: current insights and future perspectives. World J Gastroenterol. 2018;24(30):3313. doi: 10.3748/wjg.v24.i30.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-Cadherin/β-Catenin signaling via its FadA Adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steponaitiene R, Kupcinskas J, Langner C, et al. Epigenetic silencing of miR-137 is a frequent event in gastric carcinogenesis. Mol. Carcinog. 2016;55:376–386. doi: 10.1002/mc.22287. [DOI] [PubMed] [Google Scholar]
- 37.Mehta RS, Nishihara R, Cao Y, et al. Association of dietary patterns with risk of colorectal cancer subtypes classified by Fusobacterium nucleatum in tumor tissue. JAMA Oncol. 2017;3:921–927. doi: 10.1001/jamaoncol.2016.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L, Tabung FK, Zhang X, et al. Diets that promote colon inflammation associate with risk of colorectal carcinomas that contain Fusobacterium nucleatum. Clin. Gastroenterol. Hepatol. 2018;16(10):1622–1631. doi: 10.1016/j.cgh.2018.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Link J, Thon C, Schanze D, et al. Food-derived Xenomirnas: influence of diet and detectability in gastrointestinal tract—proof-of-principle study. Mol. Nutr. Food Res. 2018;63(2):1800076. doi: 10.1002/mnfr.201800076. [DOI] [PubMed] [Google Scholar]
- 40.Kupcinskas J, Steponaitiene R, Langner C, et al. LINE-1 hypomethylation is not a common event in preneoplastic stages of gastric carcinogenesis. Sci. Rep. 2017;7:4828. doi: 10.1038/s41598-017-05143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dixon MF, Genta RM, Yardley JH, et al. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am. J. Surg. Pathol. 1996;20:1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 42.Link A, Schirrmeister W, Langner C, et al. Differential expression of microRNAs in preneoplastic gastric mucosa. Sci. Rep. 2015;5:8270. doi: 10.1038/srep08270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ben Said, I., & Boujemaa, H. Cooperative localization using MUSIC algorithm and received signal strength measurements. In 2011 International Conference on Communications, Computing and Control Applications, CCCA 2011. Vol 23. 712–4723 (2011).
- 44.Boutaga K, Winkelhoff AJ, Vandenbroucke-Grauls CMJE, et al. Periodontal pathogens: a quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunol. Med. Microbiol. 2005;45:191–199. doi: 10.1016/j.femsim.2005.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.