ABSTRACT
The neonatal Fc receptor (FcRn) is a key membrane protein that plays an integral role in serum immunoglobulin (IgG) recycling, which extends the half-life of antibody. In addition, FcRn is known to traffic antigen-bound immunoglobulins (Ag-IgGs), and to interact with immune complexes to facilitate the antigen cross-presentation of peptides derived from the immune complexes in antigen-presenting cells (APCs). Studies on the IgG-FcRn molecular interactions have primarily focused on the Fc region, and only recently have shown the potential impact of the antigen-binding fragment physiochemical properties on FcRn binding. However, the effect of the antigen physiochemical properties on IgG structure as it relates to Ag-IgG-FcRn binding is not well understood. Here we used an IgG-peptide antigen complex as a model system to investigate the structural effects of the antigen’s physiochemical properties on the IgG structure, and the subsequent effects of Ag-IgG-FcRn interactions. We used hydroxyl radical footprinting–mass spectrometry to investigate the structural impact on an IgG upon antigen binding, and observed that the physicochemical properties of the antigen differentially induce conformational changes in the IgG FcRn binding region. The extent of these structural changes directly correlates to the magnitude of the affinity differences between the Ag-IgG complexes and FcRn. Moreover, the antigen’s physicochemical properties differentially induce structural differences within the Ag-IgG-FcRn ternary complex. We also provide electron microscopy data that shows corroborating Fab-FcRn interactions, and confirms the hypothesis of potential 2:1 FcRn:IgG binding stoichiometry. These data demonstrate antigen-induced Fc structural rearrangements affect both the affinity toward FcRn and the trimeric antigen-IgG-FcRn complex, providing novel molecular insights in the first steps toward understanding interactions of FcRn-containing large(r)-sized immune complex.
KEYWORDS: Immunoglobulin, Fc neonatal receptor, antigen-induced allosteric effects, hydroxyl radical footprinting-mass spectrometry, negative stain electron microscopy
Introduction
Antibody-based molecules have shown great success as treatments for various diseases. Of the immunoglobulin (Ig) subclasses, IgG is the most widely developed.1,2 IgGs contain antigen-binding fragments (Fabs), which are responsible for the specific recognition of antigen, and the Fc domain, which determines the binding to Fc gamma receptors (FcγR) and the neonatal Fc receptor (FcRn). These domains are connected by a flexible linker denoted as the hinge, which varies in length within the four IgG isoforms.
Conventionally, the Fab and Fc domains are thought to function independently due to the flexibility of the hinge region that connects the two domains. However, emerging studies have implied that there might be cross-talk between these domains. For example, IgG with the same Fc framework but different variable domain sequences showed an impact on Fc functions,3–5 and switching the Fc region from different IgG isotypes with the same variable domain results in changes in antigen-binding affinity.6,7 Moreover, studies using Protein A and Protein G, as molecule binding probes, have shown long-range allosteric effects in the Fc region upon antigen-binding.8,9 Molecular dynamic (MD) simulations showed that the dynamic distribution of IgG structure is stimulated by antigen binding, and propagates to influence Fc-receptor interaction.10 Finally, new work by Orlandi et al. have shown that antigen binding allosterically effects Fcγ-receptor binding.11 However, the role of antigen-induced structural changes on FcRn binding requires further investigation.
FcRn is a major histocompatibility complex (MHC) class I-related heterodimeric Fc receptor, best known for regulating the homeostasis of IgGs.12,13 Binding to FcRn facilitates protection of monomeric IgGs from intracellular degradation and therefore prolongs its serum half-life.14–16 As indicated, the biological role of FcRn on antigen-free IgG recycling has been well established; however, the molecular mechanism of FcRn on antigen-bound IgG (Ag-IgG) trafficking is not as well understood. It has been shown that FcRn can transport Ag-IgG complexes across epithelial cells and FcRn enhances antigen presentation (reviewed in Ref.17). In addition, FcRn-mediated “antibody buffering” can result in prolonged half-life of Ag-IgG from recycling or transcytosis, leading to novel mAb therapeutic strategies for pH-dependent antigen release.18–20 Surprisingly, however, there is little information on the structure-function relationship within the Ag-IgG-FcRn ternary complex, or how the physiochemical properties of the antigen may affect FcRn affinity.
Crystallographic data have provided information on the molecular contacts between the Fc region of an IgG and the FcRn receptor, shedding light on the pH-dependent affinity by critical histidine residues.21–24 FcRn binds to the CH2-CH3 domains of IgG Fc, and interacts through histidine residues (in particular, H310 and H435) in a pH-dependent manner. These histidine residues are protonated under low pH and form salt bridges with negatively charged residues in FcRn. In the acidic pH range (below 6.5), FcRn binds to IgG at high affinity, but with low affinity at neutral pH and above. The pH-dependent interaction enables FcRn to capture IgG in the acidified endosomal compartment and to release monomeric IgG back into circulation when exposed to the extracellular physiological pH condition.25 The Fc-FcRn interactions are well understood; but understanding of the role the Fab domain involvement in the IgG-FcRn complex is still in its infancy. Previous work has shown that antibodies with similar wild-type human Fc sequences can exhibit a wide range of half-life and clearance values,26 and changes in the complementarity-determining region (CDR) residues or swapping of Fab regions of the IgG can directly influence FcRn binding and pharmacokinetics (PK).3–5 These studies clearly demonstrate that the Fab plays a role in FcRn biology, but understanding the molecular mechanism is challenging due to the size and dynamics of a full-length antibody and the membrane association of FcRn.
Bottom-up mass spectrometry (MS) techniques have shown great promise in studying molecular interactions of large protein structures since there are virtually no size limitations. For example, hydrogen-deuterium exchange (HDX) MS analysis was able to detect the predicted Fc-FcRn interactions indicated by crystallography, and revealed potential Fab-FcRn interactions. This led to the hypothesis of a two-step process where FcRn first recognizes Fc at high affinity, and then recruits Fab in close proximity as a secondary interaction site.27,28 However, as with all bottom-up MS techniques, deciphering the difference between direct binding interactions and conformational changes due to stabilization or allostery typically requires information from orthogonal methods.
Hydroxyl radical footprinting (HRF) MS is an emerging technology orthogonal to HDX that, instead of detecting changes in the protein backbone, probes solvent accessible surface area (SASA) of protein side-chain residues. This method has successfully been used to elucidate protein-protein interface and allosteric conformational changes of biotherapeutic proteins,29 and to investigate the binding interactions of IgG/FcγR complex in solution.30 However, the structural interactions within the FcRn-IgG complex have yet to be investigated using this technology. As such, we were interested in applying HRF to investigate the IgG-FcRn complex to corroborate the putative Fab-FcRn observed for HDX. In addition, we were interested in probing if HRF can detect structural changes on a full-length IgG triggered by antigen binding, and if antigen-bound IgG (Ag-IgG) affects FcRn affinity.
Here, we performed a series of structure-function relationship studies to probe the effect of antigen physiochemical properties on an IgG1 structure and FcRn affinity. To simplify our study design, we chose an IgG1 with a peptide antigen (~2 kDa) to enable physiochemical modifications of the antigen outside the epitope sequence through a simple linker sequence. We generated a series of peptide antigens with positive, negative, and hydrophobic differences to understand how antigen properties effect IgG and IgG-FcRn interactions. We show that the physiochemical properties of the antigen differentially induce conformational changes on the Fc-receptor binding region of the IgG, in which the extent of the structural change directly correlate with Ag-IgG affinity toward FcRn. Moreover, the affinity differences are reflected in differences in receptor conformation within the Ag-IgG-FcRn ternary complex. Finally, we used electron microscopy (EM) to visualize the IgG-FcRn complex and the observed classification indicating direct Fab-FcRn interactions and the 2:1 FcRn:IgG complex. This work provides unique structural insights into the Ag-IgG-FcRn ternary complex.
Results
Analysis of IgG-FcRn interactions using hydroxyl radical footprinting
As indicated, the crystal structure of Fc-FcRn has shed light on the molecular mechanism of IgG-FcRn binding,22 and HDX has been used to further understand how the full-length IgG interacts with FcRn.27 However, the contributions from the Fab region to FcRn binding and the potential impact of antigen-binding on FcRn biology is not fully understood. To this end, we were interested in determining if HRF analysis could provide additional insight into the structural interactions within the IgG1-FcRn complex.
We first compared the HRF data from the FcRn-unbound and bound states, and mapped the data onto a homology model to visualize the results. Upon FcRn binding, we observed three tryptic peptides in the heavy chain (HC) of the Fc region of the IgG1 (residues HC 249–255, HC 302–317, and HC 417–439) with a decrease in SASA (Figure 1 highlighted in Red). All three peptides are located between CH2-CH3 domains and are consistent with the previously identified Fc-FcRn interface.22 In addition to these expected regions, three peptides on the Fab region of IgG1 from both the HC and light chain (LC) (HC 1–19, HC 148–210, and LC 67–108) also exhibit reduced SASA. The peptide regions identified by HRF corroborate the findings from the previous HDX study,27 and demonstrate a global conformational change that includes apparent interactions between FcRn and the Fab domain at LC 67–108.
Figure 1.
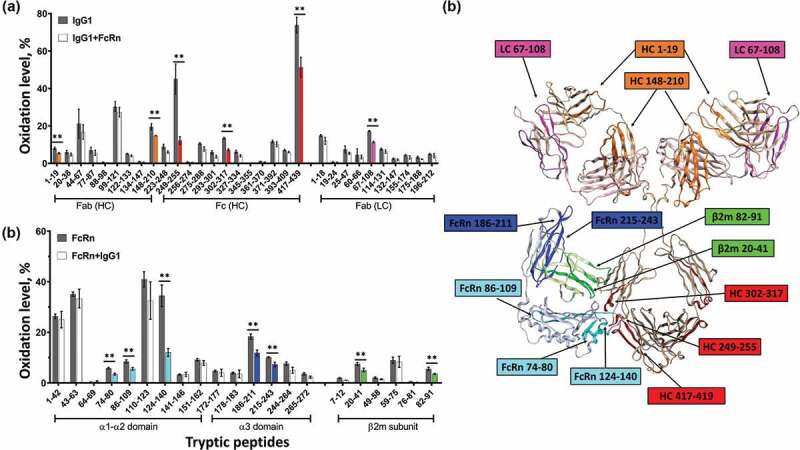
HRF structural analysis of the IgG1-FcRn complex. Oxidative footprint of the tryptic peptides from IgG (a) and FcRn (b) in the free and bound states. (c) The model structure of IgG-FcRn complex. The region color-highlighted in the structure corresponding to the color in the bar graph. **P < .01.
The receptor also experiences substantial solvent accessibility changes upon binding the IgG molecule. In particularly, we observed 3 peptides within the α1-α2 domains (residues 124–140,74-80 and 86–109) and 2 peptides within the β2 m subunit (residues 20–41 and 82–91) that exhibit reduced solvent accessibility in the bound state. All of these peptides are located close to the Fc interaction site as predicted by crystal structure analysis. In addition, we observed 2 peptides in the α3 domain (residues 186–211 and 215–243) that exhibit reduced solvent accessibility in the bound state, which are distal from the Fc interaction site and facing the direction of the Fab domain as shown in the homology model (Figure 1(c)). Our homology model is configured such that the LC of the Fab is facing down toward the bound FcRn, and suggests a potential interaction between LC peptide residues 67–108 (magenta) and FcRn peptide residues 186–211 (blue). There are also regions on Fab (HC 1–19 and HC 148–210) that are not in direct contact with FcRn based on the homology model, indicating there is likely additional conformational effect upon FcRn binding. These data are consistent with the notion that there may be direct interactions between FcRn and the Fab region of an IgG.
Visualization of IgG-FcRn complex by electron microscopy
To further assess the putative Fab and FcRn interactions, we analyzed the IgG-FcRn complex structure by negative stain electron microscopy, and observed several distinct classifications of IgG-FcRn complex (Figure 2). The Fab arms of IgG appear to be highly dynamic as previously observed,31 with multiple conformations previously predicted by Booth et al.32 These include the canonical Y-shaped conformation (Figure 2(a and d)), a T-shaped conformation (Figure 2(b and e)) or a mixed Y/T conformation (Figure 2(c and f)). In addition, we observed that the soluble form of FcRn can bind to the IgG at either 1:1 (Figure 2(b-d)) or 2:1 (Figure 2(e-f)) stoichiometry. Both of the stoichiometry conformations appear to show the Fab arm directly interacting with FcRn (Figure 2(b, c and e)). It is also interesting to note that in both of the 2:1 stoichiometry classifications the Fc region appears to be in a bent conformation (relative to the Fab arms), and not a planar conformation as is observed in the 1:1 classifications. Whether or not this is an artifact of the EM staining or a biologically relevant conformation remains unclear.
Figure 2.
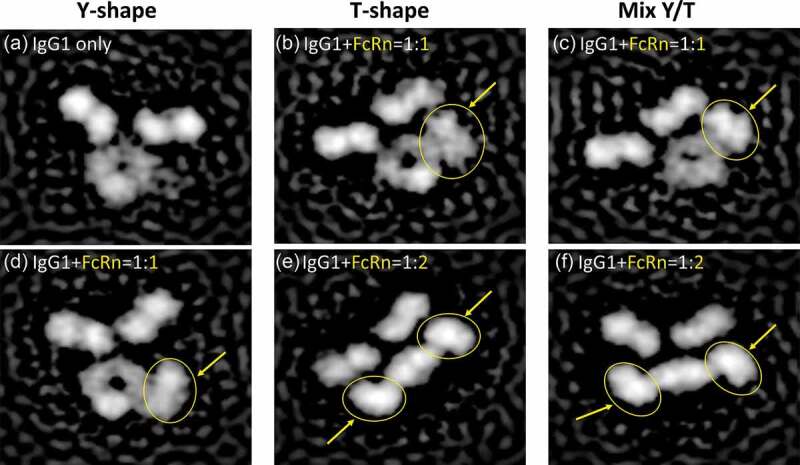
Visualization of IgG1-FcRn complex by negative stain electron microscopy. Six representative classifications of IgG1 and IgG1-FcRn complex are shown. FcRn is highlighted with yellow arrows/circles. (a) IgG1 alone in a canonical Y-shaped conformation. (b) IgG1 in a T-shaped conformation and binds to FcRn at 1:1 stoichiometry. (c) IgG1 in a Y/T mixed conformation and binds to FcRn at 1:1 stoichiometry. (d) IgG1 in a canonical Y-shaped conformation and binds to FcRn at 1:1 stoichiometry. (e) IgG1 in a T-shaped conformation and binds to FcRn at 1:2 stoichiometry. (f) IgG1 in a Y/T mixed conformation and binds to FcRn at 1:2 stoichiometry.
Antigen binding induces global conformational changes at the Fc receptor binding site
Allosteric activity between antibody Fab and Fc regions has been proposed and studied since the 1970s, but the research has remained inconclusive (reviewed in Ref. 7). With the development of novel bottom-up MS technologies such as HRF, higher order structure analysis of flexible large molecule like full-length IgG becomes possible. Therefore, we sought to utilize HRF-MS to understand the effect of antigen binding on the IgG structure. We analyzed the solvent accessibility difference in the IgG1 in the free and antigen-bound states, and observed structural changes across both the Fab and Fc regions (Figure 3). In the Fab region, the CDR-containing peptides showed the expected reduced solvent accessibility in the bound state (Figure 3 highlighted in red); however, a peptide in CH1 domain (residues 148–210) showed increased solvent accessibility upon binding to antigen. The former observation is consistent with expected changes in the Fab upon antigen binding, and the latter observation corroborates previous findings that antigen-binding induces conformational changes on the CH1-1 loop on Fab.33
Figure 3.
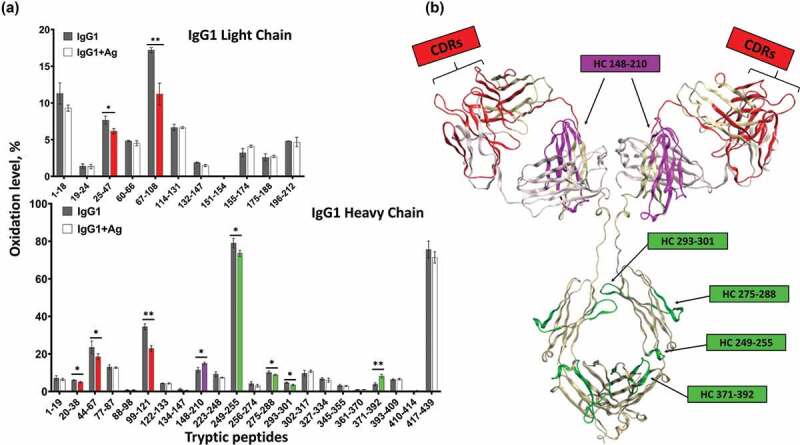
Structural changes of IgG1 induced by antigen binding. Bar graph one the left shows the oxidative labeling footprint of the tryptic peptides from IgG1 light chain (upper panel) and heavy chain (lower panel) in the free and antigen-bound states. The model structure of IgG1 antibody is shown on right. The region color-highlighted in the structure corresponding to the color in the bar graph. *P < .05, **P < .01.
In the Fc region, four peptides also showed changes in solvent accessibility, indicating antigen-binding induces conformational changes distal from the antigen binding sites. Of particular interest was the observation that an Fc peptide located at the FcRn binding interface (residues 249–255) showed reduced solvent accessibility. This peptide is remote from the antigen binding region and contains residues directly involved in FcRn binding. These data indicated that either remote conformational changes are occurring in the IgG upon antigen binding, or that antigen binding leads to conformational stability changes in the Fc region that results in reduction of the overall solvent accessibility. Since the solvent accessibility measurements from HRF are an average of the global population, these differences cannot be readily teased out. Nonetheless, our HRF data is consistent with previous literature indicating long-range conformational effects within the IgG structure upon antigen binding.
The physiochemical property of antigens affects FcRn binding affinity
To further investigate whether the observed structural changes in the Fc region could lead to functional effects, a panel of peptide antigens was designed by adding a single amino acid to glycine spacer sequences on either side of the N-terminal and C-terminal, flanking the same antigen epitopes (Table S1). First-pass analysis using surface plasmon resonance (SPR) indicated that these antigen variants retained similar binding affinity to IgG1, but they appeared to show differential effects on Ag-IgG-FcRn binding (Figure S1, S2).
We subsequently measured the binding affinity of an IgG1 to FcRn in the presence of saturating amounts of wild-type antigen (Ag-WT) and the three modified peptide antigens with the highest effect of FcRn interaction in the first-pass analysis. The antigen with arginine (Ag-R) and tryptophan (Ag-W) modifications increased IgG1-FcRn binding with the largest response, and the antigen with aspartic acid (Ag-D) modification was the only one that showed reduced binding as compared to Ag-WT. To minimize the SPR assay-induced FcRn avidity and charge effect from sensor chip,34 we used a C1 sensor chip with a flat carboxymethylated surface without dextran matrix. FcRn was immobilized at low density at 100RU, and binding of IgG1 to the FcRn surface was measured in the presence of saturating amounts of each antigen under the same experimental condition. We confirmed that the WT and three modified antigens from different physiochemical categories displayed differential influence on FcRn binding affinity (Table 1). Due to the complex nature of the possible 1:1 and/or 1:2 mab:FcRn binding, the SPR data were fitted to both a 1:1 binding model (Figure S6) and a 1:2 binding model (not shown). Both models demonstrated a similar trend in the changes in FcRn affinity from antigen binding, and therefore a 1:1 binding model was used to obtain data for Table 1. Among the antigen variants, Ag-R and Ag-W improved the affinity of IgG1 to FcRn (2.4-fold and 3.5-fold increase, respectively), while Ag-D displayed a small reduction in FcRn affinity (~20%). The effects of the Ag-R and Ag-D antigens on FcRn affinity are consistent with previous observations described for Fab charge patches (positive increase affinity, negative decreases affinity).4 However, the increase in FcRn affinity resulting from flanking tryptophan on the antigen has not been previously observed.
Table 1.
FcRn affinity to Ag-IgG1 complexes.
| Sample | Antigen Sequence | ka (1/Ms) | kd (1/s) | KD (M) | Change (%), N = 2 |
|---|---|---|---|---|---|
| IgG1 unbound | No Antigen | 6.15E+05 | 0.5798 | 9.43E-07 | 76 ± 18% |
| IgG1 + Ag-WT | GGGS-Ag-GGGS | 2.91E+05 | 0.1971 | 6.78E-07 | Ctrl (100 ± 3%) |
| IgG1 + Ag-R | RGGGS-Ag-GGGR | 3.88E+05 | 0.1099 | 2.83E-07 | 239 ± 11% |
| IgG1 + Ag-W | WGGGS-Ag-GGGSW | 4.70E+05 | 0.0917 | 1.95E-07 | 349 ± 2% |
| IgG1 + Ag-D | DGGGS-Ag-GGGD | 2.84E+05 | 0.2309 | 8.14E-07 | 83 ± 0% |
Correlating antigen-induced IgG structural changes to FcRn affinity
The structural effects of WT antigen binding to the IgG1 suggested that there are structural changes or conformational stability changes that may affect FcRn affinity. Generating peptides that differentially affect FcRn affinity enabled us to probe the correlation between FcRn affinity and antigen-induced IgG structural changes. To this end, we compared the structural changes of IgG1 when binding to the antigens that had largest effect on FcRn affinity (Ag-R and Ag-W), and then analyzed the structural difference in the Ag-IgG-FcRn ternary complexes.
In comparing the IgG1 structure in the free and antigen-bound states, the SASA pattern was very consistent between the different antigen-bound IgG1 (Figure S3). We observed that the CDR-containing peptides showed the expected reduced oxidation level in the bound states for all three antigens; however, the antigens displayed a differential effect in the Fc region. Two peptides directly involved in FcRn binding (residues 249–255 and 417–439) displayed a trending decrease in solvent accessibility (No Ag>Ag-WT>AG-R> Ag-W) that mirrored the antigen’s trending increase on FcRn binding affinity (No Ag<Ag-WT<AG-R< Ag-W) (Figure 4(a and b)). However, when bound within the antigen/IgG1/FcRn ternary complexes these Fc-peptides displayed similar SASA for all antigens tested (Figure 4(c), Figure S4). These data implied that the FcRn binding site on the IgG1 is likely fully occupied, and that antigen binding does not change the structure of Fc in the FcRn-bound state. Moreover, if the observed decrease in SASA in the Fc-region upon antigen-binding was due to the excess antigen making direct antigen-Fc surface interactions (nonspecific binding), then one would expect steric hindrance with FcRn and an associated decrease in binding affinity, not the observed affinity increase. Thus, these data are strong evidence that the SASA change in Fc-region is due to Fab-antigen binding, not nonspecific interactions in the Fc. It is unclear if this is from conformational rearrangement or conformational stability, but in either case, the SASA change in the Fc upon antigen binding directly correlate with the observed FcRn affinity.
Figure 4.
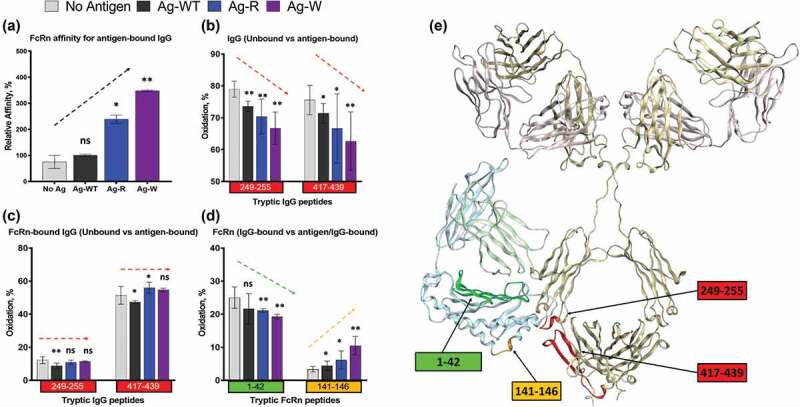
Structural changes in IgG1-FcRn complex induced by different antigens. (a) SPR analysis. The relative affinity of IC-FcRn as compared to Ag-WT/IgG1/FcRn (100%). (b-d) HRF analysis. Oxidative labeling footprint of two tryptic peptides from IgG1 Fc region in the FcRn-free antigen-bound forms (b), and FcRn-bound antigen-bound forms. (c). Oxidative labeling footprint of two tryptic peptides from FcRn in Ag-bound forms. (e) The model structure of IgG1-FcRn. The region color-highlighted in the structure corresponding to the color in the bar graph. *denotes significant difference relative to no Ag. ns: not significant, *p < .05, **p < .01.
For the FcRn receptor, antigen binding to Fab appeared to change the structure within the FcRn-bound state. We observed two distinct receptor peptides that exhibited changes in SASA that also mirrored the increasing trend of the binding affinity between Ag-IgG and FcRn. Peptide residues 1–42 and 141–146 both displayed trending effects in SASA similar to the IgG Fc-region, albeit in opposite directions (Figure 4(d), Figure S5). As the affinities of Ag-IgG to FcRn increases, the changes in the solvent accessibility for peptides residues 1–42 and 141–146 decrease and increase, respectively. While both the affinity changes and SASA changes are subtle, the direct correlation from two distinct analytical methods provides strong evidence that antigen physiochemical properties directly influence the Ag-IgG-FcRn ternary state.
Discussion
The observations that maternal IgGs are transferred to neonatal mice35 led to the discovery of the FcRn, and an extensive scientific effort to understand the molecular interactions that are responsible for the biology of this receptor. The last fifty years of scientific research on FcRn biology has enabled a detailed understanding of the pH-dependency of the molecular residues responsible for Fc-FcRn interactions,22,36 the transcytosis and recycling function (reviewed in Ref.37), and the effects of Fab charge patches on FcRn affinity.4 These discoveries have been harnessed to modulate the pharmacokinetics of monoclonal antibodies as biotherapeutics.26,38
In addition to the crucial function in IgG homeostasis, FcRn also mediates “antibody buffering”, which can result in prolonged half-life of Ag-IgG from recycling or transcytosis.18 Moreover, it is now understood that this MHC class-I related receptor has an integral role in antigen presentation (reviewed in Refs.39–41). FcRn-mediated immune function bridges the gap between humoral and cellular immunity through the simultaneous presentation of an antigen-bound antibody to both MHCII and MHCI molecules in dendritic cells (DCs). This subsequently activates CD4+ and CD8 + T cell responses, respectively. Specifically, FcRn-mediated immune complex cross-presentation has been found to be of exceptional importance in activating tumor-reactive cytotoxic T cells, and hence providing protection from tumors,42 while disruption of IC-FcRn interaction by introducing IHH mutations ((I253A, H310A, and H435A) on the Fc region results in the loss of cross-presentation ability in DCs.43 However, how antigen-bound IgGs effect FcRn structure-function relationship is poorly understood.
To further our understanding of the molecular interactions between and antigen-bound IgG and FcRn, we designed a model system using an IgG1 and peptide antigen to probe the structural consequences of antigen binding on both the Ag-IgG binary and Ag-IgG-FcRn ternary complexes. The use of a peptide antigen enabled us to perform structure-function relationship studies to investigate the role of antigen physiochemical properties on FcRn interactions. Combining the sensitivity of SPR for affinity measurements with the region-specific structural information from HRF provides a new strategy for probing these interactions.
HRF has previously been used to elucidating protein-protein interactions,29–31,44 but this is the first work that investigated IgG-FcRn interactions. Our initial HRF analysis showed all of the expected Fc-FcRn interactions and the distal structural changes that are consistent with literature (Figure 1). These results provided confidence that the SASA data from HRF in this system is accurate and interpretable. We then utilized negative stain electron microscopy to visualize the IgG-FcRn complex and observed structural conformations corroborating the HRF analysis. To date, one of the best visualization of the likely conformations the IgG must adopt comes from recent modeling work by Booth et al.32 By creating a “best-fit” model, Booth et al. describe significant steric hindrance that would occur if the IgG interacted with membrane-bound FcRn in the canonical Y-conformation. Instead, the IgG would have to adopt a T-shape conformation to enable the Fab arms to lay against the endosomal membrane. This in turn would shift the Fab arms into a position of direct interaction with the receptor.
Using negative stain electron microscopy, we observed multiple conformations demonstrating direct contact between the Fab arm and the FcRn receptor. These orientations are also consistent with the Y- and T-shaped conformations predicted by Booth et al.,32 in which the latter conformation was suggested for reduction of Fab-membrane steric hindrance. In addition, we observed the 2:1 FcRn-IgG structure, which clearly demonstrates the flexibility of the Fab arms and interactions with FcRn. This stoichiometry has been hypothesized from multiple studies, including Fc-FcRn crystal structure analysis, FcRn binding studies, in vitro transport studies and in vivo half-life investigations (reviewed in Ref. 41). However, to our knowledge this is the first data set to directly visualize the 2:1 FcRn:IgG stoichiometry.
Having established that HRF provides data consistent with our EM data and previous literature, we then used this technology to explore how antigen binding may affect the IgG structure. The HRF data indicated that the WT antigen clearly induced SASA changes within the Fc-region at peptides known to be involved in FcRn binding. Investigating the antigen’s potential effect on FcRn binding showed there was a modest increase in the IgG-FcRn affinity, leading to design of a series of antigen variants to probe the physiochemical effect on the molecular interactions of the Ag-IgG-FcRn ternary complex. All of the peptides generated displayed similar affinities toward the IgG1, and showed similar reduction in SASA in the CDR-containing peptides. However, the antigen with positive (Arginine) or aromatic (Tryptophan) properties induced further structural changes in the Fc-region. Similar to the wild-type antigen, these antigen variants effected the same 2 peptides involved in receptor binding (residues 249–255 and 417–439), but with increasing SASA responses. The trend in the effect of SASA directly correlated with an increase in affinity of the Ag-IgG complex to FcRn. These two sets of data imply that the physiochemical properties alter the Fc-region structure to affect FcRn affinity of an antigen-bound IgG. How exactly the antigen’s properties affect the Fc structure remains unclear.
Since HRF data is simply a SASA measurement of a global population of protein, deciphering whether changes are due to direct protein-protein interactions or distal conformational changes (such as allostery) remains a challenge. Moreover, the excess amount of peptide antigens may have nonspecific interactions with the Fc-region, resulting in reduced apparent SASA, which would make data interpretation challenging. However, since the Fc peptides affected by antigen binding are the same as the ones involved in FcRn binding, one would expect a negative impact on FcRn binding through steric hindrance. Since we observed an increase in FcRn affinity in the presence of excess antigen, the structural changes in the Fc region are therefore unlikely due to nonspecific binding.
While the exact nature of antigen-induced molecular events in the Fc are still unknown, comparison of the Ag-IgG binary complex to the Ag-IgG-FcRn ternary complex provides additional structural insight. For example, the decreasing trend in SASA for the Fc peptides in the Ag-IgG binary complexes is lost within the Ag-IgG-FcRn ternary complexes. These data indicate that the Ag-IgG complexes are fully bound with FcRn in the ternary complex. We do not know how much of the equilibrium population is in the 1:1 vs. 2:1 FcRn-IgG stoichiometry during HRF analysis. We added 2-fold molar excess of FcRn for complex collection, but our EM data clearly indicates both species are formed. These details are important when considering the HRF percent (%) modification calculation because of the 2-fold symmetry of an IgG. If the equilibrium stoichiometry is anything but 2:1 FcRn:IgG, then the unbound protein residues located at the interface would be more solvent accessible than the bound protein residues, obfuscating the HRF calculations. Here we observe that all of the Fc-region and FcRn peptides at the binding interface have similar SASA results in the Ag-IgG-FcRn complexes. This indicates that the samples are in a similar state of equilibrium for each antigen regardless of the stoichiometry. This is important for the interpretation of results for the FcRn peptides (residues 1–42, 141–146) that showed opposite trending in SASA data in the Ag-IgG-FcRn ternary states. Since the equilibrium states are similar for all Ag-IgG-FcRn samples, these data imply that the physiochemical properties of the antigen are directly affecting the FcRn conformation with the Ag-IgG-FcRn ternary complex. Taken together, our data indicates that antigen binding allosterically effects both the FcRn affinity and the receptor conformation within the ternary state.
This study complements the work by Jensen et al.27 demonstrating that IgG FcRn binding is a multistep mechanism. Jensen et al. proposed that FcRn binds the Fc region first, then the Fab domain interacts with FcRn. Here we demonstrate additional properties of the IgG and the antigen bound IgG need to be taken into account. First the antibody needs to be considered as a whole with all domains involved in the interactions. Second, as soon as the antibody binds its target, the new molecular environment needs to be considered. These are the first structural data of a non-Fc engineered mAb and wild-type FcRn to show that the soluble form of FcRn interacts with the Fab domain. The peptide antigen, which is relatively small by comparison to the IgG, does not sterically interfere with FcRn binding. In contrast, it strengthens the FcRn interaction as demonstrated by the affinity gain. Finally, this study provides insight into the first mechanistic steps of small antigen trafficking in the acidified endosome and the molecular details that need to be taken into account.
In summary, understanding the molecular interactions between FcRn and the antigen-bound IgG is critical for both fundamental immunology and in designing next-generation mAb therapeutics. Our work provides insight on the role of antigen binding on IgG structures, and demonstrates that physiochemical properties can influence FcRn affinity. This may have implications for antigen targeting and the subsequent trafficking of antigens with different isoforms or genetic polymorphisms. These data also provide direct evidence for Fab-FcRn interactions that modulate IgG-FcRn affinity, and provides the first structural images for the 2:1 FcRn:IgG binding stoichiometry. The structural data depicts conformations that have been hypothesized, but never before directly observed. In addition, this work supports the view that the Fab and Fc domains are not independent entities within the IgG structure, but rather involved in intricate allosteric interactions that are transmitted through the two domains and that influence Fc receptor binding. The structural data provided by HRF details the unique effect the antigen has on the structural interactions within the Ag-IgG-FcRn ternary complex, and provides novel details on the role of the physiochemical properties of the antigen.
Materials and methods
The IgG and FcRn proteins used in this study were expressed and purified in-house at Genentech, Inc. to clinical-quality drug substance. The IgG is a humanized IgG1 monoclonal antibody with kappa light chain. The antigen and its derivatives used in this study are short peptides (2.1 ~ 2.5 kDa) that are specific for IgG1. Peptide antigens were synthesized by ABclonal (Woburn, MA).
Hydroxyl radical footprinting- mass spectrometry
HRF-MS was performed as previously described.29,31 All proteins were combined in equal molar ratio and purified by size-exclusion high performance liquid chromatography into a phosphate buffer system at pH5.5 to facilitate binding.45 The equal-weight strategy previously used for protein-protein interaction studies was employed, and the total protein concentration remained equal at 1 mg/mL for unbound protein, binary and ternary complexes.29 Peptide mapping, liquid chromatography and data analysis was performed as previously described.31 All data were obtained in triplicate and error bars represent 95/99 tolerance interval. Non-overlapping error bars have p < .05 (or otherwise denoted) and are considered significant differences.
Homology model
The Molecular Operating Environment (MOE) software package from Chemical Computing Group is used to build Fab domain models. The Fab models are aligned to the left and right arms of a generic full-length IgG1 molecule to generate full-length IgG homology models. The IgG1 framework used as a template is based on a full structure of the IgG1 antibody that Brandt et al. composed from crystal structures of the fragments and equilibrated to a relaxed conformation.46 The FcRn protein used for molecular modeling was obtained from the previously characterized crystal structure (PDB: 4N0 F).
Negative Stain Electron Microscopy
Concentrated protein was diluted in 20 mM histidine chloride, pH 5.5 to 0.01 mg/ml. Then 4 µl of the diluted sample was deposited, and incubated for a minimum of 30 s on a previously glow-discharged ultra-thin carbon coated 400-mesh, copper grid (Electron Microscopy Sciences). Following incubation, the sample was negatively stained by touching the protein bound side of the grid serially onto 5, 40 µl drops and placed on a parafilm working surface, of 2% uranyl acetate (Electron Microscopy Sciences). Excess stain is allowed to sit on the grid for >30 s and then wicked away using #1 Whatman filer paper. Imaging was performed on a Talos 200 C equipped with 4 K Ceta CMOS camera, (ThermoFisher Scientific, Waltham, MA), at a magnification of 73 k. Resulting micrographs were then processed in cisTEM analysis software (cisTEM.org) to produce 2D class averages from which overall structure can be observed.
Surface Plasma Resonance (SPR)
The kinetic characteristics and binding affinities of IgG1 and antigen-bound IgG1 to FcRn were measured by Biacore 2000 instrument (GE Healthcare; Piscataway, NJ) at 25°C. It has been reported that the assay orientations may affect the IgG-FcRn binding data.34,47 To minimize variability, all the kinetic measurements in this study were run on the same chip set up.
Human FcRn protein was immobilized on a C1 sensor chip with the amine coupling procedure. In the first-pass analysis, a single dose of IgG1 (66.7 nM) were pre-incubated with various antigens (500 µM) in the running buffer (phosphate-buffered saline (PBS)/0.05% PS20, PH6.0), and the antigen-antibody complex were injected at a flow rate of 50 µl/min for 180s to reach equilibrium, and dissociate for 240 s. After each injection, the chip surface was regenerated by PBS PH8.0. The binding level of each complex was normalized by an internal standard (set as 100%). For kinetic analysis, various concentrations of IgG1 were serially diluted from 2 µM to 8 nM, and were pre-incubated with excess amount of antigen (500 µM) in the running buffer. Antigen-antibody complex were injected during multiple cycles at a flow rate of 100 μL/min for 5 s, and dissociated for 10s. After each injection, the chip surface was regenerated by PBS PH8.0. Analysis was performed after an in-line reference cell correction and followed by buffer sample subtraction. The dissociation equilibrium constant (KD), dissociation rate constant (kd), and association rate constant (ka) were calculated using the Biacore BIA evaluation software (version 4.1; GE Healthcare) with a 1:1 binding model. All binding data were measured in duplicate.
Supplementary Material
Supplemental material
Supplemental data for this article can be here.
References
- 1.Suzuki M, Kato C, Kato A.. Therapeutic antibodies: their mechanisms of action and the pathological findings they induce in toxicity studies. J Toxicol Pathol. 2015;28(3):133–9. doi: 10.1293/tox.2015-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh S, Kumar NK, Dwiwedi P, Charan J, Kaur R, Sidhu P, Chugh VK. Monoclonal antibodies: a review. Curr Clin Pharmacol. 2018;13(2):85–99. doi: 10.2174/1574884712666170809124728. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Lu P, Fang Y, Hamuro L, Pittman T, Carr B, Hochman J, Prueksaritanont T. Monoclonal Antibodies with Identical Fc Sequences Can Bind to FcRn Differentially with Pharmacokinetic Consequences. Drug Metab Dispos. 2011;39(9):1469–77. doi: 10.1124/dmd.111.039453. [DOI] [PubMed] [Google Scholar]
- 4.Schoch A, Kettenberger H, Mundigl O, Winter G, Engert J, Heinrich J, Emrich T. Charge-mediated influence of the antibody variable domain on FcRn-dependent pharmacokinetics. Proc Natl Acad Sci U S A. 2015;112(19):5997–6002. doi: 10.1073/pnas.1408766112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piche-Nicholas NM, Avery LB, King AC, Kavosi M, Wang M, O’Hara DM, Tchistiakova L, Katragadda M. Changes in complementarity-determining regions significantly alter IgG binding to the neonatal Fc receptor (FcRn) and pharmacokinetics. mAbs. 2018;10(1):81–94. doi: 10.1080/19420862.2017.1389355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janda A, Bowen A, Greenspan NS, Casadevall A. Ig Constant Region Effects on Variable Region Structure and Function. Front Microbiol. 2016;7:22–22. doi: 10.3389/fmicb.2016.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang D, Kroe-Barrett R, Singh S, Roberts CJ, Laue TM. IgG cooperativity – is there allostery? Implications for antibody functions and therapeutic antibody development. mAbs. 2017;9(8):1231–52. doi: 10.1080/19420862.2017.1367074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oda M, Kozono H, Morii H, Azuma T. Evidence of allosteric conformational changes in the antibody constant region upon antigen binding. Int Immunol. 2003;15(3):417–26. doi: 10.1093/intimm/dxg036. [DOI] [PubMed] [Google Scholar]
- 9.Sagawa T, Oda M, Morii H, Takizawa H, Kozono H, Azuma T. Conformational changes in the antibody constant domains upon hapten-binding. Mol Immunol. 2005;42(1):9–18. doi: 10.1016/j.molimm.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Zhao J, Nussinov R, Ma B. Antigen binding allosterically promotes Fc receptor recognition. mAbs. 2019;11(1):58–74. doi: 10.1080/19420862.2018.1522178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orlandi C, Deredge D, Ray K, Gohain N, Tolbert W, DeVico AL, Wintrode P, Pazgier M, Lewis GK. Antigen-Induced Allosteric Changes in a Human IgG1 Fc Increase Low-Affinity Fcγ Receptor Binding. Structure. 2020;28(5): 516–527.e5. doi: 10.1016/j.str.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat Rev Immunol. 2007;7(9):715–25. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 13.Pyzik M, Rath T, Lencer WI, Baker K, Blumberg RS. FcRn: the architect behind the immune and nonimmune functions of IgG and albumin. J Immunol 2015;194(10):4595–603. doi: 10.4049/jimmunol.1403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaccaro C, Zhou J, Ober RJ, Ward ES. Engineering the Fc region of immunoglobulin G to modulate in vivo antibody levels. Nat Biotechnol. 2005;23(10):1283–88. doi: 10.1038/nbt1143. [DOI] [PubMed] [Google Scholar]
- 15.Petkova SB, Akilesh S, Sproule TJ, Christianson GJ, Al Khabbaz H, Brown AC, Presta LG, Meng YG, Roopenian DC. Enhanced half-life of genetically engineered human IgG1 antibodies in a humanized FcRn mouse model: potential application in humorally mediated autoimmune disease. Int Immunol. 2006;18(12):1759–69. doi: 10.1093/intimm/dxl110. [DOI] [PubMed] [Google Scholar]
- 16.Dall’Acqua WF, Kiener PA, Wu H. Properties of Human IgG1s Engineered for Enhanced Binding to the Neonatal Fc Receptor (FcRn). J Biol Chem. 2006;281(33):23514–24. doi: 10.1074/jbc.M604292200. [DOI] [PubMed] [Google Scholar]
- 17.Ward ES, Ober RJ.Multitasking by exploitation of intracellular transport functions the many faces of FcRn. Adv Immunol. 2009;103:77–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devanaboyina SC, Lynch SM, Ober RJ, Ram S, Kim D, Puig-Canto A, Breen S, Kasturirangan S, Fowler S, Peng L, et al. The effect of pH dependence of antibody-antigen interactions on subcellular trafficking dynamics. mAbs. 2013;5(6):851–59. doi: 10.4161/mabs.26389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sampei Z, Haraya K, Tachibana T, Fukuzawa T, Shida-Kawazoe M, Gan SW, Shimizu Y, Ruike Y, Feng S, Kuramochi T, et al. Antibody engineering to generate SKY59, a long-acting anti-C5 recycling antibody. Plos One. 2018;13(12):e0209509. doi: 10.1371/journal.pone.0209509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuzawa T, Nezu J. A Novel Recycling Antibody for Complement-mediated Diseases. Curr Med Chem. 2020;27(25):4157–4164. doi: 10.2174/0929867326666191016115853 [DOI] [PubMed] [Google Scholar]
- 21.Burmeister WP, Gastinel LN, Simister NE, Blum ML, Bjorkman PJ. Crystal structure at 2.2 Å resolution of the MHC-related neonatal Fc receptor. Nature. 1994;372(6504):336–43. doi: 10.1038/372336a0. [DOI] [PubMed] [Google Scholar]
- 22.Burmeister WP, Huber AH, Bjorkman PJ. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature. 1994;372(6504):379–83. doi: 10.1038/372379a0. [DOI] [PubMed] [Google Scholar]
- 23.Vaughn DE, Bjorkman PJ. Structural basis of pH-dependent antibody binding by the neonatal Fc receptor. Structure (London, England: 1993). 1998;6(1):63–73. doi: 10.1016/S0969-2126(98)00008-2. [DOI] [PubMed] [Google Scholar]
- 24.Martin WL, West AP Jr., Gan L, Bjorkman PJ. Crystal structure at 2.8 Å of an FcRn/Heterodimeric Fc Complex. Mol Cell. 2001;7(4):867–77. doi: 10.1016/S0969-2126(98)00008-2. [DOI] [PubMed] [Google Scholar]
- 25.Borrok MJ, Wu Y, Beyaz N, Yu X-Q, Oganesyan V, Dall’Acqua WF, Tsui P. pH-dependent binding engineering reveals an FcRn affinity threshold that governs IgG recycling. J Biolog Chem. 2015;290(7):4282–4290. doi: 10.1074/jbc.M114.603712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Wang EQ, Balthasar JP. Monoclonal antibody pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther. 2008;84(5):548–58. doi: 10.1038/clpt.2008.170. [DOI] [PubMed] [Google Scholar]
- 27.Jensen PF, Larraillet V, Schlothauer T, Kettenberger H, Hilger M, Rand KD. Investigating the Interaction between the Neonatal Fc Receptor and Monoclonal Antibody Variants by Hydrogen/Deuterium Exchange Mass Spectrometry. Mol celL Proteom. 2015;14(1):148–61. doi: 10.1074/mcp.M114.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen PF, Schoch A, Larraillet V, Hilger M, Schlothauer T, Emrich T, Rand KD. A two-pronged binding mechanism of IgG to the neonatal fc receptor controls complex stability and IgG serum half-life. Mol celL Proteom. 2017;16(3):451–56. doi: 10.1074/mcp.M116.064675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Wecksler AT, Molina P, Deperalta G, Gross ML. Mapping the binding interface of VEGF and a monoclonal antibody Fab-1 Fragment with Fast Photochemical Oxidation of Proteins (FPOP) and mass spectrometry. J Am Soc Mass Spectrom. 2017;28(5):850–58. doi: 10.1007/s13361-017-1601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi L, Liu T, Gross ML, Huang Y. Recognition of human IgG1 by Fcγ Receptors: structural insights from hydrogen–deuterium exchange and fast photochemical oxidation of proteins coupled with mass spectrometry. Biochemistry. 2019;58(8):1074–80. doi: 10.1021/acs.biochem.8b01048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin M, Krawitz D, Callahan MD, Deperalta G, Wecksler AT. Characterization of ELISA antibody-antigen interaction using footprinting-mass spectrometry and negative staining transmission electron microscopy. J Am Soc Mass Spectrom. 2018;29(5):961–71. doi: 10.1007/s13361-017-1883-9. [DOI] [PubMed] [Google Scholar]
- 32.Booth BJ, Ramakrishnan B, Narayan K, Wollacott AM, Babcock GJ, Shriver Z, Viswanathan K. Extending human IgG half-life using structure-guided design. mAbs. 2018;10(7):1098–110. doi: 10.1080/19420862.2018.1490119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sela-Culang I, Alon S, Ofran Y. A systematic comparison of free and bound antibodies reveals binding-related conformational changes. The Journal of Immunology . 2012;189(10):4890–99. doi: 10.4049/jimmunol.1201493. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, McKay P, Yee LT, Dutina G, Hass PE, Nijem I, Allison D, Cowan KJ, Lin K, Quarmby V, et al. Impact of SPR biosensor assay configuration on antibody: neonatal Fc receptor binding data. mAbs. 2017;9(2):319–32. doi: 10.1080/19420862.2016.1261774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brambell FW. The transmission of immune globulins from the mother to the foetal and newborn young. Proc Nutr Soc. 1969;28(1):35–41. doi: 10.1079/PNS19690007. [DOI] [PubMed] [Google Scholar]
- 36.Rodewald R. pH-dependent binding of immunoglobulins to intestinal cells of the neonatal rat. J Cell Biol. 1976;71(2):666–69. doi: 10.1083/jcb.71.2.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martins JP, Kennedy PJ, Santos HA, Barrias C, Sarmento B. A comprehensive review of the neonatal Fc receptor and its application in drug delivery. Pharmacol Ther. 2016;161:22–39. doi: 10.1016/j.pharmthera.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Datta-Mannan A, Witcher DR, Tang Y, Watkins J, Wroblewski VJ. Monoclonal antibody clearance. J Biol Chem. 2007;282(3):1709–17. doi: 10.1074/jbc.M607161200. [DOI] [PubMed] [Google Scholar]
- 39.Baker K, Rath T, Pyzik M, Blumberg RS. The role of FcRn in antigen presentation. Front Immunol. 2014;5:408–408. doi: 10.3389/fimmu.2014.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Platzer B, Stout M, Fiebiger E. Antigen cross-presentation of immune complexes. Front Immunol. 2014;5:140–140. doi: 10.3389/fimmu.2014.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pyzik M, Sand KMK, Hubbard JJ, Andersen JT, Sandlie I, Blumberg RS. The Neonatal Fc Receptor (FcRn): a misnomer? Front Immunol. 2019;10:1540. doi: 10.3389/fimmu.2019.01540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker K, Rath T, Flak MB, Arthur JC, Chen Z, Glickman JN, Zlobec I, Karamitopoulou E, Stachler MD, Odze RD, et al. Neonatal Fc receptor expression in dendritic cells mediates protective immunity against colorectal cancer. Immunity. 2013;39(6):1095–107. doi: 10.1016/j.immuni.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker K, Qiao S-W, Kuo TT, Aveson VG, Platzer B, Andersen J-T, Sandlie I, Chen Z, de Haar C, Lencer WI, et al. Neonatal Fc receptor for IgG (FcRn) regulates cross-presentation of IgG immune complexes by CD8-CD11b+ dendritic cells. Proc Natl Acad Sci U S A. 2011;108(24):9927–32. doi: 10.1073/pnas.1019037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deperalta G, Alvarez M, Bechtel C, Dong K, McDonald R, Ling V. Structural analysis of a therapeutic monoclonal antibody dimer by hydroxyl radical footprinting. mAbs. 2013;5(1):86–101. doi: 10.4161/mabs.22964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wecksler AT, Yin J, Lee Tao P, Kabakoff B, Sreedhara A, Deperalta G. Photodisruption of the structurally conserved Cys-Cys-Trp triads leads to reduction-resistant scrambled intrachain disulfides in an IgG1 monoclonal antibody. Mol Pharm. 2018;15(4):1598–606. doi: 10.1021/acs.molpharmaceut.7b01128. [DOI] [PubMed] [Google Scholar]
- 46.Brandt JP, Patapoff TW, Aragon SR. Construction, MD simulation, and hydrodynamic validation of an all-atom model of a monoclonal IgG antibody. Biophys J. 2010;99(3):905–13. doi: 10.1016/j.bpj.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdiche YN, Yeung YA, Chaparro-Riggers J, Barman I, Strop P, Chin SM, Pham A, Bolton G, McDonough D, Lindquist K, et al. The neonatal Fc receptor (FcRn) binds independently to both sites of the IgG homodimer with identical affinity. mAbs. 2015;7(2):331–43. doi: 10.1080/19420862.2015.1008353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


