Abstract
The year 2020 brought the news of the emergence of a new respiratory disease (COVID-19) from Wuhan, China. The disease is now a global pandemic and is caused by a virus named SARS-CoV-2 by international bodies. Important viral transmission sources include human contact, respiratory droplets and aerosols, and through contact with contaminated objects. However, viral shedding in feces and urine by COVID-19-afflicted patients raises concerns about SARS-CoV-2 entering aquatic systems. Recently, targeted SARS-CoV-2 genome fragments have been successfully detected in wastewater, sewage sludge and river waters around the world. Wastewater-based epidemiology (WBE) studies can provide early detection and assessment of COVID-19 transmission and the growth of active cases within given wastewater catchment areas. WBE surveillance's ability to detect the growth of cases was demonstrated. Was this science applied throughout the world as this pandemic spread throughout the globe? Wastewater treatment efficacy for SARS-CoV-2 removal and risk assessments associated with treated water are reported. Disinfection strategies using chemical disinfectants, heat and radiation for deactivating and destroying SARS-CoV-2 are explained. Analytical methods of SARS-CoV-2 detection are covered. This review provides a more complete overview of the present status of SARS-CoV-2 and its consequences in aquatic systems. So far, WBE programs have not yet served to provide the early alerts to authorities that they have the potential to achieve. This would be desirable in order to activate broad public health measures at earlier stages of local and regional stages of transmission.
Keywords: Coronavirus disease, Covid-19 disease, SARS-CoV-2, Removal of SARS-CoV-2
Graphical abstract
1. Introduction
With the end of 2019, a respiratory coronavirus disease (COVID-19) outbreak emerged from Wuhan city in China (WHO, 2020a). This illness is caused by a new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (WHO, 2020a). The outbreak of COVID-19 has become a universal pandemic since March 11, 2020 (WHO, 2020a). By 18 September 2020, a total of 30,055,710 affected patients were confirmed and 943,433 deaths were reported due to COVID-19 illness worldwide (WHO, 2020b), although both these numbers are surely much higher, especially the former.
Starting from Wuhan in December 2019, the virus had spread to 216 countries by the date this paper was submitted (WHO, 2020b). Respiratory droplets along with direct contacts touching an infected person or contacting contaminated objects and then transmitting the virus from one's hand to the mouth, nose and eyes are main SARS-CoV-2 transmission routes (Chan et al., 2020b; Meselson, 2020). Recent evidence shows the presence of SARS-CoV-2 in human urine and feces (Sun et al., 2020; Tang et al., 2020; Wölfel et al., 2020; Zhang et al., 2020b). Human shedding of the virus through nose, mouth, urine and feces led to the presence of SARS-CoV-2 in the environment. SARS-CoV-2 has been reported in air, on household objects including door knobs, taps, and handles (Cai et al., 2020b; Chan et al., 2020b; Chin et al., 2020). Recently, SARS-CoV-2 was also reported in wastewater and sewage (Ahmed et al., 2020a; Arora et al., 2020; Balboa et al., 2020; Kocamemi et al., 2020a, Kocamemi et al., 2020b; Kumar et al., 2020; Medema et al., 2020; Meulemans, 1987; Nemudryi et al., 2020; Randazzo et al., 2020a; La Rosa et al., 2020b; Wurtzer et al., 2020b). The presence of viable SARS-CoV-2 in human urine and feces raises concerns about the potential spread of COVID-19 through water, soil, and other environmental compartments (Núñez-Delgado, 2020). Even, the SARS-CoV-2 RNA has been found on particulate matter (PM10) in Bergamo in Northern Italy (Setti et al., 2020).
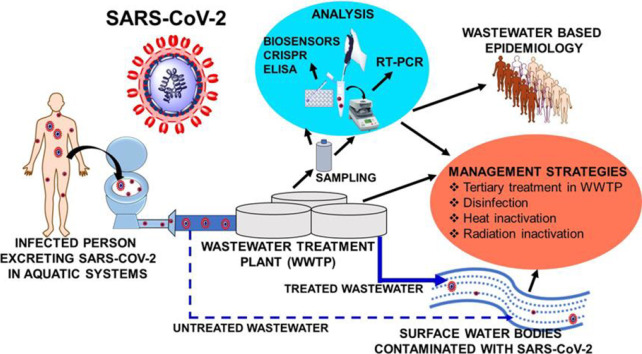
This widespread SARS-CoV-2 presence in the environment is increasing as the pandemic spreads. It needs to be properly assessed and evaluated as a possible transmission route. Viral diseases, where the infectious viruses are found in water and wastewater systems are known to cause community level transmissions (Gormley et al., 2020). The connections between water and wastewater plumbing systems with human day-to-day activities can cause spread of SARS-CoV-2 within/between buildings and even between communities. Similar high-risk transmission issues are of particular concern in hospitals and healthcare buildings (Gormley et al., 2020). Untreated sewage running directly into surface waters which are then used without any purification treatments is a special risk in low income countries, where proper sewage and wastewater disposal is often missing. With a survival of up to 21 days on various surfaces (Chin et al., 2020; Doremalen et al., 2020; Kasloff et al., 2020) and an estimated survival of 25 days at 5 °C in water and wastewater sources (Shutler et al., 2020), SARS-CoV-2 can possibly contaminate surface water sources. This could spread infections widely, as surface waters frequently serve directly for day-to-day purposes in low and some middle income countries.
Wastewater surveillance, also known as wastewater-based epidemiology, could accurately estimate SARS-CoV-2 presence in wastewater sources (Daughton, 2020b; Wigginton and Boehm, 2020). SARS-CoV-2 presence in wastewater might also estimate the extent of current COVID-19 infections in a community (Medema et al., 2020; La Rosa et al., 2020b; Wurtzer et al., 2020b). Thus, wastewater-based epidemiology could estimate the number of symptomatic, asymptomatic and presymptomatic individuals in a particular area (Daughton, 2020b; Wigginton and Boehm, 2020). Water and wastewater surveillance also limits the possibility of unnoticed spread of SARS-CoV-2 through aquatic sources (Daughton, 2020b; Wigginton and Boehm, 2020). Thus, SARS-CoV-2 presence in aquatic sources becomes an important aspect of study.
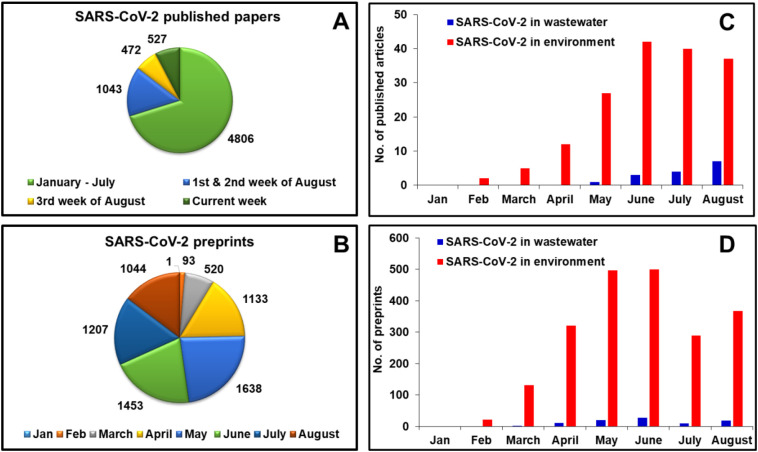
An enormous literature has already been published on SARS-CoV-2 (Fig. 1 ). But, at present little scientific evidence has appeared regarding SARS-CoV-2 transmissions through food, day-to-day objects, water and wastewater (Carraturo et al., 2020). Limited literature of SARS-CoV-2 in water and wastewater sources is available to date (Fig. 1). Coronaviruses are more easily inactivated versus non-enveloped viruses such as adenoviruses, rotaviruses and noroviruses (Carraturo et al., 2020). Therefore, this review summarizes what is currently known about SARS-CoV-2 in aquatic environments. These include its occurrence, stability, persistence, transmission, analysis and possible strategies for water and wastewater treatment. Special emphasis is given to wastewater-based epidemiology to find and evaluate COVID-19 cases in a specific region. High temperature, changes in pH, sunlight, and common disinfection agents are essential modes for the inactivation of viruses (Carraturo et al., 2020). Disinfection and other viral water treatment inactivation strategies are evaluated to provide safe water sources for human beings.
Fig. 1.
Analytical presentation of available literature on SARS-CoV-2 (A) SARS-CoV-2 published papers available as per “web of science” database, (B) SARS-CoV-2 preprints on medRxiv and bioRxiv database, (C) peer reviewed “SARS-CoV-2” articles (data available as per different mentioned keys) on “web of science” database, (D) “SARS-CoV-2” preprints (data available as per different mentioned keys) on medRxiv and bioRxiv database.
To date a large number of viewpoints, short communications and reviews have been published on SARS-CoV-2 and coronaviruses in the environment. Reviews include topics such as occurrence in water and wastewater sources (Foladori et al., 2020; Gormley et al., 2020; Núñez-Delgado, 2020), transmission (Heller et al., 2020; Hindson, 2020; Mahabee-Gittens et al., 2020), stability and persistence (Aboubakr et al., 2020; Kampf et al., 2020a; Scheller et al., 2020), analysis (Ahmed et al., 2020b; Bofill-Mas and Rusiñol, 2020; Farkas et al., 2020a; Haramoto et al., 2018; Jalandraad et al., 2020; Michael-Kordatou et al., 2020; La Rosa et al., 2020a), wastewater-based epidemiological approaches (Bivins et al., 2020; Daughton, 2020a; Daughton, 2020b; Farkas et al., 2020a; Hata and Honda, 2020; Michael-Kordatou et al., 2020; Naddeo and Liu, 2020; Sims and Kasprzyk-Hordern, 2020) and disinfection strategies (Chauhan, 2020; Kamp, 2020; Kampf et al., 2020b). None of these reviews compile all the issues to provide a complete picture of the issue of SARS-CoV-2 in the environment, with a special focus on water and wastewater. This review provides a more complete overview of SARS-CoV-2 occurrence, persistence, analysis and disinfection in water and wastewater sources. Data compiled and presented in many of these prior reviews are on postulated and derived from the studies of other coronaviruses including SARS and MERS. In contrast, this review work provides SARS-CoV-2 information available in literature prior to the manuscript submission.
Web of Science and Google scholar were used for peer reviewed literature selection. Literature from bioRvix and medRvix were used for preprint collection for writing this review. According to Web of Science, bioRxiv and medRxiv data, more than 16,500 preprints and peer reviewed articles have been published on SARS-CoV-2 up to September 2020. Only 62 peer reviewed papers and 500 preprints were focused on the keyword “SARS-CoV-2 in water” and “SARS-CoV-2 in wastewater” combined. A large number of preprints were not included in the study as these are not directly related to the topic of this review. Approximately 150 articles, preprints and reports were selected based on their relevance to SARS-CoV-2 in water and wastewater. A total of 60 peer reviewed articles/reviews together with 50 relevant preprints published in 2020 were cited. In addition, 45 other relevant articles published before 2020 were also discussed.
2. Coronavirus (SARS-CoV-2) in the environment
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a member of the Coronaviridae family and the Nidovirales order and the sub family Coronavirinae (Harapan et al., 2020). This virus belongs to genera Betacoronavirus which also includes SARS-CoV-1, HCoV-OC43, MERS-CoV and HCoV-HKU1 (Harapan et al., 2020). SARS-CoV-2 is responsible for COVID-19. Coronaviruses are enveloped single stranded RNA viruses (size 60–220 nms) with crown like structures on their surface (Naddeo and Liu, 2020; La Rosa et al., 2012). Transmission routes involve human-to-human spread that occurs mainly through aerosol droplets from mouth and nose of the infected person. High viral loads have been found in the respiratory tract of infected individuals.
Therefore, viral transmission from patient to surrounding air and onto objects is an important route of transmission. The median time of SARS-CoV-2 detection in feces (22 days) is higher than in serum samples (16 days) and in respiratory airways (18 days) in COVID-19 patients (Zheng et al., 2020). Another study reports persistent SARS-CoV-2 shedding in patient's feces for up to 33 days after being tested negative for respiratory viral RNA (Wu et al., 2020b). The large viral load in urine and feces leads to SARS-CoV-2 presence in terrestrial and aquatic sources in the environment (Ahmed et al., 2020a; Arora et al., 2020; Kocamemi et al., 2020a; Kocamemi et al., 2020b; Medema et al., 2020). According to another study, SARS-CoV-2 can remain active for up to 25 days in water sources (Shutler et al., 2020). This study also estimates that contaminated water sources (water systems, waterways and rivers) can deliver the equivalent of >100 SARS-CoV-2 genome copies with 100 mL or less water in the countries with high SARS-CoV 2 prevalence (Shutler et al., 2020). Asymptomatic persons can also disseminate coronaviruses through second-hand aerosols (SHA) and second-hand smoke (SHS) from cigarettes and combustible tobacco products (Mahabee-Gittens et al., 2020). Therefore, more comprehensive assessments of occurrence, persistence, its analysis and management strategies are necessary.
2.1. Persistence in the environment
In general, viral persistence in a given environment is essential for its transmission. However, the environmental presence of a virus is dependent upon several factors.
SARS-CoV-2 has been detected on surfaces including cell phones, door handles and many other day-to-day items (Aboubakr et al., 2020). Only limited data are available until now regarding the SARS-CoV-2 persistence on various materials in the environment (Table 1 ). Extensive literature search points toward a deficiency of available data concerning SARS-CoV-2 persistence in aquatic systems.
Table 1.
Persistence of SARS-CoV-2 in/on different environmental matrices.
| Matrix | Temp. (°C) | Relative humidity (%) | Persistence | 100% decay time | Reference |
|---|---|---|---|---|---|
| Aerosol | 21–23 | 65 | 3 ha | NR | (Doremalen et al., 2020) |
| Aerosol | RT | NR | 16 hb | NR | (Fears et al., 2020) |
| Banknote paper | 22 | 65 | 2 d | 4 d | (Chin et al., 2020) |
| Cardboard | 21–23 | 65 | 1 d | 2 d | (Doremalen et al., 2020) |
| Cloth | 22 | 65 | 1 d | 2 d | (Chin et al., 2020) |
| Copper | 21–23 | 65 | 4 h | 8 h | (Doremalen et al., 2020) |
| Cotton | 20 | 35–40 | 1 h | 4 h | (Kasloff et al., 2020) |
| Gloves (chemical) | 20 | 35–40 | 4 d | 4 d | (Kasloff et al., 2020) |
| Gloves (nitrile) | 20 | 35–40 | 7 d | 7 d | (Kasloff et al., 2020) |
| N95 mask | 20 | 35–40 | 14 d | 21 d | (Kasloff et al., 2020) |
| N100 mask | 20 | 35–40 | 14 d | 21 d | (Kasloff et al., 2020) |
| Paper | 22 | 65 | 30 min | 3 h | (Chin et al., 2020) |
| Plastic (polypropylene) | 21–23 | 65 | 3 d | 4 d | (Doremalen et al., 2020) |
| Plastic | 22 | 65 | 4 d | 7 d | (Chin et al., 2020) |
| Plastics from face shield | 20 | 35–40 | 21 d | 21 d | (Kasloff et al., 2020) |
| Stainless steel | 21–23 | 65 | 3 d | 4 d | (Doremalen et al., 2020) |
| Stainless steel | 22 | 65 | 4 d | 7 d | (Chin et al., 2020) |
| Stainless steel | 20 | 35–40 | 14 d | 21 d | (Kasloff et al., 2020) |
| Surgical mask outer layer | 22 | 65 | 7 d | NR | (Chin et al., 2020) |
| Surgical mask inner layer | 22 | 65 | 4 d | 7 d | (Chin et al., 2020) |
| Tissue paper | 22 | 65 | 30 min | 3 h | (Chin et al., 2020) |
| Tyvek | 20 | 35–40 | 14 d | 21 d | (Kasloff et al., 2020) |
| Virus transport medium | 4 | – | 14 dc | (Chin et al., 2020) | |
| 22 | – | 14 d | |||
| 37 | – | 2 d | |||
| 56 | – | 30 min | |||
| 70 | – | 5 min | |||
| Wood | 22 | 65 | 1 d | 2 d | (Chin et al., 2020) |
15.8% at the end 3 h.
55–100% at the end 16 h.
Only 10% decline in 14 d.
Stability in air and on surfaces is a prominent factor determining the efficiency of SARS-CoV-2 transmission (Doremalen et al., 2020). Contaminated dry surfaces also play an important role in SARS-CoV-2 transmission (Kamp, 2020). With the enormous number of 108 viral copies in just 1 mL of sputum, SARS-CoV-2 can rapidly infect numerous people (Rothe et al., 2020). Different coronaviruses have been reported to persist between 2 h to 9 days on different surfaces (Kampf et al., 2020a). The SARS-CoV-2 half-life in aerosol and on copper, cardboard, polypropylene and stainless steel are 2.74, 3.4, 8.45, 15.9 and 13.1 h, respectively (Doremalen et al., 2020). SARS-CoV-2 persistence in two aerosols and on several different surfaces is summarized in Table 1.
SARS-CoV-2 persisted for 3 h in aerosol at 21–23 °C and 65% relative humidity (Doremalen et al., 2020). Another study asserted that the SARS-CoV-2's prevalence and aerosol stability in ambient environmental conditions (23 °C and ~53% relative humidity and in the absence of UV) lasted >16 h (Fears et al., 2020). Infectious SARS-CoV-2 was detected for the entire 16 h period, and a minor but constant SARS-CoV-2 fraction maintained its replication-competence (Fears et al., 2020). Thus, this virus can be considered an airborne pathogen for entire 16 h. These authors also assessed the qualitative integrity of SARS-CoV-2 after longer-term aerosol experimentation through scanning electron microscopy (SEM) imaging (Fears et al., 2020). SARS-CoV-2 is heterogeneous and ovoid in shape. It maintained its shape, size and morphologies for the entire 16 h, which is consistent with the aerosol suspension stability experiment (Fears et al., 2020). The wide variations in data of these two available studies demonstrate the lack of reliability and proper statistical analysis in the available data. However, both studies concluded that SARS-CoV-2 aerosols can remain pathogenic for several hours.
Significant contamination of common household objects including remote control, mobile phones, bed table, toilets, washbasins, bed rails, window ledges, ventilation grates and floor under patient's bed by SARS-CoV-2 is reported (Cai et al., 2020a; Ong et al., 2020; Santarpia et al., 2020). SARS-CoV-2 contamination at room sites (87%), toilet sites (such as sink, toilet bowl and door handle) (60%) was reported for a mild symptomatic patient (Ong et al., 2020). Transmission of SARS-CoV-2 to humans from different contaminated surfaces is also reported (Aboubakr et al., 2020; Cahill and Morris, 2020; Núñez-Delgado, 2020), thus evaluation of viral persistence on everyday encountered common surfaces becomes essential. SARS-CoV-2 survival on some important surfaces is provided in Table 1. Plastic surfaces are common objects, often discarded in water in our society (Geyer et al., 2017; Hale and Song, 2020). Plastic bags and other plastic substances can carry bacteria and viruses with them (Hale and Song, 2020). Reusable bags and other common plastic-based products are easily contaminated and have the potential to spread coronaviruses, since SARS-CoV-2 can survive between 4 and 7 days (Chin et al., 2020; Doremalen et al., 2020). This is another reason they need to be kept out of waterways. Proper disposal of single use plastics, including PPE, gloves, gowns, syringes used by patients and their caretakers in medical facilities is necessary. SARS-CoV-2 was reported to survive for >7 days on layers of surgical masks (Chin et al., 2020). Table 1 documents SARS-CoV-2 survival studies on other household objects including paper (3 h), tissue paper (3 h), copper (8 h), cardboard (2 d), cloth (2 d), wood (2 d), banknote paper (4 d) and stainless steel (4 d). However, these studies do not mimic the real environmental situations by using higher (>104) numbers of infectious virus particles in a small study area (Goldman, 2020). Therefore, the chances of depositing highly infectious viral particle concentrations (>104) on fomite surfaces are minimal thus, the chances of viral transmission from fomites are low (Goldman, 2020).
Regardless of SARS-CoV-2 survival on various surfaces, low transmission possibilities through fomites exists (Goldman, 2020). Till date no clear evidences are available for infectious potential of SARS-CoV-2 from different surfaces (Cai et al., 2020a; Cai et al., 2020b). However, Middle East respiratory syndrome (MERS) coronavirus can remain infectious up to 60 min after aerosolization (Cai et al., 2020a). This point substantiate the possible infectious potential of SARS-COV-2 from fomites (Cai et al., 2020a). A study conducted in a shopping mall in Wenzhou, China, demonstrated the SARS-CoV-2 spread through fomites e.g. restroom taps or elevator buttons (Cai et al., 2020a). This study indicated low intensity SARS-CoV-2 transmission by indirect conveyance routes such as fomites (Cai et al., 2020a).
The persistence of various coronaviruses have been reported in both treated and untreated water (Carraturo et al., 2020; Gundy et al., 2009). The persistence of SARS-CoV-2 in aquatic systems is largely unknown and undocumented. Survival and sustainability of SARS-CoV-2 in aqueous systems is influenced by initial viral load, type of medium, temperature, organic matter, presence of biologic fluids and with the presence of organic and inorganic substances (Carraturo et al., 2020; Romano-Bertrand et al., 2020). Since coronaviruses are highly sensitive to temperature, changes can cause drastic survival time differences (Naddeo and Liu, 2020). Coronavirus (HCoV 229E), a SARS-CoV-2 surrogate, can survive (99.9% inactivation) up to 588 d at 4 °C in filtered tap water (Gundy et al., 2009). However, this survival level (99.9% inactivation) reduced to just 10.1 d at 23 °C (Gundy et al., 2009). Similarly, survival times of HCoV 229E in unfiltered tap water, filtered primary effluent, unfiltered primary effluent and secondary effluents at 23 °C are 12.1, 2.35, 3.54, and 2.77 days, respectively (Gundy et al., 2009). Although, SARS-CoV-2 inactivation studies are still lacking, based on other coronaviruses studies similar persistences are expected (Carraturo et al., 2020; Gundy et al., 2009). SARS-CoV-2 persistence in a viral transport medium (concentration 6.8 log TCID50/mL) is exponentially reduced with a rise in temperature (Chin et al., 2020). Only a 0.7 log TCID50/mL (10% approx.) reduction in SARS-CoV-2 concentration was achieved at 4 °C in 14 days, which was reduced to 14 days at 22 °C (Chin et al., 2020). Further, SARS-CoV-2 survivability progressively dropped to 2 days, 30 min and 5 min for 37, 56 and 70 °C, respectively (Chin et al., 2020).
Exposure to sunlight or UV light drastically limits coronavirus survival, as is the case for many microorganisms (Naddeo and Liu, 2020). Shielding viruses from light exposure and viral settling behavior (settling of virus in aquatic sources with time, as well as with suspended load) are both enhanced by the presence of organic matter. SARS-CoV-2 can attach itself to organic matter particles and settle quite easily (Naddeo and Liu, 2020). The presence of antagonist microorganisms can decrease the viral survival (Naddeo and Liu, 2020). Coronavirus studies suggested extremely low SARS-CoV-2 survival occurred especially in wastewater temperatures of >20 °C (Collivignarelli et al., 2020). In contrast to that report, SARS-CoV-2 has also been reported in Indian wastewaters during the peak of summer (with ambient temperatures as high as 40–45 °C) (Arora et al., 2020; Kumar et al., 2020). This raises concerns. A variety of further studies are needed on the stability and survival of SARS-CoV-2 in aqueous systems.
Recently, a computational model was developed to estimate the SARS-CoV-2 persistence in wastewater (Hart and Halden, 2020a). This model estimates the half-life of SARS-COV-2 by considering an exponential viral decay dependent on wastewater temperature (Hart and Halden, 2020a; Hart and Halden, 2020b; Hart and Halden, 2020c). Based on this model the SARS-CoV-2 half-life is estimated between 4.8 and 7.2 h at 20 °C. The 99.9% reduction time ranges between 48 and 72 h at 20 °C for SARS-CoV-2 in wastewater (Hart and Halden, 2020a).
2.2. Coronavirus (SARS-CoV-2) in water, wastewaters and sewage sludge
Viral shedding in urine and feces is likely the biggest source of viral RNA in water and wastewater systems. An estimated load of 0.056 to 11.3 billion SARS-CoV-2 genomes/infected person/per day is injected into wastewater (Hart and Halden, 2020a). Coronavirus RNA shedding has earlier been reported for SARS-CoV and MERS-CoV through feces (Corman et al., 2016; Leung et al., 2003). As many as 107 and 2.5 × 104 SARS-CoV RNA copies/mL were reported in case of diarrhea and urine respectively (Hung et al., 2004). Persistent SARS-CoV-2 RNA shedding has been also reported in 27–89% of COVID-19 patients' excreta specimens including anal/rectal swabs and feces (Cai et al., 2020b; Holshue et al., 2020; Tang et al., 2020; Wölfel et al., 2020; Zhang et al., 2020b). As many as 108 viral RNA copies per gram of feces were reported in several studies (Lescure et al., 2020; Pan et al., 2020; Wölfel et al., 2020). SARS-CoV-2 RNA fecal shedding can last up to seven weeks after onset of first symptoms has also been reported in clinical studies (Cai et al., 2020b; Wu et al., 2020b; Xiao et al., 2020). Viral RNA was shed through feces by 81.8% of cases even after patients received negative results from throat swab tests (Ling et al., 2020). Furthermore, the feces of asymptomatic patients were also found positive for SARS-CoV-2 RNA (Mizumoto et al., 2020; Nishiura et al., 2020; Tang et al., 2020).
Coronaviruses were detected previously in sewage, water and wastewater sources (Hung et al., 2004; Leung et al., 2003) as already mentioned. SARS-CoV-2 enters these waters from human urine and feces. The fecal-oral route can be important for SARS-CoV-2 transmission and future investigations on the possibilities of SARS-CoV-2 fecal–oral transmission should incorporate environmental studies to ascertain the possible conditions favoring such transmission (Yeo et al., 2020). At present none of the available studies have provided proof of fecal-oral transmission of SARS-CoV-2 (Cahill and Morris, 2020; Foladori et al., 2020; Gupta et al., 2020; Hindson, 2020; Sehmi and Cheruiyot, 2020). However, the presence of viable SARS-CoV-2 in wastewater and water sources point toward a potential transmission of the virus through contaminated aerosols (Foladori et al., 2020; Gupta et al., 2020; Heller et al., 2020; Hindson, 2020; Sehmi and Cheruiyot, 2020; Shutler et al., 2020). Three possible primary pathways were proposed for fecal-oral SARS-CoV-2 transmissions (Heller et al., 2020). The first is direct contact with contaminated water; the second is through vectors including insects; the third is through surfaces which came in contact with contaminated water or surfaces contaminated by vectors (Heller et al., 2020).
SARS coronaviruses are reported to remain infectious for up to 4 days in stool samples (Weber et al., 2016). Of particular significance, coronaviruses can also remain active and infectious in sewage and water for several days and weeks (Casanova et al., 2009; Gundy et al., 2009). Similarly, high percentages (up to 99%) of coronaviruses can remain viable for several days in tap waters and in sewage effluents at room temperature (Casanova et al., 2009; Gundy et al., 2009). No studies have performed specific SARS-CoV-2 systematic survival time determinations in water and wastewater systems up to the date of the current review. SARS-CoV-2 can remain active for a long as 14 and 2 days at 22 °C and 37 °C, respectively, in viral transport medium (Chin et al., 2020). On the basis of these viral transport results, SARS-CoV-2 significant survival times in water and wastewater systems are thought to be likely (Shutler et al., 2020). That study also concludes that SARS-CoV-2 survival is temperature driven and decreases with increases in temperature. SARS-CoV-2 can contribute to transmission above detection levels and remains active for as long as 25 days at 5 °C in wastewater (Shutler et al., 2020).
Recent studies also suggested possible SARS-CoV-2 cross-transmissions occurred between 9 patients in a bath center in Huai'an, Jiangsu province, China (Luo et al., 2020). Other possible SARS-CoV-2 transmissions through recreational and rehabilitation pools has also been suggested (Cahill and Morris, 2020; Romano-Bertrand et al., 2020). Likely pathways include direct contact and through the fecal-oral route (Romano-Bertrand et al., 2020). SARS-CoV-2 transmission through public baths and recreational pools certainly enhances the importance of careful analysis for the possible presence of SARS-CoV-2 in aquatic systems (Romano-Bertrand et al., 2020). Maintaining proper social distancing measures and meeting standard pool disinfection measures can help control possible transmissions (Romano-Bertrand et al., 2020). So far, scientists from Australia (Ahmed et al., 2020a), France (Wurtzer et al., 2020a; Wurtzer et al., 2020b), India (Arora et al., 2020; Kumar et al., 2020), Israel (Or et al., 2020), Italy (La Rosa et al., 2020b), Netherlands (Medema et al., 2020), Spain (Balboa et al., 2020; Randazzo et al., 2020a; Randazzo et al., 2020b), USA (Nemudryi et al., 2020; Wu et al., 2020a) and others have found SARS-CoV-2 present in wastewater (Table 2 ). These studies have also found correlations between the number of COVID-19 cases and the amount of SARS-CoV-2 RNA fragments present in wastewater (Medema et al., 2020). Furthermore, SARS-CoV-2 RNAs were also found in the household wastewaters of home quarantined COVID-19 affected persons (Döhla et al., 2020). These wastewater sample sources include washbasin siphons, shower siphons and toilets (Döhla et al., 2020). While these findings allow one method of mapping COVID-19 cases in a location, the key question is: what do these detected SARS-CoV-2 RNAs mean for human transmission in each location they are found? What virus loads were present?
Table 2.
Presence of SARS-CoV-2 and its viral RNA found in wastewater, sewage sludge and river water around the world.
| Country | City/state | Water/sludge type | Sample concentration | Extraction technique | Analysis methoda | Genome target region | Detection frequency | Detected viral genome copies/L | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Wastewaters | |||||||||
| Australia | Brisbane, Queensland | Untreated wastewater | RNA is extracted directly from electronegative membranes and ultrafiltration | RNeasy PowerWater Kit, RneasyPowerMicrobiome Kit; (Qiagen) | RT-qPCR | N | 2/9 (22%) | 0.19–1.2 × 102 | (Ahmed et al., 2020a) |
| Brazil | Florianopolis, Santa Catalina | Urban sewage | – | QIAamp viral RNA mini kit (Qiagen) | Real time RT-PCR | N1, S, RdRp |
4/6 (66.67%) | 5.49–6.68 log10 | (Fongaro et al., 2020) |
| Brazil | Niterói, State of Rio de Janeiro | Raw sewage samples | Ultracentrifugation | QIAamp® Viral RNA Mini kit (QIAGEN, CA, USA) and a QIAcube® automated system (QIAGEN) | RT-qPCR | N2 | 5/12 (41.6) | (Prado et al., 2020) | |
| Chile | Farfana, Santiago | WWTP influent | Ultracentrifugation | QIAamp viral RNA mini kit (Qiagen) | Taqman 2019-nCoV assay Kit v1 (ThermoFisher) | ORF1, N, S |
2/4 (50%) | 354–2304 | (Ampuero et al., 2020) |
| WWTP effluent | 2/4 (50%) | 20–167 | |||||||
| Trebal Santiago | WWTP influent | 2/4 (50%) | 628–4805 | ||||||
| WWTP effluent | ¼ (25%) | 0–10 | |||||||
| China | Wuchang Fangcang Hospital, Wuhan | Hospital wastewater | Centrifugation | EZ1 virus Mini kit (Qiagen, Germany) | AgPath-ID™ One-Step RT-PCR Kit | ORF1, N |
– | 0.05–1.87 × 104 | (Zhang et al., 2020a) |
| Czech Republic | Within Czech Republic | Untreated wastewater samples | Direct flocculation using beef extract solution in glycine buffer | RT-qPCR | 13/112 (11.6%) | (Mlejnkova et al., 2020) | |||
| France | Paris | Raw wastewater | Centrifugation | PowerFecal Pro kit (QIAGEN) on a QIAsymphony automated extractor (QIAGEN) | RT-qPCR | E | 100% | – | (Wurtzer et al., 2020a) |
| France | Paris | Untreated wastewater | Ultracentrifugation | PowerFecal Pro kit on a QIAsymphony extractor | RT-qPCR | E | 23/23 (100%) | >106.5 | (Wurtzer et al., 2020b) |
| Treated wastewater | 6/8 (75%) | ~105 | |||||||
| Japan | Ishikawa Toyama | Influent WWTPs | PEG8000 precipitation | QIAamp viral RNA mini kit (Qiagen) | qRT-PCR | – | – | 2.1 × 104–4.4 × 104 | (Hata et al., 2020) |
| Japan | Yamanashi | Secondary treated wastewater | Electronegative membrane vortex (EMV) method | RNeasy PowerWater Kit (Qiagen) | RT-qPCR | ORF1ab, S |
1/5 (20%) | 2.4 × 103 | (Haramoto et al., 2020) |
| India | Jaipur | Untreated wastewater | Adsorption | Allplex™ 2019-nCoV Assay kit (cat# RP10244Y RP10243X) | Real Time-PCR | ORF1ab, S, E, N and RdRp | 2/6 (33.34%) | – | (Arora et al., 2020) |
| India | Ahmedabad | Untreated wastewater | PEG precipitation of centrifugated supernatant | NucleoSpin® RNA Virus, Macherey-Nagel GmbH & Co. KG, Germany |
RT-PCR | ORF1, N, S |
100% | 0.78 × 102- 8.05 × 102 |
(Kumar et al., 2020) |
| Treated wastewater | Nil | – | |||||||
| Israel | Tel Aviv metropolis | Sewage Wastewater Effluent |
Polyethylene glycol (PEG) or alum (20 mg/l) precipitation followed by centrifugation | RNeasy mini kit- QIAGEN and EasyMAG -bioMerieux, France | RT-qPCR | E | 10/26 (38.5%) | (Or et al., 2020) | |
| Italy | Milan, Rome |
Untreated wastewater | Two-phase PEG-dextran method | NucliSENS miniMAG semi-automated extraction system with magnetic silica | Nested RT-PCR and real-time RT-qPCR | ORF1ab, S |
6/12 (50%) | – | (La Rosa et al., 2020b) |
| Italy | Monza/Brianza Milano |
WWTPs | Filtration | QIAMP VIRAL RNA mini kit | Real time RT-PCR | – | – | – | (Rimoldi et al., 2020) |
| Netherlands | Amsterdam, Den Haag, Utrecht, Apeldoorn, Amersfoort, Schiphol, Tilburg |
Sewage wastewater | Ultrafiltration | RNeasy PowerMicrobiome Kit, Magnetic extraction reagents of the Biomerieux Nuclisens kit in combination with the semi-automated KingFisher mL | Real-time RT-PCR | N, E |
14/24 (58%) | 2.6–2200 × 103 | (Medema et al., 2020) |
| Pakistan | Lahore | Sewage water samples | Centrifugation | Hero 32 RNA extraction system | RT-qPCR | ORF1ab, N | (Yaqub et al., 2020) | ||
| Pakistan | Several cities | Untreated wastewater samples | Polymers, dextran and polyethylene glycol (PEG) | Spin star viral nucleic acid kit 1.0 | RT-qPCR | ORF1ab | – | – | (Sharif et al., 2020) |
| Spain | Barcelona | Frozen archived wastewater samples | 20% polyethylene glycol 6000 for precipitation | NucliSENS® miniMAG® extraction system (bioMérieux) | RT-qPCR | IP2, IP4 | 1/9 (11%) | 8.3 × 102 | (Chavarria-Miró et al., 2020) |
| Spain | Murcia, Cartagena, Molina de Segura, Lorca, Cieza, Totana |
WWTP | Centrifugation | NucleoSpin RNA virus kit | RT-qPCR | N | 37/72 (51.4%) | 5.1–5.5 log10 | (Randazzo et al., 2020b) |
| Spain | Ourense | Wastewater | Ultrafiltration | STARMag 96 × 4 Universal Cartridge Kit (Seegene, Seoul, South Korea) | RT-qPCR | N, E, RdRp |
– | 7.5–15 cp/ml | (Balboa et al., 2020) |
| Spain | Valencia | Wastewater | Aluminum-driven flocculation, centrifugation | Nucleospin RNA virus Kit | RT-qPCR | N | 13/15 (86.67%) |
5.22–5.99 log10 | (Randazzo et al., 2020a) |
| Turkey | Istanbul | Wastewater | Centrifugation | QIAamp cador Pathogen Mini Kit |
RT-qPCR | RdRp | 7/9 (77.78%) | 0.0–2.6 × 104 | (Kocamemi et al., 2020a) |
| USA | Bozeman, Montana |
Raw wastewater | Ultrafiltration | RNeasy Mini Kit (Qiagen) | RT-qPCR | N | 7/7 (100%) | >3 × 104 | (Nemudryi et al., 2020) |
| USA | Louisiana | Untreated wastewater | Ultrafiltration and adsorption-elution method using electronegative membrane | ZR viral RNA Kit (Zymo Research, USA) | RT-qPCR | N1, N2 | 2/7 | For N1–3.2 log10 and for N2–2.5-3.0 log10 |
(Sherchan et al., 2020) |
| Secondary treated wastewater | 0/4 | ||||||||
| Chlorine disinfected final effluents | 0/4 | ||||||||
| USA | Massachusetts | Raw wastewater | PEG precipitation | Nucleic acid extraction | RT-qPCR | N | 10/14 (71%) | 1.2–24.0 × 103 | (Wu et al., 2020a) |
| USA | New York | Wastewater | Ultracentrifugation | AllPrep® PowerViral® DNA/RNA Kit | RT-qPCR | – | – | 1.68 × 105 |
(Green et al., 2020) |
| USA | Southeastern Virginia | Influent wastewater samples | Centrifugation Electronegative filtration |
RT-ddPCR | N1, N2, N3 | 101–104 copies/100 mL | (Gonzalez et al., 2020) | ||
| Sewage sludge | |||||||||
| Spain | Ourense | Sludge | Precipitation with polyethylene glycol (PEG) | STARMag 96 × 4 Universal Cartridge Kit (Seegene, Seoul, South Korea) | RT-qPCR | N, E, RdRp |
– | 7.5–10 cp/ml | (Balboa et al., 2020) |
| Turkey | Istanbul | Primary sludge, Waste activated sludge |
Centrifugation | Roche MagNA pure LC system (Penzberg, Germany) | RT-qPCR | RdRp | 9/9 (100%) | 1.15 × 104–4.02 × 104 | (Kocamemi et al., 2020b) |
| USA | New haven, Connecticut | Primary sewage sludge | – | RNeasey PowerSoil Total RNA kit, (Qiagen). | one-step qRT-PCR | N | 44/44 (100%) | 1.7 × 106–4.6× 108 | (Peccia et al., 2020) |
| River waters | |||||||||
| Equador | Quito | Urban river water | Skimmed milk flocculation method then centrifugation | AccuPrep® Universal RNA extraction kit - Bioneer | RT-qPCR | N1, N2 | 3/3 (100%) | 2.91 × 105–3.19 × 106 | (Guerrero-Latorre et al., 2020) |
RT-PCR = Reverse transcription polymerase chain reaction, RT-qPCR = Reverse transcription quantitative polymerase chain reaction, qRT-PCR = Quantitative reverse transcription polymerase chain reaction.
Wide variations in intact SARS-CoV-2 detection frequencies and its loads in wastewater have been observed (Table 2). Higher viral concentrations were expected and reported from the countries with high COVID-19 caseloads, such as France, Japan, Turkey and USA (Table 2). Similarly, locations with high COVID-19 cases also show higher detection frequencies for SARS-CoV-2 RNA in Brazil (Fongaro et al., 2020), France (Foladori et al., 2020; Rimoldi et al., 2020), India (Arora et al., 2020; Kumar et al., 2020), Turkey (Kocamemi et al., 2020a) and USA (Sherchan et al., 2020) (Table 2). E.g. during February and March, only 29 COVID-19 cases were reported in Milan. SARS-CoV-2 presence in wastewaters also shows a similar pattern with a sample occurrence frequency of 2/8 (25%) and this was only reported on February 24th and 28th (La Rosa et al., 2020b). However, when high numbers of COVID-19 cases occurred in Rome between March 31st and April 2nd, the SARS-CoV-2 sample detection frequency was 4/4 (100%) (La Rosa et al., 2020b). However, high numbers of COVID-19 cases in Rome, SARS-CoV-2 detection frequency translates to 4/4 (100%) samples between March 31st and April 2nd (La Rosa et al., 2020b). Similar results were also reported in other studies from Japan (Haramoto et al., 2020), Netherlands (Medema et al., 2020), Brazil (Fongaro et al., 2020) and India (Arora et al., 2020). Thus, wastewater analysis can provide accurate predictions of epidemiological cases in a regional level.
The SARS-CoV-2 presence in river waters was also examined and reported (Guerrero-Latorre et al., 2020; Haramoto et al., 2020). River water samples in Yamanshi prefecture of Japan tested negative for the presence of SARS-CoV-2 (Haramoto et al., 2020). The low COVID-19 prevalence in the studied region was the suggested explanation (Haramoto et al., 2020). However, in high COVID-19 prevalence regions, as in case of the urban rivers of Quito (Ecuador), all river water samples were tested positive for SARS-CoV-2 (Guerrero-Latorre et al., 2020). Both N1 and N2 target genome assays were utilized for SARS-CoV 2 RNA evaluations in urban river waters. Urban river water in Quito contained 2.84 × 105 to 3.19 × 106 N1 and 2.07 × 105 to 2.23 × 106 for N2 target genome copies/Liter (Guerrero-Latorre et al., 2020). This study provided correlations with COVID-19 active cases 14 days prior the sampling study (Guerrero-Latorre et al., 2020).
Like wastewater, sewage sludge is also known to host a wide variety of human viruses as well as recent strains of circulating coronavirus (Bibby and Peccia, 2013). SARS-CoV-2 RNA's presence in sewage makes sense since it is present in the stool of COVID-19 patients as well as being present in untreated wastewaters (Núñez-Delgado, 2020). The ability of SARS-CoV-2 RNA to survive primary and secondary wastewater treatment phases to accumulate in sewage sludge raises concerns. However, none of the available studies have evaluated the SARS-CoV-2 infectivity in sewage sludge of wastewaters. Application of sewage sludge in agriculture adds further concerns. Present studies do not provide sufficient data to determine and eliminate all perceived risks related to SARS-CoV-2 spread in sludge and soil (Collivignarelli et al., 2020).
SARS-CoV-2 RNA presence in sewage sludge has been identified in USA (Peccia et al., 2020), Turkey (Kocamemi et al., 2020b), and Spain (Balboa et al., 2020) so far. During a COVID-19 outbreak in Turkey, the primary sludge of 2 wastewater treatment plants (WWTPs) and the secondary sludge of 7 WWTPs were analyzed (Kocamemi et al., 2020b). SARS-CoV-2 RNA quantification in primary and waste activated sludge samples collected from WWTPs was carried out in a US authenticated real time RT-PCR diagnostic panel in Istanbul. Both, primary and secondary sewage sludge reported the presence of SARS-CoV-2 with Cq values between 33.52 and 35.86 and corresponding concentrations of 1.15 × 104–4.02 × 104 genome copies/L (Kocamemi et al., 2020b).
Viral shedding, as stated earlier, starts even before the detection of Covid-19 infections as in asymptomatic and presymptomatic cases. SARS-CoV-2 monitoring in sludge would be advantageous, as sludge is a concentrated and well-mixed sample (Peccia et al., 2020). Therefore, analyzing sewage for SARS-CoV-2 can become an early indicator for outbreak dynamics assessment within a community (Peccia et al., 2020). Primary sludge samples were collected daily from a wastewater treatment facility serving nearly 200,000 residents from March 19, 2020 to May 1, 2020 during a Covid-19 outbreak area of metropolitan area of New Haven, Connecticut (CT), USA (Peccia et al., 2020). All the tested samples show the existence of SARS-CoV-2 RNA (values vary from 1.7 × 103 to 4.6 × 105 copies per mL). Results were also quantitatively compared with new COVID-19 cases and community hospital admission data. SARS-CoV-2 RNA amounts in sludge exhibits a high correlation (R2 > 0.97) with new COVID-19 cases and hospital admissions data (Peccia et al., 2020). The SARS-CoV-2 RNA presence in primary sludge is 2–3 times higher than in wastewater with no treatment (Peccia et al., 2020). Therefore, sludge can archive the SARS-CoV-2 presence with COVID-19 infections with a high correlation (Michael-Kordatou et al., 2020; Peccia et al., 2020).
3. SARS-CoV-2 analysis in aquatic sources and wastewater based epidemiology
3.1. SARS-CoV-2 analysis in aquatic sources
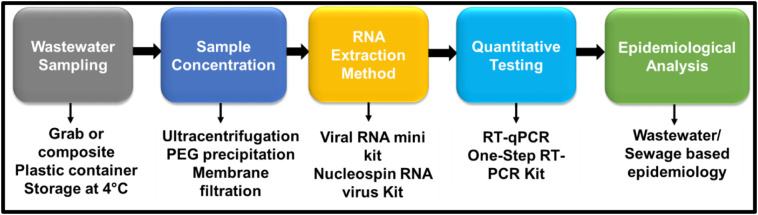
Accurate SARS-CoV-2 detection and quantification in aquatic systems is necessary for wastewater-based epidemiology (WBE). Due to very low virus concentrations, water and wastewater samples often need several pre-analysis steps to render SARS-CoV-2 RNA detectable. A typical viral analysis in wastewater involves sample collection, storage, preparation, extraction and detection (Patel et al., 2019). Sample preparation and virus concentration step(s) are necessary in untreated wastewater sample to analyze SARS-CoV-2 (Ahmed et al., 2020a, Ahmed et al., 2020b; Medema et al., 2020; Nemudryi et al., 2020; Wurtzer et al., 2020b). A diagrammatic representation of steps involved in SARS-CoV-2 analysis is presented in Fig. 2 . Matrix effect and even slight errors in these analytical steps (including sample collection, sample handling, preservation, sample concentration, RNA extraction and instrumental error) can impact proper evaluation. Recently, an excellent review was compiled by Michael-Kordatou and colleagues on analysis of SARS-CoV-2 in water and wastewater sources (Michael-Kordatou et al., 2020).
Fig. 2.
Common analytical steps required for SARS-CoV-2 RNA analysis in water and wastewater samples.
3.1.1. Sample collection and preservation
Recent studies vary in sample collection and preservation. Wastewater sampling (sampling method), container (type and volume), and storage conditions are summarized in Table 3 . Both composite and grab sampling were used for wastewater collection for SARS-CoV-2 detection (Michael-Kordatou et al., 2020). Plastic bottles are commonly used for wastewater sampling. Collected samples were commonly stored (for short duration) at 4 °C in dark. Low temperature (−20/−80 °C) is recommended for the samples to be stored for the long duration (Michael-Kordatou et al., 2020). In grab sampling, the viral concentration only represents the snapshot of that particular sample while the composite sample collection expresses the average virus RNA concentration during the collection period (Michael-Kordatou et al., 2020). Grab and composite sampling were carried out (Nemudryi et al., 2020). Composite samples provide more reliable data for SARS-CoV-2 analysis (Nemudryi et al., 2020). Large variations in sample collection volume (36 ml to 2 L) have been reported (Table 3). Other important considerations for the method development, diagnosis and quantification of SARS-CoV-2 RNA in wastewater are summarized in a review article (Michael-Kordatou et al., 2020). A standardized method for sample collection and preservation to be applied for an accurate representation of SARS-COV-2 detection in aqueous systems is the need of the hour.
Table 3.
Common sampling and storage techniques for SARS-COV-2 detection in aqueous systems.
| Sampling method | Container (volume) | Sample storage temperature (°C) | References |
|---|---|---|---|
| Grab (autosampler) | 100–200 mL | 4 | (Ahmed et al., 2020a) |
| Composite | 250 mL | 4 | (Medema et al., 2020) |
| Grab | Sterile HDPE plastic (500–1000 mL) | 4 | (Randazzo et al., 2020a) |
| Composite | 250 mL | −20 | (La Rosa et al., 2020b) |
| Grab | – | 4 | (Wurtzer et al., 2020b) |
| Composite | 1.9 L | 4 | (Green et al., 2020) |
| Composite | Plastic bottle (2 L) | −20/−80 | (Or et al., 2020) |
| Composite | 250 mL | 4 | (Balboa et al., 2020) |
| Grab | – | 4 | (Randazzo et al., 2020b) |
| Grab | 500 mL | (Nemudryi et al., 2020) | |
| Composite | 500 mL | ||
| Grab | 1 L plastic bottle | On ice | (Haramoto et al., 2020) |
| Grab | 500 mL polypropylene | – | (Rimoldi et al., 2020) |
| Composite | Polypropylene bottle | −80 | (Ampuero et al., 2020) |
3.1.2. Sample preparation and viral concentrations
Sample preparation/pre-conditioning is an important step before sample concentration can be done. Pre-conditioning steps enhances viral recoveries and overall concentration efficiencies. Common pre-conditioning steps include pre-filtration. sample pH and salinity adjustment (Bofill-Mas and Rusiñol, 2020). MgCl2, beef extract, and glycine are commonly used pre-conditioning agents to enhance viral elution (Bofill-Mas and Rusiñol, 2020).
Common sample concentration approaches prior to quantification have been recently reviewed (Bofill-Mas and Rusiñol, 2020; Haramoto et al., 2020). Several virus concentration methods with specific advantages and disadvantages can be found in Table 4 . Common methods include precipitation/flocculation (using an organic flocculant or ammonium sulfate precipitation), adsorption-elution (negatively/positively charged filters, glass powder or fiber), ultracentrifugation, ultrafiltration, lyophilisation and filtration (Bofill-Mas and Rusiñol, 2020; Bosch et al., 2006).
Table 4.
Advantages and drawbacks of viral concentration methodologies (Abdelzaher et al., 2008; Ahmed et al., 2020b; Bofill-Mas and Rusiñol, 2020).
| Concentration principle | Concentration method | Advantages | Drawbacks | References |
|---|---|---|---|---|
| Adsorption | All* | - Rapid process requires <40 m for sample processing - Effective for both solid and liquid phase viral concentration - Easy to upscale or downscale up to 200 mL - Possible field applications - Filtration unit is the main necessary equipment - Multiple sample processing using multiple filtration units |
- Cleaning and washing vessels are required - Requires pH adjustments - High turbidity can cause clogging - Requires bead beating step equipped RNA extraction kits |
(Ahmed et al., 2020a, Ahmed et al., 2020b) |
| Centrifugation-based | All* | - High recovery rates - Can be useful in turbid samples too |
- Need expensive equipment - Inapplicable in large sample volumes |
(Bofill-Mas and Rusiñol, 2020) |
| Ultracentrifugation | - No preconditioning - Single step method - Effective for both solid and liquid phase viral concentration - Low sample processing cost |
- Applicable only for small volume samples - Process consumes 3 h time - Limited number of samples can be processed at a time - Expensive ultracentrifuges are required - Inapplicable in fields |
||
| Filtration-adsorption | Dual membrane system (polyvinylidene fluoride (PVDF) + HA membrane) | - Can sequentially concentrate bacteria and virus separately - Large particles (>0.45 μm) removal by top filter |
- Large differences in viral concentrations in samples can cause differences in volume needed for filtration | (Ahmed et al., 2020a, Ahmed et al., 2020b) |
| Negative filters | All* | - Lesser costs - Facilitates direct extraction through membrane |
- Not applicable for turbid samples and large volume samples - Sample conditioning is necessary |
(Bofill-Mas and Rusiñol, 2020) |
| Positive filters | All* | - Can be applied for large sample volumes - Field deploying ability |
- Not applicable for turbid samples | |
| Virocap/ NanoCeram® cartridge filters |
- USEPA recommended process - Automatization is possible |
- Costly process | ||
| Flocculation and Precipitation | All* | - Effective for turbid samples - Low cost inputs |
- Not applicable for large sample volumes - Sample preconditioning/centrifugation might be required |
(Bofill-Mas and Rusiñol, 2020) |
| Polyethyleneglycol dextan precipitation | -WHO recommended process for poliovirus surveillance | |||
| Polyethylene glycol 8000 | -Centrifuge (upto 10,000g) is the only requirement - Effective for both solid and liquid phase viral concentration -Less expensive relatively - Can even process the large sample volumes |
- Process consumes 4–6 h - Hazardous chemical (Trizol) is required - Inapplicable for field applications - Can leave target behind as only a small sample portion is required for RNA extraction |
||
| Skimmed milk flocculation | - Can be deployed in field (Virwatest) - Single step method- Large sample volumes (up to 40 L) can be processed |
-Process is time consuming | ||
| Ultrafiltration | Tangential flow Ultrafiltration | - Efficient for large sample volumes | - Requires filter conditioning | (Ahmed et al., 2020a, Ahmed et al., 2020b; Bofill-Mas and Rusiñol, 2020) |
| Dead end ultrafiltration | - Automatization is possible - Efficient for large sample volumes - Can be coupled with Innovaprep® wet foam elution |
– | ||
| Concentrating pipette Select™ (Innovaprep®) | - Process is fast and reproducible | - Requires expensive consumables and equipment - Can only use samples up to 500 mL |
||
| Amicon® Ultra-15 centrifugal filter | - Rapid process (usually 1 h, time depends upon sample's turbidity) - Centrifuge (up to 4750 g) is the only equipment required | - Only applicable for liquid samples - Unit is quite expensive - Can only process up to 15 mL sample - Highly turbid samples requires multiple units for filtration - Turbidity can cause membrane clogging - Cannot be used for field applications - Target viruses can be absorbed onto membrane - Also concentrate PCR inhibitors along with target |
||
| Centricon Plus-70 centrifugal filter | - Rapid process (usually 1 h, time depends upon sample's turbidity) | |||
| Others | Monolyth chromatography | - High recoveries | - Requires expensive consumables and equipment | (Bofill-Mas and Rusiñol, 2020) |
* All- the overall advantages and disadvantages of a concentration principle and all the methods based upon discussed in this section.
Surrogate viruses are often used when studying uncultivable viruses (Bosch et al., 2006). The murine hepatitis virus, for example, is commonly used for persistence and recovery studies as a human corona virus surrogate due to their structural similarities (Ahmed et al., 2020b; Casanova et al., 2009; Patel et al., 2017; Ye et al., 2016). Another advantage is the non-pathogenic nature of these surrogate viruses toward humans which also reduces the need for the highest levels of biosafety precautions (Ahmed et al., 2020b). Common concentration methods (extraction-adsorption, ultrafiltration, precipitation and ultracentrifugation) were recently evaluated for a human coronavirus (SARS-CoV-2) surrogate, as a murine hepatitis virus (MHV) (Ahmed et al., 2020b). Method recovery efficiencies were quantified using RT-qPCR with mean murine hepatitis virus recoveries ranging between 26.7 and 65.7% (Ahmed et al., 2020b). Adsorption-extraction methods with MgCl2 pre-treatment (65.7 ± 23.8%) and without any pretreatment (60.5 ± 20.2%) were the most efficient concentration methods (Ahmed et al., 2020b).
Centrifugation or filtration to remove debris, electronegative membrane filtration (Ahmed et al., 2020a), ultrafiltration (Ahmed et al., 2020a; Balboa et al., 2020; Kocamemi et al., 2020a; Medema et al., 2020; Nemudryi et al., 2020; Or et al., 2020), polyethylene glycol precipitation (PEG) (Kocamemi et al., 2020a; Or et al., 2020; Wu et al., 2020a; Zhang et al., 2020a), aluminum flocculation (Randazzo et al., 2020a; Randazzo et al., 2020b) and ultracentrifugation (Wurtzer et al., 2020a; Wurtzer et al., 2020b) are common methods used for SARS-CoV-2 concentration in wastewater samples (Table 2, Table 3). Sample concentration methods increase RNA concentration by 20× to 800× in wastewater samples. For sludge samples, either RNA is directly extracted (Peccia et al., 2020) or virus is eluted by PEG precipitation from the matrix (Balboa et al., 2020). Organic compounds (such as humic substances) can interfere with downstream/in vitro viral detection via co-concentration with viral RNA (Farkas et al., 2020b). Advantages and disadvantages of various concentration procedures are provided in Table 4.
Sample volume is another important factor affecting virus detection results. In general, enteric viruses detection in untreated wastewater samples normally used <100 mL samples for concentration (Haramoto et al., 2018). In general, 200 mL raw wastewater samples were utilized for concentration in SARS-CoV-2 detection studies (Ahmed et al., 2020b; Medema et al., 2020; Nemudryi et al., 2020; Wu et al., 2020a; Wurtzer et al., 2020a, Wurtzer et al., 2020b). However, in a less prevalent COVID-19 region, large wastewater volumes would be required for SARS-CoV-2 detection (Haramoto et al., 2020).
3.1.3. Instrumental analysis and detection
Polymerase chain reactions (PCR) based techniques including quantitative PCR (qPCR) and reverse transcription quantitative PCR (RT-qPCR) are widely used for RNA and DNA viruses quantification in wastewater (Farkas et al., 2020a; Haramoto et al., 2018). These methods are useful for small viral genome segment detection. PCR based methods provide rapid, accurate and sensitive strain-level detections for up to five targets in one assay (Jiang et al., 2014). Several designed RT-qPCR assays were applied for SARS-CoV-2 detection (Chan et al., 2020a; Nalla et al., 2020; Vogels et al., 2020), which also provide satisfactory results in wastewater monitoring (Ahmed et al., 2020a; Medema et al., 2020; Wurtzer et al., 2020a; Wurtzer et al., 2020b). A SARS-CoV-2 RNA detection study conducted in Australian wastewaters used the N_Sarbeco assay (Ahmed et al., 2020a), CDC-N1, -N2, -N3 and the E_Sarbeco assays was used in a Dutch study (Medema et al., 2020) and CDC-N1, -N2 and -N3 assays in a Spanish study (Randazzo et al., 2020a; Randazzo et al., 2020b). Different SARS-CoV-2 genome sections used in these assays are provided in Table 2. However, dissimilar assays may provide varying performance for viral detection. Use of different primer/probes for quantification showed substantial differences in rates of viral detection. For example, the ‘N1’ and ‘N3’ genes were detected for SARS-CoV-2 analysis in wastewater (positive) while the ‘N2’ assay did not (Medema et al., 2020). Hence, multiple primer/probe sets are recommended for usage. The presence of organic co-contaminants limits the use of qPCR-based methods by inhibiting polymerase enzymes and reverse transcription (Farkas et al., 2020b).
Digital PCR (d-PCR) were also reported for the estimation of viruses in environmental samples (Farkas et al., 2020a). This method provides absolute target quantification and minimizes inhibition. However, digital PCR is more costly than quantitative PCR analysis. Biosensors and isothermal amplification for detection and quantification of viral DNA/RNA in environmental samples are also emerging techniques. These techniques provide results within an hour (Farkas et al., 2020a). Paper-based microfluidics devices are another easy and inexpensive platform with the potential to rapidly detect viruses in wastewaters (Mao et al., 2020). Antibody based detection techniques involving immunoassays including colloidal gold immunoassays, enzyme linked immunosorbent assays (ELISA), lateral flows immunoassays, time-resolved fluorescence immunoassays are also in development along with a variety of other antibody based detection kits commercially available (Jalandraad et al., 2020). Aptamer-based techniques, CRISPR-based techniques, electrochemical immunosensor techniques, loop-mediated isothermal amplification based techniques, microarray based techniques, and molecularly imprinted polymer based techniques are other analytical techniques under development (Jalandraad et al., 2020). However, the low sensitivity of these assays versus traditional PCR-based methods is a disadvantage. These methods were not rigorously tested in the field (Ishii et al., 2014; Jalandraad et al., 2020). Thus, RT-qPCR based techniques provide the most reliable results to date.
3.1.4. Quality control
Quality control and quality assurance (QA/QC) is important for analytical methods. Collected samples must be true representatives and proper precautions are necessary for pre-conditioning and concentration steps. Method concentration efficiencies need to be determined properly before applications with real world samples. Only methods with high recoveries should to be applied for wastewater analysis. Method repeatability and reproducibility must be assessed properly. Artificially contaminated samples are often used to evaluate detection methods and the use of surrogate viruses is a well explored practice. Surrogates are very helpful when addressing problems with uncultivable (not easily cultivable) viruses (Bosch et al., 2006). Special precaution are needed in wastewater samples because the presence of turbidity and organic matter can affect sample concentration (Bofill-Mas and Rusiñol, 2020).
The development of standard methods is required to accurately evaluate viral concentration for accurate evaluation of WBE and environmental surveillance. At present no widely accepted standard method/procedure exists for wastewater SARS-CoV-2 detection (Collivignarelli et al., 2020), however, modified standard methods have been employed. For example, WHO's standard poliovirus surveillance procedure (WHO, 2003), was modified to develop a standardized method for SARS-CoV-2 RNA analysis in Italian wastewaters (La Rosa et al., 2020b). The development of similar standardized methods for wastewater SARS-CoV-2 RNA detection could fulfill significant analytical research gaps for wastewater surveillance. Standard methods should also be able to quantify viruses in complex wastewater matrixes and thus would provide results that could be compared from samples collected around the world (Collivignarelli et al., 2020).
3.2. SARS-CoV-2 and wastewater-based epidemiological studies
Identifying symptomatic COVID-19 infected individuals is straightforward, while identification of presymptomatic and asymptomatic individuals is a bigger concern. The median COVID-19 incubation period with no symptoms is 5.1 days (Lauer et al., 2020). Between 18 and 31% of infected patients are reported to be asymptomatic (Mizumoto et al., 2020; Nishiura et al., 2020). Asymptomatic individuals also shed SARS-CoV-2 virions in their feces (Tang et al., 2020). These presymptomatic and asymptomatic patients are a major source of untraced COVID-19 transmissions (Hata and Honda, 2020). The lack of proper instrumentation and high cost limits diagnostic testing in many countries. This problem is more serious in developing countries. Thus, limited diagnostic testing coupled with the presence of presymptomatic and asymptomatic patients has led to uncertainties of the extent COVID-19 spread in many regions (Bivins et al., 2020).
Water and wastewater systems harbor numerous pathogenic microorganisms (Adriaenssens et al., 2018). Enveloped viruses, including coronaviruses, inactivate rapidly in water and wastewater without a host when compared to other viruses (Casanova et al., 2009; Casanova et al., 2010; La Rosa et al., 2020a). Nevertheless, they were present in many wastewaters due to the continuous SARS-CoV-2 influx from humans through urine and excreta (Sehmi and Cheruiyot, 2020; Singer and Wray, 2020; Sun et al., 2020) (Table 2). SARS-CoV-2 contamination of water supplies has the potential to infect whole communities (Gormley et al., 2020). A super-spreading event previously reported for a SARS spread in Hong Kong was related to faulty sewage system (Gormley, 2020; Gormley et al., 2020, Gormley et al., 2012, Gormley et al., 2014; Peiris et al., 2003; WHO, 2003). About 342 SARS cases were reported from high rise building in Amory Garden, Hong Kong. Epidemiological studies suggested the role of faulty sewage system contaminated with the excreta of “index patient” in causing this super spreading event (Gormley, 2020; Gormley et al., 2020; Peiris et al., 2003).This faulty sewage system facilitated transmission of virus-laden droplets through aerosols as well as contaminating surfaces in the bathrooms (Peiris et al., 2003; WHO, 2003). Aerosols derived from wastewaters through leakage, flushing and malfunctioning sewer plumbing facilities are identified as possible infection routes during SARS spread in 2003 (Nghiem et al., 2020). Norovirus transmission through wastewater flow and wastewater-derived aerosols was also reported (Gormley et al., 2014). Toilet flushing can generate aerosols with airborne pathogens including E. coli, Staphylococcus epidermidis, Pseudomonas alcaligenes as well as viruses (Lai et al., 2018; Li et al., 2020; Wang et al., 2020a). A recent study also reported the SARS-CoV-2 spread through wastewater plumbing systems in Guangzhou, China (Gormley, 2020; Kang et al., 2020).
The ubiquitous SARS-CoV-2 presence in human excreta offers the potential of using viral RNA sewage surveys to estimate the epidemiological status of COVID-19's prevalence in a region (Bivins et al., 2020). This epidemiological monitoring is known as wastewater-based epidemiology (WBE) or environmental surveillance (Bivins et al., 2020; Daughton, 2020a; Daughton, 2020b; Sims and Kasprzyk-Hordern, 2020).
Effective screening of suspected infectious individuals from every individual household remains a tough, highly challenging logistical task for medical professionals. It is highly labor intensive, time-consuming and costly. WBE could be an alternative and effective method for SARS-CoV-2 local and regional assessments (Daughton, 2020a; Daughton, 2020b; Hata and Honda, 2020). Temporal changes of viral concentrations in wastewater using WBE can provide information related to a specific viral absence or presence, outbreak dynamics and its demographic and human health effects (Farkas et al., 2020b). A 2013 example of silent transmissions and wild poliovirus type 1 reintroductions were observed in Israel during routine sewage samples surveillance, without any clinically reported case appearing (Manor et al., 2014). In another example, the first SARS-CoV-2 case was reported in Italy on February 21st, 2020 and the first report of SARS-CoV-2 being present in wastewater was reported shortly after, on February 24th (La Rosa et al., 2020b).
This wastewater report also suggested that SARS-CoV-2 infections might have started before the detection of the first case in Italy (La Rosa et al., 2020b). COVID-19 cases have been correlated with the SARS-COV-2 presence in wastewater from various WWTP catchment areas by several studies (Ahmed et al., 2020a; Haramoto et al., 2020; Medema et al., 2020; Randazzo et al., 2020a; Randazzo et al., 2020b; La Rosa et al., 2020b; Wurtzer et al., 2020b). These studies have successfully demonstrated the wastewater surveillance's potential to provide epidemiological dynamics assessments to better understand and design outbreak handling approaches. For example, SARS-CoV-2 analysis within sewage treatment plants of the Valencian metropolitan area (~1,200,000 inhabitants) in Spain, provided a direct correlation with declared active cases in the region (Randazzo et al., 2020a). This study illustrated that SARS-COV-2 detection in wastewaters coincided with the emergence of the first case in the region. The rise in COVID-19 cases also correlates with the rise in SARS-CoV-2 RNA's presence in wastewater (Randazzo et al., 2020a). This was illustarted in a plot of log10 gc/L of water versus the data for the Valencian metropolitan area, showing a measure of the rise of cases in this cathment area. On this same time axis was a plot of declared active COVID-19 cases detected clinically. The two plots followed the same rise and leveling off, except for a 13 day off set between the first case detected by WBE and the subsequent first clinical case detected. A subsequent time log between the rising number of declared cases and rise in genome levels found in wastewater existed. Such plots should be coordinated with health care and government leaders immediately to get a headstart on public responses.
Regions with only a few COVID-19 cases have also reported SARS-CoV-2 load in wastewaters (Haramoto et al., 2020; Randazzo et al., 2020b; Wurtzer et al., 2020b). Wastewater surveillance also provided the proof of SARS-CoV-2 RNA detection weeks before the actual confirmation of cases in the region of Murcia (Spain) had occurred (Randazzo et al., 2020b). Similarly, sewage showed the presence of SARS-CoV-2 in Barcelona, long before the first COVID-19 case was declared (Chavarria-Miró et al., 2020). Furthermore, a rise in SARS-CoV-2 genome occurrence was recorded in the archived wastewater samples (from 15 January to 4 March 2020) (Chavarria-Miró et al., 2020). SARS-CoV-2 was first detected in wastewater on January 15 (41 days before the first COVID-19 case was declared on 25th February 2020) validating the efficacy of WBE in pandemic surveillances (Chavarria-Miró et al., 2020). A sewage assessment study conducted in Santa Catalina, Brazil achieved surprising results, when SARS-CoV-2 RNA was found in sewage on 27th November 2019 (Fongaro et al., 2020). However, the region only reported its first case on 4th March 2020 (Fongaro et al., 2020). Thus, WBE can provide an early warning and helps gain time to plan prevention and mitigation efforts in disease spread. WBE can also provide warnings in the occurrence of future pandemic/epidemic waves (Chavarria-Miró et al., 2020).
This early SARS-CoV-2 detection in WWTPs also supports the idea that COVID-19 has been spreading widely and silently within the population (Chavarria-Miró et al., 2020). Advantages of environmental surveillance over clinical surveillance include, a) WBE can predict the complete status of a studied catchment region for less effort and expense than the clinical surveillance. b) a single wastewater sample has the potential to provide a clear view of an outbreak within the studied catchment, while clinical surveillance requires huge numbers of samples. Moreover, c) clinical surveillance is more time consuming and costly due to sample collection and testing compared to environmental surveillance, and d) environmental surveillance collects information about symptomatic, presymptomatic and asymptomatic patients, which clinical surveillance cannot provide (Hata and Honda, 2020). Even a single infected individual can be detected within a population of 2,000,000 individuals (Hart and Halden, 2020a). Globally, 2.1 billion people can be monitored with the existing 105,600 WWTPs (Hart and Halden, 2020a). Thus, WBE should become an essential tool for global pandemic/epidemic analysis and prevention.
4. Strategies for SARS-CoV-2 contaminated water treatment
As established above, SARS-CoV-2 contaminated water and wastewaters have the potential to spread COVID-19. Thus, assessing SARS-CoV-2 removal using existing techniques is important. These include secondary and tertiary treatments in WWTPs and advanced drinking water treatments. The use of disinfecting agents also needs to be evaluated. Heat and radiation are also important methods for microbial removal and viral removal from aqueous systems (Arora et al., 2020; Chin et al., 2020). Use of high temperature on SARS-CoV-2 inactivation and rapid SARS-CoV-2 removal using bleach, benzyl alkyl ammonium chloride, chloroxylenol, ethanol and povidine‑iodine are well known (Chin et al., 2020). Many disinfectants, including hypochlorites, quaternary ammonium salts, hydrogen peroxide, peracetic acids, mono persulfates, and chlorine dioxide are listed by United States Environmental Protection Agency for SARS-CoV-2 disinfection (USEPA, 2020). Successful SARS-CoV-2 inactivation in viral transport medium was achieved using common disinfectants including household bleach (both 1:49 and 1:99 ratios in water), benzylkonium chloride (0.1%), chloroxylenol (0.05%), chlorhexidine (0.05%), ethanol (70%) and povidone‑iodine (7.5%) within 5 min (Chin et al., 2020). However, only 54% inactivation can be achieved with hand soap solution (1:49) in 5 min and 100% inactivation is observed in 15 min (Chin et al., 2020). Thus, common disinfectants can also be applied for wastewater disinfection. For example, sodium hypochlorite was successfully applied for large scale SARS-CoV-2 loaded hospital wastewater treatment in Wuhan, China (Zhang et al., 2020a). Potential of other techniques have also been suggested (Wang et al., 2020b).
4.1. Removal in wastewater treatment plants
Coronaviruses have low survival rates in high temperature waters (Gundy et al., 2009). For example, survival (99.9% inactivation) of the human coronavirus HCoV 229E in filtered primary effluent, unfiltered primary effluent and secondary effluents at 23 °C are just 2.35, 3.54, and 2.77 days, respectively (Gundy et al., 2009). Preliminary studies also consider low SARS-CoV-2 RNA survival times in wastewaters above 20 °C (Collivignarelli et al., 2020). SARS-CoV-2 added into both raw and treated wastewater samples 48 and 72 h after inoculation displayed insignificant vitality and no cytopathic effects on Vero E6 cells (Rimoldi et al., 2020).
Several studies revealed SARS-CoV-2 RNA detection in wastewaters recently, but few of these have investigated SARS-CoV-2 RNA removal from wastewater in treatment plants (Arora et al., 2020; Collivignarelli et al., 2020; Kumar et al., 2020; Randazzo et al., 2020a, Randazzo et al., 2020b; Zhang et al., 2020a). However, some studies evaluated the presence of SARS-CoV-2 RNA in both influents and effluents of WWTPs (Arora et al., 2020; Collivignarelli et al., 2020; Kumar et al., 2020; Randazzo et al., 2020a; Randazzo et al., 2020b; Zhang et al., 2020a). Table 5 summarizes SARS-CoV-2 RNA concentrations in both influent and effluent wastewaters versus the WWTPs' treatment techniques. These studies indicated declines in SARS-CoV-2 RNA in WWTP effluents compared to the corresponding influents (Table 5). For example, 42 influents, 18 secondary and 12 tertiary treated effluents were collected and analyzed for SARS-CoV-2 (Randazzo et al., 2020b). Of these, only 35 of 42 (85%) influent samples tested SARS-CoV-2 positive compared to only 2 of 18 (11%) of the secondary-treated samples (Randazzo et al., 2020a). This provides insights into possible SARS-CoV-2 removal within WWTPs.
Table 5.
Possible SARS-CoV-2 removal from traditional wastewater treatment plants (WWTPs).
| City/country | Sampling date | SARS-CoV-2 RNA in influent wastewater (Copies/L) |
WWTPs treatment techniques | SARS-CoV-2 RNA in effluent wastewater (Copies/L) |
Removal status | Reference |
|---|---|---|---|---|---|---|
| Ahmedabad/ India |
08/05/2020 27/05/2020 |
0.78 × 102 8.05 × 102 |
UASBa treatment, Aeration pond |
0 | Yes | (Kumar et al., 2020) |
| Jaipur/ India |
03/05/2020 14/06/2020 |
Positive | MBBRb, SBRc |
0 | Yes | (Arora et al., 2020) |
| Santiago/Chile | 25/05/2020 15/06/2020 |
354–2304 | – | 20–167 | Yes | (Ampuero et al., 2020) |
| 628–4805 | 0–10 | |||||
| Paris/France | 05/03/2020 07/04/2020 |
>106.5 | – | ~105 | Yes | (Wurtzer et al., 2020b) |
| Murcia/Spain | 12/03/2020 14/04/2020 |
N1: 1.4 × 104 N2: 3.4 × 104 N3: 3.1 × 104 |
Secondary treatment, | <2.5 × 104 | Yes | (Randazzo et al., 2020b) |
| Tertiary treatment | 0 | |||||
| Valencia/ Spain |
12/02/2020 14/04/2020 |
104–105 | – | 0 | Yes | (Randazzo et al., 2020a) |
| Wuchang/ China |
– | 0.05–1.87 × 104 | Septic tank, MBBRb, Sedimentation tank, Disinfection tank |
0 at 6700 g/m3 sodium hypochlorite application | Yes | (Zhang et al., 2020a) |
UASB - Up-flow anaerobic sludge blanket.
MBBR - Moving bed biofilm reactor.
SBR - Sequencing batch Reactor.
The efficacy of Up-flow anaerobic sludge blanket (UASB) and aeration pond-based secondary treatments to remove SARS-CoV-2 RNA was demonstrated by influent and effluent sample concentration analyses conducted on 08/05/2020 and 27/05/2020 in Pirana, Ahmedabad in Gujarat, India (Kumar et al., 2020). All influent samples tested SARS-CoV-2 positive with a maximum concentration of 2.419 × 108 genome copies/L (Kumar et al., 2020). However, all effluent samples tested negative for SARS-CoV-2 RNA (Kumar et al., 2020). A study employing a moving bed biofilm reactor (MBBR) and a sequencing batch reactor (SBR) treatment also turned SARS-CoV-2 positive untreated wastewater samples into a SARS-CoV-2 negative effluents (Arora et al., 2020).
Application of treated wastewater for agricultural purposes and in gardening might have public health risks, when contaminated with SARS-CoV-2 (Arora et al., 2020). Similarly, application of sewage biosolids loaded with SARS-CoV-2 have potential health risks. Quantitative microbial risk assessment (QMRA) of SARS-CoV-2 in sewage was conducted at the entrance of a WWTP with viral load of 1.03 × 102 to 1.31 × 104 genome copies/mL (Zaneti et al., 2020). QMRA was performed to estimate the risk of infection for workers in a three-tiered approach (moderate, aggressive and extreme COVID-19 spread scenarios). The estimated risk values for aggressive and extreme scenarios are 6.5 × 10−3 and 3.1 × 10−2 respectively (Zaneti et al., 2020). Obtained QMRA values were higher than WHO benchmark for tolerable viral infection risk (10−3). Thus, sewage systems appear to be a possible route of viral transmission (Zaneti et al., 2020). Previous reports indicates viral persistence in its active state and could cause severe safety risk to the farmers indulged in irrigation process and it can also affect public health (Arora et al., 2020; La Rosa et al., 2020b). Thus, validation of the absence of SARS-CoV-2 genome in treated effluents becomes necessary (Arora et al., 2020).
The low number of wastewater treatment facilities raises serious problems for developing countries (Arora et al., 2020). Under such circumstances, validation of the SARS-CoV-2 genome presence in both treated and untreated wastewaters becomes essential. Thus, both untreated and treated wastewaters with SARS-CoV-2 load can cause large scale viral spread in developing countries, where limited medical facilities are available (Arora et al., 2020). Places where wastewater is untreated and drinking water purification plants do not operate are particular loci for transmission.
4.2. Tertiary treatment and advanced disinfection strategies
Tertiary and advanced treatment technologies feature chlorination, ozonation, photocatalysis, advanced oxidation processes, filtrations (including reverse osmosis, ultra-, micro-, nano-filtrations) and adsorption for water and wastewater treatment (Patel et al., 2019). Some of these technologies have also been evaluated for coronaviruses including SARS-CoV-2 removal from aqueous systems (Collivignarelli et al., 2020; Wang et al., 2005). Disinfection agents including bleach, benzyl alkyl ammonium chloride, chloroxylenol, ethanol, povidine‑iodine, hypochlorites, quaternary ammonium salts, hydrogen peroxide, peracetic acids, mono persulfates, and chlorine dioxide were suggested for SARS-CoV-2 disinfection (Chin et al., 2020; USEPA, 2020). They add cost to wastewater treatment.
Chlorine is one of the earliest and most used disinfection agents for wastewater treatment due to its powerful oxidizing nature (Ghernaout et al., 2018; Ghernaout et al., 2011; Yu-mei et al., 2010). Common chlorine-based disinfectants are chlorine (liquid), chlorine dioxide and sodium hypochloride (Wang et al., 2020c). Chlorine-based disinfectants destroy anabolic pathways of proteins and further neutralize microorganisms including viruses, bacteria, spores, fungi, and Clostridium botulinum. In comparison to chlorine, the chlorine dioxide is 5 times more soluble and has 2.63 times more oxidizing capability (Wang et al., 2020c). Coronaviruses are highly sensitive to chlorine and seem unstable in presence of chlorine (La Rosa et al., 2020b). SARS coronaviruses have a greater vulnerability to residual chlorine than E. coli and f2 phage (Wang et al., 2020c). Complete inactivation of SARS coronaviruses can be achieved with residual chlorine >0.5 mg/L or chlorine dioxide >2.19 mg/L in 30 min (Wang et al., 2005). SARS-CoV-2 inactivation using diluted household bleach (1:99) was performed in vitro and complete inactivation was achieved in 5 min contact time (Chin et al., 2020). 100% SARS-CoV-2 removal was also achieved through tertiary wastewater treatment equipped with both NaClO, and NaClO coupled with UV irradiation (Randazzo et al., 2020b). These studies provide insights into the potential of chlorination based process to successfully disinfect aqueous sources contaminated with SARS-CoV-2. However, present research does not clarify facts about required dosage and contact time for viral disinfection in most aqueous systems (Collivignarelli et al., 2020).
Disinfection agents including ethanol (78–95%), 2-propanol (70–100%), 2-propanolol and 1-propanol in combination (45% + 30%), formaldehyde (0.7–1%), glutardialdehyde (0.5–2.5%) and povidone iodine (0.23–7.5%) can rapidly inactivate the infectivity of coronaviruses by >4 log10 in suspensions (Kampf et al., 2020a). Sodium hypochlorite (>0.2%), hydrogen peroxide (0.5%) can also disinfect coronavirus contaminated water (Kampf et al., 2020a). These doses were based on previous studies used for inactivation of different coronaviruses strains including SARS-CoV-1, HCoV and MERS (Kampf et al., 2020a). However, detailed studies are still lacking for SARS-CoV-2 inactivation. One study from the Wuchang Cabin hospital in China provides striking revelations about challenges of hospital wastewater disinfection (Zhang et al., 2020a). Unexpectedly high (0.5–18.7) × 103 SARS-CoV-2 genome copies/L were found even after disinfection with an 800 g/m3 sodium hypochlorite dose, which is recommended by WHO and China CDC (Zhang et al., 2020a). Complete viral inactivation is only achieved with a dose of 6700 g/m3 sodium hypochlorite dose application (Zhang et al., 2020a).
4.3. Inactivation by heat and radiation
Several studies have been performed to inactivate SARS-CoV-1 in presence of high temperatures (Darnell et al., 2004; Darnell and Taylor, 2006; Kariwa et al., 2006; Scheller et al., 2020). When treated at 56 °C for different time periods, SARS-CoV-1 rate of infection was below the limit of detection (LOD) after a 20 min incubation time. However, complete inactivation is only possible after 60 min or greater incubation periods (Scheller et al., 2020). Complete SARS-CoV inactivation was also reported at 75 °C in 45 min (Scheller et al., 2020). Total inactivation of the virus (>6 Log10 decrease) was observed at 92 °C after 15 min (Pastorino et al., 2020). Similar trends were also observed in previous studies on SARS-CoV-1 and MERS-CoV (Darnell et al., 2004; Leclercq et al., 2014). The effect of a wide temperature range (37, 42, 56, and 60 °C) on SARS-CoV-2 inactivation is reported in Table 6 (Wang et al., 2020c). The virus is active at least for 24 h at 37 °C. The virus is effectively inactivated while the viral RNA was preserved in both sputum samples and human sera when heated for 30 min at 56 °C. This finding suggests that why some tropical countries have not been able to escape from the spread of SARS-CoV-2 (Wang et al., 2020c).
Table 6.
Inactivation of SARS-CoV-2 under different heat conditions (Wang et al., 2020c).
| Temperature (°C) |
Virus titers (TCID 50/mL) |
||||||
|---|---|---|---|---|---|---|---|
| 15 min | 30 min | 60 min | 90 min | 120 min | 24 h | 48 h | |
| 37 | 5.6 × 106 | 1.0 × 107 | 2.5 × 107 | 1.8 × 107 | 2.5 × 107 | 1.8 × 106 | 320 |
| 42 | 3.2 × 105 | 3.2 × 105 | 1.9 × 106 | n.d.a | 1–10 | Undetectable | |
| 56 | 2.5 × 103 | Undetectable | Undetectable | n.d.a | |||
| 56 (50% human serum) | n.d.a | Undetectable | n.d.a | ||||
| 60 | Undetectable | Undetectable | Undetectable | ||||
| Unheated | 1.4 × 107 | ||||||
n.d., not determined.
SARS-CoV-2 inactivation studies using heat in water and wastewater were almost completely absent. However, wastewater monitoring studies have successfully applied heat for SARS-CoV-2 inactivation for the safety of scientific personal (Arora et al., 2020; La Rosa et al., 2020b). For example, thermal inactivation were performed by treating wastewater samples at 56 °C for 30 min (La Rosa et al., 2020b). Other studies inactivated SARS-CoV-2 in wastewater samples using heat at 60 °C for 90 min (Arora et al., 2020; Wu et al., 2020a).
Ultraviolet (UV) light is comprised of UV-A (400–315 nm), UV-B (315–280 nm), UV-C (280–200 nm), and vacuum UV (200–100 nm) components. The disinfection of drinking water using UV was first reported in 1910 (Wang et al., 2020c). UV wavelength between 200 and 300 nm can destroy the structure of RNA and DNA of viruses, bacteria, and single-celled microorganisms. Therefore, the synthesis of protein is blocked (Ghernaout and Elboughdiri, 2020a; Ghernaout and Elboughdiri, 2020b). A wavelength of ~254 nm is considered optimum for microbial disinfection through UV (Meulemans, 1987). In comparison to chlorine disinfection, UV disinfection requires considerably smaller operation costs and investments (Wang et al., 2020b). Using UV-C has disadvantages of low depth penetration and possible personal health risks (Wang et al., 2020b). However, large scale applications have been successfully demonstrated using UV for wastewater disinfections (Hassen et al., 2000). According to a study, SARS-CoV-1 viral stocks placed in 24-well tissue culture plates (depth = 1 cm) suffers no significant effects of UV-A (365 nm emitting 2133 μW/cm2 at a distance of 3 cm) on its infectivity even after 15 min exposure. By contrast, UV-C (254 nm emitting 4016 μW/cm2 at a distance of 3 cm) partially inactivated the virus in 1 min and provided complete viral inactivation in 15 min (Darnell et al., 2004).
Gamma radiation has been used as an effective pathogen inactivation method for decades (Grieb et al., 2002). Gamma radiation acts by two mechanisms: (i) direct energy transfer by photons of the irradiations, (ii) inactivation of biological material via dislocation of electrons, covalent bond breakage, or by free radicals causing indirect damage (Grieb et al., 2002). Gamma radiation was also evaluated for the inactivation of SARS-CoV-1 (Darnell et al., 2004). A SARS-CoV-1 solution (400 μL of 106.33 TCID50 per mL) was subjected to 30, 50, 100, and 150 Gray (1 Gy = J/kg) of gamma radiation (source 60Co). But, viral infectivity did not have any significant effect even after 15 min exposure of gamma radiations (Darnell et al., 2004). A recent review of different solar energy systems for viral disinfection included 1) direct UV, and heat exposure. 2) photocatalytic/thermocatalytic/combined methods for disinfection for use in SARS-CoV-2 inactivation in water (Chauhan, 2020). Use of plasma discharge has also being suggested for SARS-CoV-2 inactivation (Ghernaout and Elboughdiri, 2020a).
5. Conclusions and recommendations
It has been 100 years since the Spanish Flu (H1N1 influenza A) virus swept around the world. In 1918, recommended defensive actions included quarantines, social distancing, banning of mass gatherings and the wearing of face masks (Tomes, 2010). There were no antiviral or antibiotics to treat the Spanish Flu virus or consequential infections. Today's world is more scientifically advanced, and scientists are focused on understanding, managing, treating, and eradicating the new COVID-19 disease. With the might of the modern scientific world mobilized, the amount of data being produced is massive and more is being added daily.
Herein we provided a literature snapshot focused on the occurrence, persistence, analysis, and possible management of coronavirus (SARS-CoV-2) in the aquatic systems. The reader will understand where the virus can be found in the environment including waters, wastewaters, and sewage sludge. We describe the range of methods used for virus sample collection, preservation, preparation, extraction, analysis, and detection. Virus eradication strategies are also addressed for wastewater treatment plants and other aquatic systems using advanced disinfection strategies, heat, and radiation. The questions and recommendations that follow are based on perceived research gaps in the exiting literature.
-
1.
A statistical study is needed to determine if contaminated surface waters, drinking water and wastewaters pose a route for human transition of COVID-19? How does this risk vary by country and region based on sewage collection and treatment processing and use of these waters with and without water treatment plants. Within these geographical areas how do the statistical risks of water transmission vary from urban to suburban to rural areas?
-
2.
Since SARS-CoV-2 has spread from bats to a variety of hosts, as well as to humans, what new risks does SARS-CoV-2 possess for humans? How will this expansion in hosts and new environments impact viral mutation rates and types of mutation?
-
3.
What new analytical methods or improvements in existing methodology or instrumentation are needed for labs in this field? How would large capacity, centralized analytical facilities versus smaller capacity, widely dispersed assets impact the study of COVID-19? What are the benefits and drawbacks to local, regional and national COVID-19 testing?
-
4.
Are there practical ways to relate virus loading detected in area water systems to known infections, their growth rates and quantitatively measure asymptomatic and presymtomatic case loads? Could this be an early warning system used to alert locals of potential outbreaks during the early phase, when most infected cases are still asymptomatic?
-
5.
Wastewater surveillance in Barcelona discovered the virus 41 days prior to the first COVID-19 case was announced (Chavarria-Miró et al., 2020). Application of wastewater based epidemiology (WBE) worldwide could help communities take immediate action including social distancing, mask wearing, hand washing, acquisition of protective gear (PPG), planning for increase load on intensive care unit (ICU) facilities and the stockpiling of associated equipment and supplies. Such planning should be a component of government agencies preparing for COVID responses worldwide.
-
6.
WBE provides a very low-cost screening of large populations which can follow a pandemic's spread and flow within defined catchments. Data can be collected rapidly with low expenditure of labor and potentially provide an estimation of the asymptomatic and pre-symptomatic individuals which more expensive clinical surveillance does not capture.
-
7.
The emergence of wastewater analysis studies in many countries to monitor SARS-CoV-2 is reported in Table 2. Still, proper WBE studies are lacking at both regional and global level to monitor pandemic situations. Several agencies from Australia, USA and European countries including Netherlands, Britain and Spain are formalizing/started national initiatives to develop early warning systems to tackle future COVID-like outbreaks. It's been >6 months since, the pandemic has spread over the entire planet and no proper WBE system has been developed and implemented to tackle situations like COVID. National and regional health agencies must develop WBE systems to monitor and warn citizens of possible hotspots and rising pandemic situations. These studies must be extended to archived samples as well. Should national health agencies and authorities have presented this data to the public to warn of emerging virus hotspots? For example, the states of Texas, Florida, Arizona, California and others (USA) are currently undergoing a drastic jump in cases as this paper is being written. But this is 5 months after outbreaks in North America were documented. Was there a large jump in SARS-CoV-2 RNA occurring in the influents to WWTPs one to three months previously, indicating a silent pre-advancing number of cases? Were all the relevant agencies monitoring this situation via wastewater surveillance? If not, why not? Going back earlier was WBE being used to monitor wastewaters in New York City, Detroit, New Orleans, etc. These same questions apply worldwide. What about New Delhi (India), Rio de Janeiro (Brazil), and other cities of Latin America, Russia etc.
Most puzzling to the authors of this review is why news media worldwide were not continuously asking questions about the application and results of WBE investigations to public officials, and government health agencies in their countries and cities. A continual news blitz, medical predictions and political commentary filled TV, radio, newspapers, magazines and social media. Individually, however, the authors did not see officials discuss results of WBE or appear to be using this data to warn or educate the public. Why one wonders wasn't WBE applied, or if it was applied, how were public officials were notified and how was the available data used? Finally, where were all branches of the omnipotent news media hiding on this topic?
Declaration of competing interest
All the Authors declarer no conflict of interests.
Acknowledgements
Financial support [DST/TM/WTI/2K15/121 (C) dated: 19.09.2016] in the project entitled “Removal and Recovery of Pharmaceuticals from water using sustainable magnetic and nonmagnetic biochars” from Department of Science and Technology, New Delhi, India is thankfully acknowledged. Authors are also thankful to University Grant Commission (UGC), New Delhi for providing the financial assistance under 21st Century Indo-US Research Initiative 2014 to Jawaharlal Nehru University, New Delhi and Mississippi State University, USA in the project “Clean Energy and Water Initiatives” [UGC No. F.194-1/2014(IC)]. One of the authors (DM) also thanks to Jawaharlal Nehru University for providing financial assistance under the Second phase of a University with Potential of Excellence (UPOEII) grant (ID 189). MP is thankful to CSIR for providing financial assistance under CSIR-SRF. Authors also acknowledge the funding support from DST PURSE, Government of India. One of the authors (DM) also thanks University Grant Commision for providing financial assistance under UGC- SAP DSA II.
Editor: Daniel CW Tsang
References
- Abdelzaher A.M., Solo-Gabriele H.M., Wright M.E., Palmer C.J. Sequential concentration of bacteria and viruses from marine waters using a dual membrane system. J. Environ. Qual. 2008;37:1648–1655. doi: 10.2134/jeq2007.0238. [DOI] [PubMed] [Google Scholar]
- Aboubakr H.A., Sharafeldin T.A., Goyal S.M. OSF Preprints. 2020. Stability of SARS-CoV2 and other coronaviruses in the environment and on common touch surfaces. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriaenssens E.M., Farkas K., Harrison C., Jones D.L., Allison H.E., McCarthy A.J. Viromic analysis of wastewater input to a river catchment reveals a diverse assemblage of RNA viruses. MSystems. 2018;3 doi: 10.1128/mSystems.00025-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739:139960. doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampuero M., Valenzuela S., Valiente-Echeverria F., Soto-Rifo R., Barriga G.P., Chnaiderman J., et al. SARS-CoV-2 detection in Sewage in Santiago, Chile-preliminary results. medRxiv. 2020 doi: 10.1101/2020.07.02.20145177. [DOI] [Google Scholar]
- Arora S., Nag A., Sethi J., Rajvanshi J., Saxena S., Shrivastava S.K., et al. Sewage surveillance for the presence of SARS-CoV-2 genome as a useful wastewater based epidemiology (WBE) tracking tool in India. medRxiv. 2020 doi: 10.1101/2020.06.18.20135277. [DOI] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodríguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., et al. The fate of SARS-CoV-2 in wastewater treatment plants points out the sludge line as a suitable spot for incidence monitoring. medRxiv. 2020 doi: 10.1101/2020.05.25.20112706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 2013;47:1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., et al. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54:7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Bofill-Mas S., Rusiñol M. Recent trends on methods for the concentration of viruses from water samples. Curr. Opin. Environ. Sci. Health. 2020;16:7–13. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A., Pintó R.M., Abad F.X. Viruses in Foods. Springer; 2006. Survival and transport of enteric viruses in the environment; pp. 151–187. [Google Scholar]
- Cahill N., Morris D. Recreational waters–a potential transmission route for SARS-CoV-2 to humans? Sci. Total Environ. 2020;740:140122. doi: 10.1016/j.scitotenv.2020.140122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Sun W., Huang J., Gamber M., Wu J., He G. Indirect virus transmission in cluster of COVID-19 cases, Wenzhou, China, 2020. Emerg. Infect. Dis. 2020;26:1343–1345. doi: 10.3201/eid2606.200412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Xu J., Lin D., Yang Z., Xu L., Qu Z., et al. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin. Infect. Dis. 2020;71(6):1547–1551. doi: 10.1093/cid/ciaa198. ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraturo F., Giudice C.D., Morelli M., Cerullo V., Libralato G., Galdiero E., et al. Persistence of SARS-CoV-2 in the environment and COVID-19 transmission risk from environmental matrices and surfaces. Environ. Pollut. 2020;265:115010. doi: 10.1016/j.envpol.2020.115010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L.M., Jeon S., Rutala W.A., Weber D.J., Sobsey M.D. Effects of air temperature and relative humidity on coronavirus survival on surfaces. Appl. Environ. Microbiol. 2010;76:2712–2717. doi: 10.1128/AEM.02291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Yip C.C.-Y., To K.K.-W., Tang T.H.-C., Wong S.C.-Y., Leung K.-H., et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Yuan S., Kok K.-H., To K.K.-W., Chu H., Yang J., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan V. Can Solar Energy take part in fighting against COVID-19 pandemic? A review on inactivation of SARS-CoV-2 in Water and Air using Solar Energy. EnerarXiv. 2020 http://www.enerarxiv.org/page/thesis.html?id=1907 [Google Scholar]
- Chavarria-Miró G., Anfruns-Estrada E., Guix S., Paraira M., Galofré B., Sáanchez G., et al. Sentinel surveillance of SARS-CoV-2 in wastewater anticipates the occurrence of COVID-19 cases. medRxiv. 2020 doi: 10.1101/2020.06.13.20129627. [DOI] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., et al. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1 doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collivignarelli M.C., Collivignarelli C., Miino M.C., Abbà A., Pedrazzani R., Bertanza G. SARS-CoV-2 in wastewater treatment plants. medRxiv. 2020;143:196–203. doi: 10.1016/j.psep.2020.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Albarrak A.M., Omrani A.S., Albarrak M.M., Farah M.E., Almasri M., et al. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin. Infect. Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell M.E.R., Taylor D.R. Evaluation of inactivation methods for severe acute respiratory syndrome coronavirus in noncellular blood products. Transfusion. 2006;46:1770–1777. doi: 10.1111/j.1537-2995.2006.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell M.E.R., Subbarao K., Feinstone S.M., Taylor D.R. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J. Virol. Methods. 2004;121:85–91. doi: 10.1016/j.jviromet.2004.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C. The international imperative to rapidly and inexpensively monitor community-wide Covid-19 infection status and trends. Sci. Total Environ. 2020;726:138149. doi: 10.1016/j.scitotenv.2020.138149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 2020;736:139631. doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhla M., Wilbring G., Schulte B., Kümmerer B.M., Diegmann C., Sib E., et al. SARS-CoV-2 in environmental samples of quarantined households. medRxiv. 2020 doi: 10.1101/2020.05.28.20114041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Hillary L.S., Malham S.K., McDonald J.E., Jones D.L. Wastewater and public health: the potential of wastewater surveillance for monitoring COVID-19. Curr. Opin. Environ. Sci. Health. 2020;17:14–20. doi: 10.1016/j.coesh.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Mannion F., Hillary L.S., Malham S.K., Walker D.I. Emerging technologies for the rapid detection of enteric viruses in the aquatic environment. Curr. Opin. Environ. Sci. Health. 2020;16:1–6. [Google Scholar]
- Fears A.C., Klimstra W.B., Duprex P., Hartman A., Weaver S.C., Plante K.C., et al. Comparative dynamic aerosol efficiencies of three emergent coronaviruses and the unusual persistence of SARS-CoV-2 in aerosol suspensions. medRxiv. 2020 doi: 10.1101/2020.04.13.20063784. [DOI] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., et al. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743:140444. doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fongaro G., Stoco P.H., Souza D.S.M., Grisard E.C., Magri M.E., Rogovski P., et al. SARS-CoV-2 in human sewage in Santa Catalina, Brazil, November 2019. medRxiv. 2020 doi: 10.1101/2020.06.26.20140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer R., Jambeck J.R., Law K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghernaout D., Elboughdiri N. Disinfecting water: plasma discharge for removing coronaviruses. Open Access Libr. J. 2020;7:1–29. [Google Scholar]
- Ghernaout D., Elboughdiri N. Urgent proposals for disinfecting hospital wastewaters during COVID-19 pandemic. Open Access Libr. J. 2020;7:1–18. [Google Scholar]
- Ghernaout D., Wahib M., Aouabed N.A. On the dependence of chlorine by-products generated species formation of the electrode material and applied charge during electrochemical water treatment. Desalination. 2011;270:9–22. [Google Scholar]
- Ghernaout D., Algahmdi A., Aichouni M., Touahmia M. The lethal water tri-therapy: chlorine, alum, and polyelectrolyte. World J. Appl. Chem. 2018;3:65–71. [Google Scholar]
- Goldman E. Exaggerated risk of transmission of COVID-19 by fomites. Lancet Infect. Dis. 2020;20:892–893. doi: 10.1016/S1473-3099(20)30561-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetk K., et al. COVID-19 surveillance in southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186:116296. doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormley M. SARS-CoV-2: the growing case for potential transmission in a building via wastewater plumbing systems. Ann. Intern. Med. 2020 doi: 10.7326/M20-6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormley M., Swaffield J.A., Sleigh P.A., Noakes C.J., Meche M.I. An assessment of, and response to, potential cross-contamination routes due to defective appliance water trap seals in building drainage systems. Build. Serv. Eng. Res. Technol. 2012;33:203–222. [Google Scholar]
- Gormley M., Templeton K.E., Kelly D.A., Hardie A. Environmental conditions and the prevalence of norovirus in hospital building drainage system wastewater and airflows. Build. Serv. Eng. Res. Technol. 2014;35:244–253. [Google Scholar]
- Gormley M., Aspray T.J., Kelly D.A. COVID-19: mitigating transmission via wastewater plumbing systems. Lancet Glob. Health. 2020;8 doi: 10.1016/S2214-109X(20)30112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H., Wilder M., Middleton F.A., Collins M., Fenty A., Gentile K., et al. Quantification of SARS-CoV-2 and cross-assembly phage (crAssphage) from wastewater to monitor coronavirus transmission within communities. medRxiv. 2020 doi: 10.1101/2020.05.21.20109181. [DOI] [Google Scholar]
- Grieb T., Forng R.-Y., Brown R., Owolabi T., Maddox E., Mcbain A., et al. Effective use of gamma irradiation for pathogen inactivation of monoclonal antibody preparations. Biologicals. 2002;30:207–216. doi: 10.1006/biol.2002.0330. [DOI] [PubMed] [Google Scholar]
- Guerrero-Latorre L., Ballesteros I., Villacres I., Granda-Albuja M.G., Freire B., Rios-Touma B. First SARS-CoV-2 detection in river water: implications in low sanitation countries. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1:10. [Google Scholar]
- Gupta S., Parker J., Smits S., Underwood J., Dolwani S. Persistent viral shedding of SARS-CoV-2 in faeces-a rapid review. Color. Dis. 2020;22:611–620. doi: 10.1111/codi.15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale R.C., Song B. Single-use plastics and COVID-19: scientific evidence and environmental regulations. Environ. Sci. Technol. 2020;54:7034–7036. doi: 10.1021/acs.est.0c02269. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., et al. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapan H., Itoh N., Yufika A., Winardi W., Keam S., HayphengTe, et al. Coronavirus disease 2019 (COVID-19): a literature review. J. Infect. Public Health. 2020;13:667–673. doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730:138875. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Modeling wastewater temperature and attenuation of sewage-borne biomarkers globally. Water Res. 2020;172:115473. doi: 10.1016/j.watres.2020.115473. [DOI] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Simulated 2017 nationwide sampling at 13,940 major US sewage treatment plants to assess seasonal population bias in wastewater-based epidemiology. Sci. Total Environ. 2020;727:138406. doi: 10.1016/j.scitotenv.2020.138406. [DOI] [PubMed] [Google Scholar]
- Hassen A., Mahrouk M., Ouzari H., Cherif M., Boudabous A., Damelincourt J.J. UV disinfection of treated wastewater in a large-scale pilot plant and inactivation of selected bacteria in a laboratory UV device. Bioresour. Technol. 2000;74:141–150. [Google Scholar]
- Hata K., Honda R. Potential sensitivity of wastewater monitoring for SARS-CoV-2: comparison with norovirus cases. Environ. Sci. Technol. 2020;54:6451–6452. doi: 10.1021/acs.est.0c02271. [DOI] [PubMed] [Google Scholar]
- Hata A., Honda R., Hara-Yamamura H., Meuchi Y. Detection of SARS-CoV-2 in wastewater in Japan by multiple molecular assays-implication for wastewater-based epidemiology (WBE) medRxiv. 2020 doi: 10.1101/2020.06.09.20126417. [DOI] [Google Scholar]
- Heller L., Mota C.R., Greco D.B. COVID-19 faecal-oral transmission: are we asking the right questions? Sci. Total Environ. 2020;729:138919. doi: 10.1016/j.scitotenv.2020.138919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson J. COVID-19: faecal–oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2020;17:259. doi: 10.1038/s41575-020-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I.F.N., Cheng V.C.C., Wu A.K.L., Tang B.S.F., Chan K.H., Chu C.M., et al. Viral loads in clinical specimens and SARS manifestations. Emerg. Infect. Dis. 2004;10:1550. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S., Kitamura G., Segawa T., Kobayashi A., Miura T., Sano D., et al. Microfluidic quantitative PCR for simultaneous quantification of multiple viruses in environmental water samples. Appl. Environ. Microbiol. 2014;80:7505–7511. doi: 10.1128/AEM.02578-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalandraad R., Yadav A.K., Verma D., Dalala N., Sharma M., Singh R., et al. Strategic and perspectives to develop SARS-CoV-2 detection methods and diagnostics. Biomed. Pharmacother. 2020;129:110446. doi: 10.1016/j.biopha.2020.110446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Fang L., Shi X., Zhang H., Li Y., Lin Y., et al. Simultaneous detection of five enteric viruses associated with gastroenteritis by use of a PCR assay: a single real-time multiplex reaction and its clinical application. J. Clin. Microbiol. 2014;52:1266–1268. doi: 10.1128/JCM.00245-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp G. Potential role of inanimate surfaces for the spread of coronaviruses and their inactivation with disinfectant agents. Infect. Prev. Pract. 2020;2:100044. doi: 10.1016/j.infpip.2020.100044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G., Voss A., Scheithauer S. Inactivation of coronaviruses by heat. J. Hosp. Infect. 2020;105:348–349. doi: 10.1016/j.jhin.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M., Wei J., Yuan J., Guo J., Zhang Y., Hang J., et al. Probable evidence of fecal aerosol transmission of SARS-CoV-2 in a high-rise building. Ann. Intern. Med. 2020 doi: 10.7326/M20-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariwa H., Fujii N., Takashima I. Inactivation of SARS coronavirus by means of povidone-iodine, physical conditions and chemical reagents. Dermatology. 2006;212:119–123. doi: 10.1159/000089211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasloff S.B., Strong J.E., Funk D., Cutts T.A. Stability of SARS-CoV-2 on critical personal protective equipment. medRxiv. 2020 doi: 10.1101/2020.06.11.20128884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocamemi B.A., Kurt H., Hacioglu S., Yarali C., Saatci A.M., Pakdemirli B. First data-set on SARS-CoV-2 detection for Istanbul wastewaters in Turkey. medRxiv. 2020 doi: 10.1101/2020.05.03.20089417. [DOI] [Google Scholar]
- Kocamemi B.A., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. SARS-CoV-2 detection in Istanbul wastewater treatment plant sludges. medRxiv. 2020 doi: 10.1101/2020.05.12.20099358. [DOI] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., et al. The first proof of the capability of wastewater surveillance for COVID-19 in India through the detection of the genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Fratini M., Sd Libera, Iaconelli M., Muscillo M. Emerging and potentially emerging viruses in water environments. Ann. Ist. Super. Sanita. 2012;48:397–406. doi: 10.4415/ANN_12_04_07. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods-a scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L., et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A.C.K., Tan T.F., Li W.S., Ip D.K.M. Emission strength of airborne pathogens during toilet flushing. Indoor Air. 2018;28:73–79. doi: 10.1111/ina.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq I., Batéjat C., Burguière A.M., Manuguerra J.-C. Heat inactivation of the Middle East respiratory syndrome coronavirus. Influenza Other Respir. Viruses. 2014;8:585–586. doi: 10.1111/irv.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure F.-X., Bouadma L., Nguyen D., Parisey M., Wicky P.-H., Behillil S., et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect. Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W.K., To K.-F., Chan P.K.S., Chan H.L.Y., Wu A.K.L., Lee N., et al. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-y., Wang J.-X., Chen X. Can a toilet promote virus transmission? From a fluid dynamics perspective. Phys. Fluids. 2020;32 doi: 10.1063/5.0013318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y., Xu S.-B., Lin Y.-X., Tian D., Zhu Z.-Q., Dai F.-H., et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020;133:1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Yao L., Zhang L., Yao M., Chen X., Wang Q., et al. Possible transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a public bath center in Huai’an, Jiangsu Province, China. JAMA Netw. Open. 2020;3:e204583. doi: 10.1001/jamanetworkopen.2020.4583. [DOI] [PubMed] [Google Scholar]
- Mahabee-Gittens E.M., Merianos A.L., Matt G.E. Letter to the editor regarding: “an imperative need for research on the role of environmental factors in transmission of novel coronavirus (COVID-19)”—secondhand and thirdhand smoke as potential sources of COVID-19. Environ. Sci. Technol. 2020;54:5309–5310. doi: 10.1021/acs.est.0c02041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor Y., Shulman L.M., Kaliner E., Hindiyeh M., Ram D., Sofer D., et al. Intensified environmental surveillance supporting the response to wild poliovirus type 1 silent circulation in Israel, 2013. Eurosurveillance. 2014;19:20708. doi: 10.2807/1560-7917.es2014.19.7.20708. [DOI] [PubMed] [Google Scholar]
- Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020;54:3733–3735. doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Meselson M. Droplets and aerosols in the transmission of SARS-CoV-2. N. Engl. J. Med. 2020;382:2063. doi: 10.1056/NEJMc2009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans C.C.E. The basic principles of UV–disinfection of water. Ozone Sci. Eng. 1987;9:299–313. [Google Scholar]
- Michael-Kordatou I., Karaolia P., Fatta-Kassinosa D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020;8:104306. doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25:2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlejnkova H., Sovova K., Vasickova P., Ocenaskova V., Jasikova L., Juranova E. Preliminary study of Sars-Cov-2 occurrence in wastewater in the Czech Republic. Int. J. Environ. Res. Public Health. 2020;17:5508. doi: 10.3390/ijerph17155508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naddeo V., Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci.: Water Res. Technol. 2020;6:1213–1216. [Google Scholar]
- Nalla A.K., Casto A.M., Huang M.-L.W., Perchetti G.A., Sampoleo R., Shrestha L., et al. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., et al. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100098. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem L.D., Morgan B., Donner E., Short M.D. Case Studies in Chemical and Environmental Engineering. Vol. 1. 2020. The COVID-19 pandemic: considerations for the waste and wastewater services sector; p. 100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Kobayashi T., Miyama T., Suzuki A., Jung Sung-mok, Hayashi K., et al. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int. J. Infect. Dis. 2020;94:154–155. doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez-Delgado A. What do we know about the SARS-CoV-2 coronavirus in the environment? Sci. Total Environ. 2020;727 doi: 10.1016/j.scitotenv.2020.138647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.X., Tan Y.K., Chia P.Y., Lee T.H., Ng O.T., YenWong M.S., et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA. 2020;323:1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Or I.B., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., et al. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: a proof-of-concept for quantitative environmental surveillance. medRxiv. 2020 doi: 10.1101/2020.04.26.20073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino B., Touret F., Gilles M., Lamballerie Xd, Charrel R.N. Evaluation of heating and chemical protocols for inactivating SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.04.11.036855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.J., Zhao G., Penna V.R., Park E., Lauron E.J., Harvey I.B., et al. A murine herpesvirus closely related to ubiquitous human herpesviruses causes T-cell depletion. J. Virol. 2017;91 doi: 10.1128/JVI.02463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M., Kumar R., Kishor K., Mlsna T., Pittman C.U., Jr., et al. Pharmaceuticals of emerging concern in aquatic systems: chemistry, occurrence, effects, and removal methods. Chem. Rev. 2019;119:3510–3673. doi: 10.1021/acs.chemrev.8b00299. [DOI] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., et al. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. medRxiv. 2020 doi: 10.1101/2020.05.19.20105999. [DOI] [Google Scholar]
- Peiris J.S.M., Chu C.M., Cheng V.C.C., Chan K.S., Hung I.F.N., Poon L.L.M., et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Maranhão A.G., Siqueira M.M., Miagostovich M.P. Preliminary results of SARS-CoV-2 detection in sewerage system in Niterói municipality, Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz. 2020;115 doi: 10.1590/0074-02760200196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuan R., Domingo-Calap P., Sanchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health. 2020;230 doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., et al. Presence and vitality of SARS-CoV-2 virus in wastewaters and rivers. Science of The Total Environment Volume. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano-Bertrand S., Glele L.-S.A., Grandbastien B., Lepelletiere D. Preventing SARS-CoV-2 transmission in rehabilitation pools and therapeutic water environments. J. Hosp. Infect. 2020;105:625–627. doi: 10.1016/j.jhin.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia J.L., Rivera D.N., Herrera V.L., Morwitzer M.J., Creager H.M., Santarpia G.W., et al. Aerosol and surface contamination of SARS-CoV-2 observed in quarantine and isolation care. Sci. Rep. 2020;10:1–8. doi: 10.1038/s41598-020-69286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller C., Krebs F., Minkner R., Astner I., Gil-Moles M., Wätzig H. Physicochemical properties of SARS-CoV-2 for drug targeting, virus inactivation and attenuation, vaccine formulation and quality control. Electrophoresis. 2020;41:1137–1151. doi: 10.1002/elps.202000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehmi P., Cheruiyot I. Presence of live SARS-CoV-2 virus in feces of coronavirus disease 2019 (COVID-19) patients: a rapid review. medRxiv. 2020 doi: 10.1101/2020.06.27.20105429. [DOI] [Google Scholar]
- Setti L., Passarini F., Gennaro G.D., Barbieri P., Grazi M., Borelli P.M., et al. SARS-Cov-2RNA found on particulate matter of Bergamo in northern Italy: first evidence. Environ. Res. 2020;188 doi: 10.1016/j.envres.2020.109754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif S., Ikram A., Khurshid A., Salman M., Mehmood N., Arshad Y., et al. Detection of SARS-Coronavirus-2 in wastewater, using the existing environmental surveillance network: An epidemiological gateway to an early warning for COVID-19 in communities. medRxiv. 2020 doi: 10.1101/2020.06.03.20121426. [DOI] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., et al. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutler J., Zaraska K., Holding T.M., Machnik M., Uppuluri K., Ashton I., et al. Risk of SARS-CoV-2 infection from contaminated water systems. medRxiv. 2020 doi: 10.1101/2020.06.17.20133504. [DOI] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A., Wray R. Detection and survival of SARS-coronavirus in human stool, urine, wastewater and sludge. Preprints. 2020 doi: 10.20944/preprints202006.0216.v2. [DOI] [Google Scholar]
- Sun J., Zhu A., Li H., Zheng K., Zhuang Z., Chen Z., et al. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg. Microbes Infect. 2020;9:991–993. doi: 10.1080/22221751.2020.1760144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z.-d., Wang H.-l., Dai Y.-x., Li K.-f., Liu J.-n., et al. Early release-detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomes N. “Destroyer and teacher”: managing the masses during the 1918–1919 influenza pandemic. Public Health Rep. 2010;125:48–62. doi: 10.1177/00333549101250S308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA . Agency USEP; 2020. List N: Disinfectants for Use Against SARS-CoV-2 (COVID-19) [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., et al. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR assays. medRxiv. 2020 doi: 10.1101/2020.03.30.20048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-W., Li J.-S., Jin M., Zhen B., Kong Q.-X., Song N., et al. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-X., Li Y.-Y., Liu X.-D., Cao X. Virus transmission from urinals. Phys. Fluids. 2020;32 doi: 10.1063/5.0021450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Lien C., Liu S., Selveraj P. Effective heat inactivation of SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.04.29.20085498. [DOI] [Google Scholar]
- Wang J., Shen J., Ye D., Yan X., Zhang Y., Yang W., et al. Disinfection technology of hospital wastes and wastewater: suggestions for disinfection strategy during coronavirus disease 2019 (COVID-19) pandemic in China. Environ. Pollut. 2020;262 doi: 10.1016/j.envpol.2020.114665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber D.J., Rutala W.A., Fischer W.A., Kanamori H., Sickbert-Bennett E.E. Emerging infectious diseases: focus on infection control issues for novel coronaviruses (Severe Acute Respiratory Syndrome-CoV and Middle East Respiratory Syndrome-CoV), hemorrhagic fever viruses (Lassa and Ebola), and highly pathogenic avian influenza viruses, A (H5N1) and A (H7N9) Am. J. Infect. Control. 2016;44:e91–e100. doi: 10.1016/j.ajic.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; Geneva: 2003. Consensus document on the epidemiology of severe acute respiratory syndrome (SARS) p. 46. [Google Scholar]
- WHO . World Health Organization; Geneva: 2020. Coronavirus disease (COVID-19) pandemic.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Google Scholar]
- WHO . WHO Coronavirus Disease (COVID-19) Dashboard. World Health Organization; Geneva: 2020. https://covid19.who.int/ [Google Scholar]
- Wigginton K.R., Boehm A.B. Environmental engineers and scientists have important roles to play in stemming outbreaks and pandemics caused by enveloped viruses. Environ. Sci. Technol. 2020;54:3736–3739. doi: 10.1021/acs.est.0c01476. [DOI] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv. 2020 doi: 10.1101/2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Maday Y., Teyssou R., Richard E., et al. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. medRxiv. 2020 doi: 10.1101/2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Moulin L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. medRxiv. 2020 doi: 10.1101/2020.04.12.20062679. [DOI] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. (e3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaqub T., Nawaz M., Shabbir M.Z., Ali M.A., Altaf I., Raza S., et al. A longitudinal survey for genome-based identification of SARS-CoV-2 in sewage water in selected lockdown areas of Lahore city, Pakistan; a potential approach for future smart lockdown strategy. medRxiv. 2020 doi: 10.1101/2020.07.31.20165126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-mei M., Wei Z., Hong-yan L. Application progress of hospital wastewater treatment methods. Occup. Health. 2010;26:1180–1182. [Google Scholar]
- Zaneti R.N., Girardi V., Spilki F.R., Mena K., Westphalen A.P.C., ERdC Colares, et al. QMRA of SARS-CoV-2 for workers in wastewater treatment plants. medRxiv. 2020 doi: 10.1101/2020.05.28.20116277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen C., Zhu S., Shu C., Wang D., Song J., et al. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Weekly. 2020;2:123–124. [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., et al. Potential spreading risks and disinfection challenges of medical wastewater by the presence of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of fangcang hospital. Sci. Total Environ. 2020;741 doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]