Abstract
Brucella ceti infections have been increasingly reported in cetaceans, although a very limited characterization of Mediterranean Brucella spp. isolates has been previously reported and relatively few data exist about brucellosis among cetaceans in Italy. To address this gap, we studied 8 cases of B. ceti infection in striped dolphins (Stenella coeruleoalba) stranded along the Italian coastline from 2012 to 2018, investigated thanks to the Italian surveillance activity on stranded cetaceans. We focused on cases of stranding in eastern and western Italian seas, occurred along the Apulia (N = 6), Liguria (N = 1) and Calabria (N = 1) coastlines, through the analysis of gross and microscopic findings, the results of microbiological, biomolecular and serological investigations, as well as the detection of other relevant pathogens. The comparative genomic analysis used whole genome sequences of B. ceti from Italy paired with the publicly available complete genomes. Pathological changes consistent with B. ceti infection were detected in the central nervous system of 7 animals, showing non-suppurative meningoencephalitis. In 4 cases severe coinfections were detected, mostly involving Dolphin Morbillivirus (DMV). The severity of B. ceti-associated lesions supports the role of this microbial agent as a primary neurotropic pathogen for striped dolphins. We classified the 8 isolates into the common sequence type 26 (ST-26). Whole genome SNP analysis showed that the strains from Italy clustered into two genetically distinct clades. The first clade comprised exclusively the isolates from Ionian and Adriatic Seas, while the second one included the strain from the Ligurian Sea and those from the Catalonian coast. Plotting these clades onto the geographic map suggests a link between their phylogeny and topographical distribution. These results represent the first extensive characterization of B. ceti isolated from Italian waters reported to date and show the usefulness of WGS for understanding of the evolution of this emerging pathogen.
Introduction
Brucella is a genus of bacteria that infects many terrestrial and aquatic vertebrates [1] and brucellosis represents a widespread zoonosis and an important economic and public health problem in many areas of the world [2].
Brucella spp. infections were first described in pinnipeds and cetaceans in the early 1990’s [3, 4], in California and Scotland, and have been reported since then in several wild marine mammal species all over the world. Since 2007, isolates of Brucella spp. from marine mammals have been classified further based on molecular investigations, differences in metabolism and host-bacteria interactions, into two species, B. ceti and B. pinnipedialis that infect cetaceans and pinnipeds, respectively [5].
The occurrence of a variety of lesions caused by brucellosis in cetaceans, such as endometritis, placentitis, abortion, orchitis, mastitis, pneumonia, myocarditis, pericarditis, osteoarthritis, spinal discospondylitis, subcutaneous abscesses, hepatic, splenic or lymph node necrosis, macrophage infiltration in liver and spleen, and meningoencephalomyelitis has been demonstrated [6–18]. However, compared with the reported high global seroprevalence of marine mammal Brucella spp. infection, clinical disease does not appear to be common [7, 11, 19, 20], suggesting that most of the infected animals overcome clinical disease, eventually remaining Brucella carriers and shedders [14].
Based on the common infection patterns of meningitis and/or meningoencephalitis, resembling “human neurobrucellosis” [21, 22], a specific susceptibility has been suggested for the striped dolphin (Stenella coeruleoalba) [9, 11, 19, 23–26]. Central nervous system (CNS) involvement in Brucella infection in man occurs in about 5–7% of the cases [1], and includes findings of meningitis, encephalitis, meningovascular disease, brain abscesses and demyelinating syndrome [21].
The dynamics of B. ceti infection in cetaceans are largely unknown, and the cell receptor(s) allowing the entry of this pathogen and the subsequent dissemination throughout cetacean host’s tissues have not yet been identified [27]. Although the role of metazoan parasites in the eco-epidemiology and pathogenesis of brucellosis in cetaceans is unclear, the localization of B. ceti in lungworms and cestoda raise the possibility that they may serve as carriers for the transmission of the infection [11, 13].
According to Multi Locus VNTR (Variable copy of Tandem Repeats) Analysis (MLVA), the marine Brucella strains can be divided into three major groups containing eight clusters [28, 29], and the Multi Locus Sequence Typing (MLST) identified 15 sequence types (STs) so far as reported at the public PubMLST repository (https://pubmlst.org/brucella/). Marine mammal Brucella strains have potential to infect and cause disease in domestic animals [30] and humans. However, only few human clinical cases have been observed to date and linked mainly to raw fish and seafood ingestion [31, 32] or to laboratory operations without proper biosafety containment [33–35].
B. ceti infections have been frequently described in dolphins from both, the Atlantic and Pacific Oceans, but to date the information about isolates from marine mammals of Mediterranean Sea is limited and relatively little data exist about brucellosis in cetaceans in Italy. The detection of anti-Brucella spp. antibodies was first demonstrated in cetaceans stranded along the Spanish coast of the Mediterranean from 1997 to 1999 [36], while no evidence of seropositivity was detected in cetaceans of Italian Seas until the beginning of 2015 [16, 37].
The isolation of B. ceti in the Mediterranean Sea was first achieved in 2009, in a striped dolphin stranded along the Spanish Catalonian coast [14]. Later, in 2012, the presence was documented in four other cetaceans, two of them, one striped dolphin and one bottlenose dolphin, stranded in Spain along the same Catalonian coastline [14], and the other two, both striped dolphins, stranded in Italy, in the Tyrrhenian and the Adriatic Sea [26, 38]. Based on MLST investigations all B. ceti strains isolated in these cases belonged to ST-26 [38].
Recently, a coinfection by Brucella spp., Listeria monocytogenes and Toxoplasma gondii was confirmed using molecular methods in a striped dolphin stranded in Italy along the Ligurian coastline, and was associated with related pathological changes in brain, blubber, liver and spleen [16]. Moreover, a B. ceti ST-27 strain, previously found only in Pacific Ocean species [11, 39], was isolated from multiple lymph nodes of one bottlenose dolphin in the Croatian part of the northern Adriatic Sea, representing the evidence of this zoonotic strain in Mediterranean waters [40, 41] and in European waters in general.
In order to gain a deeper understanding about brucellosis in cetaceans in Italy, we studied eight cases of B. ceti infection detected in striped dolphins (S. coeruleoalba) stranded along the Italian shoreline from 2012 to 2018. We focused on the pathogenic role shown by the pathogen and on the genetic constitution of the strains involved, subjected to a comparative genomic analysis in order to determine the relationship of Italian isolates to those previously described in European/Mediterranean waters [42].
Materials and methods
Ethics statement
The National Reference Centre for Diagnostic Investigations on Stranded Marine Mammals (C.Re.Di.Ma.–Istituto Zooprofilattico Sperimentale del Piemonte, Liguria e Valle d’Aosta, Torino, Italy) and the Istituti Zooprofilattici Sperimentali (IIZZSS) are public laboratories authorized by the Italian Ministry of Health to perform systematic surveys on infectious diseases of aquatic mammals stranded on the cost of Italy. This study was done through passive surveillance sampling animals found dead, therefore the procedures applied did not harm live animals.
Dolphins
We investigated 8 cases of stranding, associated with striped dolphins found stranded dead along the Apulian (Adriatic and Ionian Seas), Ligurian (Ligurian Sea) and Calabrian (Ionian Sea) coastlines, from 2012 to 2018 that resulted positive to the isolation of B. ceti from one or more tissues.
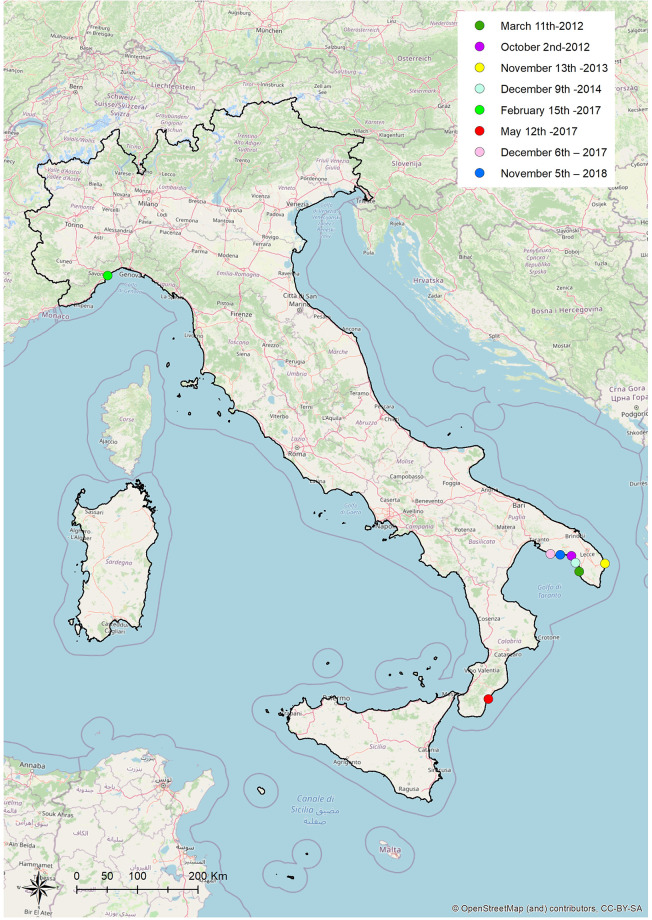
These positive cetaceans were distributed along the Apulian (N = 6), Calabrian (N = 1) and Ligurian (N = 1) coastlines (Fig 1).
Fig 1. Stranding sites of striped dolphins infected by B. ceti under study, Italy, 2012–2018.
Geographical mapping was obtained by ArcGIS® software using the geographical coordinates found from the strandings. Location data (dots): Case 1 dark green: Gallipoli Lido Pizzo, Apulian coastline, Ionian Sea (39.58 N, 18.00 E) March 11th 2012. Case 2 purple: Porto Cesareo Bacino Grande, Apulian coastline, Ionian Sea (coordinates not available) October 2nd 2012. Case 3 yellow: Alimini, Otranto, Apulian coastline, Adriatic Sea (coordinates not available) November 13th 2013. Case 4 light blue: Porto Cesareo Bacino Grande, Apulian coastline, Ionian Sea (coordinates not available) December 9th 2014. Case 5 green: Savona, on the Ligurian coastline, Ligurian sea (44.94 N, 8.18 E) February 15th 2017. Case 6 red: Ardore, on the Calabrian coastline, Ionian Sea (38.11 N, 16.12 E) May 12th 2017. Case 7 pink: Maruggio, on the Apulian coastline, Ionian Sea (40.17 N, 17.35 E) December 6th 2017. Case 8 blu: Manduria, on the Apulian coastline, Ionian Sea (40.18 N, 17.40 E) November 5th 2018. The Italy map was used under a CC BY-SA copyright from OpenStreetMap contributors (https://www.openstreetmap.org/copyright/en) (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Necropsy and diagnostic investigations
At the time of stranding, the carcasses were submitted to the diagnostic laboratories, belonging to the network of Istituti Zooprofilattici Sperimentali laboratories, coordinated by the C.Re.Di.Ma, where a detailed post-mortem examination was performed according to standard protocols [43], depending on the carcasses’ preservation status.
Individual data, including date and location, sex, age class categories (based on total body length-TBL), decomposition code [43] and body condition [44] were recorded. The presence of helminths was estimated by macroscopic and microscopic examination of tissues. Endoparasites were preserved in 70% alcohol for microscopic identification, according to established morphological characteristics [45, 46]. Coronal sections from different brain regions (telencephalon, diencephalon, mesencephalon, pons, cerebellum, medulla) [47], as well as from all major organs, were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned at 4 μm, stained with hematoxylin and eosin, and finally examined under a light microscope.
The presence of relevant pathogens like DMV and T. gondii was investigated by PCR methods as previously reported [48, 49]. Frozen CNS tissue samples of all animals and additional organs for cases 5, 6 and 7 were tested for both pathogens.
In one case (case 5), the presence of DMV and T. gondii was also explored through immunohistochemical investigation (IHC) [50], on CNS and bladder sections, for DMV, and CNS sections, for T. gondii, respectively.
Serological investigations to estimate the presence of specific antibodies against DMV and T. gondii were also performed in two cases (5, 6) [50], specifically on serum, cerebrospinal fluid (CSF) and aqueous humor of case 5, and on serum of case 6.
Macroscopical and microscopical findings of all cases, except case 3, were recorded.
Selected tissues and/or fluids were collected for microbiological, biomolecular and serological investigations focused on Brucella infection diagnosis.
Brucella isolation and identification
The primary isolation of Brucella spp. was performed from CNS samples of all animals, and from other tissues available for cases 1, 5, 6, 7 and 8.
Specifically, the isolation of Brucella spp. was also attempted from spleen of Case 1, spleen, lung, prescapular lymph node and cerebrospinal fluid (CSF) of case 5, spleen, lung, lymph nodes and liver of case 6, spleen, lung, liver, kidney and testes of Case 7, and spleen, lung, mesenteric lymph nodes, liver and kidney of case 8.
The CNS of case 6 was submitted for bacterial isolation subsequently to the observation of microscopic lesions suggestive of neurobrucellosis, thanks to the histopathological analysis performed retrospectively on CNS tissue of the animal under study.
Cultures were performed according to the technique described in the OIE Manual of Diagnostic Tests and Vaccines [51], using both selective and non-selective solid media and enrichment broths to enhance the chance of isolating, except for tissues other than CNS of case 6.
For cases 1, 2, 3, 4, we used Farrell’s and Columbia blood Agar media, for cases 7 and 8 we added modified Thayer Martin and CITA media, while for cases 5 and 6 we used a combination of Farrell’s and CITA media. The solid media were incubated at 37°C, aerobically and in a microaerophilic atmosphere containing 5% CO2, for at least 10 days. Enrichment cultures were carried out in Brucella enrichment broth, supplemented with fetal horse serum and modified Brucella selective supplement, and incubated at 37°C in a microaerophilic atmosphere containing 5% CO2, while for cases 7 and 8 we added Thayer Martin broth. Weekly for six times or up until isolation, a loopful of the enrichment broth was streaked to Farrell’s Agar medium. Suspect colonies (circular, convex, shiny, 1–2 mm in diameter after 48–72 h) were seeded onto blood agar medium and incubated for a further 2 days before re-examination. When Brucella spp. was suspected based on the Gram’s staining [52], the colonies were tested for catalase, oxidase and urease activities [52]. Motility and slide agglutination tests with Brucella anti-A and anti-M antisera were also performed for cases 1, 2, 3, 4, 7, 8, together with nitrate reduction, H2S production and growth in the presence of CO2 for cases 5 and 6 [52].
For DNA extraction, all B. ceti isolates were subcultured in Brucella medium base (BAB; Oxoid, Hampshire, UK) and incubated in a 5–10% CO2 atmosphere at 37°C for 48 h to assess the purity of cultures and the absence of dissociation. Bacterial DNA was extracted from single colonies using Maxwell® 16 Tissue DNA Purification Kit using Maxwell® 16 Instrument (Promega, Madison, WI, USA) or High Pure DNA Template Preparation kit (Roche Diagnostics, France) according to the manufactures’ instructions.
All strains isolated from the striped dolphins under study were identified as B. ceti using the PCR-RFLP method [53] and then subjected to genomic analysis at the National and OIE Reference Laboratory for Brucellosis, Istituto Zooprofilattico Sperimentale dell’Abruzzo e del Molise, Teramo, Italy.
Molecular detection of Brucella spp. from tissues
Frozen tissue samples of cases 5, 6 and 7 were submitted to PCR for the detection of Brucella spp. by hemi-nested PCR targeting an outer membrane protein gene of B. abortus [54].
The reactions were loaded as previously reported using B. suis bv 2 strain Thomsen as positive control and no template control as negative control.
Specifically, molecular detection was attempted on CNS, spleen, lung, liver, pre-scapular, tracheobronchial, pulmonary lymph nodes, tongue and skin ulcers, laryngeal tonsil and CSF of case 5; on CNS of case 6; on CNS, spleen, liver, lung and testes of case 7. For DNA extraction, tissue samples (30–50 mg) were physically disrupted using a TissueLyser II homogenizer (Qiagen, Hilden, Germany) by high-speed shaking in plastic tubes with stainless steel beads (5 mm in diameter). Genomic DNA was then extracted from the disrupted tissues with an AllPrep DNA/RNA Mini kit (Qiagen) according to the manufacturer’s instructions.
The PCR products were analyzed by electrophoresis on 2% agarose gel containing GelRed (Biotium, Fremont, California, USA), compared with molecular weight markers and subsequently photographed on a Gel-Doc UV transilluminator system (Bio-Rad, Hercules, California, USA).
Serological tests for brucellosis
Serum, cerebrospinal fluid and aqueous humor from case 5, and serum from cases 6 and 8, were tested by rapid serum agglutination (Rose Bengal plate test, RBT) using RBT antigen produced from B. abortus strain S99 [24, 50] to detect anti-smooth Brucella spp. antibodies. For case 8, the test was performed on a fresh sample, while for cases 6 and 8 was carried on thawed sera.
Whole genome sequencing and bioinformatics
Total genomic DNA of eight samples, from the cases studied with the following ID numbers 31957, 2780, 3838, 17753, 1259 and 25153, was sequenced using the Illumina NextSeq 500 platform. Briefly, the quantity of total DNA was measured with the Qubit fluorometer (QubitTM DNA HS assay; Life Technologies, Thermo Fisher Scientific, Inc.). The libraries were prepared using the Nextera XT library preparation kit (Illumina Inc., San Diego, CA) following the manufacturer’s instructions and the libraries were sequenced using NextSeq 500/550 Mid Output Reagent Cartridge v2 with 300 cycles generating 150 bp paired-end reads. Reads shorter than 70 bp and average Phred mean quality < 24 were automatically discarded. Raw reads were quality assessed using FastQC and trimmed to remove nucleotides with quality score less than 20 from 5′ end and 3′ end. Read coverage ranged from 29X to 113X, with an average of 63X.
Genomes were assembled using SPAdes version 3.11.1. and the scaffolds were used to assign MLST profiles using mlst tool (https://github.com/tseemann/mlst) which incorporates components of PubMLST database (https://pubmlst.org).
Sequence reads were deposited in Sequence Read Archive (SRA) database under NCBI Bioproject PRJNA623338.
Two samples, 10759 and 28753, were sequenced using IonTorrent platform and full genomes were assembled with Velvet 1.1.0 and deposited in NCBI with RefSeq Accession Numbers GCF_000590795.1 and GCF_000590815.1, respectively [42]. Additional 51 B. ceti and B. pinnipedialis WGS sequences available from the public database GenBank or Sequence Read Archive (SRA) were also included in the analysis. The dataset was limited to non-identical sequences that mapped to the B. ceti reference genome with less than 500 ambiguous matches (GenBank Accession Numbers NC_022905.1; NC_022906.1). SNP analysis was performed using In Silico Genotyper (ISG) version 0.16.10–3[55] using BWA-MEM (version 0.7.12-r1039) [56] as the aligner and GATK (version 3.9) [57] as a SNP caller. B. ceti genome (GenBank Accession Numbers NC_022905.1; NC_022906.1) was used as a reference. Default filters were applied to remove SNPs from duplicated regions, from regions with read coverage of less than 10X and with base call proportion less than 90%. Concatenated unique variants were used to generate maximum likelihood tree using IQ-TREE (version 1.6.9) [58, 59]. Ascertainment bias correction option was used to correct the branch lengths for the absence of constant sites in the SNP alignment. ModelFinder was used to select the best fit model and based on Bayesian Information Criterion (BIC) value and TVM+F+R2 model was chosen for phylogenetic reconstruction. Branch support was assessed using non-parametric bootstrap with 500 bootstrap replicates.
The population structure was assessed using a Bayesian approach implemented in BAPS 6.0 software with the module hierarchical BAPS (hierBAPS), which examines the existence of subgroups within a population and predicts the placement of individual sequences into specific clusters [60–62].
Results
Post-mortem and histopathological investigations were performed on seven of the eight animals with positive culture for B. ceti (cases 1, 2, 4, 5, 6, 7, 8). The main gross findings included a moderate parasitic infection by Phyllobothrium delphini and/or Monorygma grimaldi plerocercoids (5/7; 71,4%), lymphadenomegaly (3/7; 42,8%), parasitic bronchopneumonia (3/7; 42,8%) and meningeal hyperaemia (3/7; 42,8%), which is regarded as the only macroscopic Brucella spp.-related lesion (cases 1, 4, 8) (Table 1).
Table 1. Brucella ceti-infected striped dolphins: stranding data, body condition, most significant findings (gross and microscopic), bacteriological, molecular and serological Brucella spp. investigations results, along with coinfections and the most probable cause of death.
| Case no. | ID strain | Place, coordinates, date and stranding condition | DC | Sex | Age | NS | Main pathological findings (macro/micro) ** | B. ceti isolation*** | Molecular detection of Brucella spp *** | RBT | Coinfections | Most probable cause of death |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 10759 | Gallipoli, (39.58 N, 18.00 E) March 11th-2012 found dead | 2 | Ma | Ju | Mo | moderate parasitic infection by Phyllobothrium delphini and Monorygma grimaldi plerocercoids; meningeal hyperaemia; cerebral edema; pulmonary edema; non-suppurative leptomeningitis | CNS-spleen | NP | NP | severe cerebral impairment, associated to a primary B. ceti infection. | |
| 2 | 28753 | Porto Cesareo*, October 2nd-2012 found dead | 3 | Fe | Ju | Go | hepatocyte vacuolar degeneration; generalized lymphoid depletion; non-suppurative meningitis | CNS | NP | NP | severe cerebral impairment, associated to a primary B. ceti infection. | |
| 3 | 31957 | Otranto*, November 13th -2013 found dead | ND | ND | ND | ND | data not available | CNS | NP | NP | ND | |
| 4 | 2780 | Porto Cesareo*, December 9th -2014 found dead | ND | Ma | Ju | Mo | meningeal hyperemia; cerebral edema; marked lymphomonocytic meningitis (+++ medulla oblongata), lymphomonocytic plexocoroiditis, perivascular mononuclear cuffing; non-suppurative meningoencephalitis | CNS | NP | NP | severe cerebral impairment by a B. ceti infection | |
| 5 | 3838 | Savona, 44.94 N, 8.18 E), February 15th -2017 found dead | 2 | Ma | Ju | Po | moderate parasitic infection by Phyllobothrium delphini and Monorygma grimaldi plerocercoids; ulcerative glossitis; skin ulcers; splenic and prescapular lymphadenomegaly; bronchointerstitial pneumonia; multicentric lymphoid necrosis (spleen, PSC and PUL lymph nodes, laryngeal tonsil); multifocal necrotizing hepatitis; interstitial nephritis; non-suppurative meningoencephalitis | CNS-spleen- lung- PSC ln-CSF | CNS-spleen-lung, liver, PSC-TB-PUL ln tongue and skin ulcers- laryngeal tonsil-CSF | Neg (CSF, AH, S) | DMV T. gondii | severe cerebral impairment, associated to a coinfection (DMV, T. gondii, B. ceti) |
| 6 | 17753 | Ardore, (38.11 N, 16.12 E) May 12th -2017 found dead | 2 | Fe | Ad | Go | moderate parasitic infection by Phyllobothrium delphini and Monorygma grimaldi plerocercoids; severe parasitic bronchopneumonia; parasitic gastritis; non-suppurative meningoencephalitis | CNS, spleen, LNs, liver, lung | CNS | Neg (S) | DMV | severe cerebral impairment, associated to a coinfection (DMV, B. ceti) |
| 7 | 1259 | Maruggio, (40.17 N, 17.35 E) December 6th– 2017 found dead | 2 | Ma | Ju | Po | moderate parasitization by Phyllobothrium delphini and Monorygma grimaldi plerocercoids; moderate parasitic bronchopneumonia; multicentric reactive lymphadenopathy; moderate hepatocyte vacuolar degeneration; moderate multifocal bronchointerstitial pneumonia; mild multifocal mononuclear myocardial infiltrate; cerebral haemorrhages; severe non-suppurative meningoencephalitis | CNS-spleen-liver-lung-kidney-testes | CNS, spleen, liver, lung, kidney, testes | NP | DMV | severe cerebral impairment, associated to a coinfection (DMV, B. ceti) |
| 8 | 25153 | Manduria, (40.18 N, 17.0 E) November 5th– 2018 found dead | 2 | Ma | Ju | Mo | moderate parasitization by Phyllobothrium delphini plerocercoids; hemothorax and hemoperitoneum; generalized congestion; multicentric reactive lymphadenopathy; epicardial petechiae; mild parasitic bronchopneumonia; hemorrhagic parasitic gastritis; severe necrotizing enteritis; meningeal hyperaemia; non-suppurative meningoencephalitis | CNS-lung-liver-kidney-spleen-MES ln | NP | Pos (S) | DMV | severe cerebral impairment, associated to a coinfection (DMV, B. ceti) |
*: coordinates not available. DC, decomposition code (2, fresh; 3, moderate autolysis); Ma, male; Fe, female; Ad: adult; Ju: juvenile; NS, nutritional status; Mo, moderate; Po, poor; ND, not determined; NP, not performed; Neg, negative; Pos, positive; CNS: central nervous system; PSC ln, prescapular lymph node; TB ln, tracheo-bronchial lymph node; PUL ln, pulmonary lymph node; MES ln, mesenteric lymph node; LNs, lymph nodes; CSF, cerebrospinal fluid; AH, aqueous humor; S, serum; RBT, rosa bengala test; DMV, Dolphin Morbillivirus.
**Brucella-associated pathological features are shown in bold.
***Tissues positive are shown in bold.
The main microscopic findings were: non-suppurative meningoencephalitis (5/7; 71,4%); non-suppurative meningitis/leptomeningitis (2/7; 28,5%); multicentric lymphoid reactive hyperplasia (2/7; 28,5%); bronchointerstitial pneumonia (2/7; 28,5%); hepatocyte vacuolar degeneration (2/7; 28,5%); parasitic gastritis (2/7; 28,5%). Non-suppurative meningitis and/or meningoencephalitis, involving a mononuclear cell infiltration, observed in cases 1, 2, 4, 6, 7, 8, along with lymphoid necrosis and necrotizing hepatitis in case 5, were considered Brucella-type lesions (Table 1).
Moreover, protozoan cysts were apparent in the brain tissue of case 5 (S1 Table).
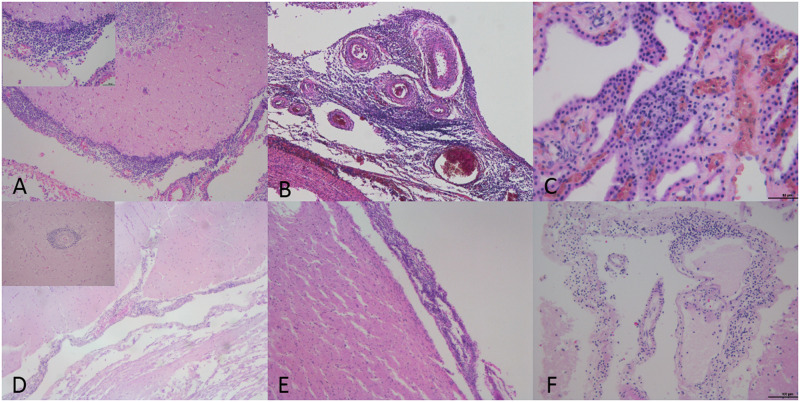
The microscopical features of B. ceti-associated CNS lesions are shown in Fig 2.
Fig 2. B. ceti-associated lesions in central nervous system of striped dolphins (S. coeruleoalba).
(A) Severe non-suppurative meningitis. Cerebellar meninges are infiltrated by mononuclear cells (case 2). 10x. H&E. Left upper inset: detail of the lympho-monocytic inflammatory infiltrate. 40x. H&E. (B) Severe non-suppurative meningitis (case 4). Meninges at the level of medulla oblongata are infiltrated by lympho-monocytic cells. 10x. H&E. (C) Mild non-suppurative meningoencephalitis (case 5). Chorioid plexuses are infiltrated by lympho-monocytic cells. 40x. H&E. (D) Mild non-suppurative meningoencephalitis (case 6). cerebellar meninges are infiltrated by mononuclear cells. 10x. H&E. Left upper inset: perivascular cuff characterized by the presence of lympho-monocytic cells. 20x. H&E. (E) Non-suppurative meningitis (case 7). Cerebral cortex meninges are infiltrated by mononuclear cells. 10x. H&E. (F) Non-suppurative meningitis (case 8). Meninges at the level of parietal cortex are infiltrated by mononuclear cells. 20x. H&E.
B. ceti was isolated from CNS of all the studied dolphins, while spleen was positive only in cases 1 and 5, and lung in case 5. The PCR for Brucella spp. detection directly from tissues resulted positive from CNS in case 5 and 6, in two out the three animals investigated, and from six different body sites in only one case (Table 1). On the contrary, the case 7 showed negative results from all the tissues tested.
The RBT test did not detect anti-Brucella antibodies in samples from cases 5 and 6, while resulted positive in the serum of the case 8 (Table 1).
Cases 6, 7 and 8 showed the coinfection by B. ceti and DMV and case 5 demonstrated the coinfection by B. ceti, DMV and T. gondii. Specifically, the animal showed anti-T.gondii and anti-DVM antibodies along with positive IHC and PCR tests from different tissues.
A full description of gross and microscopic findings observed, along with complete analytical data and considerations about the cause of death and the pathogenic role of B. ceti infection for each case considered are available in the S1 Table.
The genotyping of B. ceti using MLST classified the 8 strains as sequence type 26 (ST-26). The publicly available B. ceti and B. pinnipedialis genomes were assigned to the STs 23, 24, 25, 26 and 27.
The SNP analysis revealed 6,320 putative polymorphisms. The mapping of sequences to the reference ranged between 92% and 99%, with an average of 98%. The constructed phylogeny revealed that the population was divided into two major clades. As shown in Fig 3, the BAPS analysis split our dataset further into five groups based on the secondary level of clustering. Groups were monophyletic, confirming the robustness of the major branches of the SNP phylogeny, which were also supported by 100% bootstrap scores.
Fig 3. Maximum likelihood tree of B. ceti and B. pinnipedialis.
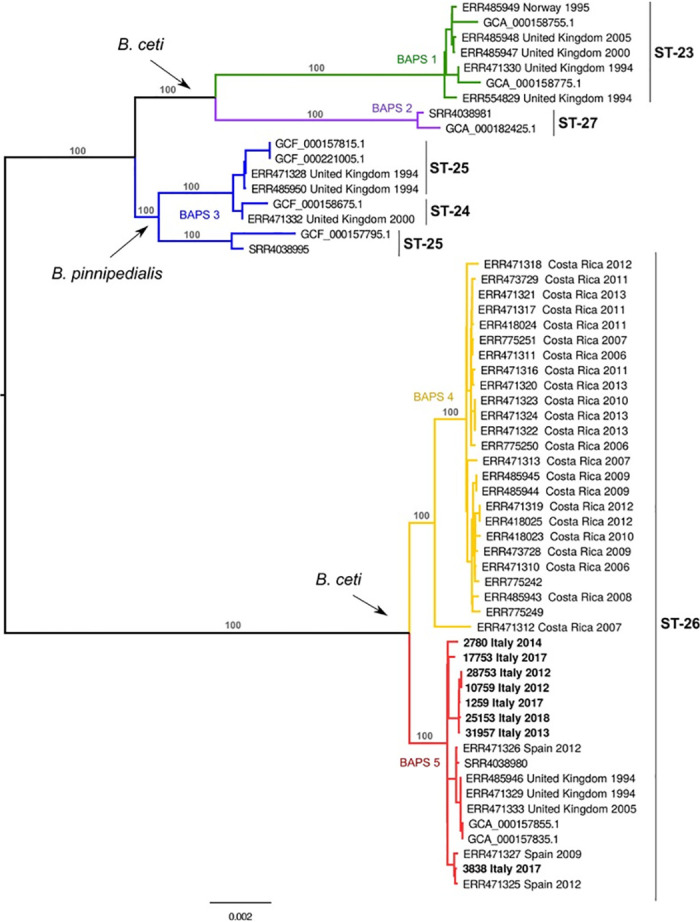
The tree was constructed using concatenated SNP sequences of 59 isolates and mid-point rooted. The branch colours correspond to BAPS populations and the major branches are labelled with bootstrap values.
The SNP analysis demonstrated that the clades comprising the ST-23, ST-27, corresponded to BAPS1 and BAPS2 groups while the ST-24 and ST-25 were included in the BAPS3 and the ST-26 was further subdivided into 2 groups (BAPS4 and BAPS5). The BAPS1 group comprised strains from the B. ceti species which contained mostly strains isolated from porpoises. BAPS2 group was composed of two strains from B. ceti ST-27, one of which was isolated from a human in New Zealand and the second from a dolphin in Croatia. The BAPS3 was composed of strains from the B. pinnipedialis species. The isolates from ST-26 were split into two subpopulations belonging to two different geographical areas with BAPS4 isolated in Costa Rica and BAPS5 isolated in the European Atlantic Sea and in the Mediterranean Sea. Interestingly, BAPS4 and BAPS5 contained mostly isolates from dolphins.
Whole genome SNP analysis showed that the strains from Italy were divided further into two genetically distinct subclades. The first subclade comprised exclusively the isolates from Ionian and Adriatic seas, while the second one included the strains from the Ligurian Sea and the Catalonian coast (Fig 3). Plotting these clades onto the geographic map suggests a link between their phylogeny and topographical distribution.
Discussion
To our knowledge, this study represents the first survey of B. ceti infection in striped dolphins from Italian waters and the first extensive characterization of B. ceti isolates reported to date.
The isolation of B. ceti was achieved from the CNS of all the dolphins under investigation. Only in two cases the isolation was obtained also from the other tissues (specifically spleen for case 1 and spleen and lung for case 5).
Although we observed several not pathognomonic signs, we detected specific gross pathological findings associated with Brucella infection, represented by hyperaemia of the meninges [9, 11] in three animals (cases 1, 4, 8).
No lesions were detected in the reproductive organs, and no signs associated with a potential abortion were found in the only adult female sampled (case 6).
A correlation between the infection and the pathological microscopic changes was observed in all cases submitted to histopathological investigations (neurobrucellosis, associated in one case with hepatic and lymph node necrosis). Specifically, non-suppurative meningitis or meningoencephalitis were detected in the CNS of 7 B. ceti-infected animals, thereby recapitulating the features of neurobrucellosis observed in humans [21, 22], as well as in striped dolphins elsewhere [9, 11, 19, 23–26].
We detected severe coinfections in half of the animals investigated (cases 5, 6, 7, 8), involving DMV in all cases and T. gondii in case 5, as reported before for several cetacean species infected by Brucella spp. and Cetacean Morbillivirus [63] or T. gondii [26].
Considering the role of B. ceti infection in the striped dolphins under study, in cases 1, 2 and 4 the stranding could have resulted from a severe cerebral impairment, associated with severe brain inflammation caused by B. ceti infection.
Moreover, in case 2, the finding of a generalized lymphoid depletion, described before in dolphins with brucellosis [18, 64], suggests an immunocompromised host response, though the evidence of hepatocyte vacuolar degeneration could additionally make the effects of toxic environmental pollutants and/or an undisclosed viral infection other than DMV plausible.
The cause of death could not be hypothesized for case 3, considering the limited data available.
In cases 5, 6, 7 and 8 the stranding could have resulted from a severe cerebral impairment, associated with a coinfection by B. ceti and DMV, and, for case 5, also T. gondii. Noteworthy, in case 5, a striped dolphin in a poor body condition, the systemic spread of B. ceti infection, the evidence of additional Brucella-type lesions (multicentric lymphoid necrosis and multifocal necrotizing hepatitis), with the absence of anti-Brucella spp. antibodies and negative bacteriological and biomolecular results in CSF, suggests an acute fatal brucellosis infection that appears to be the most likely contributing cause of death. DMV and T. gondii infections, associated with typical pathological findings, represented respectively by bronchointerstitial pneumonia and protozoan cysts at cerebral level, along with specific immunopositivity and the presence of antibodies for both agents, may indeed support the potentially relevant role played by DMV and T. gondii in initiating the animal’s decline.
Interestingly, the anti-Brucella spp. antibodies observed in case 8 represent the only positive result among the three tested animals. The limited number of samples hampers any conclusion on the use of RBT for the detection of Brucella infection in dolphins. Nevertheless, the fact that the positive result was obtained by testing a fresh serum sample collected from an animal in a good conservation code, supports a true positive result [11]. Moreover, the negative serological findings are supported by the simultaneous detection of other antibodies in case 5 and a supposed immunocompromised host response in case 6, in presence of a widespread DMV infection. RBT test, while generally considered consistent, may produce false results due to variety of factors [65]. Therefore, in order to screen the immunological status of the examined animals, other serological tests such as ELISA could be used, as previously shown [66]. Although some discrepancies between results of RBT and ELISA tests have been reported, ELISA tests, such as iELISA have been successfully used to detect anti-Brucella antibodies in odontocetes and arctic wildlife [65, 66] and could therefore serve as a complementary method serological response to B. ceti in dolphins. The highest frequency of B. ceti infection was confirmed in juveniles. This observation seems to be in discordance with a study performed in striped dolphins stranded in Costa Rica [64], in which meningoencephalomyelitis was revealed in the same number of juveniles and adults that displayed neurological syndromes before death, as well as with a previous study in marine mammals stranded in Brazil [18]. Specifically, in a population made up of several cetacean species, the highest frequency of Brucella spp. infection was confirmed in the newborn calves, whereas, within the genus Stenella, the most commonly infected age-group were the adults.
None of the cases considered in this report stranded alive, so it was not possible to observe neurological symptoms at the time of stranding.
The genomic analysis of the B. ceti strains isolated from the stranded dolphins grouped them into the common ST-26 in agreement with the previous reports [28]. The whole genome SNP analysis showed that the population was divided into 5 main groups, one of which included the B. pinnipedialis species while the remaining four were assigned to B. ceti. It is interesting to note how these groups identify specific STs in accordance with the MLST which therefore allows us to use previous data as a legacy to give consistency to the host spectrum as well as to geographic location. According to these data, we observed that the BAPS1 from B. ceti and BAPS3 from B. pinnipedialis, although not exclusively, are mainly found in porpoises and seals. Similar cluster partition was observed previously with MLVA data, which demonstrated the existence of the B. ceti group, predominantly associated with porpoises [28]. Their geographical location, instead, appeared to be linked to the Atlantic Ocean, even though the real distribution could be wider.
On the other hand, the BAPS 2, 4 and 5 populations seemed to be mainly observed in dolphins. The more consistent groups, BAPS4 and BAPS5, which belonged to the ST-26, were further divided from a geographical point of view and split the of Costa Ricans strains from those of the Mediterranean and the Atlantic Ocean. Further analysis revealed that the BAPS5 population was divided into geographically separated subgroups. The phylogeographic division found for these strains resembled the phylogeographic evolution of the striped dolphin [67]. The evolution of the striped dolphin suggests that its population is divided between the eastern and western Mediterranean, thus confirming the genomic division between B. ceti strains from the Ligurian Sea and the Adriatic and Ionian Seas observed in our analysis.
The data obtained by whole genome SNP analysis suggest an interesting relationship between phylogeny and geographical distribution of B. ceti strains in Italy, therefore providing new insights into the phylogenetic structure of B. ceti in the Mediterranean. Nevertheless, further studies from the Mediterranean Sea are required to elucidate the molecular evolution of B. ceti and its actual distribution.
In summary, our results provide novel data and pathological evidence of B. ceti infection in cetacean species in Italy, and the geographic distribution range of this agent in Italian waters. Considering the results of this survey and the other data available [16, 26], the occurrence of B. ceti infection in cetaceans stranded, along the Italian coastline appears to be limited to specific areas (Liguria, Tuscany, Apulia, Calabria), with the highest occurrence of B. ceti infected cetaceans along the Ionian coastline, which suggests consistent circulation of the bacterial pathogen in that area. Our results highlight the need for continuous surveillance and monitoring studies to better understand the pathogen, host and environmental factors involved in cetacean Brucella spp. infection’s epidemiology, in tight agreement with the “One Health” concept. WGS typing proved to be useful for molecular classification of B. ceti strains and allowed the typing of large populations. The genetic clustering based on SNP analysis was in agreement with all previously reported methods and additionally it provided a much higher discriminatory power.
The severity of B. ceti-associated lesions reported in the present study supports the role of this microbial agent as a primary neurotropic pathogen in striped dolphins, as well as a probable cause of stranding events and death, as previously described elsewhere [14]. In this regard, our results corroborate previous reports indicating striped dolphins as highly susceptible hosts for developing neurobrucellosis in comparison with the other cetaceans [64], thus confirming neurobrucellosis as one of the most significant lesions’ pattern associated to B. ceti infection [26, 64, 68].
Additional studies are required to identify the mechanisms involved in the crossing of the blood-brain barrier and the pathogen and host-related factors driving B. ceti neuroinvasion, colonization and persistence in the CNS [27]. Moreover, a detailed understanding of the effects of pollutant-related immunotoxicity on pathogenicity of B. ceti, as suggested by some case reports [16, 69], is required, particularly in light of conflicting result obtained using ex vivo model [70].
Surveillance of strandings in Italy involves organisations from governmental and academic institutions with different areas of expertise such as public health, animal health and environment. Such network made our study possible, and our findings highlight the importance of the multidisciplinary approach in the monitoring of stranded cetaceans, with epidemiological data and laboratory information truly shared across sectors in the perspective of the one-health approach.
Finally, based on the demonstrated zoonotic capability of B. ceti [31, 32, 33], proper handling of stranded animals, together with an ad hoc adoption of all necessary biosafety and biosecurity measures and protocols during post mortem and diagnostic investigations on stranded cetaceans are strongly recommended to avoid the risk of transmission to humans of this and other zoonotic pathogens.
Supporting information
(DOCX)
Data Availability
The WGS data is submitted at NCBI GenBank with the following accession number: PRJNA623338.
Funding Statement
This work was supported by the Italian Ministry of Health. WM received funding from the Italian Ministry of Health under gran agreement code IZSPLV 09/18. GG received funding from the Italian Ministry of Health under gran agreement code IZSAM 02/17 RC. The funder had no role in study design, date collection and analysis, decision to publish, or preparation of the manuscript. There was no additional external funding received for this study.
References
- 1.Pappas G. The changing Brucella ecology: novel reservoirs, new threats. Int J Antimicrob Agents. 2010. November 36 Suppl 1: S8–11. 10.1016/j.ijantimicag.2010.06.013 [DOI] [PubMed] [Google Scholar]
- 2.Corbel MJ. Brucellosis in humans and animals. World Health Organization. 2006. https://apps.who.int/iris/handle/10665/43597
- 3.Ewalt DR, Payeur JB, Martin BM, Cummins DR, Miller WG. Characteristics of a Brucella species from a bottlenose dolphin (Tursiops truncatus) J Vet Diagn Invest. 1994; 6:448–452 10.1177/104063879400600408 [DOI] [PubMed] [Google Scholar]
- 4.Ross HM, Foster G, Reid RJ, Jahans KL, MacMillan AP. Brucella species infection in sea-mammals. Vet Rec. 1994; 134(14): 359 10.1136/vr.134.14.359-b [DOI] [PubMed] [Google Scholar]
- 5.Foster G, Osterman BS, Godfroid J, Jacques I, Cloeckaert A. Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int J Syst Evol Microbiol. 2007. November;57(Pt 11):2688–93. 10.1099/ijs.0.65269-0 [DOI] [PubMed] [Google Scholar]
- 6.Miller WG, Adams LG, Ficht TA, Cheville NF, Payeur JP, Harley DR, et al. Brucella-induced abortions and infection in bottlenose dolphins (Tursiops truncatus). J Zoo Wildl Med. 1999. March; 30(1):100–10. [PubMed] [Google Scholar]
- 7.Foster G, MacMillan AP, Godfroid J, Howie F, Ross HM, Cloeckaert A, et al. A review of Brucella sp. infection of sea mammals with particular emphasis on isolates from Scotland. Vet Microbiol. 2002. December 20; 90(1–4):563–80. 10.1016/s0378-1135(02)00236-5 [DOI] [PubMed] [Google Scholar]
- 8.Ohishi K, Zenitani R, Bando T, Goto Y, Uchida K, Maruyama T, et al. Pathological and serological evidence of Brucella-infection in baleen whales (Mysticeti) in the western North Pacific. Comp Immunol Microbiol Infect Dis. 2003. March; 26(2):125–36. 10.1016/s0147-9571(02)00036-x [DOI] [PubMed] [Google Scholar]
- 9.González-Barrientos R, Morales JA, Hernández-Mora G, Barquero-Calvo E, Guzmán-Verri C, Chaves-Olarte E, et al. Pathology of striped dolphins (Stenella coeruleoalba) infected with Brucella ceti. J Comp Pathol. 2010. May;142(4):347–52. 10.1016/j.jcpa.2009.10.017 [DOI] [PubMed] [Google Scholar]
- 10.Nymo HI, Tryland M, Godfroid J. A review of Brucella infection in marine mammals, with special emphasis on Brucella pinnipedialis in the hooded seal (Cystophora cristata). Vet Res. 2011; 42(1): 93 10.1186/1297-9716-42-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzmán-Verri C, González-Barrientos R, Hernández-Mora G, Morales JA, Baquero-Calvo E, Chaves-Olarte E, et al. Brucella ceti and Brucellosis in Cetaceans Front Cell Infect Microbiol. 2012; 2: 3 10.3389/fcimb.2012.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sierra E, Sánchez S, Saliki JT, Blas-Machado U, Arbelo M, Zucca D, et al. Retrospective study of etiologic agents associated with nonsuppurative meningoencephalitis in stranded cetaceans in the Canary Islands. J Clin Microbiol. 2014. July;52(7):2390–7. 10.1128/JCM.02906-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsen SC, Palmer MV. Advancement of knowledge of Brucella over the past 50 years. Vet Pathol. 2014. November;51(6):1076–89. 10.1177/0300985814540545 [DOI] [PubMed] [Google Scholar]
- 14.Isidoro-Ayza M, Ruiz-Villalobos N, Pérez L, Guzmán-Verri C, Muñoz PM, Alegre F, et al. Brucella ceti infection in dolphins from the Western Mediterranean sea. BMC Vet Res. 2014. September 17; 10:206 10.1186/s12917-014-0206-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colegrove KM, Venn-Watson S, Litz J, Kinsel MJ, Terio KA, Fougeres E, et al. Fetal distress and in utero pneumonia in perinatal dolphins during the Northern Gulf of Mexico unusual mortality event. Dis Aquat Organ. 2016. April 12;119(1):1–16. 10.3354/dao02969 [DOI] [PubMed] [Google Scholar]
- 16.Grattarola C, Giorda F, Iulini B, Pintore MD, Pautasso A, Zoppi S, et al. Meningoencephalitis and Listeria monocytogenes, T. gondii and Brucella spp. coinfection in a dolphin in Italy. Dis Aquat Organ. 2016. February 25;118(2):169–74. 10.3354/dao02957 [DOI] [PubMed] [Google Scholar]
- 17.Davison NJ, Perrett LL, Dawson C, Dagleish MP, Haskins G, Muchowski J, et al. Brucella ceti Infection in a Common Minke Whale (Balaenoptera acutorostrata) with Associated Pathology. J Wildl Dis. 2017. July;53(3):572–576. 10.7589/2016-08-200 [DOI] [PubMed] [Google Scholar]
- 18.Sánchez-Sarmiento AM, Carvalho VL, Díaz-Delgado J, Ressio RA, Fernandes NCCA, Guerra JM, et al. Molecular, serological, pathological, immunohistochemical and microbiological investigation of Brucella spp. in marine mammals of Brazil reveals new cetacean hosts. Transbound Emerg Dis. 2019. July;66(4):1674–1692. 10.1111/tbed.13203 [DOI] [PubMed] [Google Scholar]
- 19.Bossart GD. Marine Mammals as Sentinel Species for Oceans and Human Health. Vet Pathol. 2011; 48(3): 676–690. 10.1177/0300985810388525 [DOI] [PubMed] [Google Scholar]
- 20.Hernández-Mora G, Palacios-Alfaro JD, González-Barrientos R. Wildlife reservoirs of brucellosis: Brucella in aquatic environments. Rev Sci Tech. 2013. April;32(1):89–103. 10.20506/rst.32.1.2194 [DOI] [PubMed] [Google Scholar]
- 21.Shakir RA, Al-Din AS, Araj GF, Lulu AR, Mousa AR, Saadah MA. Clinical categories of neurobrucellosis. A report on 19 cases. Brain. 1987. February;110 (Pt 1):213–23. [DOI] [PubMed] [Google Scholar]
- 22.Obiako OR, Ogoina D, Danbauchi SS, Kwaifa SI, Chom ND, Nwokorie E. Neurobrucellosis—a case report and review of literature. Niger J Clin Pract. 2010. September;13(3):347–50. [PubMed] [Google Scholar]
- 23.González L, Patterson IA, Reid RJ, Foster G, Barberán M, Blasco JM, et al. Chronic meningoencephalitis associated with Brucella sp. Infection in live-stranded striped dolphins (Stenella coeruleoalba). J Comp Pathol. 2002. Feb-Apr;126(2–3):147–52. 10.1053/jcpa.2001.0535 [DOI] [PubMed] [Google Scholar]
- 24.Hernández-Mora G, González-Barrientos R, Morales JA, Chaves-Olarte E, Guzmán-Verri C, Barquero-Calvo E, et al. Neurobrucellosis in stranded dolphins, Costa Rica. Emerg Infect Dis. 2008. September;14(9):1430–3. 10.3201/eid1409.071056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davison NJ, Cranwell MP, Perrett LL, Dawson CE, Deaville R, Stubberfield EJ, et al. Meningoencephalitis associated with Brucella species in a live-stranded striped dolphin (Stenella coeruleoalba) in south-west England. Vet Rec. 2009. July 18;165(3):86–9. 10.1136/vetrec.165.3.86 [DOI] [PubMed] [Google Scholar]
- 26.Alba P, Terracciano G, Franco A, Lorenzetti S, Cocumelli C, Fichi G, et al. The presence of Brucella ceti ST26 in a striped dolphin (Stenella coeruleoalba) with meningoencephalitis from the Mediterranean Sea. Vet Microbiol. 2013. May 31;164(1–2):158–63. 10.1016/j.vetmic.2013.01.023 [DOI] [PubMed] [Google Scholar]
- 27.Di Guardo G, Centelleghe C, Mazzariol S. Cetacean Host-Pathogen Interaction(s): Critical Knowledge Gaps. Front Immunol. 2018. November 28; 9:2815 10.3389/fimmu.2018.02815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maquart M, Le Flèche P, Foster G, Tryland M, Ramisse F, Djønne B, et al. MLVA-16 typing of 295 marine mammal Brucella isolates from different animal and geographic origins identifies 7 major groups within Brucella ceti and Brucella pinnipedialis. BMC Microbiol. 2009. July 20;9:145 10.1186/1471-2180-9-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suárez-Esquivel M, Baker KS, Ruiz-Villalobos N, Hernández-Mora G, Barquero-Calvo E, González-Barrientos R et al. Brucella Genetic Variability in Wildlife Marine Mammals Populations Relates to Host Preference and Ocean Distribution, Genome Biol Evol. 2017. July 9; 7: 1901–1912, 10.1093/gbe/evx137 10.1093/gbe/evx137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhyan JC, Gidlewski T, Ewalt DR, Hennager SG, Lambourne DM, Olsen SC. Seroconversion and abortion in cattle experimentally infected with Brucella sp. isolated from a Pacific harbor seal (Phoca vitulina richardsi). J Vet Diagn Invest. 2001. September;13(5):379–82. 10.1177/104063870101300502 [DOI] [PubMed] [Google Scholar]
- 31.Sohn AH, Probert WS, Glaser CA, Gupta N, Bollen AW, Wong JD, et al. Human neurobrucellosis with intracerebral granuloma caused by a marine mammal Brucella spp. Emerg Infect Dis. 2003. April;9(4):485–8. 10.3201/eid0904.020576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald WL, Jamaludin R, Mackereth G, Hansen M, Humphrey S, Short P, et al. Characterization of a Brucella sp. strain as a marine-mammal type despite isolation from a patient with spinal osteomyelitis in New Zealand. J Clin Microbiol. 2006. December;44(12):4363–70. 10.1128/JCM.00680-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brew SD, Perrett LL, Stack JA, MacMillan AP, Staunton NJ. Human exposure to Brucella recovered from a sea mammal. Vet Rec. 1999. April 24;144(17):483 [PubMed] [Google Scholar]
- 34.Whatmore AM, Perret LL, MacMillan AP. Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol, 2007. April 20; 7: 34 10.1186/1471-2180-7-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whatmore AM, Dawson CE, Grossaud P, Koylass MS, King AC, Shankster SJ et al. Marine Mammal Brucella genotype associated with zoonotic infection. Emerg Infect Dis. 2008. March; 14 (3):517–8. 10.3201/eid1403.070829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Bressem MF, Van Waerebeek K, Raga JA, Godfroid J, Brew SD, MacMillan AP. Serological evidence of Brucella species infection in odontocetes from the south Pacific and the Mediterranean. Vet Rec. 2001. May 26;148(21):657–61. 10.1136/vr.148.21.657 [DOI] [PubMed] [Google Scholar]
- 37.Profeta F, Di Francesco CE, Marsilio F, Mignone W, Di Nocera F, De Carlo E, et al. Retrospective seroepidemiological investigations against Morbillivirus, T. gondii and Brucella spp. in cetaceans stranded along the Italian coastline (1998–2014). Res Vet Sci. 2015. August;101:89–92. 10.1016/j.rvsc.2015.06.008 [DOI] [PubMed] [Google Scholar]
- 38.Garofolo G, Zilli K, Troiano P, Petrella A, Marotta F, Di Serafino G, et al. Brucella ceti from two striped dolphins stranded on the Apulia coastline, Italy. J Med Microbiol. 2014. February;63(Pt 2):325–9. 10.1099/jmm.0.065672-0 [DOI] [PubMed] [Google Scholar]
- 39.Whatmore AM, Dawson CE, Grossaud P, Koylass MS, King AC, Shankster SJ, et al. Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect Genet Evol. 2009. December;9(6):1168–84. 10.1016/j.meegid.2009.07.001 [DOI] [PubMed] [Google Scholar]
- 40.Cvetnić Ž, Duvnjak S, Đuras M, Gomerčić T, Reil I, Zdelar-Tuk M et al. Evidence of Brucella strain ST27 in bottlenose dolphin (Tursiops truncatus) in Europe. Vet Microbiol. 2016. November 30; 196:93–97. 10.1016/j.vetmic.2016.10.013 [DOI] [PubMed] [Google Scholar]
- 41.Duvnjak S, Špičić S, Kušar D, Papić B, Reil I, Zdelar-Tuk M, et al. Whole-Genome Sequence of the First Sequence Type 27 Brucella ceti Strain Isolated from European Waters. Genome Announc. 2017. September 14;5(37). pii: e00988-17. 10.1128/genomeA.00988-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ancora M, Marcacci M, Orsini M, Zilli K, Di Giannatale E, Garofolo G, et al. Complete Genome Sequence of a Brucella ceti ST26 Strain Isolated from a Striped Dolphin (Stenella coeruleoalba) on the Coast of Italy. Genome Announc. 2014. March 6;2(2). pii: e00068-14. 10.1128/genomeA.00068-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geraci JR & Lounsbury VJ. 2005. Marine Mammals Ashore: A Field Guide for Strandings (Second Edition) National Aquarium in Baltimore, Baltimore, MD. [Google Scholar]
- 44.Pugliares KR, Bogomolni AL, Touhey KM, Herzig SM, Harry CT, Moore MJ. Small cetacean necropsy technique and anatomy. In: Marine mammal necropsy: An introductory guide for stranding responders and field biologists. Woods hole Oceanographic Institution Technical Report WHOI-2007-06, Woods Hole, Massachussets; 2007. pp. 53–55
- 45.Anderson RC. Keys to the genera of the superfamily Metastrongyloidea In: CIH keys to the nematode parasites of vertebrates. No. 5. Anderson RC, Chabaud AG, Willmott S editors. Commonwealth Agricultural Bureaux, Farnham Royal, UK; Vol.5, 1978. pp.1–40 [Google Scholar]
- 46.Khalil LF, Jones A, Bray RA. Keys to the Cestode Parasites of Vertebrates. CAB International, Wallingford, UK; 1994. 752 pp [Google Scholar]
- 47.Pintore MD, Mignone W, Di Guardo G, Mazzariol S, Ballardini M, Florio CL et al. Neuropathologic findings in cetaceans stranded in Italy (2002–14). J Wildl Dis. 2018. April;54(2):295–303. 10.7589/2017-02-035 [DOI] [PubMed] [Google Scholar]
- 48.Verna F, Giorda F, Miceli I, Rizzo G, Pautasso A, Romano A, et al. Detection of morbillivirus infection by RT-PCR RFLP analysis in cetaceans and carnivores. J Virol Methods. 2017. September;247:22–27. 10.1016/j.jviromet.2017.05.009 [DOI] [PubMed] [Google Scholar]
- 49.Vitale M, Galluzzo P, Currò V, Gozdzik K, Schillaci D, Di Marco, et al. A high sensitive nested PCR for T. gondii detection in animal and food samples. J Microb Biochem Technol. 2013; 5: 39–41. [Google Scholar]
- 50.Di Guardo G, Proietto U, Di Francesco CE, Marsilio F, Zaccaroni A, Scaravelli D, et al. Cerebral toxoplasmosis in striped dolphins (Stenella coeruleoalba) stranded along the Ligurian Sea coast of Italy. Vet Pathol. 2010. March;47(2):245–53. 10.1177/0300985809358036 [DOI] [PubMed] [Google Scholar]
- 51.OIE Terrestrial Manual. Brucellosis (Brucella abortus, B. melitensis and B. suis) (infection with B. abortus, B. melitensis and B. suis. In: Manual of Diagnostic Tests and Vaccines for terrestrial animals. 2019. online version. Part 3. Chapter 3.1.4
- 52.Alton GG, Jones ML, Angus RD, Verger JM. Techniques for the Brucellosis Laboratory. INRA: Paris; 1988
- 53.Cloeckaert A, Verger J M, Grayon M, Grepinet O. Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer membrane proteins of Brucella. Microbiology. 1995. September; 141: 2111–2121 10.1099/13500872-141-9-2111 [DOI] [PubMed] [Google Scholar]
- 54.Baily GG, Krahn JB, Drasar BS, Stoker NG. Detection of Brucella melitensis and Brucella abortus by DNA amplification. J Trop Med Hyg. 1992. August;95(4):271–5. [PubMed] [Google Scholar]
- 55.Sahl JK, Beckstrom-Sternberg SM, Babic-Sternberg JS, Gillece JD, Hepp CM, Auerbach RK et al. The In Silico Genotyper (ISG): an open-source pipeline to rapidly identify and annotate nucleotide variants for comparative genomics applications. 2015. bioRxiv preprint 10.1101/015578 [Google Scholar]
- 56.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013 preprint arXiv:1303.3997
- 57.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013; 43:11.10.1–11.10.33. 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015. January;32(1):268–74. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017. June;14(6):587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng L., Connor TR, Sirén J, Aanensen DM, Corander J. Hierarchical and Spatially Explicit Clustering of DNA Sequences with BAPS Software. Mol Biol Evol. 2013. May; 30(5): 1224–1228. 10.1093/molbev/mst028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corander J, Waldmann P, Sillanpää MJ. Bayesian analysis of genetic differentiation between populations. Genetics. 2003. January;163(1):367–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muñoz PM, Mick V, Sacchini L, Janowicz A, De Miguel M J, Cherfa MA et al. Phylogeography and epidemiology of Brucella suis biovar 2 in wildlife and domestic swine. Vet Microbiol. 2019; 233: 68–77. 10.1016/j.vetmic.2019.04.025 [DOI] [PubMed] [Google Scholar]
- 63.West KL, Levine G, Jacob J, Jensen B, Sanchez S, Colegrove K, et al. Coinfection and vertical transmission of Brucella and Morbillivirus in a neonatal sperm whale (Physeter macrocephalus) in Hawaii, USA. J Wildl Dis. 2015. January;51(1):227–32. 10.7589/2014-04-092 [DOI] [PubMed] [Google Scholar]
- 64.González-Barrientos R, Morales JA, Hernández-Mora G, Barquero-Calvo E, Guzmán-Verri C, Chaves-Olarte E, et al. Pathology of striped dolphins (Stenella coeruleoalba) infected with Brucella ceti. J Comp Pathol. 2010. May;142(4):347–52. 10.1016/j.jcpa.2009.10.017 [DOI] [PubMed] [Google Scholar]
- 65.Hernández-Mora G, Manire CA, González-Barrientos R, Barquero-Calvo E, Guzmán-Verri C, Staggs L et al. Serological diagnosis of Brucella infections in odontocetes. Clin Vaccine Immunol. 2009. June;16(6):906–15. 10.1128/CVI.00413-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nymo IH, Godfroid J, Åsbakk K, Larsen AK, da Neves CG, Rødven R et al. A protein A/G indirect enzyme-linked immunosorbent assay for the detection of anti-Brucella antibodies in Arctic wildlife. J Vet Diagn Invest. 2013. May;25(3):369–75. 10.1177/1040638713485073 [DOI] [PubMed] [Google Scholar]
- 67.Gaspari S, Azzellino A, Airoldi S, Hoelzel AR. Social kin associations and genetic structuring of striped dolphin populations (Stenella coeruleoalba) in the Mediterranean Sea. Mol Ecol. 2007. July;16(14):2922–33. 10.1111/j.1365-294X.2007.03295.x [DOI] [PubMed] [Google Scholar]
- 68.Muñoz M, García-Castrillo G, López-García P, González-Cueli JC, De Miguel MJ, Marín CM, et al. Isolation of Brucella species from a live-stranded striped dolphin (Stenella coeruleoalba) in Spain. Vet Rec. 2006. 158: 450–451. 10.1136/vr.158.13.450 [DOI] [PubMed] [Google Scholar]
- 69.Davison NJ, Perrett LL, Law RJ, Dawson CE, Stubberfield EJ, Monies RJ et al. Infection with Brucella Ceti and High Levels of Polychlorinated Biphenyls in Bottlenose Dolphins (Tursiops Truncatus) Stranded in South-West England. Vet Rec. 2011. July; 2;169(1):14 10.1136/vr.d2714 [DOI] [PubMed] [Google Scholar]
- 70.Nymo IH, das Neves CG, Tryland M, Bårdsen BJ, Santos RL, Turchetti AP et al. Brucella pinnipedialis hooded seal (Cystophora cristata) strain in the mouse model with concurrent exposure to PCB 153. Comp Immunol Microbiol Infect Dis. 2014. 37: 195–204. 10.1016/j.cimid.2014.01.005 [DOI] [PubMed] [Google Scholar]