Protein crystallization of 3CL Mpro from SARS-CoV-2 is described along with neutron and X-ray data collection.
Keywords: neutron diffraction, X-ray diffraction, SARS-CoV-2, 3CL Mpro
Abstract
The replication of SARS-CoV-2 produces two large polyproteins, pp1a and pp1ab, that are inactive until cleavage by the viral chymotrypsin-like cysteine protease enzyme (3CL Mpro) into a series of smaller functional proteins. At the heart of 3CL Mpro is an unusual catalytic dyad formed by the side chains of His41 and Cys145 and a coordinated water molecule. The catalytic mechanism by which the enzyme operates is still unknown, as crucial information on the protonation states within the active site is unclear. To experimentally determine the protonation states of the catalytic site and of the other residues in the substrate-binding cavity, and to visualize the hydrogen-bonding networks throughout the enzyme, room-temperature neutron and X-ray data were collected from a large H/D-exchanged crystal of ligand-free (apo) 3CL Mpro.
1. Introduction
COVID-19 is a global pandemic that is causing worldwide disruption to travel, economic activity and social life (Zhou et al., 2020 ▸; Wu et al., 2020a ▸,b ▸; Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020 ▸). It is caused by the SARS-CoV-2 coronavirus, an enveloped positive-sense single-stranded RNA virus with a genome of around 30 kb. The virus is composed of around 28 proteins that can be broken down into two main classes: nonstructural proteins (NSPs) that are typically involved in viral replication and structural proteins that are involved in forming the structure of the virus. NSP5 or 3CL Mpro is a cysteine protease that is essential for viral replication. It performs a critical function by cleaving the two large viral polyproteins pp1a and pp1ab into smaller functional proteins (Gorbalenya & Snijder, 1996 ▸). The lack of a homologous human protein makes 3CL Mpro one of the most promising targets for the design of specific protease inhibitors (Zhang et al., 2020 ▸; Kneller et al., 2020 ▸; Jin et al., 2020 ▸). The global crystallographic community has rapidly been exploring chemical space in the search for drug candidates that can block the action of 3CL Mpro (Zhang et al., 2020 ▸). To date, no FDA-approved inhibitors (Pillaiyar et al., 2016 ▸) have been developed against the closely related 3CL Mpro from the 2003 SARS-CoV that shares 96% amino-acid homology with the 3CL Mpro from SARS-CoV-2.
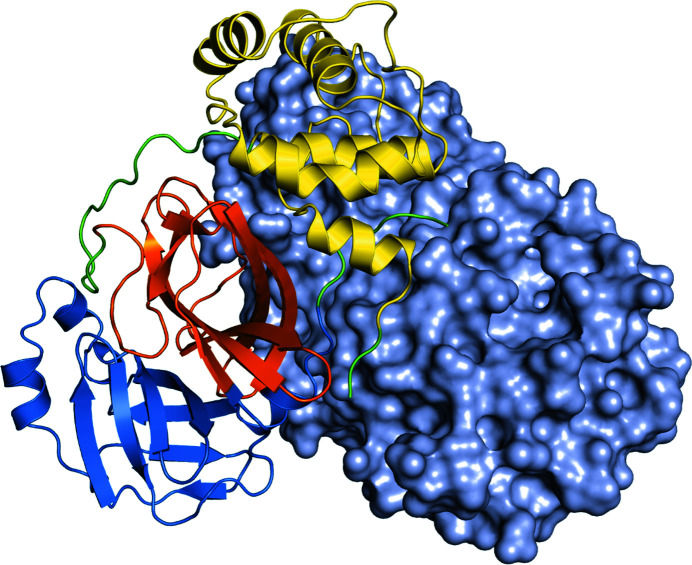
3CL Mpro is active as a homodimer that is composed of two amino-acid chains of 306 amino acids (Kneller et al., 2020 ▸; Zhou et al., 2020 ▸; Jin et al., 2020 ▸). Each chain is folded into three domains. Domain I is composed of residues 8–101, while domain II is composed of residues 102–184. Domains I and II are formed from antiparallel β-barrel structures and are the catalytic domains. Domain III is formed from residues 201–303 and consists mainly of five α-helices where the two polypeptide chains interact to form a dimer (Fig. 1 ▸). Based on earlier studies of SARS-CoV 3CL Mpro, domain III plays an essential role in the protease function as the monomeric enzyme is not catalytically active (Hsu et al., 2005 ▸).
Figure 1.
The structure of 3CL Mpro from SARS-CoV-2. One polypeptide chain of the dimer is shown as a cartoon, where domain I is colored blue, domain II is colored orange and domain III is colored yellow, whereas the loops connecting the domains are colored green. The other polypeptide chain is shown as a light blue surface.
To improve the drug design of SARS-CoV-2 3CL Mpro-specific protease inhibitors, knowledge of the complete enzyme structure, including the locations of H atoms, and the direct visualization of hydrogen-bonding networks is of paramount importance. The presence or absence of H atoms on ionizable chemical groups changes the electrical charges of amino-acid side chains and therefore can modulate the electrostatics of the drug-binding cavity. The only technique that is capable of the direct determination of H-atom positions is neutron crystallography. Neutron protein crystallography (NPC) can provide the locations of H and D atoms experimentally rather than by inference based on general chemical knowledge. The atomic nucleus scatters neutrons, and those scattered from deuterium, an isotope of hydrogen, are scattered with the same magnitude as the heavier elements of the protein such as carbon, nitrogen and oxygen. Thus, with NPC the positions of D atoms can be determined directly, even at moderate resolutions of around 2 Å (Bacik et al., 2017 ▸; Vandavasi et al., 2016 ▸; Kovalevsky et al., 2010 ▸; Gerlits et al., 2016 ▸; Kumar et al., 2020 ▸), compared with X-rays, which typically require much higher resolutions of below 1 Å to begin to observe some H-atom positions (Erskine et al., 2003 ▸; Coates et al., 2001 ▸).
Here, we report the crystallization of, and X-ray and neutron diffraction data collection from, a large SARS-CoV-2 3CL Mpro crystal of 0.3 mm3 in volume. We demonstrate that neutron diffraction-quality crystals of this enzyme can be grown successfully using a microseeding crystallization technique. The crystal of hydrogenous H/D-exchanged 3CL Mpro diffracted X-rays to a resolution of 2.3 Å and neutrons to a resolution of 2.5 Å at room temperature.
2. Materials and methods
2.1. 3CL Mpro expression and purification
The 3CL Mpro (NSP5 Mpro) gene from SARS-CoV-2 was cloned into the pD451-SR plasmid harboring kanamycin resistance (ATUM, Newark, California, USA). The 3CL Mpro gene was surrounded on either side by an upstream maltose-binding protein (MBP) and a downstream His6 tag. This plasmid was transformed into Escherichia coli BL21(DE3) cells and sequence-verified. MBP fusion is normally used to improve the solubility of proteins during overexpression, and kanamycin resistance is the preferred selectable marker for future expression in deuterated media. In this construct, MBP is followed by the the SARS-CoV-2 NSP4-NSP5 autoprocessing site SAVLQ↓SGFRK and 3CL Mpro is followed by the human rhinovirus 3C protease (HRV 3C) cleavage site SGVTFQ↓GP. MBP and the His6 tag can be cleaved by Mpro in vivo autoprocessing and by in vitro HRV 3C protease treatment, respectively, allowing clarification of the authentic 3CL Mpro by nickel-affinity chromatography.
Expression of 3CL Mpro was performed in Luria–Bertani broth with 1 g l−1 glucose using the antibiotic kanamycin (50 mg l−1). When the culture had grown to an OD600 of 0.8 at 37°C, overexpression was induced with 0.2 mM isopropyl β-d-1-thiogalactopyranoside for 18 h at 18°C. The harvested cells were lysed by sonication in 20 mM Tris pH 8.0, 150 mM NaCl, 40 mM imidazole, 1 mM tris(2-carboxyethyl)phosphine (TCEP) and centrifuged at 30 000g for 30 min. The soluble lysate was loaded onto a HisTrap FF column and eluted as a single peak using a linear gradient of 20 mM Tris pH 8.0, 150 mM NaCl, 500 mM imidazole, 1 mM TCEP. The His6 tag was removed by treating the pooled eluate with HRV 3C protease also containing a His6 tag (Sigma–Aldrich, St Louis, Missouri, USA) at a ratio of 500 mg 3CL Mpro to 1 mg HRV 3C protease. The sample was then dialyzed in 20 mM Tris pH 8.0, 150 mM NaCl, 1 mM TCEP for 18 h at 4°C. When this mixture was applied onto a HisTrap FF column, HRV 3C, uncleaved 3CL Mpro and cleaved His6 tags were retained and authentic 3CL Mpro was collected as the flowthrough. The final production yield of purified protein was ∼15 mg per litre of expression culture. The protein was kept in 20 mM Tris pH 8.0, 150 mM NaCl and fresh 1 mM TCEP baseline buffer conditions. Each 3CL Mpro molecule features 12 cysteine residues, including the catalytic Cys145, which may be prone to oxidation in the absence of a reducing agent such as TCEP. Protein samples were concentrated to 4–6 mg ml−1 (∼120–180 µM based on monomers) for crystallization using polyethersulfone membrane centrifugal concentrators (Sartorius, Göttingen, Germany). Mpro is expected to be primarily dimeric at this concentration as the homodimer dissociation constant has previously been measured to be 2.5 µM by analytical ultracentrifugation (Zhang et al., 2020 ▸).
2.2. 3CL Mpro crystallization
High-throughput crystal screening of 3CL Mpro was initially conducted at the Hauptman–Woodward Medical Research Institute (HWI; Luft et al., 2003 ▸) in order to explore new conditions that are capable of growing high-quality crystals for neutron diffraction at room temperature. This identified initial crystallization conditions for further optimization efforts onsite at Oak Ridge National Laboratory (ORNL). 3CL Mpro crystals were grown at ORNL using the sitting-drop vapor-diffusion method with a 1:1 protein:well solution ratio. Initial crystal aggregates appeared in 20 µl drops with 25% PEG 3350, 0.1 M Bis-Tris pH 6.5 as the reservoir solution using microbridges (Hampton Research, Aliso Viejo, California, USA). Multiple crystal clusters (Fig. 2 ▸) were harvested to generate microseeds with Hampton Research Seed Beads. The microseeding technique provides the reliable reproduction of single crystals, allowing optimization trials for precipitant concentration and pH using grid searching to be carried out (Budayova-Spano et al., 2020 ▸). It is also important to determine whether the freeze–thaw process of concentrated protein samples and microseed stocks has deleterious effects on crystallization. This is especially true for proteases that are capable of autoproteolysis. 3CL Mpro samples can grow crystals after storage at −30°C, but preparing aliquots for crystallization avoiding multiple freeze–thawing cycles is highly recommended.
Figure 2.
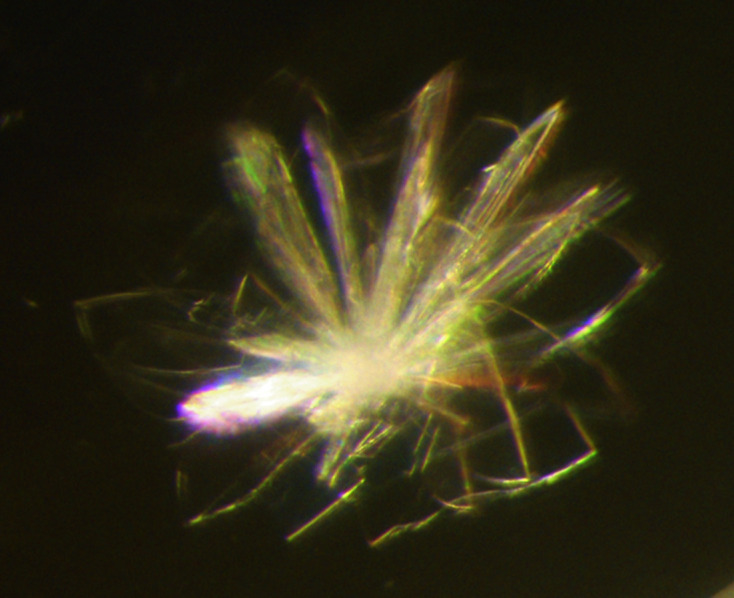
A small cluster consisting of many 3CL Mpro crystals with a maximum length of 200 µm. Several clusters were used to generate the seed stocks necessary to nucleate the large single crystal grown for neutron protein crystallography studies at ORNL.
Large-volume sitting-drop vapor diffusion for neutron diffraction-quality crystals of 3CL Mpro was performed in 200 µl drops of freshly prepared hydrogenated protein mixed with 18% PEG 3350, 0.1 M Bis-Tris pH 6.0, 3% DMSO seeded with 1 µl microseeds (1:500 dilution) in a Hampton Research 9-well plate and sandwich-box setup with 50 ml well solution.
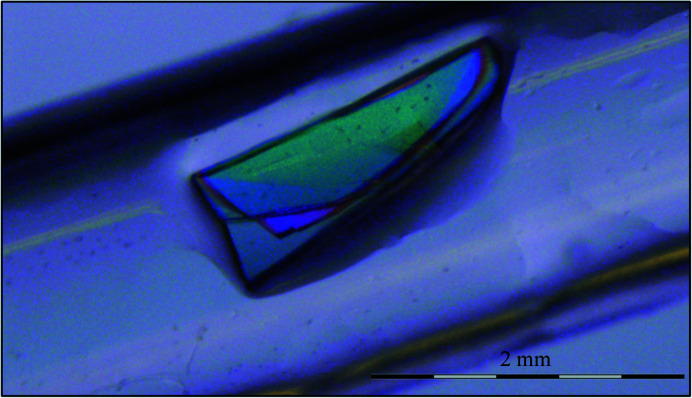
In this condition, we observed that individual crystals as large as ∼0.8 × 0.2 × 0.05 mm in size appeared if left undisturbed for 3–5 days at 18°C and continued to increase in size for 2–3 weeks until growth ceased. To encourage continued crystal growth, the temperature can be gradually reduced at this point to decrease the solubility of the protein in the crystallization drops. 3CL Mpro follows the traditional trend of fewer nucleation sites, microseeds in this case, and a larger crystallization drop volume driving increased crystal size. The largest crystal (Fig. 3 ▸) grew to ∼2 × 0.8 × 0.2 mm (∼0.3 mm3) after incubation at 18°C gradually reduced to 10°C over five weeks in a drop with one other crystal. This hydrogenated crystal was then mounted in a capillary with mother liquor prepared with D2O and allowed to exchange with deuterium for one week before starting data collection. A pH of 6.6 for the final crystallization drop was measured using a microelectrode, corresponding to a pD of 7.0 (pD = pH + 0.4).
Figure 3.
A large protein crystal of 3CL Mpro of roughly 2 × 0.8 × 0.2 mm in size, giving a total crystal volume of around 0.3 mm3, was used for neutron data collection on the MaNDi instrument at the Spallation Neutron Source.
3. Results and discussion
3.1. Neutron and X-ray room-temperature data collection
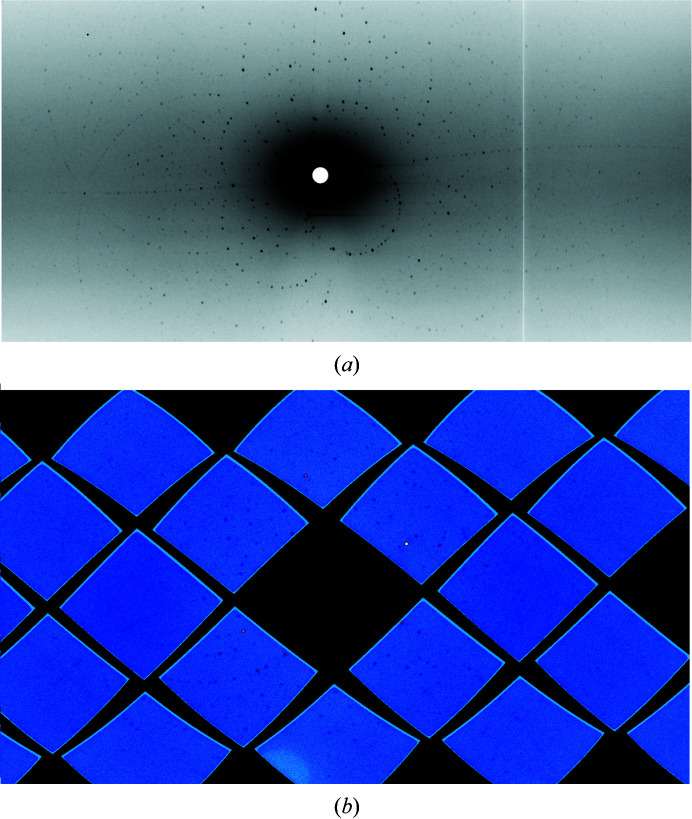
Crystals were initially screened for diffraction quality using a broad-bandpass Laue configuration using neutrons from 2.8 to 10 Å wavelength at the IMAGINE instrument (Coates et al., 2018 ▸; Schröder et al., 2018 ▸) at the High Flux Isotope Reactor (HFIR) at Oak Ridge National Laboratory (Fig. 4 ▸ a). The observed strong neutron diffraction indicated that the crystal was single and worthy of complete data collection. Neutron diffraction data were then collected using the time-of-flight (TOF; Langan et al., 2008 ▸) technique on the Macromolecular Neutron Diffractometer (MaNDi) instrument (Coates et al., 2010 ▸, 2015 ▸, 2018 ▸) at the Spallation Neutron Source (SNS) because the geometry of the MaNDi diffractometer provides better reciprocal-space coverage for the low-symmetry monoclinic space group. The crystal was held stationary at room temperature, and diffraction data (Fig. 4 ▸ b) were collected for 24 h using all neutrons between 2 and 4.16 Å wavelength. Following this, the crystal was rotated by Δφ = 10° and a subsequent data frame was collected. A total of 23 data frames were collected to form the final neutron data set. The diffraction data were reduced using the Mantid package (Arnold et al., 2014 ▸), with integration being performed by three-dimensional TOF profile fitting (Sullivan et al., 2018 ▸). Wavelength-normalization of the Laue data was performed using LAUENORM from the LAUEGEN suite (Campbell et al., 1998 ▸; Helliwell et al., 1989 ▸).
Figure 4.
Neutron diffraction images obtained from a 3CL Mpro crystal. (a) A 12 h exposure on the IMAGINE instrument using all neutrons between 2.8 and 10 Å wavelength. (b) A 24 h exposure on the MaNDi instrument using neutrons between 2 and 4.16 Å wavelength.
Using the same crystal, X-ray diffraction images were recorded at ORNL using a Rigaku MicroMax-007 HF X-ray generator coupled with a Dectris EIGER R 4M detector and were reduced using the CrysAlisPro software suite (Rigaku, The Woodlands, Texas, USA). The diffraction data were then scaled using AIMLESS (Evans & Murshudov, 2013 ▸) from the CCP4 suite (Cowtan et al., 2011 ▸; Winn et al., 2011 ▸). The neutron and X-ray data-collection and processing statistics are given in Table 1 ▸.
Table 1. Room-temperature neutron and X-ray data-collection statistics.
| X-ray | Neutron | |
|---|---|---|
| Unit-cell parameters (Å, °) | a = 114.93, b = 54.69, c = 45.22, α = 90, β = 101.46, γ = 90 | |
| Space group | C2 | |
| Wavelength(s) (Å) | 1.54 | 2–4.16 |
| No. of unique reflections | 12092 (1155) | 7995 (765) |
| Resolution range (Å) | 28.16–2.30 (2.38–2.30) | 14.95–2.50 (2.59–2.50) |
| Multiplicity | 4.30 (3.70) | 5.67 (4.68) |
| 〈I/σ(I)〉 | 8.6 (1.7) | 13.0 (3.70) |
| CC1/2 | 0.993 (0.480) | 0.961 (0.551) |
| R p.i.m. | 0.069 (0.544) | 0.078 (0.156) |
| Data completeness (%) | 98.20 (96.60) | 83.93 (83.33) |
The resolution of the diffraction data that we obtained with neutrons was limited by the size and the morphology of the hydrogenated crystal that could be produced in a limited time frame. It is envisioned that larger crystal volumes and deuterium labeling of the protein by expression in deuterated medium will allow us to collect neutron data sets to higher resolutions.
Acknowledgments
This research used resources at the Spallation Neutron Source and the High Flux Isotope Reactor, which are DOE Office of Science User Facilities operated by Oak Ridge National Laboratory. The Office of Biological and Environmental Research supported research at ORNL’s Center for Structural Molecular Biology (CSMB), a DOE Office of Science User Facility. Initial crystallization screening at HWI was supported through NSF grant 2029943. This research used resources at the Second Target Station, which is a DOE Office of Science User Facilities Construction Project at Oak Ridge National Laboratory.
Funding Statement
This work was funded by Office of Science, National Virtual Biotechnology Laboratory grant .
References
- Arnold, O., Bilheux, J. C., Borreguero, J. M., Buts, A., Campbell, S. I., Chapon, L., Doucet, M., Draper, N., Ferraz Leal, R., Gigg, M. A., Lynch, V. E., Markvardsen, A., Mikkelson, D. J., Mikkelson, R. L., Miller, R., Palmen, K., Parker, P., Passos, G., Perring, T. G., Peterson, P. F., Ren, S., Reuter, M. A., Savici, A. T., Taylor, J. W., Taylor, R. J., Tolchenov, R., Zhou, W. & Zikovsky, J. (2014). Nucl. Instrum. Methods Phys. Res. A, 764, 156–166.
- Bacik, J.-P., Mekasha, S., Forsberg, Z., Kovalevsky, A. Y., Vaaje-Kolstad, G., Eijsink, V. G. H., Nix, J. C., Coates, L., Cuneo, M. J., Unkefer, C. J. & Chen, J. C.-H. (2017). Biochemistry, 56, 2529–2532. [DOI] [PubMed]
- Budayova-Spano, M., Koruza, K. & Fisher, Z. (2020). Methods Enzymol. 634, 21–46. [DOI] [PubMed]
- Campbell, J. W., Hao, Q., Harding, M. M., Nguti, N. D. & Wilkinson, C. (1998). J. Appl. Cryst. 31, 496–502.
- Coates, L., Cao, H. B., Chakoumakos, B. C., Frontzek, M. D., Hoffmann, C., Kovalevsky, A. Y., Liu, Y., Meilleur, F., dos Santos, A. M., Myles, D. A. A., Wang, X. P. & Ye, F. (2018). Rev. Sci. Instrum. 89, 092802. [DOI] [PubMed]
- Coates, L., Cuneo, M. J., Frost, M. J., He, J., Weiss, K. L., Tomanicek, S. J., McFeeters, H., Vandavasi, V. G., Langan, P. & Iverson, E. B. (2015). J. Appl. Cryst. 48, 1302–1306.
- Coates, L., Erskine, P. T., Wood, S. P., Myles, D. A. A. & Cooper, J. B. (2001). Biochemistry, 40, 13149–13157. [DOI] [PubMed]
- Coates, L., Stoica, A. D., Hoffmann, C., Richards, J. & Cooper, R. (2010). J. Appl. Cryst. 43, 570–577.
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (2020). Nat. Microbiol. 5, 536–544.
- Cowtan, K., Emsley, P. & Wilson, K. S. (2011). Acta Cryst. D67, 233–234. [DOI] [PMC free article] [PubMed]
- Erskine, P. T., Coates, L., Mall, S., Gill, R. S., Wood, S. P., Myles, D. A. A. & Cooper, J. B. (2003). Protein Sci. 12, 1741–1749. [DOI] [PMC free article] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Gerlits, O., Wymore, T., Das, A., Shen, C. H., Parks, J. M., Smith, J. C., Weiss, K. L., Keen, D. A., Blakeley, M. P., Louis, J. M., Langan, P., Weber, I. T. & Kovalevsky, A. (2016). Angew. Chem. Int. Ed. 55, 4924–4927. [DOI] [PMC free article] [PubMed]
- Gorbalenya, A. E. & Snijder, E. J. (1996). Perspect. Drug Discov. Des. 6, 64–86. [DOI] [PMC free article] [PubMed]
- Helliwell, J. R., Habash, J., Cruickshank, D. W. J., Harding, M. M., Greenhough, T. J., Campbell, J. W., Clifton, I. J., Elder, M., Machin, P. A., Papiz, M. Z. & Zurek, S. (1989). J. Appl. Cryst. 22, 483–497.
- Hsu, W.-C., Chang, H.-C., Chou, C.-Y., Tsai, P.-J., Lin, P.-I. & Chang, G.-G. (2005). J. Biol. Chem. 280, 22741–22748. [DOI] [PMC free article] [PubMed]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., Yang, X., Bai, F., Liu, H., Liu, X., Guddat, L. W., Xu, W., Xiao, G., Qin, C., Shi, Z., Jiang, H., Rao, Z. & Yang, H. (2020). Nature, 582, 289–293. [DOI] [PubMed]
- Kneller, D. W., Phillips, G., O’Neill, H. M., Jedrzejczak, R., Stols, L., Langan, P., Joachimiak, A., Coates, L. & Kovalevsky, A. (2020). Nat. Commun. 11, 3202. [DOI] [PMC free article] [PubMed]
- Kovalevsky, A. Y., Chatake, T., Shibayama, N., Park, S.-Y., Ishikawa, T., Mustyakimov, M., Fisher, Z., Langan, P. & Morimoto, Y. (2010). J. Mol. Biol. 398, 276–291. [DOI] [PubMed]
- Kumar, M., Mandal, K., Blakeley, M. P., Wymore, T., Kent, S. B. H., Louis, J. M., Das, A. & Kovalevsky, A. (2020). ACS Omega, 5, 11605–11617. [DOI] [PMC free article] [PubMed]
- Langan, P., Fisher, Z., Kovalevsky, A., Mustyakimov, M., Sutcliffe Valone, A., Unkefer, C., Waltman, M. J., Coates, L., Adams, P. D., Afonine, P. V., Bennett, B., Dealwis, C. & Schoenborn, B. P. (2008). J. Synchrotron Rad. 15, 215–218. [DOI] [PMC free article] [PubMed]
- Luft, J. R., Collins, R. J., Fehrman, N. A., Lauricella, A. M., Veatch, C. K. & DeTitta, G. T. (2003). J. Struct. Biol. 142, 170–179. [DOI] [PubMed]
- Pillaiyar, T., Manickam, M., Namasivayam, V., Hayashi, Y. & Jung, S.-H. (2016). J. Med. Chem. 59, 6595–6628. [DOI] [PMC free article] [PubMed]
- Schröder, G. C., O’Dell, W. B., Myles, D. A. A., Kovalevsky, A. & Meilleur, F. (2018). Acta Cryst. D74, 778–786. [DOI] [PubMed]
- Sullivan, B., Archibald, R., Langan, P. S., Dobbek, H., Bommer, M., McFeeters, R. L., Coates, L., Wang, X. P., Gallmeier, F., Carpenter, J. M., Lynch, V. & Langan, P. (2018). Acta Cryst. D74, 1085–1095. [DOI] [PMC free article] [PubMed]
- Vandavasi, V. G., Weiss, K. L., Cooper, J. B., Erskine, P. T., Tomanicek, S. J., Ostermann, A., Schrader, T. E., Ginell, S. L. & Coates, L. (2016). J. Med. Chem. 59, 474–479. [DOI] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z.-W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y., Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E. C. & Zhang, Y.-Z. (2020a). Nature, 579, 265–269. [DOI] [PMC free article] [PubMed]
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z.-W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y., Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E. C. & Zhang, Y.-Z. (2020b). Nature, 580, E7. [DOI] [PMC free article] [PubMed]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K. & Hilgenfeld, R. (2020). Science, 368, 409–412. [DOI] [PMC free article] [PubMed]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., Zheng, X.-S., Zhao, K., Chen, Q.-J., Deng, F., Liu, L.-L., Yan, B., Zhan, F.-X., Wang, Y.-Y., Xiao, G.-F. & Shi, Z.-L. (2020). Nature, 579, 270–273.