Food animal reservoirs contribute to Shiga toxin-producing Escherichia coli (STEC) evolution via the acquisition of horizontally acquired elements like Shiga toxin bacteriophages that enhance pathogenicity. In cattle, persistent fecal shedding of STEC contributes to contamination of beef and dairy products and to crops being exposed to contaminated water systems. Hence, identifying factors important for STEC persistence is critical. This longitudinal study enhances our understanding of the genetic diversity of STEC types circulating in a cattle herd and identifies genotypic and phenotypic traits associated with persistence. Key findings demonstrate that multiple STEC types readily persist in and are transmitted across cattle in a shared environment. These dynamics also enhance the persistence of virulence genes that can be transferred between bacterial hosts, resulting in the emergence of novel STEC strain types. Understanding how pathogens persist and diversify in reservoirs is important for guiding new preharvest prevention strategies aimed at reducing foodborne transmission to humans.
KEYWORDS: Escherichia coli, Shiga toxins, biofilms, cattle, genomics
ABSTRACT
Shiga toxin-producing Escherichia coli (STEC) is a leading cause of foodborne infections. Cattle are an important STEC reservoir, although little is known about specific pathogen traits that impact persistence in the farm environment. Hence, we sought to evaluate STEC isolates recovered from beef cattle in a single herd in Michigan. To do this, we collected fecal grabs from 26 cattle and resampled 13 of these animals at 3 additional visits over a 3-month period. In all, 66 STEC isolates were recovered for genomics and biofilm quantification using crystal violet assays. The STEC population was diverse, representing seven serotypes, including O157:H7, O26:H11, and O103:H2, which are commonly associated with human infections. Although a core genome analysis of 2,933 genes grouped isolates into clusters based on serogroups, some isolates within each cluster had variable biofilm levels and virulence gene profiles. Most (77.8%; n = 49) isolates harbored stx2a, while 38 (57.5%) isolates formed strong biofilms. Isolates belonging to the predominant serogroup O6 (n = 36; 54.5%) were more likely to form strong biofilms, persistently colonize multiple cattle, and be acquired over time. A high-quality single nucleotide polymorphism (SNP) analysis of 33 O6 isolates detected between 0 and 13 single nucleotide polymorphism (SNP) differences between strains, indicating that highly similar strain types were persisting in this herd. Similar findings were observed for other persistent serogroups, although key genes were found to differ among strong and weak biofilm producers. Together, these data highlight the diversity and persistent nature of some STEC types in this important food animal reservoir.
IMPORTANCE Food animal reservoirs contribute to Shiga toxin-producing Escherichia coli (STEC) evolution via the acquisition of horizontally acquired elements like Shiga toxin bacteriophages that enhance pathogenicity. In cattle, persistent fecal shedding of STEC contributes to contamination of beef and dairy products and to crops being exposed to contaminated water systems. Hence, identifying factors important for STEC persistence is critical. This longitudinal study enhances our understanding of the genetic diversity of STEC types circulating in a cattle herd and identifies genotypic and phenotypic traits associated with persistence. Key findings demonstrate that multiple STEC types readily persist in and are transmitted across cattle in a shared environment. These dynamics also enhance the persistence of virulence genes that can be transferred between bacterial hosts, resulting in the emergence of novel STEC strain types. Understanding how pathogens persist and diversify in reservoirs is important for guiding new preharvest prevention strategies aimed at reducing foodborne transmission to humans.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) is a foodborne pathogen that can cause a wide range of disease outcomes from hemorrhagic colitis to hemolytic uremic syndrome (HUS) (1). Since the first STEC O157:H7 outbreak in 1982 linked to ground beef (2), numerous outbreaks have been associated with cattle products, such as beef, milk, and cheese (3–5), or with cattle directly (6). In the United States, six non-O157 serogroups, namely, O26, O45, O103, O111, O121, and O145, which are denoted the “big six,” have increased in frequency (7) and predominate among non-O157 infections (8). Although cattle have been implicated as an important reservoir for STEC, other ruminants and farm animals can harbor STEC (9, 10); contact with the farming environment has also been reported as a risk factor (11). The high prevalence of STEC in cattle and agriculture settings supports the need to understand the environmental niche that is occupied in order to develop new strategies to minimize the risk of transmission to humans.
Environmental factors and farm practices may impact the prevalence of STEC within a farm regardless of the strain serotype and genetic composition. Warmer temperatures in the summer months have been linked to higher frequencies of STEC (12, 13). Variation in STEC frequencies has also been observed across herds and geographic locations. Indeed, STEC prevalence was reported in 44.4% and 4.1% to 10.5% of beef feedlots and in 12.6% and 9.2% to 18.3% of dairy farms in South Korea and Nebraska, respectively (14, 15). Our prior study in Michigan also showed that beef feedlot farms had a higher prevalence of STEC than dairy farms (13), although this distribution was contrary to findings from Washington state (16). In addition, a prior study of Midwestern cattle farms reported a higher prevalence of non-O157 serogroups (19.3%) than O157 serogroups (12.9%) (12), which was similar to our data from Michigan cattle (13).
Variation in STEC prevalence estimates across cattle populations as well as the identification of hundreds of serogroups suggests that there is a considerable degree of bacterial genetic variation among isolates recovered in these environments. Genetic variation also contributes to differences in STEC phenotypes, including biofilm formation, which is an important survival mechanism against antibiotics, bacteriophages, and environmental stressors. Biofilm formation in STEC has been hypothesized to contribute to persistence and has been shown to facilitate survival in cattle water troughs (17) and on food processing equipment (18, 19). The ability to persist within a biofilm has also been shown to enhance the likelihood of horizontal gene transfer across strains (20). To form a biofilm, STEC has been shown to utilize different classes of adhesins, including type 1 fimbriae and curli, to initiate attachment to surfaces (21, 22), while surface proteins, such as the lipopolysaccharide (LPS), were also found to be important (23, 24). Moreover, production of the exopolysaccharide matrix was deemed necessary for biofilm maturation (25). Although it is logical to assume that biofilm formation may play a role in STEC persistence in the farm environment, prior studies have not examined the ability of different serotypes to form biofilms in cattle or assessed the role that genetic variation plays on biofilm production in a given environment or reservoir.
Despite cattle being implicated as an important reservoir for STEC, little is known about the transmission and persistence of specific strain types within this reservoir host. Characterizing STEC strains from cattle collected at multiple time points is critical for identifying virulence attributes that are important for persistence within a herd. Additionally, persistent STEC populations can contribute to the emergence of novel or antibiotic-resistant pathogens via the transfer of Shiga toxin-encoding bacteriophages or resistance genes from STEC or other bacterial species. Through this study, we aimed to examine the genotypic and phenotypic profiles of longitudinally collected STEC strains from cattle to give insight on persistent strain types in the farm environment and identify genetic similarities between cattle-derived and clinical isolates.
RESULTS
STEC virulence characteristics.
Among the 66 STEC isolates recovered for characterization, 7 serogroups were identified with serogroup O6 (n = 36; 57.1%) predominating, followed by O26, O168, and O103 serogroups (Table 1). Three additional isolates were classified as O157, O8, and O185, while three were nontypeable (NT) because of incomplete wzy/wzx sequences. Although these genes were present, incomplete sequencing prevented in silico classification of the O-antigen type or serogroup. The same was true for the H antigen gene fliC in two isolates. All but 1 of the 11 O26 isolates had complete whole-genome sequencing (WGS) data, leaving a total of 63 STEC isolates available for genomic analyses.
TABLE 1.
Serotypes and virulence gene profiles of Shiga toxin-producing Escherichia coli isolates recovered from beef cattle
| Serogroupa | Total no. of isolates (%) | stx profile | eae variant | ehxA subtype |
|---|---|---|---|---|
| O6:H34 | 36 (57.1) | 2c | ||
| O8:H19 | 1 (1.6) | 2a | A | |
| O26:H11 | 10 (15.9) | 1a | Beta | C |
| O103:H2 | 5 (7.9) | 1a (n = 4), 1a,2a (n = 1) | Epsilon | Cb |
| O157:H7 | 1 (1.6) | 1a,2a | Gamma | B |
| O168:H8 | 8 (12.7) | 2a | ||
| O185:H7 | 1 (1.6) | 2a | ||
| ONT:H8 | 1 (1.6) | 2a | A |
Virulence gene alleles were not evaluated for two nontypeable (NT) isolates and one O26 isolate because they lacked whole-genome sequencing data.
One O103:H2 isolate with stx1a lacked ehxA.
The 63 isolates had diverse virulence gene profiles that varied by serogroup. The gene encoding Stx2 (stx2) was detected in 77.8% (n = 49) of the isolates. All O168 isolates (n = 8) as well as the O8 (n = 1), O185 (n = 1), ONT (n = 1), and O157 (n = 1) isolates contained the stx2a variant, while the 36 O6 isolates had stx2c. Only three serogroups, namely, O26 (n = 10, 100%, eae: beta), O103 (n = 5, 100%, eae: epsilon), and O157 (n = 1, 100%, eae: gamma), harbored stx1a. All but one O103 isolate from these three serogroups had distinct eae alleles and were positive for ehxA. Only two isolates belonging to serogroups O157 and O103 had both stx1 and stx2.
Biofilm formation.
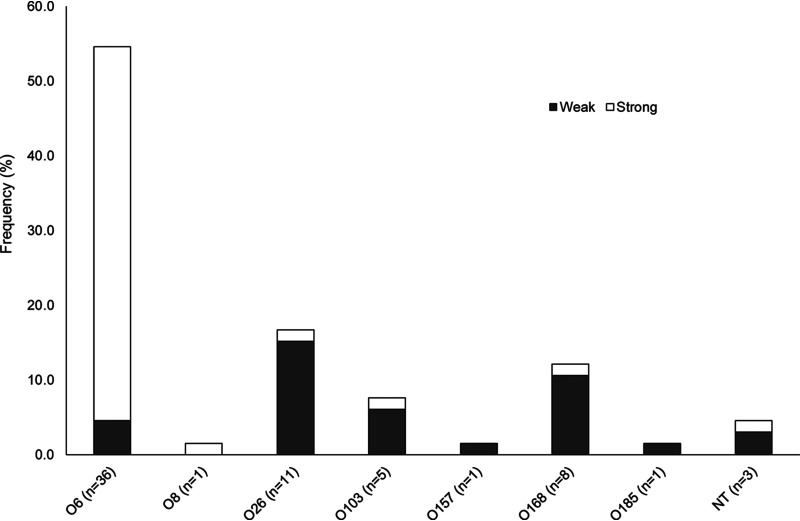
Static biofilm assays performed on all 66 STEC isolates resulted in a range of absorbance values from 0.15 to 5.93. Plotting the absorbance values identified a distinct break in the distribution of data points at an absorbance of 2.0, which is close to the average of 2.48. Therefore, high/strong biofilm production was classified by absorbance at 595 nm (A595) values greater than 2.0, and low/weak production was classified by A595 values less than 2.0. Roughly 38 (57.6%) isolates were classified as strong biofilm formers, while the remaining 28 (42.4%) were classified as weak. In all, there was a range in biofilm production across and within the serogroups (Fig. 1). Isolates representing serogroups O26 (n = 11), O103 (n = 5), and O168 (n = 8) were mainly weak biofilm formers, although 1 isolate in each of the 3 serogroups was classified as a strong biofilm former. Notably, the serogroup O6 isolates were significantly more likely to form strong biofilms relative to all other serogroups (Fisher’s exact test, P ≤ 0.0001). Variation in biofilm production, however, was observed among these O6 isolates, as three (4.5%) were classified as weak biofilm formers with values of 0.25, 1.20, and 1.90. The remaining 33 O6 strains had strong biofilm production ranging between 2.48 and 4.91.
FIG 1.
Level of biofilm production in Shiga toxin-producing Escherichia coli isolates recovered from beef cattle by serogroup.
Genetic diversity of cattle-derived STEC.
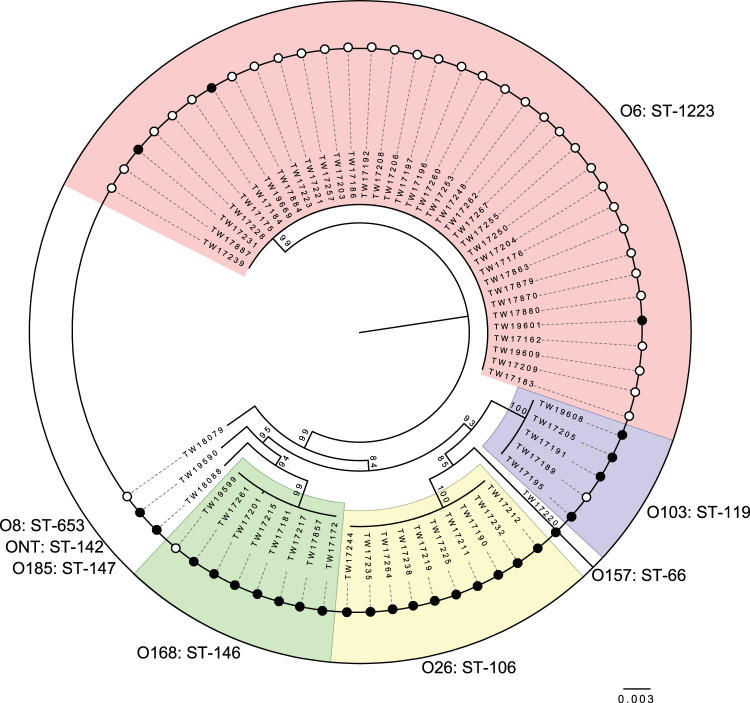
Multilocus sequence type (MLST) loci sequences were first extracted to classify isolates into sequence types (STs). Eight STs were identified among the 62 isolates with sequencing data available; one additional O6 isolate was excluded from the MLST analysis due to missing loci sequences. Interestingly, all O6 isolates were classified as a new genotype, ST-1223, due to a G-T SNP present in mdh at nucleotide 300 that generated a new mdh allele. In all cases, the STs were identical among isolates of the same serogroup. The 5 O103 isolates, for example, comprised ST-119, the 8 O168 isolates represented ST-146, and the 10 O26 isolates were classified as ST-106. The ONT isolate along with those representing serogroups O8, O157, and O185 had unique STs.
To better understand the genetic relatedness of the cattle-derived isolates, a reference-free pangenome analysis was performed; all but the two NT isolates lacking WGS data were included in this analysis. Construction of a maximum likelihood phylogeny of 2,933 core concatenated genes, which were shared across the 64 isolates, identified 4 clades, or clusters, with significant (>99%) bootstrap support (Fig. 2). Each cluster was comprised of strains representing the same STs and serogroups. Although one O26 and one O6 isolate could not be evaluated by MLST due to a missing gene sequence, both isolates clustered with strains of the same ST and serogroup in the core genome analysis. Consequently, the ST was inferred for these two isolates. No clustering was observed among the four isolates with unique serogroups, which were represented as singletons in the phylogeny. Although the long branch length for the O157 isolate (TW17220) indicates that it is more distantly related to the other non-O157 STEC isolates in this analysis, it grouped together (93% bootstrap support) with the two big six serogroup clusters. In addition, specific virulence gene profiles were identical among strains in each cluster, although one exception was noted. One of the five O103 isolates, TW17191, differed slightly in that it possessed both stx1 and stx2, unlike the rest of the isolates, which only harbored stx1. Moreover, the variation in biofilm production observed among isolates among three of the four serogroups did not affect the clustering in the phylogeny.
FIG 2.
Maximum likelihood phylogeny of 2,933 concatenated genes in 62 Shiga toxin-producing Escherichia coli isolates from beef cattle. The phylogeny was constructed with 1,000 bootstrap replications. Serogroup and multilocus sequence type (ST) designations are indicated for each cluster and high (open circles) and low (colored circles) biofilm formation. Clusters containing multiple isolates belonging to the same ST and serogroup, indicated as groups sharing the same colored shading, had ≥99% bootstrap support.
STEC prevalence and persistence in beef cattle.
The prevalence of STEC varied across samplings, and some animals were colonized with multiple strain types representing unique serogroups, biofilm levels, and virulence gene profiles. Among the 26 cattle sampled at the first visit, 19 (35.2%) were positive for STEC, although 20 unique isolates were recovered. Indeed, animal 776 had an O103:H2 as well as an O26:H11 isolate, and both isolates harbored stx1 and eae. Animal 776 along with 12 others, however, were not included in the longitudinal sampling, although their isolate profiles were included to evaluate the possibility of transmission to other animals sampled longitudinally. The following serotypes were recovered from these 12 cattle sampled only at visit 1: O6:H34 (n = 7), O8:H19 (n = 1), O168:H8 (n = 2), ONT:HNT (n = 1), and ONT:H8 (n = 1).
At visit 2, 8 of the 13 (61.5%) cattle followed longitudinally were culture positive for STEC; 16 isolates were recovered (Table 2). More than one serotype was isolated from the fecal samples of four animals. Three animals also possessed isolates belonging to the same serotype but had different levels of biofilm production. Animal 752, for example, had four isolates with various virulence gene profiles or biofilm levels representing only two serotypes, whereas animals 753 and 767 had two distinct serotypes. Animal 763 had two O6:H32 isolates with various biofilm levels and one ONT:HNT isolate with a distinct stx profile. At visits 3 and 4, 12 (92.0%) and 9 (69.2%) animals were STEC positive, respectively. Four of the cattle sampled at visit three had more than one unique STEC isolate as did two animals sampled at the fourth and final visit. In all, 10 of the 13 cattle had more than one isolate with a unique serogroup, virulence gene, and/or biofilm level profile recovered from one sample at one visit. Only animals 751, 758, and 759 had one isolate recovered per sample.
TABLE 2.
Dynamics of Shiga toxin-producing Escherichia coli shedding in beef cattle by visit, serotype, and biofilm productiona
The serotype, Shiga toxin gene profile (e.g., stx1, stx2, and stx1/2), and presence of eae are indicated for each isolate. Isolates belonging to the same serotype are represented by the same color. ONT, nontypeable O antigen; HNT, nontypeable H antigen. Isolates with strong biofilm production are indicated with an asterisk (*).
Animal 752 had 2 O6:H34 isolates recovered at visit 2; however, they differed slightly in the level of biofilm production even though both were above the cutoff for strong biofilm production.
Importantly, all seven (100%) of the STEC-negative animals sampled at visit 1 acquired STEC in at least one follow-up visit, whereas three of the six (50.0%) STEC-positive animals sampled at all four visits lost STEC in at least one subsequent visit. Several animals were also found to have persistent colonization, which was defined by the recovery of isolates with identical serogroups, biofilm levels, and virulence gene profiles at multiple samplings. Over the entire sampling period, eight (61.5%) cattle were persistently colonized by a single serogroup and profile. Isolates classified as O6:H34 were most commonly found (n = 7; 87.5%) among these eight animals with persistent shedding; animal 760 was negative for O6:H34 but was colonized with O168:H8 at three of the four samplings. Some animals also acquired unique STEC isolates that were not detected at prior visits but were found in at least one other animal at a prior visit. Indeed, 11 of the 13 (84.6%) cattle acquired a unique strain of STEC that was previously found in at least 1 other animal. The most commonly acquired serogroup was O26 (n = 9; 69.2%) followed by O6 (n = 8; 61.5%), O103 (n = 3; 23.1%), and O168 (n = 2; 15.4%). A unique O157:H7 isolate was acquired by animal 761; however, this isolate was not detected in any of the other animals sampled.
High biofilm formers were significantly more likely to persist among the 13 animals followed over the four samplings (Fisher's exact test, P = 0.0048). Specifically, 13 of the 28 (46.4%) high biofilm formers were recovered from cattle at more than 1 sampling period compared with only 2 of the 23 (8.7%) low biofilm formers. This association is likely due to serogroup O6, which was found in two or more phases in 7 of the 13 animals (53.8%) and was independently associated with strong biofilm formation. Three animals that were negative for O6:H34 isolates at the first sampling acquired a persistent, high biofilm-forming isolate in one of the three follow-up visits. Cow 760 was persistently colonized with O168:H8 at three consecutive samplings and was the only cow with persistence that did not belong to serogroup O6.
Genomic analysis of isolates belonging to specific serogroups.
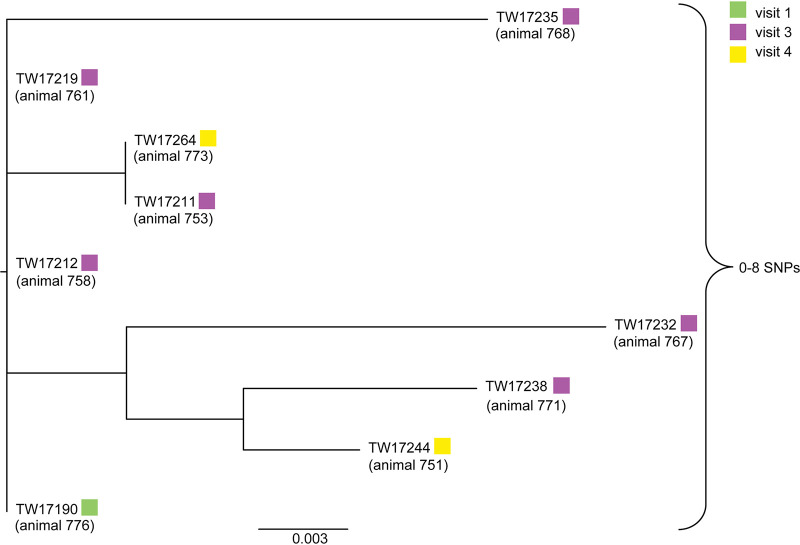
A high-quality SNP (hqSNP) analysis was performed to examine the genetic relatedness of the specific serogroups found within the herd and to determine whether identical strains were circulating throughout the sampling periods or if subtle changes (e.g., SNPs) could be detected. For O26, isolates recovered from nine cattle at three different sampling periods were evaluated; isolate TW17255 was removed from this analysis due to low-quality sequencing and lack of supported informative sites. Among the remaining isolates, the SNP analysis indicated that they are highly similar, differing by between 0 and 8 SNPs (Fig. 3). No distinct clustering of the O26 isolates was observed across visits. Although serogroup O103 isolates were recovered from multiple cattle and samplings, a phylogenetic tree could not be generated due to a lack of informative sites that met the confidence standards (95% of reads agree with SNP designation and 20 read depth at a site).
FIG 3.
High-quality SNP analysis of nine O26:H11 STEC isolates recovered from nine cattle at different visits. All isolates were positive for stx1, eae, and ehxA and formed weak biofilms. Sampling periods (visits) are indicated by the colored boxes; no O26:H11 isolates were recovered from sampling period 2.
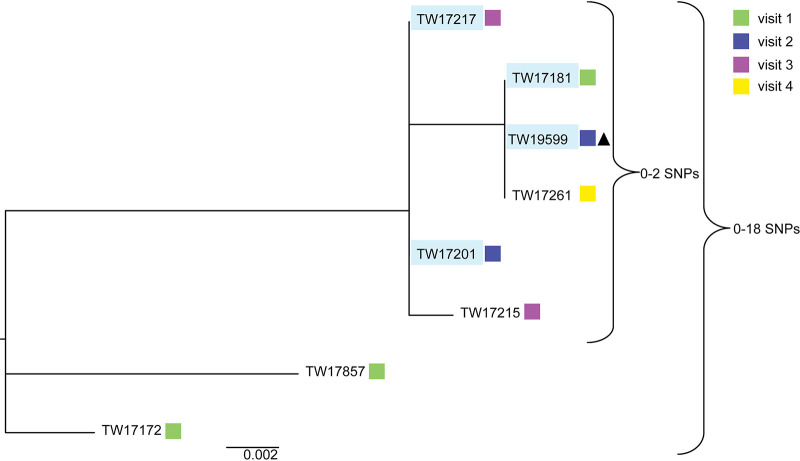
Among the eight O168:H8 serogroup isolates recovered, the hqSNP analysis demonstrated that four isolates from the same animal (760) were almost identical even though they were isolated at three different samplings and had differing biofilm levels (Fig. 4). These four isolates differed by between 0 and 2 SNPs. Interestingly, the four isolates from animal 760 also clustered with two isolates recovered from two different cattle at visits three and four. All six of these isolates clustered separately from two other isolates recovered from animals 728 and 729 at the first sampling, differing by between 0 and 18 SNPs in all.
FIG 4.
High-quality SNP analysis of eight O168:H8 STEC isolates recovered from five cattle over multiple samplings. All isolates recovered from the same animal (760) are outlined in light blue; all other isolates are from different cattle. The sampling number is indicated by colored boxes, and the one isolate with strong biofilm production is shown next to the black triangle.
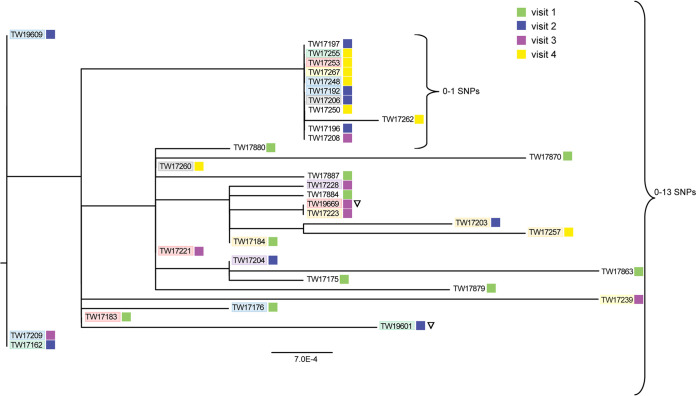
It is noteworthy that the hqSNP analysis of the 34 O6:H34 isolates indicated that all of the isolates were highly similar, although TW17231 and TW17186 could not be examined due to poor sequencing quality. In all, the O6 isolates differed by between 0 and 13 SNPs, indicating that genetically similar strains were circulating through the herd at each of the 4 sampling periods (Fig. 5). There was no clustering by animal or sampling period, although most of the visit 1 isolates were located on separate branches within the phylogeny. Interestingly, a small cluster was detected within the tree that included 11 highly similar isolates with no more than one SNP difference. Isolates in this cluster were recovered from 10 animals across 3 of the 4 sampling periods; 2 isolates from the same animal (752) were recovered at visits 2 and 4.
FIG 5.
High-quality SNP analysis of 34 O6:H34 STEC isolates from cattle over four sampling periods. All isolates possess stx2c and formed strong biofilms except for two isolates with low biofilm production (inverted open triangle). Isolates from the same animal (752, blue; 761, red; 763, green; 764, orange; 767, purple; 768, gray; 773, yellow) have similar shading, and sampling periods are indicated by the colored squares.
The four O6 isolates collected from animals 761 and 763 at the same visits were located on different branches of the phylogeny (Fig. 5). Interestingly, each pair of isolates varied in the level of biofilm production. Isolates TW19669 from animal 761 and TW19601 from animal 763 formed weak biofilms compared with the other two isolates, TW17221 (761) and TW17162 (763), recovered from both animals and formed strong biofilms. A direct genomic comparison of TW19669 with the three strong biofilm formers TW17221, TW17183, and TW17253 recovered from the same animal (761) identified differences in two genes. The first difference was in icsA, a putative outer membrane autotransporter barrel, and the second was in vgrG, a gene encoding a type VI secretion protein. Similarly, TW19601 was compared with the other two isolates, TW17162 and TW17255, with strong biofilm production from the same animal (763). Five mutations were identified in the following genes: those encoding a putative d-alanyl-d-alanine endopeptidase (F7D04_07630), a putative paraquat-inducible protein A (FPV29_09865), DNA repair (radC), a putative phage tail protein, and a type VI secretion protein (vgrG). Extraction of the vgrG gene from all 34 O6 isolates showed that it is heterozygous, with each genome containing more than one copy.
DISCUSSION
In this analysis of 66 STEC isolates recovered from cattle in a single beef herd, we identified a range of non-O157 serogroups and evidence for persistence and transmission of specific strain types. Although one O157:H7 isolate was recovered, higher frequencies of O103 and O26, which represent two of the most common non-O157 serogroups associated with human infections (8), were observed. Multiple virulence gene profiles were also detected, which can contribute to variation in pathogenicity. Indeed, different stx subtypes were found to contribute to various degrees of cytotoxicity and differences in clinical outcomes. Isolates with stx2a, for example, were more lethal in mice and have caused more severe human infections than isolates with stx1a only (26, 27). Isolates with stx2c were shown to have reduced cytotoxicity in Vero cells in vitro (28) despite having the ability to cause human infections. In the study herein, most STEC isolates possessed stx2c and were classified as serotype O6:H34. Strains belonging to O6:H34 have been linked to human infections (29), but they appear to be uncommon in the United States. Regardless, all isolates possessing Stx-encoding prophages can serve as reservoirs for the transfer of Stx genes to other E. coli populations residing in a shared environment.
The emergence of an O157:H7 isolate possessing stx1 and stx2 as well as eae and ehxA, two mobile genetic elements encoding key STEC virulence factors, highlights the importance of horizontal gene transfer. Similarly, most O103 and O26 isolates (n = 14; 93.3%) harbored stx1 along with eae and ehxA, thereby enhancing the pathogenic potential of the isolates circulating among cattle in this herd. The intimin gene eae, which is found on the locus of enterocyte effacement pathogenicity island, is responsible for attaching and effacing lesion formation on intestinal epithelial cells (30). Prior studies have shown that possession of eae influences the ability of a strain to effectively colonize cattle in addition to enhancing pathogenicity in human infections (31, 32). The ehxA gene, however, encodes an enterohemolysin found on different plasmids that vary by serotype and has been linked to virulence in humans (33). Most of our cattle isolates possessed ehxA subtypes A and C, consistent with findings in a prior study (34), although this study also reported an association between subtype C and clinical isolates. Only the single O157:H7 isolate recovered herein possessed ehxA subtype B, which has been shown to predominate among all O157 isolates regardless of the source (34).
The ability to form biofilms also allows for an increased potential for transmission and persistence of bacterial pathogens in the external environment. In this study, the predominant STEC serotype O6:H34 was more commonly acquired by other cattle in the herd and persisted in higher frequencies over the entire sampling period than other serotypes. It was therefore interesting to observe that most of these O6:H34 isolates could form strong biofilms in vitro, which is consistent with findings from other studies. A persistent bovine O157:H7 strain (MC2) found in French farms, for example, was shown to have an enhanced ability to form biofilms in specific environmental conditions and possessed unique genes that could impact adherence (35). Similarly, our genomic comparison of weak and strong biofilm-forming O6:H34 isolates identified multiple differences, including mutations in vgrG (valine-glycine repeat protein G). VgrG is part of a type VI secretion system, which was detected in the persistent O157:H7 strain MC2 genome as well (35) and has been shown to impact adherence in other pathogens. In Acinetobacter baumannii, for instance, vgrG mutants exhibited increased susceptibility to certain antibiotics, a lower growth rate, and decreased adhesion to eukaryotic cells in vitro (36). In the O6:H34 isolate examined herein, a nonsynonymous SNP in vrgG was identified, which resulted in a change from a tyrosine (aromatic) to a histidine (basic). While such a change could contribute to transcriptional or regulatory differences that influence biofilm production or enhanced survival in the farm environment, the examination of a larger sample of isolates with variable phenotypes is needed for confirmation. Other factors such as autotransporters and specific fimbrial adhesins, which have been linked to biofilm production in STEC previously (37), may also be important but were not examined in our study. We also did not have closed assemblies for each genome, and hence, we could not fully investigate the association between the number of vgrG copies and biofilm levels. Such studies would be worthwhile in the future.
Compared to the O6 isolates, some serogroups were more transient and less likely to persist in the same animal over time. O103 isolates, for example, were present only at the first and second samplings in the subset of cattle examined. Serogroup O26 isolates were recovered throughout the sampling period, and similar to the O103 isolates, they were never found in the same animal at a subsequent sampling. These findings suggest that these two serogroups are readily transmitted between animals, which is likely attributable to the high level of comingling that occurred among animals within this herd. These two serogroups, however, were less likely to persist over time. While differences between transient and persistent colonizers could be due to the presence or absence of specific bacterial traits or protective host responses, such differences could also be due to variation in the cattle gut microbiome. Indeed, specific members of the intestinal microbiome could prevent prolonged colonization and shedding of some microbes via competitive exclusion (38).
In summary, this study has enhanced our understanding of genetic profiles as well as transmission and colonization dynamics of STEC in a subset of beef cattle. Although a larger sample size of cattle should be studied in the future, the use of genomics to characterize STEC in only 13 animals over 4 sampling periods has shown high rates of transmission and persistence within these animals. Specific bacterial factors linked to biofilm production and/or persistent colonization were also identified. Such studies are important for developing novel strategies aimed at minimizing STEC transmission between animals and reducing shedding levels of persistent strain types in the farm environment. Reducing STEC diversity also has the potential to decrease the likelihood of virulence gene acquisition by resident commensal E. coli populations, thereby preventing the emergence of new STEC pathogens.
MATERIALS AND METHODS
Bacterial strains.
A total of 66 STEC isolates were examined. All isolates were recovered in 2012 from one beef cattle herd in Michigan that was previously found to have a high prevalence of STEC (13). Cattle in this herd were pasture raised with a high likelihood of comingling, thereby making it a useful population to study STEC shedding dynamics. The 66 STEC isolates were recovered from fecal grab samples of 26 animals, of which 13 were sampled over the course of 4 sampling periods that were ∼3 to 4 weeks apart. As part of our original study involving 11 herds (13), a larger percentage of cattle were sampled at the first visit in each herd, and only a subset of STEC-negative (n = 7) and STEC-positive (n = 6) cattle was selected for the longitudinal analyses. Approval to conduct the study was obtained by the Michigan State University Institutional Animal Care and Use Committee (AN12/10-223-00).
STECs were detected by incubating 5 grams of feces in 2× EC broth (Oxoid Ltd., Waltham, MA) containing novobiocin (8 mg/liter), rifampin (2 mg/liter), and potassium tellurite (1 mg/liter) at 42°C for 24 h (13). Subculture to CHROMagar STEC (Paris, France) and sorbitol MacConkey (SMAC) agar enabled identification of suspect colonies that were confirmed to be STEC based on the presence of Shiga toxin genes stx1 and/or stx2, using multiplex PCR. The PCR primers and cycling conditions were described previously (10). In some animals, multiple colonies had identical stx and serogroup profiles. One colony representing each unique profile was selected at random from each animal for characterization to avoid duplicates.
Biofilm assays.
Isolates were grown in Luria-Bertani (LB; BD Diagnostics) broth and incubated at 37°C overnight with shaking. Overnight cultures were diluted 1:10 in prewarmed LB no-salt (LB-NS) broth before 30 μl was added to a 96-well microtiter plate (TPP; Techno Plastic Products AG) with 100 μl LB-NS broth and incubated at 25°C for 48 h. The 96-well plate was washed thrice with phosphate-buffered saline (PBS) to remove any unattached cells that had not formed a biofilm. Biofilms were fixed by adding 100% methanol and incubating for 10 minutes. Following removal of the methanol and air drying, the biofilms were stained with crystal violet (CV) and incubated for 15 min at room temperature. PBS was used to wash the biofilms and to remove any CV that had not been absorbed by the biofilm. After air drying, 200 μl of 33% glacial acetic acid was used to solubilize the CV and the A595 was determined using a plate reader. The data were normalized to the blank control of LB-NS broth for each biological replicate. At least four technical replicates were averaged on a plate and repeated for at least three biological replicates.
DNA isolation and WGS.
DNA was extracted from the STEC isolates using the DNeasy kit (Qiagen, Valencia, CA, USA). Sequencing libraries were prepared using the Nextera XT library prep kit (Illumina, San Diego, CA, USA) and sequenced at 2 × 250 bp on the MiSeq platform (Illumina) at the Michigan State University Research Technology Support Facility (RTSF) and the Michigan Department of Agriculture and Rural Development.
Bioinformatic analysis.
Sequences were preprocessed with Trimmomatic to remove sequencing adapters, sequences <100 nucleotides, and reads with a Phred quality score less than 20 (Q20) (39). FastQC was used to quality check the reads prior to assembly and analysis (40). Among all isolates, the average genome size was 5.19 × 106, while the minimum and maximum genome sizes were 4.98 × 106 and 5.5 × 106, respectively.
Assemblies were generated with SPAdes 3.10.1 (kmers 21, 33, 55, 77, 99, and 127) (41), gene extraction for molecular serotyping (wzy/wzx, O-antigen; fliC, H-antigen) and stx profiling were performed using Abricate (https://github.com/tseemann/abricate), and databases were curated by the Center for Genomic Epidemiology (www.genomicepidemiology.org) as described previously (42). In-house bioinformatic pipelines using a Basic Local Alignment Search Tool (BLAST) (43) were used to extract and concatenate the seven housekeeping genes (aspC, clpX, fadD, icdA, lysP, mdh, and uidA) for multilocus sequence typing (MLST) via the Whittam scheme (44) as well as virulence genes eae and ehxA encoding intimin and enterohemolysin, respectively (30, 45). STs were classified using the EcMLST v1.2 database (http://shigatox.net/ecmlst/cgi-bin/index).
Assemblies were annotated using Prokka v1.14.0, and the pangenomes were extracted from annotated genomes using Roary v3.11.2 as described (46, 47). Lyve-SET was used to generate high-quality single nucleotide polymorphisms (hqSNPs) to further examine strains of the same serogroup and determine whether identical strains were persisting in cattle. Informative sites were included in the analysis if they met the confidence standards for Lyve-SET previously applied to foodborne pathogens (48). These parameters required that the SNP had ≥95% identity to the reference sequence, and that the region containing the SNP had at least a 20-read depth and 2 forward and 2 reverse reads.
Data analysis.
Concatenated MLST alleles were aligned with ClustalW, and the phylogeny was constructed using the neighbor-joining algorithm with 1,000 bootstrap replication in MEGA X (49). RAxML v8 was used to infer maximum likelihood phylogenies for both SNP profiles determined by Lyve-SET and pangenome concatenated gene sequences generated by Roary (50). A reference-free phylogeny was constructed for each serogroup to avoid missing SNPs unique to the cattle strains that may have been absent from an unrelated reference strain genome. As described previously, use of an unrelated strain to serve as an outgroup could cause a lower number of SNP differences to be detected and skew the distance between isolates (51, 52). Visualization of the phylogenetic trees used MEGA X, TreeGraph2, or FigTree (http://tree.bio.ed.ac.uk/software/figtree) (49, 53). Associations between the molecular characteristics and shedding status were identified using the chi-square (χ2) test, while Fisher’s exact test was used for sample sizes less than five in a cell; P values of ≤0.05 were considered significant.
Data availability.
All sequencing reads used in the analysis were deposited in the NCBI database under BioProject accession number PRJNA635222, and the associated BioSample numbers are SAMN15029623 to SAMN15029685.
ACKNOWLEDGMENTS
We thank Cristina Venegas-Vargas, Paul Bartlett, Bo Norby, and Steven Rust for overseeing the cattle sampling efforts in our initial study.
Funding for this project was provided by the United States Department of Agriculture (USDA; number 2011-67005-30004) and the Michigan Department of Agriculture and Rural Development. Additional salary support was provided by the USDA (2019-67017-29112 and MICL02475), the Michigan State University (MSU) Foundation, and AgBioResearch at MSU. Student support to H.M.B. was provided by MSU through a University Enrichment Fellowship, a College of Natural Science Dissertation Continuation Fellowship, and the Bertina Wentworth Scholar Award from the Department of Microbiology and Molecular Genetics.
REFERENCES
- 1.Karmali MA, Petric M, Lim C, Fleming PC, Arbus GS, Lior H. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis 151:775–782. doi: 10.1093/infdis/151.5.775. [DOI] [PubMed] [Google Scholar]
- 2.Riley LW, Remis RS, Helgerson SD, Mcgee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen ML. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 3.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg Infect Dis 11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2000. Outbreak of Escherichia coli O157:H7 infection associated with eating fresh cheese curds—Wisconsin, June 1998. Morb Mortal Wkly Rep 49:911–913. [PubMed] [Google Scholar]
- 5.Denny J, Bhat M, Eckmann K. 2008. Outbreak of Escherichia coli O157:H7 associated with raw milk consumption in the Pacific Northwest. Foodborne Pathog Dis 5:321–328. doi: 10.1089/fpd.2007.0072. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2001. Outbreaks of Escherichia coli O157:H7 infections among children associated with farm visits—Pennsylvania and Washington, 2000. Morb Mortal Wkly Rep 50:293–296. [PubMed] [Google Scholar]
- 7.Gould LH, Mody RK, Ong KL, Clogher P, Cronquist AB, Garman KN, Lathrop S, Medus C, Spina NL, Webb TH, White PL, Wymore K, Gierke RE, Mahon BE, Griffin PM, Emerging Infections Program Foodnet Working Group. 2013. Increased recognition of Non-O157 Shiga toxin-producing Escherichia coli infections in the United States during 2000–2010: epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog Dis 10:453–460. doi: 10.1089/fpd.2012.1401. [DOI] [PubMed] [Google Scholar]
- 8.Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Strockbine NA. 2005. Non‐O157 Shiga toxin–producing Escherichia coli infections in the United States, 1983–2002. J Infect Dis 192:1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- 9.Zschock M, Hamann HP, Kloppert B, Wolter W. 2000. Shiga toxin-producing Escherichia coli in faeces of healthy dairy cows, sheep and goats: prevalence and virulence properties. Lett Appl Microbiol 31:203–208. doi: 10.1046/j.1365-2672.2000.00789.x. [DOI] [PubMed] [Google Scholar]
- 10.Singh P, Sha Q, Lacher DW, Del Valle J, Mosci RE, Moore JA, Scribner KT, Manning SD. 2015. Characterization of enteropathogenic and Shiga toxin-producing Escherichia coli in cattle and deer in a shared agroecosystem. Front Cell Infect Microbiol 5:29. doi: 10.3389/fcimb.2015.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien SJ, Adak GK, Gilham C. 2001. Contact with farming environment as a major risk factor for Shiga toxin (Vero cytotoxin)-producing Escherichia coli 0157 infection in humans. Emerg Infect Dis 7:1049–1051. doi: 10.3201/eid0706.010626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barkocy-Gallagher GA, Arthur TM, Rivera-Betancourt M, Nou X, Shackelford SD, Wheeler TL, Koohmaraie M. 2003. Seasonal prevalence of Shiga toxin-producing Escherichia coli, including O157:H7 and Non-O157 serotypes, and Salmonella in commercial beef processing plants. J Food Prot 66:1978–1986. doi: 10.4315/0362-028X-66.11.1978. [DOI] [PubMed] [Google Scholar]
- 13.Venegas-Vargas C, Henderson S, Khare A, Mosci RE, Lehnert JD, Singh P, Ouellette LM, Norby B, Funk JA, Rust S, Bartlett PC, Grooms D, Manning SD. 2016. Factors associated with Shiga toxin-producing Escherichia coli shedding by dairy and beef cattle. Appl Environ Microbiol 82:5049–5056. doi: 10.1128/AEM.00829-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong HJ, Lee S, Kim W, An JU, Kim J, Kim D, Cho S. 2017. Prevalence, virulence potential, and pulsed-field gel electrophoresis profiling of Shiga toxin-producing Escherichia coli strains from cattle. Gut Pathog 9:22. doi: 10.1186/s13099-017-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renter DG, Morris JG, Sargeant JM, Hungerford LL, Berezowski J, Ngo T, Williams K, Acheson DWK. 2005. Prevalence, risk factors, O serogroups, and virulence profiles of Shiga toxin-producing bacteria from cattle production environments. J Food Prot 68:1556–1565. doi: 10.4315/0362-028x-68.8.1556. [DOI] [PubMed] [Google Scholar]
- 16.Cobbold RN, Rice DH, Szymanski M, Call DR, Hancock DD. 2004. Comparison of shiga-toxigenic Escherichia coli prevalences among dairy, feedlot, and cow-calf herds in Washington State. Appl Environ Microbiol 70:4375–4378. doi: 10.1128/AEM.70.7.4375-4378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeJeune JT, Besser TE, Hancock DD. 2001. Cattle water troughs as reservoirs of Escherichia coli O157. Appl Environ Microbiol 67:3053–3057. doi: 10.1128/AEM.67.7.3053-3057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silagyi K, Kim SH, Martin Lo Y, Wei CI. 2009. Production of biofilm and quorum sensing by Escherichia coli O157:H7 and its transfer from contact surfaces to meat, poultry, ready-to-eat deli, and produce products. Food Microbiol 26:514–519. doi: 10.1016/j.fm.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Wang R, Bono JL, Kalchayanand N, Shackelford S, Harhay DM. 2012. Biofilm formation by Shiga toxin-producing Escherichia coli O157:H7 and non-O157 strains and their tolerance to sanitizers commonly used in the food processing environment. J Food Prot 75:1418–1428. doi: 10.4315/0362-028X.JFP-11-427. [DOI] [PubMed] [Google Scholar]
- 20.Maeda S, Ito M, Ando T, Ishimoto Y, Fujisawa Y, Takahashi H, Matsuda A, Sawamura A, Kato S. 2006. Horizontal transfer of nonconjugative plasmids in a colony biofilm of Escherichia coli. FEMS Microbiol Lett 255:115–120. doi: 10.1111/j.1574-6968.2005.00072.x. [DOI] [PubMed] [Google Scholar]
- 21.Saldaña Z, Xicohtencatl-Cortes J, Avelino F, Phillips AD, Kaper JB, Puente JL, Girón JA. 2009. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ Microbiol 11:992–1006. doi: 10.1111/j.1462-2920.2008.01824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 23.Genevaux P, Bauda P, DuBow MS, Oudega B. 1999. Identification of Tn 10 insertions in the rlaG, rfaP, and gaLU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch Microbiol 172:1–8. doi: 10.1007/s002030050732. [DOI] [PubMed] [Google Scholar]
- 24.Puttamreddy S, Cornick NA, Minion FC. 2010. Genome-wide transposon mutagenesis reveals a role for pO157 genes in biofilm development in Escherichia coli O157:H7 EDL933. Infect Immun 78:2377–2384. doi: 10.1128/IAI.00156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danese PN, Pratt LA, Kolter R. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol 182:3593–3596. doi: 10.1128/jb.182.12.3593-3596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tesh VL, Burris JA, Owens JW, Gordon VM, Wadolkowski EA, O'Brien AD, Samuel JE. 1993. Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect Immun 61:3392–3402. doi: 10.1128/IAI.61.8.3392-3402.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostroff SM, Tarr PI, Neill MA, Lewis JH, Hargrett-Bean N, Kobayashi JM. 1989. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli 0157:H7 infections. J Infect Dis 160:994–998. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt CK, McKee ML, O'Brien AD. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H- strain. Infect Immun 59:1065–1073. doi: 10.1128/IAI.59.3.1065-1073.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin X, Klein EJ, Galanakis E, Thomas AA, Stapp JR, Rich S, Buccat AM, Tarr PI. 2015. Real-Time PCR assay for detection and differentiation of Shiga toxin-producing Escherichia coli from clinical samples. J Clin Microbiol 53:2148–2153. doi: 10.1128/JCM.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci U S A 87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naylor SW, Roe AJ, Nart P, Spears K, Smith DGE, Low JC, Gally DL. 2005. Escherichia coli O157:H7 forms attaching and effacing lesions at the terminal rectum of cattle and colonization requires the LEE4 operon. Microbiology 151:2773–2781. doi: 10.1099/mic.0.28060-0. [DOI] [PubMed] [Google Scholar]
- 32.Sheng H, Lim JY, Knecht HJ, Li J, Hovde CJ. 2006. Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect Immun 74:4685–4693. doi: 10.1128/IAI.00406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorenz SC, Monday SR, Hoffmann M, Fischer M, Kase JA. 2016. Plasmids from Shiga toxin-producing Escherichia coli strains with rare enterohemolysin gene (ehxA) subtypes reveal pathogenicity potential and display a novel evolutionary path. Appl Environ Microbiol 82:6367–6377. doi: 10.1128/AEM.01839-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorenz SC, Son I, Maounounen-Laasri A, Lin A, Fischer M, Kase JA. 2013. Prevalence of hemolysin genes and comparison of ehxA subtype patterns in Shiga toxin-producing Escherichia coli (STEC) and Non-STEC strains from clinical, food, and animal sources. Appl Environ Microbiol 79:6301–6311. doi: 10.1128/AEM.02200-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segura A, Auffret P, Bibbal D, Bertoni M, Durand A, Jubelin G, Kérourédan M, Brugère H, Bertin Y, Forano E. 2018. Factors involved in the persistence of a Shiga toxin-producing Escherichia coli O157: H7 strain in bovine feces and gastrointestinal content. Front Microbiol 9:375. doi: 10.3389/fmicb.2018.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Zhou Z, He F, Ruan Z, Jiang Y, Hua X, Yu Y. 2018. The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PLoS One 13:e0192288. doi: 10.1371/journal.pone.0192288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogeleer P, Tremblay YDN, Mafu AA, Jacques M, Harel J. 2014. Life on the outside: role of biofilms in environmental persistence of Shiga-toxin producing Escherichia coli. Front Microbiol 5:317. doi: 10.3389/fmicb.2014.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Callaway TR, Anderson RC, Edrington TS, Genovese KJ, Harvey RB, Poole TL, Nisbet DJ. 2013. Novel methods for pathogen control in livestock pre-harvest: an update, p 275–304. In Sofos J (ed), Advances in microbial food safety. Woodhead Publishing Limited, Cambridge, UK. [Google Scholar]
- 39.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrews S. 2010. FASTQC, a quality control tool for the high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 41.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blankenship HM, Mosci RE, Phan Q, Fontana J, Rudrik JT, Manning SD. 2020. Genetic diversity of Non-O157 Shiga toxin-producing Escherichia coli recovered from patients in Michigan and Connecticut. Front Microbiol 11:529. doi: 10.3389/fmicb.2020.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 44.Qi W, Lacher DW, Bumbaugh AC, Hyma KE, Ouellette LM, Large TM, Tarr CL, Whittam TS. 2004. EcMLST: an online database for multi locus sequence typing of pathogenic Escherichia coli, p 520–521. In Proceedings—2004 IEEE Computational Systems Bioinformatics Conference, CSB 2004. [Google Scholar]
- 45.Bauer ME, Welch RA. 1996. Characterization of an RTX toxin from enterohemorrhagic Escherichia coli O157:H7. Infect Immun 64:167–175. doi: 10.1128/IAI.64.1.167-175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 47.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katz LS, Griswold T, Williams-Newkirk AJ, Wagner D, Petkau A, Sieffert C, Van Domselaar G, Deng X, Carleton HA. 2017. A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Front Microbiol 8:375. doi: 10.3389/fmicb.2017.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oakeson KF, Wagner JM, Mendenhall M, Rohrwasser A, Atkinson-Dunn R. 2017. Bioinformatic analyses of whole-genome sequence data in a public health laboratory. Emerg Infect Dis 23:1441–1445. doi: 10.3201/eid2309.170416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olson ND, Lund SP, Colman RE, Foster JT, Sahl JW, Schupp JM, Keim P, Morrow JB, Salit ML, Zook JM. 2015. Best practices for evaluating single nucleotide variant calling methods for microbial genomics. Front Genet 6:235. doi: 10.3389/fgene.2015.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stöver BC, Müller KF. 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequencing reads used in the analysis were deposited in the NCBI database under BioProject accession number PRJNA635222, and the associated BioSample numbers are SAMN15029623 to SAMN15029685.