Using stimuli-responsive hydrogels, we 3D print composite microscaffolds to analyze mechanobiology on the single-cell level.
Abstract
Many essential cellular processes are regulated by mechanical properties of their microenvironment. Here, we introduce stimuli-responsive composite scaffolds fabricated by three-dimensional (3D) laser lithography to simultaneously stretch large numbers of single cells in tailored 3D microenvironments. The key material is a stimuli-responsive photoresist containing cross-links formed by noncovalent, directional interactions between β-cyclodextrin (host) and adamantane (guest). This allows reversible actuation under physiological conditions by application of soluble competitive guests. Cells adhering in these scaffolds build up initial traction forces of ~80 nN. After application of an equibiaxial stretch of up to 25%, cells remodel their actin cytoskeleton, double their traction forces, and equilibrate at a new dynamic set point within 30 min. When the stretch is released, traction forces gradually decrease until the initial set point is retrieved. Pharmacological inhibition or knockout of nonmuscle myosin 2A prevents these adjustments, suggesting that cellular tensional homeostasis strongly depends on functional myosin motors.
INTRODUCTION
In cell biological research, more and more attention is drawn to biophysical cues that influence cellular behavior in addition to biochemical cues (1). For example, adherent cells have been found to be more spread, more polarized, and more contractile in stiffer environments, they migrate differently, and stem cells differentiate into different cell types (2). Cells are able to recognize and transduce mechanical stress and strain patterns by mechanosensitive modules such as ion channels, cell adhesion sites, and the cytoskeleton (3). The mechanical input is ultimately converted to biochemical signals that guide not only the dynamic rearrangement of actin stress fibers and the actin cortex (4) but also gene expression and the response to soluble ligands. Although several approaches have been established to stretch cells, it remains challenging to monitor the cell response to mechanical stimuli.
Mechanical stimulation of cells is most commonly performed by pneumatic, piezoelectric, or electromagnetic stretching of deformable polydimethylsiloxane substrates or thin membranes (5). Here, cells are typically adhering to two-dimensional (2D) substrates in random morphologies. Other approaches such as optical tweezers, atomic force microscopy (6), microplates (7), or micromanipulators (8) offer precise displacements in 3D, but these techniques are hardly scalable to study a large number of cells. In addition, a fixation of cells in the stretched state is not possible. Moreover, all available techniques require complex setups to trigger and control the stimulation. A detailed description and comparison of these methods can be found in several reviews (9, 10). Here, we propose a different approach using 3D scaffolds based on stimuli-responsive, supramolecular polymers to simultaneously stretch a large number of single cells in tailored 3D microenvironments.
In the past, 3D laser lithography has successfully been used to manufacture cell scaffolds with tailored geometry and spatially functionalized surfaces (11). However, the transition from passive to active systems requires responsive materials that can be stimulated on demand (12). In recent years, a large number of these material systems with numerous applications in the macroscopic (13) and microscopic (14, 15) regime have been investigated and extensively reviewed (16, 17). One crucial constraint for the application in cell biology is a specific physiological stimulus of the material that does not influence or alter the behavior of the cells. Despite a number of studies demonstrating the dynamic control of cells using hydrogels responsive to temperature (18), pH (19), enzymes (20), or illumination with ultraviolet light (21), these applications are limited because the formation and cleavage of bonds are often performed under harsh conditions. Supramolecular polymers (22, 23) could provide an advantage over the abovementioned materials, if appropriate host or guest molecules are selected for the stimulation (24).
In the following, we first present a stimuli-responsive photoresist containing cross-links formed by noncovalent, directional interactions between β-cyclodextrin (host) and adamantane (guest) moieties. The resulting hydrogel microstructures fabricated by 3D laser lithography exhibit large volume changes by stimulation with the soluble low–molecular weight guest molecules under physiological conditions. Next, we combine this material with conventional photoresists to fabricate composite scaffolds consisting of protein-repellent base structures, protein-adhesive parts, and the stimuli-responsive hydrogel. These fabricated samples can easily be mounted on a microscope and only require a simple microfluidic chamber to exchange the adequate solutions for stimulation. We further use numerical calculations to model the scaffolds and correlate displacements to forces, which has enabled us to optimize the design to the expected force values of cells.
Using this approach, we stretch single cells in a spatially and temporally well-defined manner and precisely track their response as a function of time via digital image correlation. Our approach is inherently scalable and enables to simultaneously study a large number of individual cells in well-defined microenvironments. Furthermore, this fabrication technique allows us to integrate nonresponsive scaffolds as controls in all experiments. Another major advantage of this system is the ability to chemically fix the cells and the scaffolds at any point during the experiment, thus allowing us to stain the cells and their subcellular compartments while keeping them in the stretched state. We highlight this by visualizing a strong reorganization of the actin cytoskeleton as a response to the external displacement. We further show that this reorganization crucially depends on the activity of myosin II molecular motors. In the last part, we emphasize the flexibility of our approach to changing and adapting the geometrical design of the scaffolds according to the specific application. We further demonstrate this aspect by modified structures, which asymmetrically stretch cells on one side only, and we study the response to this localized stimulation.
RESULTS
Stimuli-responsive microstructures based on host-guest complexes
To create 3D microscaffolds for the manipulation of single cells, we require a material system with a large stimuli response under physiological conditions that can be processed by 3D laser lithography. A host-guest hydrogel based on β-cyclodextrin acrylamide (βCD-AAm) and adamantane acrylamide (Ad-AAm) fulfills all these requirements.
In the first step, we developed a photoresist that can be used for 3D laser lithography. Figure 1A shows a schematic representation of the composition and of the photopolymerization procedure. The two main components are βCD-AAm and Ad-AAm. Both molecules are equipped with an acrylamide group, which enables their incorporation in polymer networks. β-cyclodextrin and adamantane form a host-guest complex in aqueous environments. The hydrophobic adamantane (guest) fits nicely in the hydrophilic cavity of β-cyclodextrin (host), forming an inclusion complex with an association constant of Ka > 104 M−1 (25). The host-guest interactions are in a dynamic equilibrium and can be manipulated by adding or removing competitive adamantane molecules in solution, leading to control over this stimuli-responsive system on demand. By including additional acrylamide in the photoresist, the polymerized network consists of polyacrylamide chains, which are cross-linked by the host-guest complexes. Thus, no additional covalent cross-linker is needed to form stable 3D microstructures. This aspect circumvents the typical challenge in printing stimuli-responsive 3D hydrogels, namely, that more cross-links are required to stabilize the network, which hinder its response at the same time (16). In our system, we were able to increase the number of cross-links and the functionality simultaneously. To induce the radical polymerization, the water soluble and biocompatible photoinitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) was added (26). All components were dissolved in a mixture of 2-propanol and water (see Materials and Methods for details). The cross-linked materials were not soluble in water or in other organic solvents. Therefore, the insoluble bulk material was characterized using 1H field gradient magic angle spinning nuclear magnetic resonance spectroscopy on a model photoresist, polymerized by a one-photon process (λ = 365 nm). Figure S1 shows the successful polymerization resulting in formation of polymer networks containing host and guest moieties in the side chain.
Fig. 1. Characterization of host-guest structures.
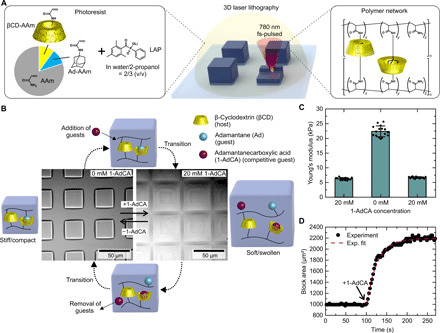
(A) Chemical structure of components and products of the 3D laser lithography. (B) Array of hydrogel blocks fabricated by 3D laser lithography with schematic depiction of the host-guest dynamics. (C) Young’s modulus of a hydrogel block in 0 and 20 mM 1-adamantanecarboxylic acid (1-AdCA) in phosphate-buffered saline (PBS). (D) Block area as a function of time. After 100 s, the solution was exchanged to 20 mM 1-AdCA in PBS.
To study the stimuli-responsive properties of the resulting material, we fabricated an array of hydrogel blocks of 25 μm by 25 μm by 10 μm by 3D laser lithography and developed them in water. These blocks were attached to the glass substrate by a prior silanization treatment of the surface (see Materials and Methods for details). At the beginning of the experiment, the polymer network of the resulting structures was in the stiff/compact state, since the host-guest moieties form stable complexes in aqueous environments (see Fig. 1B). Next, we imaged the blocks in an optical microscope during the addition of free adamantane derivatives in solution (see also movie S1). In all experiments, we used 1-adamantanecarboxylic acid (1-AdCA) because pure adamantane is poorly soluble in water. The dissolved 1-AdCA guests are also hydrophobic and start to compete with the guests in the polymer chain for the limited number of available host molecules. This exchange can happen because the host-guest complexation is a dynamic equilibrium. Hence, host-guest complexes can be dissociated and recovered spontaneously. However, only the inclusion complexes formed by the βCD-AAm and Ad-AAm units contribute to the cross-linking of polymer chains and, hence, the stability of the overall polymer network. After a transition period, a new equilibrium is formed with complexes between bound βCD-AAm units and free 1-AdCA. As a result, the number of cross-links in the polymer network is substantially reduced, and the material transitioned to the soft/swollen state. This effect can be clearly observed in the optical micrographs of the larger blocks. The fabricated blocks undergo a strong swelling when the 1-AdCA is added, since the network with less cross-links takes up more water. In addition, the image contrast is reduced because the refractive index difference to the surrounding media becomes smaller when more and more water is taken up by the material. This effect can be studied in the block area as a function of time as plotted in Fig. 1D. The addition of 1-AdCA leads to an increase in block area by more than a factor of 2 and exponential fits of the swelling yield a time constant of 28.0 ± 1.7 s (mean and SD) for an average of multiple blocks. However, note that this process is diffusion driven and the time constant strongly depends on the size of the structures. The area was calculated by tracking the edges of the block, which appear as local minima in the corresponding averaged intensity profile of the optical bright-field images.
One important aspect of this system is its complete reversibility. If the competitive guest molecules are washed out, then only the βCD-AAm and Ad-AAm units linked to the polymer chains remain. In another transition period the equilibrium shifts, the cross-links will form again and the material returns to the stiff/compact state.
The same transition can also be observed in the mechanical properties of the material (Fig. 1C). We used an atomic force microscope to determine the stiffness of hydrogel blocks by nanoindentation (see Materials and Methods for details). In a solution consisting of 20 mM 1-AdCA in phosphate-buffered saline (PBS), we measured a Young’s modulus of 7 kPa (open cross-links), which increased to 23 kPa when the solution was substituted with pure PBS (closed cross-links). As the system is reversible, we were able to retrieve the initial value by adding again a new solution containing 20 mM 1-AdCA.
Composite scaffolds as platforms for single-cell manipulation
To manipulate single cells in a 3D environment, it is necessary to first guide them to the desired positions in the scaffolds and, second, to apply a controlled external stimulus. Therefore, several materials with different surface properties have to be combined with the stimuli-responsive hydrogel in a sequential fabrication procedure.
A schematic design of the microscaffolds along with the underlying principle is depicted in Fig. 2A. The four walls (gray) consist of a photoresist based on trimethylolpropane ethoxylate triacrylate (TPETA), which is protein repellent. The four beams on top (orange), on the other hand, are fabricated with a photoresist based on pentaerythritol triacrylate (PETA) and adsorb proteins on the surface. In this manner, it is possible to precisely guide the adhesion of cells in the scaffolds. The stimuli-responsive host-guest hydrogel (yellow) is enclosed in the center. By addition of solved 1-AdCA molecules in solution, the hydrogel expands and pushes on the walls. Because they are fixed to the substrate, they start to bend and pull the four beams away from the center. Thus, a cell adhered to these beams will be substantially stretched on all sites. Fixation of the hydrogel to the substrate by silanization prevents an isotropic swelling, leading to the depicted shape with a stronger lateral expansion on the top and a bulged upper surface.
Fig. 2. Stimuli-responsive composite microscaffolds.
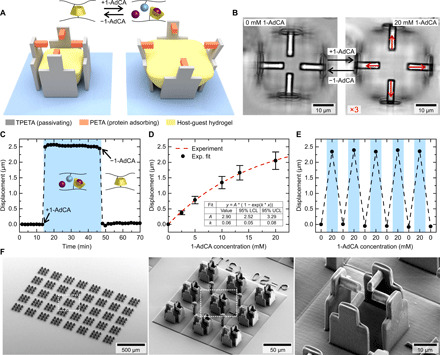
(A) Schematic image of the microscaffolds, fabricated by 3D laser lithography with three different materials. Addition of 1-AdCA leads to a swelling of the hydrogel and displacement of the beams. (B) Optical micrographs of the two states with and without 1-AdCA. The red arrows indicate the displacement of the individual beams scaled by a factor of 3. (C) Average beam displacement relative to the center for an exemplary scaffold as a function of time. Addition of 1-AdCA leads to a strong positive displacement of the beams away from the center. Rinsing with pure PBS returns the scaffold into the original state. (D) Average displacement as a function of 1-AdCA concentration for multiple scaffolds. The table lists the resulting fit values and the lower and upper confidence limits LCL and UCL. (E) Average beam displacements for multiple cycles of solution exchange demonstrate complete reversibility of the system. (F) Scanning electron microscopy (SEM) micrographs of a typical sample with 360 scaffolds (without the host-guest hydrogel) with increasing magnifications from left to right.
This indirect transmission of forces has two key advantages as compared to direct cultivation of cells on the expanding host-guest hydrogels (24). First, the change in stiffness of the material and the expansion are decoupled. Since the change in Young’s modulus can have an additional substantial impact on cellular behavior, it is necessary to isolate the response due to the mechanical stimulation. Second, our scaffold design enables us to guide the cells to defined adhesion points in a complex environment and force them in the desired shape. To ensure that the hydrogel and the cell will not contact each other, we performed 3D laser scanning microscope (LSM) reconstructions by adding fluorescent dyes to the photoresists (see fig. S2).
We monitor displacements of the beams by recording a time series and performing a digital image correlation analysis on the resulting images (see Materials and Methods for details). This technique enables us to achieve subpixel localization errors (27). This precision is mandatory to track even small cellular responses with high accuracy. Figure 2C depicts the average beam displacement of all four beams of one scaffold relative to the center over time. A positive value represents a displacement away from the center and a negative value a displacement toward the center of the scaffold. After 10 min, we exchanged the solution to 20 mM 1-AdCA in PBS, which leads to a swelling of the hydrogel and a positive displacement of about 2.5 μm per beam. This displacement is stable until we change back to pure PBS at 40 min to remove all solved 1-AdCA molecules. The hydrogel shrinks again and the beams return to their initial position. The time constants for these transitions were obtained from exponential fits as depicted for a single scaffold in fig. S3. We analyzed nine scaffolds from three independent experiments and obtained time constants of 3.1 ± 0.5 s for swelling and 4.5 ± 2.2 s for shrinking of the hydrogels. Two optical micrographs of the time series depicting these two states are shown in Fig. 2B. The red arrows correspond to the individual beam displacements, for clarity scaled by a factor of 3. With this 3D scaffold design, it is possible to exert a substantial mechanical stretch on a cell that is attached on top of the four beams.
One important feature of the system is the ability to precisely control the stimulus by the 1-AdCA concentration as can be observed in Fig. 2D. Since the host-guest complexation is a dynamic equilibrium, the number of cross-links with free 1-AdCA depends on the available molecules in solution. This dynamic equilibrium enables us to control the beam displacement and, thus, the cellular stretch by variation of the 1-AdCA concentration. In addition, the composite scaffolds demonstrate a complete reversibility, which has previously been discussed for the single hydrogel blocks (see Fig. 2E).
The reaction of all scaffolds on one sample can be directly controlled by exchanging the solution, which makes this system inherently scalable. Figure 2F shows three scanning electron micrographs of a typical fabricated sample with scaffolds with increasing magnification. Here, the central hydrogel is not included because the sample has to be dried for scanning electron microscopy (SEM) preparation. In our design, we arranged 3 by 3 scaffolds together on a protein repellent TPETA layer. The resulting size of around 150 μm by 150 μm corresponds to one writing field during the fabrication and a respective viewing field during the image acquisition. Five by eight of this fields result in a total number of 360 scaffolds per sample, making high throughput and quantitative analysis possible.
Correlating beam displacements to cellular forces
Next, we applied this system to mechanically stimulate single cells and study their resulting response. To achieve this, we used numerical calculations to correlate displacements to expected cellular forces and adapted the scaffold design accordingly.
The expected cellular behavior is schematically depicted in Fig. 3A. A cell first invades the scaffold (1) and starts to spread on the four PETA beams, which have been coated with the extracellular matrix protein fibronectin (FN). During this process, the cell builds up tension. From previous experiments, we expected the cellular forces to be on the order of 100 to 300 nN (28, 29). As a result, the cell pulls the four beams toward the center until a steady-state condition is reached in which the beams (and thus the walls) are bend inward (2). The displacements of the walls by the cellular forces are indicated by blue arrows. At this point, the external stimulation can be triggered by the addition of 1-AdCA to stretch the cells by expansion of the hydrogel (3). The external displacement of the walls due to the stimuli-responsive hydrogel is indicated by red arrows. From previous publications, we expected that displacements on the order of 1 to 3 μm per beam that can be easily achieved with this system will lead to a cellular response (6, 8).
Fig. 3. Cellular forces in the microscaffolds.
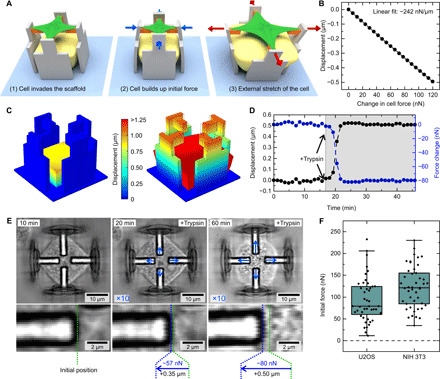
(A) Schematic images of a cell that first invades the scaffold (1), builds up initial force in the process (2), and is later stretched by the scaffold (3). (B) Linear relationship of changes in displacement and changes in cellular traction forces, obtained by numerical calculations in COMSOL Multiphysics. Negative displacements are defined to point toward the center of the scaffold. (C) Exemplary images of displacements from numerical calculations before (left) and after (right) the swelling of the hydrogel. (D) Reaction of an exemplary cell in a scaffold as a function of time after addition of trypsin at minute 17. The black data points depict the displacement change, and the blue data points depict the change in traction force. (E) Optical micrographs of a cell before, during, and after addition of trypsin. The blue arrows indicate the displacement of the individual beams scaled by a factor of 10. The images below show a magnification of the left beam, with the green dashed line denoting the beam position of the initial frame and the blue dashed line denoting the position of the current frame. The numbers next to the arrow show the cellular reaction as a change in displacement and traction force relative to the first image. (F) Quantification of initial forces in the scaffolds for U2OS and NIH 3T3 cells. Each data point corresponds to one cell in a scaffold.
The analysis of the cellular response required the correlation of beam displacements to changes in cell force. Therefore, we used numerical finite element method calculations using the commercial software COMSOL Multiphysics. We modeled our scaffolds and the expanding hydrogel and calculated the forces that are necessary to displace the beams by defined distances (Fig. 3C and see Materials and Methods for details). Thereby, we effectively calibrated the walls as Hooke’s springs with a given spring constant and found a linear relation between the displacement and the resulting force that has to be exerted (see Fig. 3B).
These calculations were also used to optimize our scaffold design according to the experimental conditions. Here, we mainly considered three aspects: first, confining the hydrogel to achieve a large displacement of the beams, resulting in a substantial stretch of the cell; second, achieving distinct displacements via the cellular forces, which can be tracked with high accuracy; and third, a robust design that allows reliable fabrication to minimize sample-to-sample variations.
Initial traction force of cells in the scaffolds
At this point, we had all the necessary tools available to study cellular forces in the scaffolds under different conditions. In the first experiment, we evaluated the equilibrium forces of cells inside the scaffolds.
After spreading on the scaffolds, cells build up an initial force in analogy to traction forces observed on 2D samples via traction force microscopy (28) or micropost assays (29). However, in our scaffolds, the cells are in a more complex environment with four restricted peripheral adhesion sites and an elevated cell body. To quantify the force in our system, we recorded a time series of cells in scaffolds without the host-guest hydrogel until they reached an equilibrium state. At this point, we added trypsin to detach all cells from the scaffolds and simultaneously recorded the beam displacements. Since the correlation between forces and displacements depends on the presence of the hydrogel, we calculated the corresponding relation without the hydrogel by numerical calculations (fig. S4). Figure 3D shows an exemplary track corresponding to the three optical micrographs in Fig. 3E. Here, the displacements are colored in black, and the corresponding calculated force change is colored in blue (see also movie S2). After addition of trypsin, the cell is quickly detached from the beams. As a result, the beams return toward their initial positions before the cell invaded the structure. The difference between these two levels is used to calculate the initial force of the cells. The position of the beams can also be observed in the magnified segment on the bottom, where the green dashed line depicts the initial position before addition of trypsin and the blue dashed line the current position on the displayed frame.
We performed this experiment with U2OS and NIH 3T3 cells with the results summarized in Fig. 3F. Here, each data point depicts one cell in a scaffold with a corresponding track as shown previously. Both cell types show a positive initial force, so they actively start to pull on the beams when they spread on the scaffolds. The average contraction force is 79 ± 50 nN for U2OS cells and 121 ± 48 nN for NIH 3T3 cells (mean values with SD). These values are lower compared to cellular contraction forces measured on either flat substrates (28, 30) or on dense micropillars (29, 31). A direct comparison, however, is difficult since we measure forces exerted by the whole cell, whereas, in the above cited publications, forces are given per contact site. Our results also agree well with recent experiments using the same NIH 3T3 cells in rigid 3D scaffolds created with 3D laser lithography (32). Here, the force scale is set by the 1D modulus of the peripheral stress fibers, which has been estimated from 3D cell shape to be around 70 nN. For the geometry studied here, with two arcs pulling on one adhesion platform, one would expect a result less than twice this number, exactly as observed. The deviation between 2D and 3D can be explained by the restricted number of adhesions sites, which are limited to the four FN-coated beams in our case. Another important factor is the roundish 3D cell shape, which leads to more distributed forces than the strongly pinned and flattened cell shapes in 2D (32). Thus, if compared to cells adhering to a substrate and an atomic force microscope cantilever, our values for NIH 3T3 cells are in good agreement with the resulting force of 117 ± 21 nN measured by Webster et al. (6). In the following, we focused on U2OS cells and included additional data with NIH 3T3 cells in figs. S5 and S6.
Cellular reaction to stretches
After analyzing the initial forces, we investigated the cellular response to mechanical stimulations. We seeded the cells and kept them for 2 hours in the incubator until the cells adhered on the structures and established contacts to all four beams. We imaged the cells for another 20 min in this steady state and applied the external stretch by adding medium with 20 mM 1-AdCA. After 30 min under the stretched condition, we changed to medium without 1-AdCA to release the cells from the external stimulation. The reaction of the cells was studied by tracking the beam displacements over the whole course of the experiment.
Figure 4A shows optical micrographs 1 to 5 of an exemplary cell in a scaffold during the different phases of the experiment. The arrows correspond to the individual beam displacements as a result of the external stimulation (in red) and the cellular response (in blue) with the respective scaling. The complete track is depicted in Fig. 4B with the numbers that correspond to the optical micrographs (see also movie S3).
Fig. 4. Cellular reaction to external stimulation.
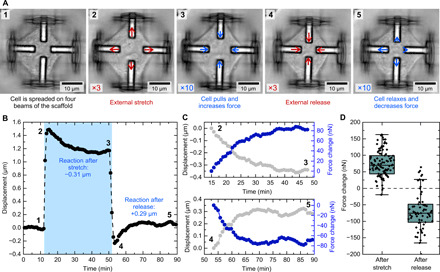
(A) Optical micrographs from different time points during a stretching experiment. The arrows indicate the individual beam displacements due to the external stimulus (red) (scaled by factor of 3) and the cellular response (blue) (scaled by factor of 10). (B) Corresponding average beam displacement as a function of time for a scaffold with an attached cell during a stretch-release cycle. The individual numbers in the plot correspond to the optical micrographs in (A). (C) Zoom in of the reaction after the stretch/after the release respectively with the displacements in black and traction force changes in blue. (D) Quantification of the cellular response 30 min after the stretch and after the release. Each data point represents one cell during the depicted cycle in (B).
In the first phase, the cell is in a steady state on the scaffold and has established an initial force (1). Next, the external stimulus is activated and the cell is stretched by 1.5 μm on each beam (2). Now, the cell starts to actively counteract this external deformation and pulls back the beams, thus increasing its contraction force substantially. After around 30 min, the displacement stabilized again on a new level, which is 0.31 μm lower than after the stretch (3). This behavior can be observed in more detail in Fig. 4C (top), which displays a zoom in of the reaction phase after the stretch. The connection to the cell force (blue dots) reveals that the cell immediately starts to increase its contraction force after the stretch to stabilize again at a value that is about 80 nN higher than the initial value. We chose 30 min as a time interval for all the experiments, because most of the cells reached a new steady-state level during this time (see fig. S7 for details).
In the next step, the cell is released from the external stretch, and the beams move 1.5 μm back toward the center. Immediately afterward, the displacement is lower compared to the initial value at the beginning of the track, since the cell is still exerting the additional contraction force (4). Over the course of the next 30 min, however, the cell relaxes and stabilizes again around the initial set point (5). The section shown in Fig. 4C (bottom) displays how the contraction force decreased by about 70 nN after the external stimulation is removed. As a result, the total contraction force of the cell is now close to the initial value at the start of the experiment.
Figure 4D shows the quantification of this effect. Each data point corresponds to one cell in a scaffold, which undergoes a cycle as shown in Fig. 4B. The displacements 30 min after the external stretch and 30 min after the external release were measured and converted to changes in cell force. In summary, cells counteract the external stretch by increasing their tension and stabilize on a new equilibrium level. Compared to the initial level, contraction forces are increased by 74 ± 41 nN. These results support previous findings that tensional homeostasis does not necessarily cause the cells to return to their initial state (6, 33). Different amounts of applied stress between 0.5- and 3-μm displacement in all directions corresponding to 4 to 24% equibiaxial stretch did not induce different force responses (see fig. S8). For large displacements, cells stabilize around a tunable force set point adjusted to the new situation, which has been termed tensional buffering in the past (6). This new set point can be maintained for at least 1 hour (see fig. S7 for details). When the external stimulation is released, the new set point initially led to a lower displacement compared to the beginning of the experiment. In the following 30 min, cells relaxed and decreased their contraction forces by −69 ± 57 nN. Consequently, the initial set point before stimulation is recovered. We performed preliminary experiments where we stretched NIH 3T3 cells three times over the course of 200 min. We found that these cells actively respond to the stimulations in the scaffolds over long periods of time (fig. S9). These experiments further emphasize the high throughput of this technique, which is an important aspect for the quantitative analysis of cellular effects. In addition, we performed control experiments with cells in scaffolds not containing a host-guest hydrogel, showing that the addition of 1-AdCA does not significantly influence the cellular behavior (fig. S10.). We further investigated the cell viability in a live/dead staining over the course of 24 hours in medium containing 1-AdCA and found no significant differences to the control sample with almost 100% cell survival (fig. S11).
Inhibition effects on cellular behavior
In the next step, we investigated whether the observed effects primarily arise from actomyosin contractility, from passive elements such as cytoskeletal cross-linkers, or a combination of both (34). Therefore, we studied the effect of Rho-associated protein kinase (ROCK) inhibition and additionally used a U2OS–knockout (KO) cell line, which lacks the nonmuscle myosin 2A (NM2A) protein. Both, ROCK and its downstream target NM2A, are known to be key generators of cellular contractility (35). However, the KO of NM2A solely affects actomyosin contractility by blocking the motor activity, while ROCK signaling acts bidirectional, affecting both actomyosin contractility and the actin stability (36, 37). If the observed effects depend on both, actomyosin contractility and passive actin stability, then ROCK inhibition and NM2A-KO should lead to a different cellular response.
Figure 5B shows the quantification of the initial force, compared to the wild-type (WT) scenario that has been discussed previously (see Fig. 3 for details). For the ROCK inhibition, we waited until cells obtained a steady state on the scaffolds and treated them with 50 μM Y-27632 for 15 min. After adding the inhibitor, we obtained a time versus displacement curve similar to Fig. 3D, where the cellular contraction force is substantially reduced. However, in contrast to the trypsin treatment, the adhesions were still maintained, and the cells stayed attached to the four beams (see Fig. 5A). For the NM2A-KO cells, we performed the experiment similar to the WT cells. Here, the cells adhered on the four beams but only generated a very small force compared to the WT scenario. In both cases, ROCK inhibition and NM2A-KO, we observed a substantial impact on the behavior and the initial forces were markedly reduced.
Fig. 5. Inhibition effects and actin filament reorganization.
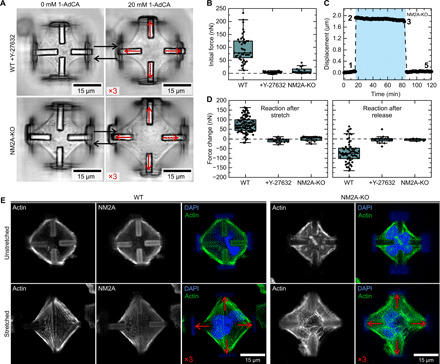
(A) Optical micrographs of ROCK inhibited and NM2A-KO cells in the unstretched and stretched state. Red arrows indicate the displacement of the individual beams scaled by a factor of 3. (B) Comparison of initial traction forces of WT cells, ROCK inhibitor–treated cells, and the NM2A-KO cell line. (C) Exemplary track of a NM2A-KO cell in a stretch-release cycle. The numbers correspond to the respective time points in Fig. 4B for comparison. (D) Quantification of the reaction after the stretch and after the release of the two cases with the WT situation. (E) Comparison between fixed and stained cells in the unstretched and the stretched state. For both cell types shown, actin and the cell nucleus are stained (the walls also show autofluorescence in the blue channel). For the U2OS WT cells, NM2A was stained additionally. The red arrows indicate the displacement of the individual beams, scaled by a factor of 3. DAPI, 4′,6-diamidino-2-phenylindole.
Next, we investigated whether the cells still react to the external stimulation. Figure 5A displays optical micrographs of the two conditions in the unstretched and the stretched state. Although NM2A is not expressed in KO cells, they still invade the scaffolds, spread on the four beams, and show a similar morphology as compared to WT cells. Force measurements, however, reveal substantial differences between the two cell types. Figure 5C depicts a track of a NM2A-KO cell during the same stretch-release cycle that has previously been used to analyze the cellular response. Here, it becomes immediately clear that both, the reaction after the stretch and the reaction after the release, are strongly reduced. As shown in the quantification in Fig. 5D, the ability to react to the external stimulation is markedly reduced for both, ROCK inhibited and NM2A-KO cells.
Reorganization of the actomyosin machinery
The strongly reduced response after mechanical stimulation in ROCK-inhibited and NM2A-KO cells indicates that mechanical homeostasis strongly depends on actomyosin contractility. We next asked how the mechanical stimulation affects the morphology of the actomyosin cytoskeleton by performing immunocytochemical stainings. By simply adding 1-AdCA to all the solutions required for immunostaining protocols, the hydrogel, the scaffold, and the cell are kept in the stretched state. Thus, the sample can be either stored or transferred and imaged in any microscope without further need of complex setups.
Figure 5E shows images of cells fixed 30 min after the applied external stretch labeled for actin, NM2A, and the nucleus. A comparison between unstretched and stretched WT cells reveals the strong emergence of actin stress fibers after external stimulation. The fibers display a high degree of organization and are well colocalized with NM2A. Furthermore, stretched WT cells also form actin stress fibers not only along the cell contour but also in the cell interior, thereby connecting opposing beams. In contrast, NM2A-KO cells show a substantially weaker response. The mechanical stimulation still leads to the induction of actin stress fibers; however, these fibers are much less organized. These results support our previous finding that NM2A-KO cells are no longer able to react to the stretch, suggesting that the cellular response upon mechanical stimulation is primarily generated by actomyosin contractility. Passive elements such as elastic cytoskeletal cross-linkers are apparently not sufficient to generate the counteracting intracellular forces for tensional homeostasis.
Asymmetric stimulation
In the last set of experiments, we exploited the inherent flexibility of the fabrication by 3D laser lithography to change the scaffold design. In the particular case shown here, we increased the thickness of three of the four walls substantially, as can be seen in the scheme in the first column of Fig. 6A. As a result, the thick walls are too stiff to be substantially bent, and the swelling hydrogel is solely displacing the remaining thin wall on the left side. This altered design enabled us to analyze how the cellular response and the cytoskeleton remodeling are affected if the stretch is only applied to one of the four beams.
Fig. 6. Asymmetric stretch.

(A) Left: Schematic images of a scaffold with three thick walls to enable an asymmetric stretch to the left side. Right: Comparison of immunocytochemical stainings of actin and the cell nucleus between an unstretched cell and a cell that has been stretched only to the left side. (B) Exemplary individual beam displacements as a function of time. (C) Quantification of force changes after stretching on the left beam.
Figure 6B shows the individual beam displacements of a cell that has only been stretched on the left side (see also movie S4). Here, the different colors refer to the individual beams. Only the left beam is substantially displaced when the 1-AdCA solution is added after 10 min. Following this stretch, the cell started to counteract the external stimulation and stabilized on a new level. Consequently, the cellular response is similar to the previously studied case of a stretch in all four directions but is now focused solely on the one displaced side. The quantification in Fig. 6C further shows how cells increased their contraction force by about 23 nN after the stretch to counteract the imposed displacement. This corresponds to about the same force per beam that we obtained for an equibiaxial stretch. Consequently, this particular asymmetry does not lead to a notable increase in the cellular contraction force, although it is focused on only one displaced side.
To study the remodeling of the cytoskeleton in this configuration, we fixed cells 30 min after the stimulation and compared them with unstretched control cells on the same sample (see Fig. 6A). We stained for actin and cell nucleus and included red arrows to denote the displacement with the respective scaling. The images clearly show the emergence of additional, organized stress fibers in the stretched state that are predominantly localized on the single displaced side. These results demonstrate how the scaffolds can be easily adapted to generate a variety of different experimental conditions. Moreover, the fabrication by 3D laser lithography allows us to place several different designs along with respective controls on a single coverslip, thus effectively parallelizing the experimental workflow.
DISCUSSION
In this work, we present a previously unknown stimuli-responsive material system for 3D laser lithography based on host-guest complexes. In combination with conventional material systems, we created and characterized composite scaffolds to simultaneously stretch a large number of single cells in tailored 3D microenvironments. Calibration of the system by numerical calculations allowed us to directly connect the cellular reaction to forces and optimize the scaffolds accordingly.
We applied this system to study the mechanoresponse of single cells in 3D microenvironments. Observation of steady-state cells in the scaffolds revealed an initial contraction force on the order of 80 and 125 nN for U2OS and NIH 3T3 cells, respectively. Inhibiting the ROCK pathway or KO of NM2A led to a marked reduction of the measured forces, which is in accordance to conventional methods such as traction force microscopy (38, 39). This inhibition demonstrated the sensitivity of our system to analyze cellular forces and to differentiate between force regimes of different cell types.
We further exploited our system to investigate the dynamic response of U2OS and NIH 3T3 cells upon mechanical stress and stress release. We find that cells have the ability to counteract mechanical stimulations by actomyosin generated contractility. This process was previously described as tensional homeostasis (40, 41). Different studies showed a similar trend concerning the general cell response but differences in the amplitude. While Weng et al. (42) observed that cells completely return to their initial default state, Webster et al. (6) showed that cells plateau at a new set point within 20 to 30 min. In our system, both NIH 3T3 and U2OS cells also plateau at a new set point, supporting the findings of Webster and coworkers (6). However, we cannot exclude that different cell types or different force regimes, which act on the cells, lead to differences in the amplitude of the cell response. Furthermore, the amount of applied stress might be a crucial variable, leading to differences in the response amplitude. While Weng et al. (42) only applied 8% equibiaxial stretch, we applied an equibiaxial stretch between 4 and 24% corresponding to a beam displacement of 0.5 to 3 μm in all directions. However, these different amounts of applied stretch did not induce different force responses.
Another interesting cellular behavior was observed when the stretch was released. In a time frame of around 30 min, cells adapted their contractility by again setting a new steady state, which was remarkably close to the initial set point before the stretch. This behavior highlights the ability of cells to rapidly adapt their force exerting machinery until an ideal configuration is reached. Similar trends were observed by Ezra et al. (33). Since our system allows quantitative image analysis in a higher throughput than cantilever-based/other stretch systems, further studies can analyze in more detail how cells “remember” their initial state and how the transition back to this default state occurs. Immunocytochemical staining of actin fibers in stretched and unstretched cells revealed notable actin filament reorganizations. A reorganization of the cytoskeleton could, in principle, lead to delayed cellular reactions including apoptosis (43). However, we did neither observe cell death nor obvious blebbing behavior during our experiments. Instead, as a response to the stretch, cells formed numerous actin stress fibers between the adhesion sites that reinforced the cell contour and the cell center. Additional actin stress fibers were observed in both, U2OS WT and NM2A-KO cells. However, WT cells showed a higher degree of fiber organization/alignment upon the stretch compared to NM2A-KO cells. Together with the finding that NM2A-KO cells are unable to counteract the mechanical stretch, we propose that tensional homeostasis is a process that heavily relies on active tension generation via NM2 motor proteins. It was previously shown for smooth muscle myosins that these molecular motors adapt their filament size in a load-dependent manner (44). Since the different NM2 isoforms are known to be the key generators of intracellular forces in nonmuscle cells and thus have a big influence on actin stress fiber organization and dynamics, similar properties could arise from NM2 proteins, driving the cellular response upon mechanical stimulation (45). The systematic investigation of NM2B or NM2C could therefore contribute to a better understanding of the molecular mechanisms that cells use to regulate tensional homeostasis.
Although our data suggest that actomyosin contractility is the main driving force during tensional homeostasis, the actin architecture might still contribute to this process in a more subtle manner. It was previously shown in theoretical models that the balanced ratio of active motors and passive cross-linkers might be necessary to generate an optimal force configuration (34). Therefore, the specific investigation of cross-linkers such as α-actinin might lead to an even more precise understanding of the regulating mechanisms during tensional homeostasis.
In the last section, we demonstrated the design flexibility of the 3D laser lithography and analyzed the respective cell behavior on an asymmetric stretch by deflecting only one of the adhesion sites. By analyzing the displacement change over time and by staining the actin cytoskeleton in fixed cells, an asymmetric cell response was observed. This is represented by the displacement of only the stretched beam via intracellular force generation and the reinforcement of actin stress fibers along the stretch site. The flexibility in design and geometry of the scaffolds enables great possibilities to address various cell biological questions that deal with the response of single cells to mechanical stress. The dynamic assembly and disassembly of actin stress fibers in response to mechanical stress are a long-standing topic of interest. It is still an ongoing debate whether mesenchymal-like cells form stress fibers in their physiological 3D environments and how these fibers might differ from their counterpart on flat, rigid substrates such as glass coverslips (46). Using fluorescent actin markers such as F-tractin or Lifeact, our system cannot only be used to investigate the assembly of stress fibers in a 3D environment but also to track their dynamics in living cells using conventional fluorescence microscopes. From a physiological point of view, interesting questions such as the induction of stress fibers in dermal myofibroblasts during wound healing could be addressed. By combining the asymmetric stretch approach with tension sensors encoding for the different RhoGTPases (RhoA, Rac1, and CDC42), the processes involved in stress fiber formation can be further dissected by observing the asymmetric localization and redistribution of these upstream effectors in subcellular compartments upon mechanical stress. The list of possible target molecules and cellular structures is not restricted to the cytoskeleton itself but can be extended to most mechanoresponsive molecules and extracellular structures, such as the influence of different extracellular matrix molecules on force transduction.
MATERIALS AND METHODS
Materials
Acrylamide (Sigma-Aldrich; >98%), N,N′-methylenebis(acrylamide) (Sigma-Aldrich; >99%), LAP (Tokyo Chemical Industry; >98%), 1-AdCA (Sigma-Aldrich; 99%), ethylene glycol (Sigma-Aldrich; >99%), 3-(trimethoxysilyl)propyl methacrylate (Sigma-Aldrich; >97%), PETA (Sigma-Aldrich; >97%), TPETA (number-average molecular weight, ~692; Sigma-Aldrich; >97%), Irgacure 819 (BASF), 7-diethylamino-3-thenoylcoumarin (J&K Scientific; 97%), 2-propanol (Carl Roth; >99.5%), and methyl isobutyl keton (MIBK; Roth; >99%) were used. All chemicals and solvents were used as received without further purification.
βCD-AAm and Ad-AAm were synthesized as described elsewhere (47). Chemicals for the synthesis are the following: β-cyclodextrin (Junsei Chemical), adamantanamine hydrochloride (FUJIFILM Wako Pure Chemical), sodium hydroxide (Nacalai Tesque), acryloyl chloride (Tokyo Chemical Industry), triethylamine (Nacalai Tesque), tetrahydrofuran (FUJIFILM Wako Pure Chemical), and dichloromethane (FUJIFILM Wako Pure Chemical).
Photoresist formulations
The resists were prepared by dissolving 20 mg of Irgacure 819 in 980 mg of TPETA for TPETA resist and in 980 mg of PETA for PETA resist. In host-guest resist, the starting mixture was prepared by dissolving 36 mg of βCD-AAm with 6.6 mg of Ad-AAm in 200-μl water and 300 μl of 2-propanol. After 1 hour of ultrasonic bath at 60°C, the mixture was cooled down to room temperature, and 68 mg of AAm and 5 mg of LAP were added. The final resist was obtained after another 5 min in the ultrasonic bath at room temperature.
Fabrication of 3D microstructures
A commercial direct laser writing system (Photonic Professional GT, Nanoscribe GmbH) equipped with a 63×, numerical aperture (NA) = 1.4 oil immersion objective was used for the fabrication. To increase the adhesion of microstructures to the glass surface, we used plasma-cleaned coverslips, treated with 3-(trimethoxysilyl)propyl methacrylate (1 mM in toluene) for 1 hour and rinsed them afterward with acetone and water.
The fabrication of cell scaffolds consisted of three consecutive writing steps. First, the TPETA resist was drop-cast on the silane-treated coverslip, and the walls along with several alignment markers were written. A laser power of 40 mW at the back focal plane and a scanning speed of 2 mm/s were used for the fabrication. After a development step in a 1:1 mixture of MIBK and 2-propanol, the samples were dried.
Second, the PETA resist was drop-cast on the structures. By lateral positioning with the alignment markers and axial positioning with the built-in interface finder, the beams were fabricated at the desired position on the walls. We typically used a laser power of 30 mW at the back focal plane and a scanning speed of 2 mm/s to fabricate the structures. Again, the sample was developed in a 1:1 mixture of MIBK and 2-propanol.
In the third step, the host-guest resist was drop-cast on the existing structures and put on a hot plate for 2 min at 50°C to reduce the amount of solvent. For the remaining writing process, the sample was contained in a sealed environment to prevent further evaporation of solvents. After alignment and fabrication of the hydrogel blocks in the center of the scaffolds, the sample was developed in a 1:1 mixture of water and 2-propanol and lastly stored in water. In this fabrication step, we used a laser power of 40 mW at the back focal plane and a scanning speed of 5 mm/s. The final concentration of monomers is 2 mol/kg containing 3 mole percent of βCD-AAm and Ad-AAm monomers. If one assumes that the yield of polymerization is 100% and the swelling is negligible, then this yields the concentration of the inclusion complex to be about 60 mM.
Before the sample was used for further experiments, it was immersed for 2 days in water with 20 mM 1-AdCA. This solution triggered the swelling of the hydrogel that helped to remove unpolymerized residues from the material network. In addition, we observed a slightly altered swelling behavior in the first contact of the hydrogel with 1-AdCA, which we attribute to tensions in the polymer that are released after one cycle. As previously reported by Harada et al. (47), the association constants of “free” host/guest molecules in solution are more than 10 times larger than the association constants between polymers. Therefore, the treatment of host-guest hydrogels with 20 mM 1-AdCA is sufficient to induce swelling.
Mechanical characterization
The mechanical analysis was performed by an atomic force microscope (NanoWizard, JPK Instruments). Soft silicon nitride cantilevers (MLCT, Bruker) with a nominal spring constant of 0.03 N/m were used to perform the indentation in an aqueous environment.
Cell culture
U2OS and NIH 3T3 cells were obtained from American Type Culture Collection and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (PAN-Biotech) supplemented with 10% bovine growth serum (HyClone) at 37°C under a humidified atmosphere containing 5% CO2.
Live-cell imaging
Live-cell imaging was performed in a LSM (LSM 800, Zeiss) with an incubation chamber at 37°C. We used a 40×, NA =1.2 water immersion objective and the motorized mechanical stage to sequentially move to all the positions in during the time series. The sample was fixed in a self-built fluidic chamber to enable solvent exchange during the experiment. At the beginning of the live-cell experiments, cells were kept in phenol red-free high-glucose DMEM containing 25 mM Hepes (Thermo Fisher Scientific) and supplemented with 10% bovine growth serum. To induce the mechanical stretch, the medium was completely replaced by a medium containing 20 mM 1-AdCA using a microfluidic chamber and syringes. Since the addition of 1-AdCA leads to a substantially decreased pH value, we adjusted the pH to 7.0 with NaOH before the experiments. Releasing the stretch was obtained by replacing the media with normal imaging medium.
Fluorescence imaging
Cells were fixed in PBS containing 4% paraformaldehyde and 20 mM
1-AdCA. Cells were then washed and permeabilized in PBS containing 0.1% Triton-X 100 and 20 mM 1-AdCA. All following washing steps were also carried out using this PBS mixture. Following antibodies and affinity probes were used for immunocytochemical labeling: rabbit anti–NMHC-IIA (BioLegend), goat anti-rabbit Cy3 (Jackson ImmunoResearch), phalloidin–Alexa Fluor 488 (Thermo Fisher Scientific), and 4′,6-diamidino-2-phenylindole (DAPI) (Carl Roth). All staining incubation steps were carried out in 1% bovine serum albumin in PBS containing 20 mM 1-AdCA. Samples were stored in PBS containing 20 mM 1-AdCA until imaging.
We used a confocal LSM (LSM 800 Airyscan, Zeiss), equipped with a 40×, NA = 1.2 water immersion objective.
Image analysis of cellular response
The images were analyzed by digital image cross-correlation based on MATLAB (MathWorks) code. All images of a time series were compared to a reference image at t = 0. In these images, regions of interest with a specified size were defined on the four beams. For each image, the calculation of the maximum cross-correlation function resulted in the 2D local displacement vector. Four different positions per beam were tracked and averaged to obtain a mean displacement per beam as a function of time. Potential offsets of the sample during the injection or after the stage movement were corrected by additional tracking of solid marker structures next to the scaffolds. In addition, we tracked reference scaffolds without cells to attribute for small displacements of the beams as a result of the swelling properties. We subtracted the reference tracks from the parts where we observed the cellular response to correct for these potential deviations.
Finite element calculations
We performed the numerical analysis by a finite-element approach using the commercial software COMSOL Multiphysics to solve the linear elastic Cauchy continuum mechanics equations. Swelling of the hydrogel was modeled as a boundary load with a fixed bottom surface to account for the silanization of the glass substrate. Geometrical nonlinearities have been accounted for.
The mechanical properties of the used materials were measured via atomic force spectroscopy (see fig. S12 for details). The Young’s moduli of PETA EPETA = 3 × 106 kPa and TPETA ETPETA = 12.7 × 103 kPa were kept constant. For the stimuli-responsive hydrogel, we used the values EHydrogel = 22 kPa in the stiff state and EHydrogel = 6.5 kPa in the soft state. All geometrical parameters were extracted from scanning electron micrographs.
Statistical analysis
All box plots in this manuscript display the median value as a solid line with the lower and upper quartile in the box around the median value. The whiskers are drawn to the last points that are still within 1.5 times the interquartile range away from the respective lower and upper quartiles to mark possible outliers.
Supplementary Material
Acknowledgments
We thank S. Bertels, T. Frenzel (both KIT), and U. Schwarz (Heidelberg University) for discussions and V. Hahn (KIT) for the SEM imaging. Funding: We acknowledge the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) under Germany’s Excellence Strategy—EXC-2082/1—390761711, JSPS (JP20H00661 and JP19H05719), Nakatani Foundation, German-Japanese University Alliance (HeKKSaGOn Alliance), and the Carl Zeiss Foundation for support. We acknowledge additional support by the Karlsruhe Nanostructure Service Laboratory (NSL) and by the Helmholtz programs Science and Technology of Nanosystems (STN) and BioInterfaces in Technology and Medicine (BIFTM). M.H. has been supported by the Karlsruhe School of Optics and Photonics (KSOP). M.H. and K.R. have been supported by the Heidelberg Karlsruhe Strategic Partnership (HEiKA) graduate school Functional Materials. The work of E.D.L. has been supported by a postdoctoral research fellowship of the Alexander von Humboldt Foundation. C.B.-K. acknowledges the Australian Research Council (ARC) for funding in the context of an Australian Laureate Fellowship. Author contributions: M.H., B.R., M.N., M.T., and M.B. conceived and initiated the study. M.N., Y.T., and A.H. synthesized and provided the stimuli-responsive materials. M.H. and E.D.L. developed the photoresist and performed the 3D laser lithography experiments and the studies of the stimuli response. M.H. carried out the numerical analysis. M.H., K.W., and K.R. performed the cell culture and cell experiments. M.H. analyzed all the experimental and numerical data. M.T., C.B.-K., Y.T., A.H., E.B., M.W., and M.B. motivated and supervised the research program. M.H., K.W., and M.B. wrote the first draft of the manuscript. All authors discussed the results and worked on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the corresponding authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/39/eabc2648/DC1
REFERENCES AND NOTES
- 1.Baker B. M., Chen C. S., Deconstructing the third dimension – How 3D culture microenvironments alter cellular cues. J. Cell Sci. 125, 3015–3024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruprecht V., Monzo P., Ravasio A., Yue Z., Makhija E., Strale P. O., Gauthier N., Shivashankar G. V., Studer V., Albiges-Rizo C., Viasnoff V., How cells respond to environmental cues – Insights from bio-functionalized substrates. J. Cell Sci. 130, 51–61 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Matthews B. D., Overby D. R., Mannix R., Ingber D. E., Cellular adaptation to mechanical stress: Role of integrins, Rho, cytoskeletal tension and mechanosensitive ion channels. J. Cell Sci. 119, 508–518 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., Piccolo S., Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Kamble H., Barton M. J., Jun M., Park S., Nguyen N.-T., Cell stretching devices as research tools: Engineering and biological considerations. Lab Chip 16, 3193–3203 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Webster K. D., Ng W. P., Fletcher D. A., Tensional homeostasis in single fibroblasts. Biophys. J. 107, 146–155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thoumine O., Ott A., Time scale dependent viscoelastic and contractile regimes in fibroblasts probed by microplate manipulation. J. Cell Sci. 110, 2109–2116 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Scheiwe A. C., Frank S. C., Autenrieth T. J., Bastmeyer M., Wegener M., Subcellular stretch-induced cytoskeletal response of single fibroblasts within 3D designer scaffolds. Biomaterials 44, 186–194 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Desmaële D., Boukallel M., Régnier S., Actuation means for the mechanical stimulation of living cells via microelectromechanical systems: A critical review. J. Biomech. 44, 1433–1446 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Brown T. D., Techniques for mechanical stimulation of cells in vitro: A review. J. Biomech. 33, 3–14 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Richter B., Hahn V., Bertels S., Claus T. K., Wegener M., Delaittre G., Barner-Kowollik C., Bastmeyer M., Guiding cell attachment in 3D microscaffolds selectively functionalized with two distinct adhesion proteins. Adv. Mater. 29, 1604342 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Hippler M., Lemma E. D., Bertels S., Blasco E., Barner-Kowollik C., Wegener M., Bastmeyer M., 3D scaffolds to study basic cell biology. Adv. Mater. 31, 1808110 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Han D., Lu Z., Chester S. A., Lee H., Micro 3D printing of a temperature-responsive hydrogel using projection micro-stereolithography. Sci. Rep. 8, 1963 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hippler M., Blasco E., Qu J., Tanaka M., Barner-Kowollik C., Wegener M., Bastmeyer M., Controlling the shape of 3D microstructures by temperature and light. Nat. Commun. 10, 232 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng H., Martella D., Wasylczyk P., Cerretti G., Lavocat J.-C. G., Ho C.-H., Parmeggiani C., Wiersma D. S., High-resolution 3D direct laser writing for liquid-crystalline elastomer microstructures. Adv. Mater. 26, 2319–2322 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Spiegel C. A., Hippler M., Münchinger A., Bastmeyer M., Barner-Kowollik C., Wegener M., Blasco E., 4D printing at the microscale. Adv. Funct. Mater. 30, 1907615 (2019). [Google Scholar]
- 17.Zhang Y. S., Khademhosseini A., Advances in engineering hydrogels. Science 356, eaaf3627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okano T., Yamada N., Okuhara M., Sakai H., Sakurai Y., Mechanism of cell detachment from temperature-modulated, hydrophilic-hydrophobic polymer surfaces. Biomaterials 16, 297–303 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Frank V., Kaufmann S., Wright R., Horn P., Yoshikawa H. Y., Wuchter P., Madsen J., Lewis A. L., Armes S. P., Ho A. D., Tanaka M., Frequent mechanical stress suppresses proliferation of mesenchymal stem cells from human bone marrow without loss of multipotency. Sci. Rep. 6, 24264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guvendiren M., Burdick J. A., Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat. Commun. 3, 792 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Yang C., Tibbitt M. W., Basta L., Anseth K. S., Mechanical memory and dosing influence stem cell fate. Nat. Mater. 13, 645–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada A., Takashima Y., Nakahata M., Supramolecular polymeric materials via cyclodextrin–guest interactions. Acc. Chem. Res. 47, 2128–2140 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Schmidt B. V. K. J., Barner-Kowollik C., Dynamic macromolecular material design—The versatility of cyclodextrin-based host-guest chemistry. Angew. Chem. Int. Ed. 56, 8350–8369 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Hörning M., Nakahata M., Linke P., Yamamoto A., Veschgini M., Kaufmann S., Takashima Y., Harada A., Tanaka M., Dynamic mechano-regulation of myoblast cells on supramolecular hydrogels cross-linked by reversible host-guest interactions. Sci. Rep. 7, 7660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rekharsky M. V., Inoue Y., Complexation thermodynamics of cyclodextrins. Chem. Rev. 98, 1875–1918 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Fairbanks B. D., Schwartz M. P., Bowman C. N., Anseth K. S., Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: Polymerization rate and cytocompatibility. Biomaterials 30, 6702–6707 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan B., Qian K., Xie H., Asundi A., Two-dimensional digital image correlation for in-plane displacement and strain measurement: A review. Meas. Sci. Technol. 20, 062001 (2009). [Google Scholar]
- 28.Balaban N. Q., Schwarz U. S., Riveline D., Goichberg P., Tzur G., Sabanay I., Mahalu D., Safran S., Bershadsky A., Addadi L., Geiger B., Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3, 466–472 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Tan J. L., Tien J., Pirone D. M., Gray D. S., Bhadriraju K., Chen C. S., Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc. Natl. Acad. Sci. 100, 1484–1489 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soiné J. R. D., Brand C. A., Stricker J., Oakes P. W., Gardel M. L., Schwarz U. S., Model-based traction force microscopy reveals differential tension in cellular actin bundles. PLOS Comput. Biol. 11, e1004076 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trichet L., Le Digabel J., Hawkins R. J., Vedula S. R. K., Gupta M., Ribrault C., Hersen P., Voituriez R., Ladoux B., Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc. Natl. Acad. Sci. 109, 6933–6938 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brand C. A., Linke M., Weißenbruch K., Richter B., Bastmeyer M., Schwarz U. S., Tension and elasticity contribute to fibroblast cell shape in three dimensions. Biophys. J. 113, 770–774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ezra D. G., Ellis J. S., Beaconsfield M., Collin R., Bailly M., Changes in fibroblast mechanostat set point and mechanosensitivity: An adaptive response to mechanical stress in floppy eyelid syndrome. Invest. Ophthalmol. Vis. Sci. 51, 3853–3863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belmonte J. M., Leptin M., Nédélec F., A theory that predicts behaviors of disordered cytoskeletal networks. Mol. Syst. Biol. 13, 941 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vicente-Manzanares M., Ma X., Adelstein R. S., Horwitz A. R., Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura K., Ito M., Amano M., Chihara K., Fukata Y., Nakafuku M., Yamamori B., Feng J., Nakano T., Okawa K., Iwamatsu A., Kaibuchi K., Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273, 245–248 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Shao J., Welch W. J., DiProspero N. A., Diamond M. I., Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol. Cell. Biol. 28, 5196–5208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shutova M. S., Asokan S. B., Talwar S., Assoian R. K., Bear J. E., Svitkina T. M., Self-sorting of nonmuscle myosins IIA and IIB polarizes the cytoskeleton and modulates cell motility. J. Cell Biol. 216, 2877–2889 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beningo K. A., Hamao K., Dembo M., Wang Y.-l., Hosoya H., Traction forces of fibroblasts are regulated by the Rho-dependent kinase but not by the myosin light chain kinase. Arch. Biochem. Biophys. 456, 224–231 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banes A. J., Tsuzaki M., Yamamoto J., Fischer T., Brigman B., Brown T., Miller L., Mechanoreception at the cellular level: The detection, interpretation, and diversity of responses to mechanical signals. Biochem. Cell Biol. 73, 349–365 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Brown R. A., Prajapati R., McGrouther D. A., Yannas I. V., Eastwood M., Tensional homeostasis in dermal fibroblasts: Mechanical responses to mechanical loading in three-dimensional substrates. J. Cell. Physiol. 175, 323–332 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Weng S., Shao Y., Chen W., Fu J., Mechanosensitive subcellular rheostasis drives emergent single-cell mechanical homeostasis. Nat. Mater. 15, 961–967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desouza M., Gunning P. W., Stehn J. R., The actin cytoskeleton as a sensor and mediator of apoptosis. BioArchitecture 2, 75–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veigel C., Molloy J. E., Schmitz S., Kendrick-Jones J., Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat. Cell Biol. 5, 980–986 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Hundt N., Steffen W., Pathan-Chhatbar S., Taft M. H., Manstein D. J., Load-dependent modulation of non-muscle myosin-2A function by tropomyosin 4.2. Sci. Rep. 6, 20554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livne A., Geiger B., The inner workings of stress fibers – From contractile machinery to focal adhesions and back. J. Cell Sci. 129, 1293–1304 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Harada A., Kobayashi R., Takashima Y., Hashidzume A., Yamaguchi H., Macroscopic self-assembly through molecular recognition. Nat. Chem. 3, 34–37 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/39/eabc2648/DC1


