Abstract
Lung cancer (LC) gene methylation detected in sputum assesses field cancerization and predicts LC incidence. Hispanic smokers have higher LC susceptibility compared to non-Hispanic Whites (NHW). We aimed to identify novel dietary nutrients affecting LC gene methylation and determine the degree of ethnic disparity in methylation explained by diet. Dietary intakes of 139 nutrients were assessed using a validated Harvard food frequency questionnaire in 327 Hispanics and 1502 NHWs from the Lovelace Smokers cohort (LSC). Promoter methylation of twelve LC genes was assessed in sputum DNA. A global association was identified between dietary intake and gene methylation (Ppermutation=0.003). Seventeen nutrient measurements were identified with magnitude of association with methylation greater than that seen for folate. A stepwise approach identified B12, manganese, sodium, and saturated fat as the minimally correlated set of nutrients whose optimal intakes could reduce the methylation by 36% (Ppermutation<0.001). Six protective nutrients included vitamin D, B12, manganese, magnesium, niacin, and folate. Approximately 42% of ethnic disparity in methylation was explained by insufficient intake of protective nutrients in Hispanics compared to NHWs. Functional validation of protective nutrients showed an enhanced DNA repair capacity towards double-strand DNA breaks, a mechanistic biomarker strongly linked to acquisition of LC gene methylation in smokers. Dietary intake is a major modifiable factor for preventing promoter methylation of LC genes in smokers’ lungs. Complex dietary supplements could be developed based on these protective nutrients for LC chemoprevention in smokers. Hispanic smokers may benefit the most from this complex for reducing their LC susceptibility.
Keywords: Nutrient, Ethnic disparity, Promoter methylation, Tumor suppressor gene, Sputum
INTRODUCTION
Lung cancer (LC) remains the leading cause of cancer death in men and women in the United States. Silencing of tumor suppressor genes (TSG) through promoter hypermethylation plays a causal role in LC etiology (1,2). The detection of promoter methylation of TSGs in exfoliated lung cells collected in sputum provides an assessment of field cancerization in the lungs (1,3). Integration of the methylation status of a validated panel of 12 TSGs in sputum into risk assessment models containing basic demographic and clinical factors can increase the prediction accuracy for LC by 11 to 16% which also does not differ between Hispanic and non-Hispanic white (NHW) smokers (4–6).
New Mexico (NM) has the highest percentage (47%) of Hispanics of any state with the majority born in the United States (7). NM Hispanics mainly include descendants of Spanish colonists and are mainly comprised of 63% European and 35% Native American ancestry (NAA) (6,8). Although NM Hispanics have lower age standardized LC incidence, previous studies have implied that the differences in LC incidence between NM Hispanics and NHWs are largely explained by the patterns of cigarette smoking of these two groups (9–11). Paradoxically, our recent study of Hispanic and NHW smokers in the Lovelace Smokers Cohort (LSC) identified that Hispanic smokers on average had an 18% increased risk for gene methylation compared to NHWs and this ethnic disparity remained with strict control of confounding effects of age, sex and smoking status (6), suggesting that Hispanic smokers may have a higher LC susceptibility. Furthermore, we and others have shown that Hispanic LC cases smoke fewer cigarettes (by 6 cigarettes per day) and Hispanic smokers have a more rapidly increasing risk for LC as a function of cumulative exposure to cigarettes compared to NHWs (6,9,10). Thus, these findings together with the greater promoter methylation of TSGs in Hispanic smokers provide provocative evidence supporting a greater susceptibility to LC for NM Hispanic smokers.
Accumulating evidence (12) has shown that specific dietary components may affect methylation status of specific regions in the human genome. Folate is a key nutritional factor in one-carbon metabolism that supplies the methyl groups for methylation reactions and is also involved in DNA repair through de novo synthesis of purines and pyrimidines (13). Extensive studies have consistently shown that folate insufficiency results in promoter hypermethylation of TSGs and/or hypomethylation of repetitive genomic elements (e.g., LINE1 and Alu), a hallmark of cancer (13,14). Our previous study assessed 21 candidate composite food variables and identified folate, leafy green vegetables, and multi-vitamin use as three protective factors against acquisition of gene methylation (8-gene panel) in the lungs of NHW smokers (14). Elucidation of the associations between dietary nutrients, ethnicity, and promoter methylation of TSGs could provide a scientific basis for designing complex dietary supplements for chemoprevention of LC in smokers and for the alleviation of ethnic disparity of LC susceptibility.
The current study took a “nutrientomics” approach that assessed dietary intake of 139 nutrients using a validated Harvard food frequency questionnaire (FFQ) in 327 Hispanics and 1502 NHWs from the Lovelace Smokers cohort (LSC) to identify dietary nutrients associated with methylation of 12 TSGs in sputum. The mediation effect of dietary nutrients on the ethnic disparity in methylation was also assessed. The functional relevance of the protective nutrients identified was assessed by their effects on double strand DNA repair capacity, a major determinant for propensity for acquisition of aberrant gene promoter methylation during smoking induced lung carcinogenesis (15–22).
MATERIALS AND METHODS
Lovelace Smokers cohort
The LSC enrollment started in 2001 with the goal to conduct longitudinal studies on biomarkers of lung carcinogenesis in biospecimens from moderate and heavy smokers (20). Enrollment was restricted to current and former smokers age 40 to 74 y with a minimum of 10 pack-years of smoking at baseline and mainly targeted residents living in the greater Albuquerque area. A detailed questionnaire written in English was used to collect information on demographics, medical, smoking, and exposure history, socioeconomic status, and quality of life at study entry. Cohort members returned approximately every 18 months to update smoking status and medical conditions. Sputum collection by induction and pulmonary functional test (PFT) were conducted at every visit. The LSC is still actively recruiting new smokers with a total number of 2500 enrolled by the end of 2015. Hispanics in the LSC (>95%) and their parents (>95%) and grandparents (>85%) were almost entirely born in the United States and recent immigrants from Latin America consist of a very small portion of the LSC (http://www.pewhispanic.org/states/state/nm/). Self-reported ethnicity was verified using ancestry informative markers in the LSC (6). The study was conducted in accordance with the Declaration of Helsinki and approved by the Western Institutional Review Board. All participants signed an informed consent form written in English.
Gene promoter methylation
Twelve TSGs with diverse cellular functions (e.g., cell cycle, DNA repair, and apoptosis) were selected for promoter hypermethylation analysis in cytologically adequate sputum samples based on our previous studies establishing their association with risk for LC, their specificity to methylation in lung epithelial cells, their functional consequences for gene expression and tumor phenotypes, and their timing of silencing in the carcinogenesis if known (1,2,4,5). The methylation of these genes was acquired during lung carcinogenesis and their detection in exfoliated lung cells collected in sputum provides an assessment of field cancerization in the lungs of smokers (1,3). In addition, enlarging this 12-gene panel by adding more TSGs did not further improve the prediction accuracy for LC risk due to the moderate to high correlations seen between any added genes and genes already in the panel (5). Methodology for detection of promoter methylation is described in Supplementary Methods.
Baseline dietary assessment
Cohort members completed at study entry the English version of the validated Harvard semi-quantitative FFQ, a self-administered instrument that includes ~150 food items distributed within the eight major dietary categories (23). The FFQ collects consumption frequency and serving size of each specified food item during the past 12 months and has excellent coverage for food items of the US Southwestern style (24). Daily estimates of the nutrient intakes are derived by summing over all foods, the products of the reported frequency of each food by the amount of nutrient in a specified (or assumed) serving of that food based primarily on US Department of Agriculture publications (23). The FFQ also has open-ended questions that collect use of food items consumed at least once per week, but not listed in the eight major dietary categories. The applicability of this FFQ in New Mexicans was further supported by the fact that the estimated consumption of vitamin A and C was highly comparable to the data from a study that itemized all chile containing traditional southwestern foods commonly consumed in New Mexico (24). In addition, serum vitamin B12, C, and folate levels were reported to be significantly lower in Hispanics versus NHWs in a previous study of an elderly New Mexican population (25). These three vitamins also showed an ethnic difference in the dietary assessment in the current study (Supplementary Table 1). A previous study compared the energy and nutrient source of elderly Hispanics and NHWs in New Mexico and identified no exclusive pattern for consumption of southwestern regional foods in New Mexico Hispanics (24). Because the number of nutrients in the output varied overtime (from 104 to 254 nutrients from 2005 to 2014), a total of 139 nutrients with <40% missing rate were included in this study. Cohort members with either extremely low or extremely high total caloric intake were excluded (14). In addition, none of the FFQs had >70 missing items. Finally, a total of 1968 LSC cohort members completed FFQs that passed the quality check. The number of unanswered food items (out of ~150) was very low in both ethnic groups (Table 1). This study focused on 139 nutrients with <40% missing rate in 1829 cohort members who were either Hispanic or NHW, the two major ethnic groups in the LSC.
Table 1.
Characteristics of LSC cohort members by ethnicity
| Variable | Self-identified ethnicity | P value | |
|---|---|---|---|
| Hispanic | NHW | ||
| n | 327 | 1502 | |
| Age (years, mean ± SD) | 54.7 ± 8.8 | 57.7 ± 9.5 | 1.5 × 10−7a |
| Gender (male, %) | 24.5 | 22.0 | 0.34b |
| Current smokers (%) | 73.7 | 51.8 | 5.0 × 10−13b |
| Packyears (pys, median (Q1-Q3)) | 30.5 (23.0 – 40.2) | 36.0 (26.8 – 50.2) | <0.0001c |
| 10–26 (%) | 33.0 | 21.9 | 3.8 × 10−6b |
| 26–37 (%) | 33.0 | 30.8 | |
| 37–145 (%) | 33.9 | 47.3 | |
| BMI (kg/m2, mean ± SD) | 29.1 ± 6.1 | 27.9 ± 6.0 | 0.0027a |
| 16–25 (%) | 25.1 | 34.2 | 0.0063b |
| 25–30 (%) | 40.4 | 36.0 | |
| 30–61 (%) | 34.6 | 29.8 | |
| Education level (%) | |||
| Less than college | 53.9 | 24.2 | 2.5 × 10−26b |
| Some college or above | 46.2 | 75.8 | |
| Ancestry component (mean ± SD) | |||
| European ancestry | 0.64 ± 0.08 | 0.97 ± 0.03 | NCd |
| Native American ancestry | 0.34 ± 0.08 | 0.03 ± 0.02 | NCd |
| African ancestry | 0.02 ± 0.02 | 0.00 ± 0.01 | NCd |
| Promoter methylation in sputum (%)e | |||
| P16 | 19.4 | 20.2 | 0.77 |
| MGMT | 26.5 | 25.3 | 0.69 |
| RASSF1A | 0.4 | 0.9 | 0.41 |
| DAPK | 19.8 | 15.5 | 0.091 |
| GATA4 | 41.8 | 35.0 | 0.036 |
| GATA5 | 19.8 | 14.2 | 0.021 |
| PAX5α | 12.3 | 14.8 | 0.30 |
| PAX5β | 8.6 | 8.0 | 0.77 |
| SULF2 | 42.9 | 33.3 | 0.0028 |
| PCDH20 | 41.8 | 34.4 | 0.022 |
| DAL1 | 7.8 | 6.7 | 0.49 |
| JPH3 | 21.3 | 22.3 | 0.72 |
| Number of unanswered food items (median (Q1-Q3)) | 0 (0 – 3) | 0 (0 – 2) | 0.0064c |
| Total calorie intake (kcal) (median (Q1-Q3)) | 1782.4 (1361.1 – 2353.5) | 1714.9 (1339.9 – 2148.9) | 0.067c |
| rs1801133 genotypef | |||
| CC | 0.34 | 0.45 | 0.0069b |
| CT | 0.51 | 0.43 | |
| TT | 0.16 | 0.12 | |
Student’s t-test.
x2 test.
Wilcoxon rank sum test.
Not calculated. Ancestry analysis included 307 Hispanics and 1390 NHW.
Methylation data is complete for 268 Hispanics and 1158 NHW.
CC, CT, and TT genotype of rs1801133 were coded as 0, 1, and 2 in the statistical analysis.
Double strand DNA repair capacity in lung epithelial cell lines
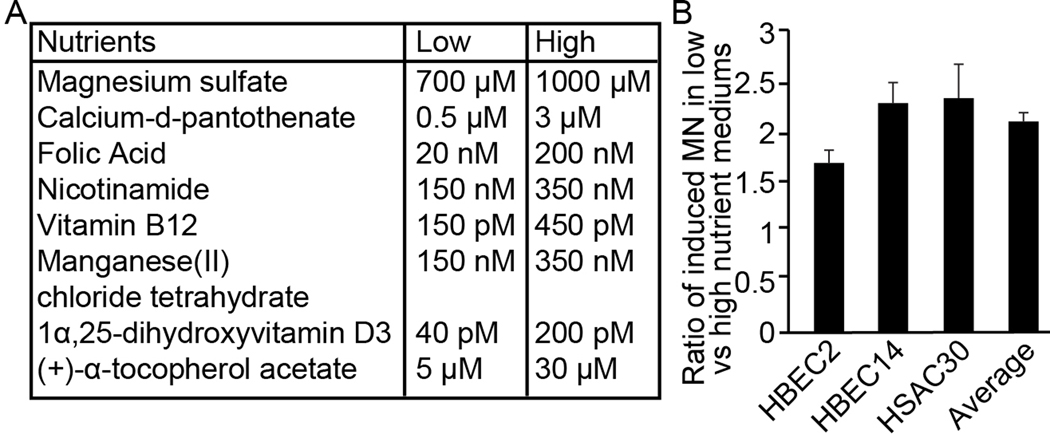
Three immortalized human lung epithelial cell lines (HBEC2, HBEC14, and HSAEC30) were obtained from Drs. Shay and Minna, Southwestern Medical Center (26). Cell lines were maintained in ACL-4 medium supplemented with bovine pituitary extract and nicotinic acid for a maximum of 6 months (24 passages). RPMI1640 as the base medium of ACL-4 is the only source for the eight protective nutrients, thus customized RPMI1640 depleted for these eight nutrients was used to prepare the low and high nutrient medium with the final concentrations of these eight nutrients determined based on the human physiological range in blood (Figure 1A). The crystal violet assay was used to assess the effect of the nutrient supplementation on cell proliferation. The cytokinesis-block micronucleus assay that specifically scores the micronuclei in cells that only divide once and minimizes the effect of different cell cycling due to nutrient levels was used to assess the DNA repair capacity in these three lung epithelial cell lines treated with 0.05U/L bleomycin, a radiomimetic drug specifically inducing double strand DNA breaks (27). Both experiments were repeated for a second time and identified same results.
Figure 1.
Protective nutrients enhanced DNA repair capacity towards double strand breaks. A. The levels of nutrients in low and high nutrient mediums were lower and upper limits of physiological levels seen in humans. B. The ratio of micronuclei frequency induced by bleomycin treatment in low nutrient versus high nutrient medium were 1.68 ± 0.12 in HBEC2, 2.28 ± 0.21 in HBEC14, and 2.33 ± 0.33 in HSAEC30 with an average of 2.10 ± 0.08 (P= 0.00077).
Statistical analysis
A rigorous analytical approach that incorporated a global test, false discovery rate (FDR) calculation, and hypothesis driven testing for B12 – methylenetetrahydrofolate reductase (MTHFR) SNP interaction was taken to minimize the chance of false positive findings (see Supplementary Methods). A global test based on principal component analysis was first employed to assess whether the total variance of nutrients was associated with risk for gene methylation. The associations between dietary intake quantified as principal components or individual nutrients and risk for gene methylation were assessed using generalized estimating equations (GEE) implemented in PROC GENMOD procedure (28) with a vector of the methylation status of 12 genes (1 for methylated status and 0 for unmethylated status for each individual gene) for each individual smoker as the outcome as described in our previous studies (6). Mediation analysis was conducted to assess a hypothesis that the effect of ethnicity on methylation was mediated by intake of dietary nutrients (29). All statistical analyses were conducted in SAS 9.3.
RESULTS
Global association between dietary nutrients and risk for methylation
Descriptive statistics for studied variables were summarized by ethnicity (Table 1). A significant global association (Ppermutation=0.003) was identified between dietary nutrients as represented by the four top-ranking principal components (Supplementary Figure) and methylation using the quasi-likelihood under the independence model criterion (QIC) statistic. In addition, component 2 which provided an integrative summary of more consumption of vitamin A, folate, and vitamin D and less consumption of saturated fat (Supplementary Table 2) was significantly associated with reduced risk for gene methylation (per standard deviation, OR=0.72, 95% CI=0.58–0.89, P=0.0031).
Effect of folate intake from different dietary sources on methylation
Among seven folate measurements (Supplementary Table 3), total folate and folate equivalents including supplements and fortified food, folic acid from supplements and fortified food, and folate from supplements were significantly associated with reduced methylation. Three major sources of folate in diet including natural food folate, folate from fortified food, and folate from supplements were assessed in one model to better understand which source of folate was most influential for methylation. Interestingly, only folate from supplements was significantly associated with methylation with 400 μg folate (the difference between lower and upper quartile in the LSC) reducing methylation by 9% (95% CI=0.84–0.99, P=0.028). Multi-vitamins and individual vitamin supplement are the two major sources for folate from supplements. Folate from supplements remained significantly associated with methylation (per 400 μg of folate, OR=0.89, 95% CI=0.81–0.98, P=0.016) when multi-vitamin use was included in GEE for adjustment, suggesting that the effect for folate from supplements was unlikely to be affected by other components in the multi-vitamins. This premise is further supported by the finding that individual folate supplement was protective against acquisition of methylation (per 400 μg of folate, OR=0.74, 95% CI=0.61–0.88, P=0.00083, n=609) in LSC members not currently taking multi-vitamins.
Other nutrients and methylation
Further analysis was conducted using the greatest significance (P=0.021 for total folate intake) seen among seven dietary folate measurements as the threshold to guide the identification of nutrients with the strongest associations with methylation (Supplementary Table 4). Seventeen nutrient measurements were identified with six (vitamin B12, manganese, manganese without supplements, magnesium, vitamin D, and niacin) protective against methylation (Table 2). Due to the moderate to high correlations observed between some of the 18 nutrients (including folate), a stepwise approach based on the QIC statistic was employed to select a set of relatively independent nutrients for assessing their contribution to methylation (see details in Supplementary methods). Two protective nutrients (vitamin B12 and manganese) and two risk nutrients (sodium and saturated fat) were identified in the final model. A combination of higher dietary intake of vitamin B12 and manganese (from lower to upper quartile) and lower intake of sodium and saturated fat (from upper to lower quartile) reduced methylation by 36% (95% CI=0.27–0.45, Ppermutation<0.001).
Table 2.
Individual nutrients associated with risk for methylation in 268 Hispanics and 1158 NHWsa
| Nutrient (unit/d) | OR (95% CI)b | Praw | PFDR | Mean ± SD | Median (Q1 - Q3) |
|---|---|---|---|---|---|
| Vitamin B12 (μg) | 0.98 (0.97 – 0.99) | 0.00035 | 0.050 | 27.17 ± 78.43 | 10.54 (5.86 – 19.67) |
| Manganese (mg) | 0.89 (0.82 – 0.96) | 0.0015 | 0.095 | 4.54 ± 2.59 | 3.94 (2.70 – 5.91) |
| Proline (g)c | 1.25 (1.08 – 1.44) | 0.002 | 0.095 | 4.89 ± 1.93 | 4.64 (3.51 – 5.97) |
| Total Trans/Cis Trans Linoleic (g)c | 1.18 (1.06 – 1.31) | 0.0032 | 0.095 | 0.44 ± 0.23 | 0.39 (0.28 – 0.55) |
| Sodium (g) | 1.22 (1.07 – 1.40) | 0.0034 | 0.095 | 2.19 ± 0.86 | 2.07 (1.58 – 2.68) |
| Saturated fat (g) | 1.19 (1.06 – 1.35) | 0.0045 | 0.095 | 23.80 ± 10.40 | 22.03 (16.38 – 29.59) |
| Animal fat (g) | 1.15 (1.04 – 1.27) | 0.0047 | 0.095 | 35.14 ± 17.02 | 32.72 (22.60 – 44.31) |
| Palmitic fatty acid (g) | 1.21 (1.06 – 1.38) | 0.0056 | 0.099 | 13.31 ± 5.65 | 12.41 (9.22 – 16.62) |
| Manganese without supplements (mg) | 0.88 (0.80 – 0.96) | 0.0064 | 0.101 | 3.39 ± 1.58 | 3.20 (2.22 – 4.22) |
| c9, t11 conjugate diene isomer 18:2 Linoleic (mg)c | 1.17 (1.04 – 1.31) | 0.0072 | 0.102 | 105.85 ± 57.76 | 96.15 (64.14 – 135.3) |
| Stearic fatty acid (g) | 1.17 (1.04 – 1.32) | 0.0084 | 0.109 | 5.99 ± 2.70 | 5.58 (4.04 – 7.42) |
| Magnesium (mg) | 0.88 (0.80 – 0.97) | 0.010 | 0.119 | 345.95 ± 137.42 | 328.79 (248.52 – 423.16) |
| Vitamin D (IU) | 0.91 (0.84 – 0.98) | 0.011 | 0.120 | 477.13 ± 373.18 | 455.08 (154.12 – 623.01) |
| Myristic fatty acid (g) | 1.12 (1.02 – 1.22) | 0.014 | 0.136 | 1.94 ± 1.06 | 1.74 (1.17 – 2.48) |
| Palmitelaidic trans fatty acid (g)c | 1.18 (1.03 – 1.34) | 0.014 | 0.136 | 0.09 ± 0.05 | 0.08 (0.05 – 0.12) |
| Total Trans (g)c | 1.17 (1.03 – 1.32) | 0.016 | 0.140 | 2.37 ± 1.18 | 2.16 (1.5 – 2.97) |
| Niacin (mg) | 0.96 (0.92 – 1.00) | 0.020 | 0.140 | 40.18 ± 36.86 | 33.56 (21.26 – 46.05) |
| Total folate intake (μg) | 0.91 (0.83 – 0.99) | 0.021 | 0.140 | 637.34 ± 343.80 | 603.57 (351.12 – 829.65) |
Methylation data is complete for 268 Hispanics and 1158 NHW.
Unit for calculating the OR is the difference between Q3 and Q1. Age, sex, ethnicity, smoking history (smoking status and packyears), BMI, and total calorie intake were included in the GEE models for covariate adjustment.
38% study subjects have missing data for these nutrients.
Interaction between B12 and MTHFR variant for risk for methylation
MTHFR catalyzes the irreversible reduction of 5,10-methylene tetrahydrofolate (THF) to 5-methylTHF that provides the methyl group for the remethylation reaction of homocysteine to methionine which is catalyzed by methionine synthase with B12 as the cofactor. SNP rs1801133 results in an Ala to Val change at codon 222 of exon 4, the folate binding site of MTHFR. Each copy of the T allele of rs1801133 leads to about a 30% reduction of MTHFR enzyme activity in vitro (30). Thus, we tested whether the association between B12 or folate intake and methylation could be modified by rs1801133. Genotype data for rs1801133 was acquired from our previous studies (19,31). Excitingly, a significant interaction was identified between B12 and rs1801133 (CC versus CT and TT) for methylation (P=0.0042). Stratification analysis by genotype identified a significant association for B12 only in LSC members with CC genotype (OR=0.97, 95% CI=0.95–0.98, P=0.000090, n=592), but not in those with T allele (OR=1.0, 95% CI=0.99–1.00, P=0.36, n=776). The interaction between folate and rs1801133 was not statistically significant (P=0.61).
Dietary nutrients and ethnic disparity in methylation
Individual nutrient analysis using multivariate linear regression identified 71 nutrients that had significant difference in their average levels between Hispanics and NHWs (Supplementary Table 1, FDR<0.05). The average difference of these 71 nutrients between the two ethnic groups was about 10.6% of the values seen in NHWs. Interestingly, Hispanics had higher dietary intake for only three nutrients (i.e., wheat germ, maltose, and copper without supplement).
Mediation analysis was conducted for the 101 nutrients without missing data to ensure optimal power. Twenty-two nutrients were identified to be nominally significantly associated with ethnicity and with methylation (Supplementary Table 1 and 4). Twelve nutrients were identified to reduce the coefficient for ethnicity by 3–23% and the delta changes of the coefficients for ethnicity for each of the 12 nutrients were all statistically significant (Ppermutation<0.05, Table 3). The mediation effect was further supported by showing no interaction between ethnicity and each of the individual 12 nutrients for affecting methylation (Ps for interaction>0.26). Inclusion of all 12 nutrients in the model reduced the coefficient for ethnicity by 42% (Ppermutation<0.001, Table 3).
Table 3.
Nutrients explaining the ethnic disparity in risk for methylationa
| Nutrient (unit/d)b | Nutrient not adjusted | Nutrient adjusted | Ratio | Ppermc | ||||
|---|---|---|---|---|---|---|---|---|
| Estimate | SE | P | Estimate | SE | P | |||
| Manganese (mg) | 0.144 | 0.069 | 0.036 | 0.112 | 0.069 | 0.106 | 0.77 | 0.001 |
| Vitamin D (IU) | 0.144 | 0.069 | 0.036 | 0.121 | 0.069 | 0.081 | 0.84 | 0.006 |
| Magnesium (mg) | 0.144 | 0.069 | 0.036 | 0.123 | 0.069 | 0.075 | 0.85 | 0.006 |
| Folate Equivalents (μg) | 0.144 | 0.069 | 0.036 | 0.127 | 0.069 | 0.068 | 0.88 | 0.023 |
| Manganese without supplements (mg) | 0.144 | 0.069 | 0.036 | 0.127 | 0.069 | 0.066 | 0.88 | 0.009 |
| Folic Acid (μg) | 0.144 | 0.069 | 0.036 | 0.128 | 0.069 | 0.066 | 0.88 | 0.026 |
| Total Folate intake (μg) | 0.144 | 0.069 | 0.036 | 0.128 | 0.069 | 0.066 | 0.88 | 0.021 |
| Niacin (mg) | 0.144 | 0.069 | 0.036 | 0.130 | 0.069 | 0.061 | 0.89 | 0.023 |
| Total Vitamin E atoco (mg) | 0.144 | 0.069 | 0.036 | 0.131 | 0.069 | 0.056 | 0.91 | <0.001 |
| Pantothenic acid (mg) | 0.144 | 0.069 | 0.036 | 0.132 | 0.069 | 0.056 | 0.92 | 0.036 |
| Alcohol (g) | 0.144 | 0.069 | 0.036 | 0.133 | 0.069 | 0.053 | 0.92 | 0.038 |
| Vitamin B12 (μg) | 0.144 | 0.069 | 0.036 | 0.140 | 0.069 | 0.041 | 0.97 | 0.002 |
| All above | 0.144 | 0.069 | 0.036 | 0.084 | 0.070 | 0.231 | 0.58 | <0.001 |
Adjusted for age, sex, current smoking status, packyears, and total calorie intake in the GEE models.
Assessment of total folate intake and folate equivalents includes all sources (i.e., natural food, supplements, and fortified foods). Folic acid includes supplements and fortified foods.
The methylation data for 12-gene was permuted randomly for 1000 times with delta change of beta coefficient for ethnicity for each individual nutrient calculated in each permutated database that formed the null distribution. The observed delta value was compared to this null distribution and P value was calculated as the percentage of permutations that resulted in delta values smaller than the observed value.
The effect of nutrients on double strand DNA repair capacity
Combinational effect of eight protective nutrients that also explained 42% of the ethnic disparity in methylation (Table 2 and 3) was assessed for the effect on double strand DNA repair capacity in 114 LSC subjects with dietary and double strand DNA repair capacity data available (20). Principal component 1 was extracted from the calorie adjusted dietary intake of the eight nutrients and explained 59% of the total variance. One inter-quartile range of principal component 1 was associated with a reduction of 1.43 cells with chromosomal aberration with adjustment of covariates, suggesting that these nutrients in combination were associated with a better double strand DNA repair capacity (P=0.02). However, this difference may be underestimated because of the in vitro culture period (72 hours) with standard medium for primary lymphocytes. Thus, we replicated these results using three immortalized human lung epithelial cell lines cultured in customized medium with high versus low levels of these eight nutrients. Although the low nutrient medium only slightly reduced the cell exponential proliferation rate by 22% to 30% (P=0.015, not shown), it dramatically increased the micronucleus frequency by 68 to 133% compared to high nutrient medium (Figure 1B). Overall, the data suggest that these eight protective nutrients for methylation in combination provide a better DNA repair capacity towards double strand DNA breaks, a mechanistic biomarker tightly linked to the acquisition of aberrant gene promoter methylation during lung carcinogenesis.
DISCUSSION
This is the first study that takes an unbiased nutrientomics approach to assess dietary intake of 139 nutrients estimated using a well validated FFQ in 1829 current and former smokers from NM. A global association analysis based on principal components was conducted first to minimize the chance for false positive results and identified a significant association between dietary intake and risk for gene methylation. This finding strongly supports dietary intake as a major modifiable factor affecting acquisition of promoter methylation of multiple TSGs in the lungs of smokers at risk for LC.
The current study replicated the protective effect of folate for methylation from our previous study of 21 candidate composite food variables (14) by using an enlarged sample size (n=1426), a larger panel of genes (n=12), and a more sophisticated analytical approach (GEE). Furthermore, among different dietary folate sources, folate from supplements was significantly associated with methylation and this association was not confounded by other components in the multi-vitamins. In addition, among all the nutrients assessed, vitamin B12 was found to be most significantly associated with methylation and this association was more prominent in LSC members carrying the wild type MTHFR SNP (rs1801133) associated with normal enzymatic activity of MTHFR, suggesting that normal physiological effect of vitamin B12 in one-carbon metabolism requires proper input of 5-methylTHF as substrate for homocysteine remethylation. Furthermore, a positive association of borderline significance was also identified between methionine and methylation (P=0.064, Supplementary Table 4), consistent with our previous finding that plasma methionine level was associated with increased methylation in 149 LSC members (32). When total folate intake, vitamin B12, and methionine as independent variables with minimal correlations (Pearson correlation coefficients range from 0.04 to 0.33, not shown) were included in one GEE model, all three nutrients were significantly associated with methylation (Ps ≤ 0.043, not shown). Thus, these findings emphasize the intricate mechanisms that potentially regulate gene-specific promoter methylation via one-carbon metabolism.
Individual nutrient association analysis further identified vitamin B12, manganese, magnesium, vitamin D, and niacin as being protective against methylation with effects stronger than seen for folate. More importantly, a combination of higher intake of vitamin B12 and manganese and lower intake of sodium and saturated fat that captured the variance carried by all 18 top-ranking nutrients could reduce risk for gene methylation by 36%. Silencing of TSGs through promoter hypermethylation plays a causal role in LC etiology (1,2). Because the detection of promoter methylation of TSGs in sputum samples provides an assessment of field cancerization in the lungs and is a validated biomarker for identifying smokers at higher risk for LC incidence, identification of dietary nutrients protective against methylation may have strong translational significance for LC chemoprevention in high risk populations. To begin to support this hypothesis, we conducted ex vivo and in vitro analyses to assess the effect of the eight protective nutrients on double strand DNA repair capacity, a mechanistic biomarker strongly linked to acquisition of gene promoter hypermethylation in lung carcinogenesis in smokers (15–22). Significant association was identified between these protective nutrients and better DNA repair capacity in cultured primary human lymphocytes. This finding was further replicated in human lung epithelial cells showing that supplementation of these nutrients at higher human physiological levels dramatically enhanced the DNA repair capacity towards double strand DNA breaks. These findings provided a functional insight that the effect of protective nutrients on methylation may be through the modulation of DNA repair towards cigarette smoke induced double strand DNA breaks.
The protective effect of vitamin D and niacin on the acquisition of methylation of TSGs in the lungs of smokers has biological probability. Similar protection has been observed in human normal rectal mucosa and early stage colorectal tumors where serum 25-D3 level or dietary vitamin D intake were associated with reduced promoter methylation of adenomatous polyposis coli or Wnt signaling genes, respectively (33,34). Furthermore, several in vitro studies provided evidence for the demethylation effect of 1,25-D3 on key TSGs whose re-expression was shown to suppress several oncogenic phenotypes in breast and prostate cancer cells (35–37). The underlying mechanism has been linked to the transcriptional activating effect of liganded vitamin D receptor through interaction with chromatin modifiers and remodelers, a well-established mechanism for demethylation or preventing methylation (38,39). Furthermore, a number of genes encoding for chromatin modifiers and remodelers such as histone demethylase are primary targets of liganded vitamin D receptor as well (38). Niacin is required for the synthesis of NAD and NADP, the cofactors in many redox reactions in cellular metabolism. The protective effect of niacin on the acquisition of methylation in the lungs of smokers is supported by findings from an in vitro experiment that nicotinamide treatment could reactivate an epigenetically silenced CMV promoter driving green fluorescent protein expression in colon cancer cells (40). This effect may be mediated through a mechanism in which higher levels of niacin act as substrates for reactions that favor direct poly(ADP-ribosyl)ation of histones and histone shuttling causing chromatin relaxation over SIRT1-catalyzed deacetylation resulting in chromatin compaction (41).
Few studies have assessed the effect of manganese and magnesium on gene methylation and the molecular mechanism remains unexplored. In a cohort of 61 mother-infant pairs, genome-wide DNA methylation status of placenta-derived DNA from placental samples was globally associated with infant toenail manganese levels which largely fell within a reference range seen in healthy individuals (42). In a transgenerational feeding study, magnesium deficiency in pregnant rats increased methylation of specific cytosines in the hepatic hydroxysteroid dehydrogenase-2 promoter of the offspring, a mechanism potentially underlying changes in glucocorticoid homeostasis of offspring caused by magnesium deficiency (43).
Although blood nutrient biomarkers were not measured in this study, previous studies have identified significant positive correlations between dietary intakes for folate (44), vitamin B12 (44,45), E (46) and D (47–49) using the Harvard FFQ or its modified forms and for pantothenic acid using 4-day diet records (50) and their levels in blood. Plasma magnesium concentrations are under tight homeostatic regulation by a variety of mechanisms, most notably by renal excretion; therefore, plasma magnesium is a poor surrogate for magnesium intake (51,52). Thus, the Harvard FFQ should provide a good to excellent assessment of dietary intakes for these nutrients that also correlate well with blood status.
NM populations provided a unique opportunity to reliably assess how the quantitative difference in dietary intake would affect the ethnic disparity in risk for gene methylation. NM Hispanics are distinct from Hispanic or Latino populations living in other states because they mainly include descendants of Spanish colonists who have settled the area of NM and Southern Colorado since the 1600s. Over the past 200 years, intense admixture of Spanish colonists with indigenous populations (Native Americans) and later (after 1912) with Anglophone Americans define the culture (e.g., diet of Southwestern style) and genetics of today’s NM Hispanics. Thus, our study is unlikely to be affected by immigration related factors such as acculturation status, healthy migrant effect, and salmon bias (53) which are common concerns in other studies involving Latino immigrants (7). This also explains why no exclusive pattern for consumption of southwestern regional foods or any ethnic foods exists in NM Hispanics versus NHWs (24). The assessment of ethnic disparity in dietary intake identified a large number of nutrients (n=71) with levels different between Hispanics and NHWs, although the average magnitude of difference is relatively small (about 10.6% of levels seen in NHWs). Mediation analysis identified that up to 42% of ethnic disparity in gene methylation could be explained by nine dietary nutrients that include manganese, vitamin D, magnesium, folate, niacin, vitamin E, pantothenic acid, alcohol, and vitamin B12, further supporting that greater risk for gene methylation in Hispanics may be due to the insufficient intake of these protective nutrients in their diet compared with NHWs (6). Thus, Hispanic smokers may benefit the most from supplements containing these nine protective nutrients for reducing their risk for gene methylation and LC susceptibility. Caution needs to be taken when generalizing our findings of ethnic disparity to other Hispanic populations living in different geographic areas because of the reasons mentioned above, and the heterogeneity of environmental and dietary exposures, admixture levels, and the source of NAA.
Supplementary Material
Acknowledgements
We thank the staff from Lovelace Scientific Resources for recruiting and enrolling study subjects and collecting clinical samples and data. We also thank the New Mexico residents who participate in this study.
Sources of Support This study was mainly funded by National Cancer Institute grant R01 CA097356. The State of New Mexico as a direct appropriation from the Tobacco Settlement Fund and NIH/NCI P30 CA118100 provided additional support for the establishment of the Lovelace Smokers cohort.
Abbreviations:
- AIM
ancestry informative marker
- GEE
generalized estimating equation
- LSC
Lovelace Smokers cohort
- NAA
Native American ancestry
- NHW
non-Hispanic White
Footnotes
The authors declare no potential conflicts of interest.
Note: Supplementary data for this article are available at Cancer Prevention Research Online (http://cancerpreventionresearch.aacrjournals.org/)
References
- 1.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nature reviews Cancer 2004;4(9):707–17. [DOI] [PubMed] [Google Scholar]
- 2.Baylin SB. The cancer epigenome: its origins, contributions to tumorigenesis, and translational implications. Proceedings of the American Thoracic Society 2012;9(2):64–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kadara H, Scheet P, Wistuba II, Spira AE. Early Events in the Molecular Pathogenesis of Lung Cancer. Cancer prevention research 2016;9(7):518–27. [DOI] [PubMed] [Google Scholar]
- 4.Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer research 2006;66(6):3338–44. [DOI] [PubMed] [Google Scholar]
- 5.Leng S, Do K, Yingling CM, Picchi MA, Wolf HJ, Kennedy TC, et al. Defining a gene promoter methylation signature in sputum for lung cancer risk assessment. Clinical cancer research : an official journal of the American Association for Cancer Research 2012;18(12):3387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leng S, Liu Y, Thomas CL, Gauderman WJ, Picchi MA, Bruse SE, et al. Native American ancestry affects the risk for gene methylation in the lungs of Hispanic smokers from New Mexico. American journal of respiratory and critical care medicine 2013;188(9):1110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harhay MO. The Hispanic paradox and chronic obstructive pulmonary disease. American journal of respiratory and critical care medicine 2012;185(11):1246, author reply 46–7. [DOI] [PubMed] [Google Scholar]
- 8.Bruse S, Sood A, Petersen H, Liu Y, Leng S, Celedon JC, et al. New Mexican Hispanic smokers have lower odds of chronic obstructive pulmonary disease and less decline in lung function than non-Hispanic whites. American journal of respiratory and critical care medicine 2011;184(11):1254–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humble CG, Samet JM. Smoking and lung cancer in New Mexico. American journal of public health 1986;76(11):1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humble CG, Samet JM, Pathak DR, Skipper BJ. Cigarette smoking and lung cancer in ‘Hispanic’ whites and other whites in New Mexico. American journal of public health 1985;75(2):145–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trinidad DR, Perez-Stable EJ, White MM, Emery SL, Messer K. A nationwide analysis of US racial/ethnic disparities in smoking behaviors, smoking cessation, and cessation-related factors. American journal of public health 2011;101(4):699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods in molecular biology 2012;863:359–76. [DOI] [PubMed] [Google Scholar]
- 13.Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Advances in nutrition 2012;3(1):21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stidley CA, Picchi MA, Leng S, Willink R, Crowell RE, Flores KG, et al. Multivitamins, folate, and green vegetables protect against gene promoter methylation in the aerodigestive tract of smokers. Cancer research 2010;70(2):568–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuozzo C, Porcellini A, Angrisano T, Morano A, Lee B, Di Pardo A, et al. DNA damage, homology-directed repair, and DNA methylation. PLoS genetics 2007;3(7):e110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Esteve PO, Chin HG, Pradhan S. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proceedings of the National Academy of Sciences of the United States of America 2005;102(4):1000–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esteve PO, Chin HG, Pradhan S. Molecular mechanisms of transactivation and doxorubicin-mediated repression of survivin gene in cancer cells. The Journal of biological chemistry 2007;282(4):2615–25. [DOI] [PubMed] [Google Scholar]
- 18.Le Gac G, Esteve PO, Ferec C, Pradhan S. DNA damage-induced down-regulation of human Cdc25C and Cdc2 is mediated by cooperation between p53 and maintenance DNA (cytosine-5) methyltransferase 1. The Journal of biological chemistry 2006;281(34):24161–70. [DOI] [PubMed] [Google Scholar]
- 19.Leng S, Liu Y, Weissfeld JL, Thomas CL, Han Y, Picchi MA, et al. 15q12 variants, sputum gene promoter hypermethylation, and lung cancer risk: a GWAS in smokers. Journal of the National Cancer Institute 2015;107(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leng S, Stidley CA, Willink R, Bernauer A, Do K, Picchi MA, et al. Double-strand break damage and associated DNA repair genes predispose smokers to gene methylation. Cancer research 2008;68(8):3049–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proceedings of the National Academy of Sciences of the United States of America 2005;102(25):8905–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS genetics 2008;4(8):e1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rimm EB, Giovannucci EL, Stampfer MJ, Colditz GA, Litin LB, Willett WC. Reproducibility and validity of an expanded self-administered semiquantitative food frequency questionnaire among male health professionals. American journal of epidemiology 1992;135(10):1114–26; discussion 27–36. [DOI] [PubMed] [Google Scholar]
- 24.Pareo-Tubbeh SL, Romero LJ, Baumgartner RN, Garry PJ, Lindeman RD, Koehler KM. Comparison of energy and nutrient sources of elderly Hispanics and non-Hispanic whites in New Mexico. Journal of the American Dietetic Association 1999;99(5):572–82. [DOI] [PubMed] [Google Scholar]
- 25.Lindeman RD, Romero LJ, Koehler KM, Liang HC, LaRue A, Baumgartner RN, et al. Serum vitamin B12, C and folate concentrations in the New Mexico elder health survey: correlations with cognitive and affective functions. Journal of the American College of Nutrition 2000;19(1):68–76. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer research 2004;64(24):9027–34. [DOI] [PubMed] [Google Scholar]
- 27.Leng S, Dai Y, Niu Y, Pan Z, Li X, Cheng J, et al. Effects of genetic polymorphisms of metabolic enzymes on cytokinesis-block micronucleus in peripheral blood lymphocyte among coke-oven workers. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2004;13(10):1631–9. [PubMed] [Google Scholar]
- 28.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics 1986;42(1):121–30. [PubMed] [Google Scholar]
- 29.Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. Journal of personality and social psychology 1986;51(6):1173–82. [DOI] [PubMed] [Google Scholar]
- 30.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nature genetics 1995;10(1):111–3. [DOI] [PubMed] [Google Scholar]
- 31.Leng S, Stidley CA, Liu Y, Edlund CK, Willink RP, Han Y, et al. Genetic determinants for promoter hypermethylation in the lungs of smokers: a candidate gene-based study. Cancer research 2012;72(3):707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flores KG, Stidley CA, Mackey AJ, Picchi MA, Stabler SP, Siegfried JM, et al. Sex-specific association of sequence variants in CBS and MTRR with risk for promoter hypermethylation in the lung epithelium of smokers. Carcinogenesis 2012;33(8):1542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tapp HS, Commane DM, Bradburn DM, Arasaradnam R, Mathers JC, Johnson IT, et al. Nutritional factors and gender influence age-related DNA methylation in the human rectal mucosa. Aging cell 2013;12(1):148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawson JB, Sun Z, Dicks E, Daftary D, Parfrey PS, Green RC, et al. Vitamin D intake is negatively associated with promoter methylation of the Wnt antagonist gene DKK1 in a large group of colorectal cancer patients. Nutrition and cancer 2012;64(7):919–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doig CL, Singh PK, Dhiman VK, Thorne JL, Battaglia S, Sobolewski M, et al. Recruitment of NCOR1 to VDR target genes is enhanced in prostate cancer cells and associates with altered DNA methylation patterns. Carcinogenesis 2013;34(2):248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopes N, Carvalho J, Duraes C, Sousa B, Gomes M, Costa JL, et al. 1Alpha,25-dihydroxyvitamin D3 induces de novo E-cadherin expression in triple-negative breast cancer cells by CDH1-promoter demethylation. Anticancer research 2012;32(1):249–57. [PubMed] [Google Scholar]
- 37.Vanoirbeek E, Eelen G, Verlinden L, Carmeliet G, Mathieu C, Bouillon R, et al. PDLIM2 expression is driven by vitamin D and is involved in the pro-adhesion, and anti-migration and -invasion activity of vitamin D. Oncogene 2014;33(15):1904–11. [DOI] [PubMed] [Google Scholar]
- 38.Fetahu IS, Hobaus J, Kallay E. Vitamin D and the epigenome. Frontiers in physiology 2014;5:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leng S, Bernauer AM, Hong C, Do KC, Yingling CM, Flores KG, et al. The A/G allele of rs16906252 predicts for MGMT methylation and is selectively silenced in premalignant lesions from smokers and in lung adenocarcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research 2011;17(7):2014–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raynal NJ, Lee JT, Wang Y, Beaudry A, Madireddi P, Garriga J, et al. Targeting calcium signaling induces epigenetic reactivation of tumor suppressor genes in cancer. Cancer research 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirkland JB. Niacin status impacts chromatin structure. The Journal of nutrition 2009;139(12):2397–401. [DOI] [PubMed] [Google Scholar]
- 42.Maccani JZ, Koestler DC, Houseman EA, Armstrong DA, Marsit CJ, Kelsey KT. DNA methylation changes in the placenta are associated with fetal manganese exposure. Reproductive toxicology 2015;57:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takaya J, Iharada A, Okihana H, Kaneko K. Magnesium deficiency in pregnant rats alters methylation of specific cytosines in the hepatic hydroxysteroid dehydrogenase-2 promoter of the offspring. Epigenetics : official journal of the DNA Methylation Society 2011;6(5):573–8. [DOI] [PubMed] [Google Scholar]
- 44.Zhang SM, Willett WC, Selhub J, Hunter DJ, Giovannucci EL, Holmes MD, et al. Plasma folate, vitamin B6, vitamin B12, homocysteine, and risk of breast cancer. Journal of the National Cancer Institute 2003;95(5):373–80. [DOI] [PubMed] [Google Scholar]
- 45.Tucker KL, Rich S, Rosenberg I, Jacques P, Dallal G, Wilson PW, et al. Plasma vitamin B-12 concentrations relate to intake source in the Framingham Offspring study. The American journal of clinical nutrition 2000;71(2):514–22. [DOI] [PubMed] [Google Scholar]
- 46.Tangney CC, Bienias JL, Evans DA, Morris MC. Reasonable estimates of serum vitamin E, vitamin C, and beta-cryptoxanthin are obtained with a food frequency questionnaire in older black and white adults. The Journal of nutrition 2004;134(4):927–34. [DOI] [PubMed] [Google Scholar]
- 47.Bunyavanich S, Rifas-Shiman SL, Platts-Mills TA, Workman L, Sordillo JE, Camargo CA Jr., et al. Prenatal, perinatal, and childhood vitamin D exposure and their association with childhood allergic rhinitis and allergic sensitization. The Journal of allergy and clinical immunology 2016;137(4):1063–70 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giovannucci E, Liu Y, Rimm EB, Hollis BW, Fuchs CS, Stampfer MJ, et al. Prospective study of predictors of vitamin D status and cancer incidence and mortality in men. Journal of the National Cancer Institute 2006;98(7):451–9. [DOI] [PubMed] [Google Scholar]
- 49.Ananthakrishnan AN, Khalili H, Higuchi LM, Bao Y, Korzenik JR, Giovannucci EL, et al. Higher predicted vitamin D status is associated with reduced risk of Crohn’s disease. Gastroenterology 2012;142(3):482–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eissenstat BR, Wyse BW, Hansen RG. Pantothenic acid status of adolescents. The American journal of clinical nutrition 1986;44(6):931–7. [DOI] [PubMed] [Google Scholar]
- 51.Chiuve SE, Sun Q, Curhan GC, Taylor EN, Spiegelman D, Willett WC, et al. Dietary and plasma magnesium and risk of coronary heart disease among women. Journal of the American Heart Association 2013;2(2):e000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiuve SE, Korngold EC, Januzzi JL Jr., Gantzer ML, Albert CM. Plasma and dietary magnesium and risk of sudden cardiac death in women. The American journal of clinical nutrition 2011;93(2):253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turra CM, Elo IT. The Impact of Salmon Bias on the Hispanic Mortality Advantage: New Evidence from Social Security Data. Population research and policy review 2008;27(5):515–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.