Abstract
The crosstalk between host immunity and the external environment in the mucous membranes of the gastrointestinal and respiratory tracts in bronchial asthma has recently been scrutinized. There is compelling evidence that the microbiota at these sites may play an important role in the pathogenesis of this chronic airway disease. The appearance of bacteria early in life in the gut before dissemination to the airways plays a pivotal role in shaping mucosal immunity. Loss of microbial diversity or dysbiosis can result in aberrant immune-mediated inflammation and mucosal barrier disruption, which coincides clinically with the successive development of the “allergic march” in asthma. Microbial manipulation may be effective in curbing asthma development by indirectly preserving homeostatic epithelial barrier functions. The protective effects and mechanisms of immunity-microbiome crosstalk at mucosal sites require further investigation to identify therapeutic and preventive measures in asthma. This topical review aims to highlight new evidence that compromised epithelial barrier function, which results in deregulated crosstalk between the microbiome and host mucosal immune system, is an important disease mechanism in asthma. In the light of current COVID-19 pandemic, the collective findings on the impact of mucosal microbiota on the suceptibility to SARS-CoV-2 infection and severity of COVID-19 is explored. The possible therapeutic implications to target these abnormalities are further discussed.
Keywords: asthma, COVID-19, SARS-CoV-2, dendritic cells, innate immunity, microbiome, barrier dysfunction
Background
Asthma is an airway disease that currently affects more than 300 million people worldwide.1 This chronic respiratory condition is characterized by a spectrum of clinical phenotypes and a range of underlying molecular mechanisms called endotypes.2 Consequently, this heterogeneous group of clinical presentations should not be thought of as a single disease but instead a spectrum of conditions with some overlapping characteristics. At the molecular level, asthma is divided into two categories: atopic and non-atopic asthma.3 Atopic asthma is characterized by type 2 inflammation, driven by IgE hypersensitivity to aeroallergen, chemoattraction of granulocytes, hyper-activation of airway epithelial cells and subsequent remodeling of the epithelium.4 Besides these well-established mechanisms, recent evidence has implicated developmental microbial exposure in the pathogenesis of asthma.5
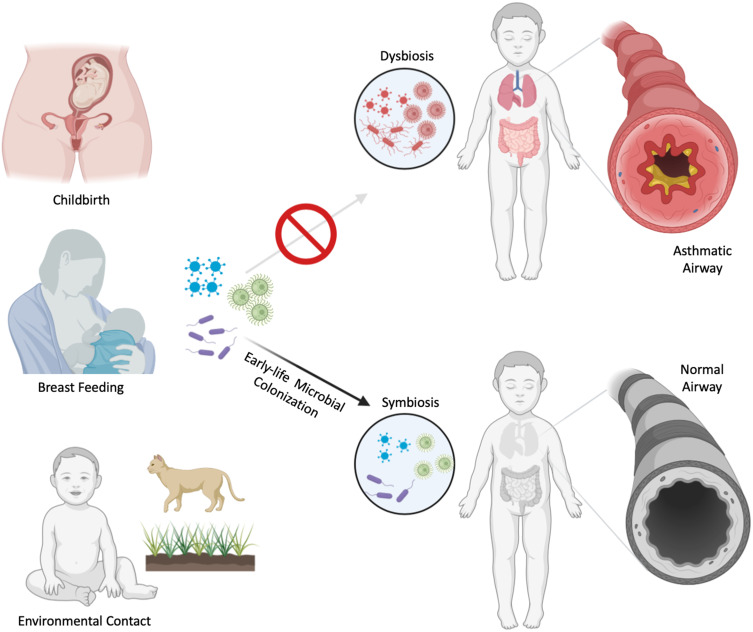
The “hygiene-hypothesis”, which was first proposed in 1989,6 suggested that while exposure to pathogens educates the immune system to avoid invoking allergic reactions, an environment that is too clean increases the risk of developing atopic conditions. Pathogens are naturally acquired during development in several ways. For example, neonates are exposed to maternal vaginal and intestinal flora during childbirth, which are important for priming the early immune system (Figure 1).7 Furthermore, contact with soil and animals can increase exposure to pathogens during the early years of life.8 Unfortunately, the increased number of cesarean section deliveries and the loss of contact with soil and animals have interrupted these early exposures. Additionally, the use of antibiotics and acid-suppressing agents during the first 6 months of infancy has been associated with a significantly increased risk of developing allergic diseases and asthma.9
Figure 1.
Early shaping of bronchial asthma by microbial exposure. Early-life microbial transfer occurs in utero and during childbirth (maternal intestinal and vaginal flora), during breast feeding (maternal milk microbiome) and contact with animals and soil (environmental microorganisms). These mechanisms allow colonization of commensals during early life. Failure to properly establish early symbiotic relationship with commensals results in dysbiosis and predisposes the host to allergic inflammation and bronchial asthma.
In congruence with this long-standing “hygiene hypothesis”, recent studies emphasize the novel role of the mucosal microbiome in the education of the immature immune system and, subsequently, in the development or prevention of allergic sensitization.10,11 The term microbiome refers to the collective genomes of all micro-organisms symbiotically existing within the human body. These organisms consist of bacteria, viruses, fungi and protozoa that take residence on the outer (skin, hair, nail) as well as inner mucosal surfaces (gastrointestinal and respiratory tracts) of the body. While previously thought to be sterile, our intestinal tracts are in fact replete with bacteria that have been acquired via early-life pathogen exposure.12,13 Similarly, the previously accepted idea of “sterile lungs” has been challenged by the discovery of the respiratory tract microbiome, demonstrated by the introduction of the lung tissues into the Human Microbiome Project.13,14 Disruption of homeostatic microbial colonization at these sites results in an imbalanced microbiota and a loss of microbial diversity, termed dysbiosis, and is a shared etiology of emerging hypotheses about the role of microbiota in asthma development.15 This dysbiosis results in mucosal barrier dysfunction, which is a postulated cause and/or consequence of inflammatory processes in childhood asthma.16 Mechanistically, abnormal microbial movement through those disrupted mucosal barriers and the subsequent aberrant interactions with the host immune system dictate, at least in part, susceptibility to focal as well as systemic inflammatory responses and asthma development.17,18 A better appreciation of how these mechanisms interconnect is critical for enhancing our understanding of the pathogenesis of asthma and aiding the development of early intervention methods.
This topical review outlines the role of mucosal barrier dysfunction and details the mechanisms of crosstalk between the gastrointestinal-airway microbiome and mucosal immunity in the pathogenesis of asthma. Finally, their potential therapeutic implications in asthma are discussed.
The Mucosal Epithelial Barrier
Gastrointestinal and Respiratory Epithelium
The intestinal mucosa is one of the largest surface areas in the body and comes into direct contact with the external environment.19 It serves the dual function of being semipermeable to water and nutrients while always preventing access of potentially harmful microbial organisms and large molecules from the lumen. Structurally, it consists of multiple layers that form physical barriers (epithelial and mucosal layers) and immunological defenses (lamina propria). Each layer and their cellular components play different but complimentary roles in host barrier defenses (Figure 2).
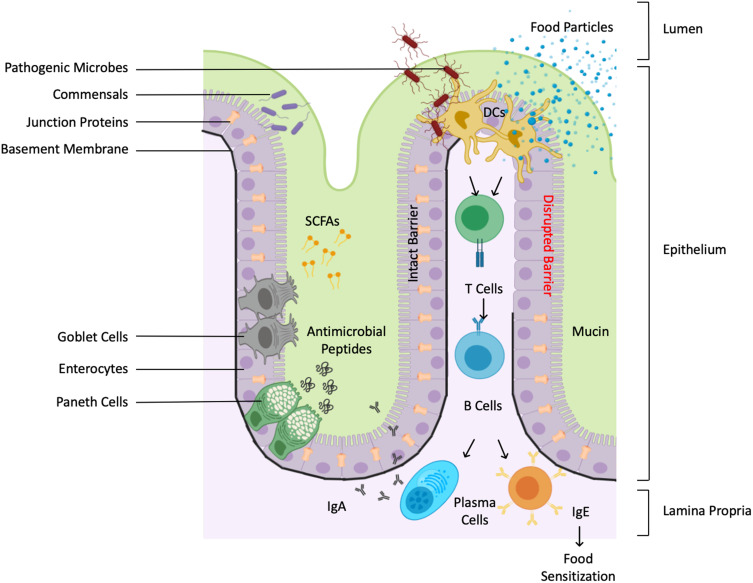
Figure 2.
Immunity-microbiome crosstalks at gastrointestinal tract. Schematic diagram depicting the gut mucosa illustrated with the various structural and immunological barriers and defenses. The intestinal mucosa consists of an epithelial layer lined by an inner mucus layer and an underlying lamina propria. The epithelial layer forms a physical barrier, facilitated by intracellular tight junctions, various cell types and their secretory products. Paneth cells produce protective antimicrobial peptides, while goblet cells produce mucin, the major component of the mucus coating. The mucus layer is rich in microbial colonies, which can synthesize immunomodulatory molecules such as SCFAs. The lamina propria are rich in immune cells that play a fundamental role in immune surveillance. Dendritic cells transmit environmental signals to activated plasma cells to produce protective secretory immunoglobulin A (sIgA). A leaky intestinal barrier can lead to displaced food antigens in the lamina propria, which in turn can activate Th2 Cell and class-switch B cells to produce specific immunoglobulin E (sIgE) production, causing sensitization to food allergens.
On the luminal side, the epithelial layer is lined by a gel-like mucus coating that consists primarily of antimicrobial molecules (antimicrobial peptides secreted from Paneth cells and other glycosylated mucin proteins from goblet cells), secretory immunoglobulin A (sIgA), and nutrient metabolites such as short-chain fatty acids (SCFAs) (Figure 2). The mucus layer protects the delicate linings of the gut, and along with the microbial colonies residing here, it provides an interface between the epithelial layer and the exterior environment.20 The inner part of the mucus layer keeps the epithelium and intestinal crypts germ-free and in complete isolation from non-self antigens, while commensal organisms in the outer coat compete with pathogenic organisms for nutrients and space.21
Similar to the gastrointestinal mucosa, the lung performs the dual function of blocking potential pathogens and harmful substances while simultaneously allowing the exchange of gases across its barrier. The integrity of the environmental barrier is maintained by various factors that differ significantly in structure and function from one compartment of the airway to the other. These factors include a combination of airway epithelial cells, surfactants, and structural proteins. The surfactant system consists of different types of secretory products of type II alveolar epithelial cells. They are formed by a distinct mixture of phospholipids (90%) and protein (10%), which significantly contributes to the regulation of airway fluid balance, airway clearance, resistance to inhaled agents, and other immunomodulatory functions in asthma (Table 1).22,23
Table 1.
Surfactants Involved in the Regulation of Airway Barrier Function
| Molecules | Types | Function | Regulator/Influencer (Effect) |
|---|---|---|---|
| SP-A | Collectins | Immunomodulatory | Binding IgE, pollen grain/Suppressing IL-8, histamine, T cell proliferation |
| SP-B | Saposins | Fluid balance, airway clearance | Downregulated by IL-4 |
| SP-C | Saposins | Fluid balance, airway clearance | Downregulated by IL-5 |
| SP-D | Collectins | Immunomodulatory | Binding IgE, pollen grain, mite allergens/Suppressing T cell proliferation |
Abbreviation: SP, surfactant protein.
Besides surfactants, airway junctional structures consist of tight junctions (TJs) and an underlying layer, which further supports the barrier function, called the adherens junctions (Table 2).24,25
Table 2.
Intercellular Structural Proteins Involved in the Regulation of Airway Barrier Function
| Class of Structural Protein | Types | Function | Regulator/Influencer (Effect) |
|---|---|---|---|
| Tight junctions | Claudin Occludin |
Epithelial junction | Oncostatin M gene (Disrupting TJ junction) |
| Adherens junction | E-cadherin Nectin |
Epithelial junction | Caveolin-1 (Stabilizing E-cadherin, increasing TSLP) |
| Cytosolic plaques | α-catenin β-catenin ZO-1/2/3 |
Epithelial junction | Caveolin-1 (Stabilizing β-catenin) |
Abbreviations: TJs, tight junctions; ZO, zonula occludens.
The Mucosal Immune System
The chief mediator of mucosal immunity in the gastrointestinal tract is the lamina propria, a specialized tissue that is rich in innate immune cells, notably dendritic cells (DCs), which reside in gut-associated lymphoid tissues such as the Peyer’s patch (PPs) of the small intestine.26 DCs patrol the mucosa and extend between epithelial cells via their elongated pseudopods to reach remote areas. The subepithelial layer in the lungs is also rich in DCs and other immune cells that perform immune surveillance functions.
The host immune response recognizes various types of organisms and dictates the immune response according to the type or virulence of the organism.27 Despite the engagement of immune cells and the constant exposure to a large inoculum of commensal microbes, the mucous membranes are not usually inflamed.28 This innate immune response also provides the host with protection against pathogenic microbial invasions through the recognition of pathogen-associated molecular patterns (PAMPs), such as double-stranded DNA in viruses, or bacterial cell surface structures, including lipopolysaccharides.29 Sensing for invading microbes is mediated by pattern-recognition receptors (PRRs), such as Toll-like receptors (TLR), NOD-like receptors, RIG-I-like-receptors and C-type lectin receptors (CLR), located on the surface of both intestinal epithelial cells and innate immune cells located in the lamina propria.29,30 Under basal conditions, PRRs are downregulated to prevent aberrant inflammatory responses; however, upon recognition of PAMPs, PRRs trigger a cascade of intracellular signaling pathways mediated by NF-κB, resulting in the production of cytokines and chemokines, which activate neighboring DCs and macrophages.29,30 Stimulation of these innate immune cells activates plasma cells and provokes antibody responses, including mucosal sIgA production, which are transported to the mucus layer and engage in the neutralization and expulsion of unwanted microorganisms.31
Epithelial Barrier Dysfunction in Asthma
While mucosal barriers in healthy guts and lungs serve as impervious protective layers for the host, they can be damaged by inflammation, chemicals, and trauma, allowing the infiltration of environmental stimuli through leaky membranes, creating an inflammatory response and subsequent allergic ailments.32,33
Intestinal barrier disruption is characterized by paracellular influx of intestinal bacteria-derived products, chiefly, lipopolysaccharides (LPS), into the systemic circulation. Direct contact with these microbial products triggers the activation of lamia propria DCs via TLR4-dependent signaling pathways, causing the release of proinflammatory mediators that perpetuate barrier disruption.34 Sustained disruption of the intestinal barriers might allow the translocation of ingested allergens to the airway through a gut-lung communication pathway, and subsequent development of allergic/asthmatic responses.35 While also marked by a general increase in epithelial permeability, the pathological hallmark of airway barrier disruption is the infiltration of inhaled allergens. This translocation causes allergic sensitization and inflammation via activation of airway DCs.36 DCs recognize and transport these stimuli to adjacent lymph nodes, where they interact with T cells, resulting in T helper 2 (Th2)-skewed immunity. Th2 CD4+ T cells are responsible for directing B cells to switch to IgE production, resulting in the recruitment of mast cells and eosinophils and subsequent induction of an allergic response (Figure 3).
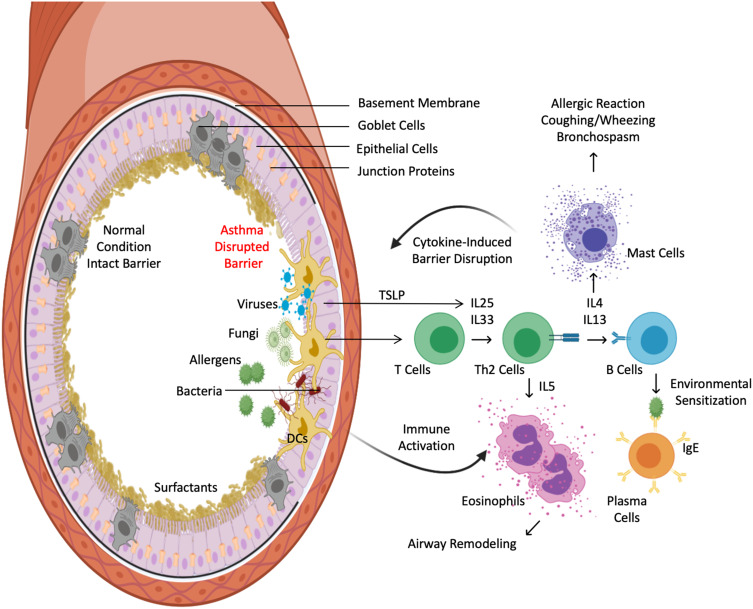
Figure 3.
Immunity-microbiome crosstalks at respiratory tract. Airway epithelial barriers are protected by junctional structures and a family of surfactant proteins. Intact barriers and homeostatic interplay between commensals and innate immunity prevent the entrance of allergens and pathogens (left). In contrast, aberrant interaction between microbes, including viruses and fungi, with mucosal dendritic cells elicits an allergic inflammatory milieu that causes barrier dysfunction (right). This further allows translocation of inhaled allergens and pathogens, heightening Th2 inflammation. Microbes and TLSP from pathogen-activated epithelium primes mucosal dendritic cells to orchestrate T-cell production of Th2 cytokines, namely, IL-4, IL-5, and IL-13, via IL-25 and IL-33 signaling. IL-5 recruits eosinophils to the airway, while IL-4 and IL-13 activate B cells to develop into plasma cells that produce allergen-specific IgE and recruit mast cells to the tissues. These events ultimately perpetuate a vicious cycle of epithelial barrier dysfunction.
Besides environmental allergens, viruses and fungi are important stimuli that play a critical role in the development of barrier disruption in asthma.37,38 During respiratory viral infection, interferons and the cytokine thymic stromal lymphopoietin (TSLP) can be produced by lung epithelial cells and lung fibroblasts through TLR3/7/8 signaling.38 In response to TSLP, IL-25 and IL-33 are produced in abundance and DCs are instructed to activate naïve T cells, causing Th2 polarization (Figure 3). Additionally, TLR3 signaling disrupts barrier function through TIR-domain-containing adapter-inducing interferon-ß (TRIF) activation, which loosens epithelial tight junctions.25 Consequently, the airway suffers from hyperresponsiveness and airway remodeling. This interaction is believed to be the causative link between early childhood respiratory infections, such as respiratory syncytial virus, influenza and human rhinovirus, and subsequent asthma development. Unlike viruses, fungi predominantly interact with mucosal immune cells by engagement of CLRs and RAGE (receptor for advanced glycation end products) as well as recognition of fungal proteases via PARs (protease-activated receptors).37 Activation of CLR results in the production of inflammatory and Th-2 polarizing cytokines, while stimulation of RAGE results in IL-33 production.39–41 Lastly, PAR activation on eosinophils and epithelial cells results in the release of granule proteins and IL-33/TSLP, respectively.42,43 Collectively, these diverse immunomodulatory mechanisms allow microbes to hijack airway immune cells and instruct them to produce allergic mediators in order to perpetuate barrier dysfunction and predispose the host to asthma development.
The Mucosal Microbiome
Microbial communities are vital for maintaining human health. These organisms not only protect the host from diseases through competing against pathogenic organisms and preventing their invasion and growth but they also support other physiological and immunological functions of the host via their endogenous synthesis of nutrient metabolites and vitamins.44,45 Pathogenic microbes can be distinguished from commensal organisms as they carry specific adhesive and invasive molecules, such as adhesins and invasins, which enable them to adhere to and invade tissues, causing harm to the host.46 In contrast, the inability of commensals to adhere to or invade the sterile epithelium makes the host tissue tolerant to their presence, as they can be easily washed off by mucus and expelled from the host through peristaltic movements in the gut or ciliary movements of the airway epithelium.46
Gastrointestinal Commensals
The gut is home to the largest microbial community. Although the importance of the gut microbiome in early infancy is well established, the timing of microbial assembly in the infant gut remains elusive. Microbial colonization in fetal membranes had only been observed in the presence of infection, and, thus, it was believed to be a phenomenon that occurred due to infection, and not during normal pregnancies.47 These early analyses, which employed culture-based techniques and microscopy, showed that the meconium (earliest stool sample from infants) is free of bacteria in the majority of studied subjects.48–50 Subsequent studies of the amniotic fluid also showed no evidence of bacterial growth in over 90% of the tested samples, suggesting that fetal development may occur in a bacteria-free environment.51,52
However, this hypothesis is now being challenged, as microbial assembly has recently been suggested to occur during the in utero phase.53 In this regard, populations of the Proteobacteria phylum, particularly the Enterobacter and Escherichia/Shigella genera, were found in the amniotic fluid, placenta and colostrum of a small cohort of 15 mothers and their babies.54 Consistent with this observation, other studies confirmed the presence of many bacterial species, such as Lactobacillus, Bifidobacterium, and Clostridium leptum, in placental tissues.54–57 Furthermore, studies that examined the meconium of newborns delivered via both vaginal birth and C-section found no differences in the composition of the gut microbiome, suggesting that colonization of the gut occurs prior to delivery and is independent of the birthing method.58,59 However, while these causal relationships appear to provide evidence that the fetal gut is not sterile, large scale and direct evidence of microbial assembly in utero is lacking and the developmental mechanisms of microbial colonization remain unclear. In fact, the largest single study of 320 placental samples to date has suggested that the presence of a microbiome in this tissue may be associated with antepartum infection in the mothers, rather than via a homeostatic mechanism.60
Besides maternal vaginal and intestinal flora, breast milk also helps to seed the infant microbiome, as it is heavily enriched with microbes. In fact, a longitudinal study of mother-infant pairs reported that 27.7% of beneficial bacteria present in the intestinal tracts of infants is obtained from the mother’s breast milk.61 The origin of the breast milk microbiome has been the subject of much debate. It was believed that the milk microbiome was a result of contamination from the mother’s skin during infant suckling, and many studies note the similarities between the adult skin microbiome and milk microbiome, particularly among the genera Staphylococcus and Corynebacterium.62 Furthermore, the discovery of anaerobic species, which are exclusively associated with the gut environment, in breast milk suggested that live bacteria from the maternal gut may travel to the mammary gland via interactions between epithelial cells, immune cells, and bacteria.63,64 Results regarding the composition of breast milk microbiota have been varied. Culture-based analyses isolated bacterial species belonging to 5 families: Micrococcaceae, Streptococcaceae, Corynebacteriaceae, Lactobacillaceae, and Neisseriaceae.65 In contrast, studies with more advanced sequencing technologies discovered 9 core genera in breast milk, including Staphylococcus, Streptococcus, Serratia, Pseudomonas, Corynebacterium, Ralstonia, Propionibacterium, Sphingomonas, and Bradyrhizobiaceae.66 Nevertheless, Staphylococcaceae and Streptococcaeceae have been reported as dominant species within breast milk.67
Notably, breast milk feeding appears to significantly dictate the diversity of the infant gut microbiome. Consistent breastfeeding appears to favor Bifidobacteria and Bacteroides as the dominant species. In contrast, infants fed formula have an increased abundance of anaerobes, such as Enterobacteriaceae, for long periods of time.68,69 Besides transferring microbes, breast milk provides other prebiotics that support beneficial bacterial colonization.70 For example, Bifidobacteria, Bacteroides, and Lactobacillus are adapted to utilize human milk oligosaccharides (HMOs), while these complex glycans inhibit the growth of other harmful bacteria.71,72 Furthermore, HMOs can prevent pathogen adhesion to intestinal epithelium by serving as soluble glycan receptor decoys. Mechanistically, HMOs could resemble the structures of viral receptors and prevent adherence to cells, therefore preventing infection.73–75 Lastly, breast milk also contains immunoglobulins, which provide passive immunity to newborn infants. The prominent antibodies present in breast milk are IgA (90–95%), IgM (2–5%) and IgG (<1%).44,76 Along with microbes, the presence of these antibodies helps to shape the composition of the infant gut microbiota and promotes a symbiotic relationship within the host.77
Airway Microbiome
Similar to the gut, it is now widely accepted that lungs are not sterile, as was previously thought.78 Distinct airway microbial profiles begin to cluster immediately after birth.79 The sequence of events may start with bacteria colonizing the intestine early in life, prior to its appearance in the airways.80,81 Alternatively, it has been hypothesized that the lower airway microbiome is derived from that of the upper airway via either microbial aspiration or direct inhalation, to a lesser extent.82–84 In healthy subjects, the respiratory microbiome has a low density and modest growth rate. With the use of new technologies such as next-generation sequencing of the 16S ribosomal RNA gene, more than 2000 bacterial genomes per cm2 were detected in bronchoalveolar lavage specimens from normal healthy lungs.85 Although there is still a relative lack of research performed on the microbiota in the lungs compared with that at other sites such as the gut, there is growing evidence of the uniqueness of the microbial community in the respiratory tract. Nevertheless, some types of bacteria in the airways and intestine do overlap, with the two most abundant phyla detected in both being Firmicutes and Bacteroidetes.80,81
Gastrointestinal-Airway Microbial Communication
Overlapping pathologies in intestinal and respiratory diseases suggest that a gut-lung communication pathway may exist. Furthermore, patients with chronic gastrointestinal diseases have a higher prevalence of pulmonary diseases.86 However, the mechanisms by which the microbiota in the gut and lung interact are not fully understood. Russell and coworkers demonstrated that a disturbance of the intestinal microbiota by vancomycin administration, which reduces gut flora diversity, early in life induced an increase in susceptibility to sensitization and asthma later in life.87 Interestingly, treatment of age-matched mice with streptomycin did not lead to reduce microbial diversity, and, ultimately, these mice were no more sensitive to asthma induction later in life than untreated controls. This result suggests that temporary disturbance of the microflora by the use of an antibiotic is not harmful unless it leads to a lasting perturbation of flora diversity. However, disturbances of the airway microbiota may also result in alterations of the intestinal microbial composition. In a study by Vital et al, allergic airway sensitization in a mouse model led to a shift in the intestinal microbial composition, demonstrating that the gut microbiota is dynamic and can be altered in response to airway insults.88 In support of this view, Wang et al found alterations in the gut microbial composition of mice after respiratory infection with influenza.89 Although the airway microbiota was not examined in these studies, the changes that occurred in the lungs in response to the gut or vice versa suggest that gut-lung axis communication works in both directions and may constitute a positive feedback loop.
Furthermore, the connection between food allergies and asthma development also supports the presence of a gut-lung mucosal crosstalk. In this regard, the leaky intestinal barrier may lead to mislocation of food antigens into the laminal propria, causing inflammation and sensitization to food allergens.90 These antigens can be translocated via leaky membranes into the lung interstitium, resulting in cross-susceptibility to food allergies and asthma, or even occurring in succession, starting with food allergies and progressing later to asthma.91,92
Mucosal Immunity-Microbiome Crosstalk
Homeostatic Interaction During Development
Early microbial colonization in the GIT and other mucosal sites, such as the respiratory tract and skin, occurs in tandem with the development of the immune system. During early microbial assembly, the immune system is amenable to the colonization of organisms because of its immaturity. The relative lack of cytokine responses results in blunted inflammatory responses, allowing for the settlement and expansion of the microbiome in various niches.93 Moreover, during this early period of rapid development of the immune system, maternally derived IgA protects the newborn from infections.94 Interestingly, in mice, IgA production is influenced by the gut microbial environment, as the microbiome contributes to the development and function of mucosal lymphoid structures, such as the PPs, which is the site for IgA production in the GIT.95,96 Indeed, mice raised in germ-free conditions have underdeveloped PPs and a reduced number of CD4+ T cells and IgA-producing plasma cells.96 Conversely, the homeostatic role of IgA in regulating gut commensals has also been observed in humans. In this regard, IgA deficiency has been associated with an altered composition of gut microbiota and increased microbial translocation, leading to a depletion of anti-inflammatory species, such as Faecalibacterium, and overexpansion of the pro-inflammatory species Gammaproteaobacteria and Prevotella.97 Furthermore, intestinal DCs also play a critical role in microbial imprinting via mechanisms that involve sampling of microbes for antigen presentation and induction of protective IgA.98
Although IgA is the dominant antibody in the gut, IgM and IgG are also present. Their role is, however, less well characterized. While IgA and IgM are both produced locally by B cells, IgG is a circulating antibody that can gain entry into the gut through the binding of Fc receptors.99 Consequently, IgG induction is a result of systemic immune activation in response to non-self antigens. As a result, IgG has been shown to target distinct microbial species, in contrast to IgA and IgM, which were shown to target the same microbial populations.100 Commensal bacteria also provide supporting nutrient metabolites for the development of the immune system, notably via their production of SCFAs. SCFAs exert anti-inflammatory activity on gastrointestinal membranes, boost antibody production and play a key role in the growth of the intestinal lining and the maintenance of its integrity.45 Furthermore, SCFAs can influence the differentiation of certain immune cell populations and, therefore, have immunomodulatory properties. For example, SCFAs can skew T helper differentiation away from the allergenic Th2 phenotype and, thus, prevent the induction of allergic sensitization.45
Aberrant Interaction During Dysbiosis and Barrier Disruption
While perturbation of the intestinal microbiome has been well established as a cardinal feature of obesity and metabolic disorders,101 early gut dysbiosis has been linked to the development of asthma. For example, infants with a reduced Bifidobacterial to Clostridium difficile (C. difficile) ratio have a significantly increased risk for asthma development in later childhood.102 This finding was mirrored in the KOALA birth cohort study of 957 infants in the Netherlands, which demonstrated that colonization of C. difficile in the feces at one month of age was significantly associated with the subsequent development of eczema, wheezing and asthma, as assessed by parent interviews and by the determination of specific immunoglobulin E (sIgE) levels from blood samples collected at 2 years of age.103 Likewise, in a study of 117 children, colonization of Bacteroides fragilis at the age of 3 weeks was associated with a significantly increased risk for developing asthma at the age of 3 years.104 Furthermore, diminished intestinal microbial diversity in asthmatic children has been documented.105 The genera Lachnospira, Veillonella, Faecalibacterium, and Rothia were significantly decreased in the guts of infants at risk for asthma in the first 100 days of life.106 Changes in the abundance of Lachnospira and Clostridium neonatale in the first 3 months of life were also linked to asthma development during the pre-school period.107 In addition, children with asthma presented a significantly lower abundance of the genera Faecalibacterium and Roseburia.108 Similar to the gastrointestinal tract, the airway microbiome also differs between healthy and diseased individuals.109 In asthmatic children, nasal secretion samples showed a distinct microbiota composition dominated by the genus Moraxella.110 Proteobacteria (Hemophilus, Moraxella and Neisseria) were also more frequent in the asthmatic bronchial tree than in that of healthy adults.85,111 Conversely, Bacteroidetes, particularly Prevotella, were significantly reduced in asthmatics.85 In addition to bacteria, the presence of human rhinovirus in the nasopharynx during early life can impact asthma development.112,113
Several studies have linked airway dysbiosis to asthma development through microbe-mediated alteration of the balance between Th2 and other immune signals. For instance, resident airway microbes have been shown to play a critical role in the regulation of basophil activation and IgE production,114 while Actinomycetaceae and Moraxella catarrhalis are able to induce eosinophilia, epithelial damage and subsequent inflammatory Th2 cytokine expression (IL-33, IL-8).110 Streptococcus, Prevotella, and Neisseria species are also associated with a Th2 cytokine profile; however, this allergic inflammatory response is driven by macrophages and T cells.115,116 Other studies have also shown that Enterococcus faecalis can suppress Th17 immune responses and elevate symptoms of allergic asthma.117
Interestingly, the airway microbiota may also drive specific asthma endotypes and phenotypes.118 In this regard, the presence of eosinophilia during asthma has been shown to be related to changes in the microbiome. For instance, elevated eosinophil counts were documented in a study of 321 neonates, whose airways are predominantly colonized by Streptococcus pneumoniae, Haemophilus influenzae, or Moraxella catarrhalis.119 Conversely, eosinophil abundance in bronchial biopsies were found to be negatively associated with airway colonization of Proteobacteria, Neisseria, Bacteroides, and Rothia.120–122 Neutrophilia has also been linked to airway microbial alterations, characterized by a general loss of diversity.121,122 A detailed dissection of the microbiome in these subjects revealed specific reduction in abundance of Streptococcus, Gemella, and Porphyromonas. Furthermore, Th2 inflammation, the signature of asthma, has been recently linked to fungal dysbiosis with specific enrichment of Trichoderma and Penicillium species.123
Collectively, the studies above provide evidence for the need to properly establish an early microbial colony in the mucosae during development so that homeostatic interaction between the microbiome and mucosal immune system is maintained. Otherwise, a disrupted immune-microbiome crosstalk could generate inflammatory pathogenic factors that contribute to epithelial barrier dysfunction and asthma development. Notably, asthmatic Th2 cytokines, such as IL-4, IL-5, IL-13, and TSLP, can negatively impact airway epithelial barrier functions.124 In this regard, Xu et al demonstrated that IL-13 could suppress surfactant D expression in the airway epithelium.125 Similarly, IL-4 and IL-5 have been shown to inhibit the expression of other secretory airway barrier factors, such as surfactant B and C, two critical regulators of respiratory functions.126,127 Ahdieh et al also demonstrated through an in vitro study on human lung adenocarcinoma cells that both IL-4 and IL-13 decreased the expression of epithelial junction proteins, such as ZO-1 and occludin, resulting in barrier leakage.128 Furthermore, decreased caveolin-1 in adult asthma patients was correlated with increased expression of the pro-inflammatory cytokine TSLP.129
Of current interest, in the pathophysiology of COVID-19, microbiome alterations have been associated with significant involvement of the respiratory and gastrointestinal tracts.130,131 In this regard, COVID-19 patients exhibit reduced bacterial diversity in the gut and reduction of healthy symbionts, in addition to an increase in the abundance of pathogenic Streptococcus, Rothia, Veillonella, and Actinomyces.132 Consistent with this report, the gut microbiome of COVID-19 patients was documented with an enrichment of opportunistic bacteria (Clostridium, Actinomyces, and Bacteroides) and fungi (Candida albicans, Candida auris, and Aspergillus flavus) and depletion of beneficial commensals, such as Eubacterium, Faecalibacterium prausnitzii, and Lachnospiraceae.133 More importantly, the severity of the symptoms related to COVID-19 was linked to an expansion of pathogenic Clostridium and reduction of Alistipes and Bacteroides commensals. Limited information with regard to respiratory microbes have been documented in COVID-19 with the general consensus that opportunistic fungal and bacterial infections are widespread in the lungs of patients suffering from this disease.134,135 Specifically, Acinetobacter, Chryseobacterium, Burkholderia, Brevundimonas, Sphingobium, and Enterobacteriaceae are the predominant bacteria while Cutaneotrichosporon, Issatchenkia, Wallemia, Cladosporium, Alternaria, Dipodascus, Mortierella, Aspergillus, Naganishia, Diutina, and Candida are the prevalent fungi in the airways of COVID-19 patients.135 These collective findings highlight the impact of mucosal microbiota on the susceptibility to SARS-CoV2 infection and severity of COVID-19.
Therapeutic Implications
Currently, the mainstay of bronchial asthma treatment is to control airway inflammation through the use of inhaled corticosteroids (ICSs). Although the mechanism is thought to occur by reducing inflammatory cytokine levels, newer evidence points to the role of ICSs in stabilizing the airway epithelium and in promoting epithelial barrier synthesis.25 Interestingly, an increase in Proteobacteria in bronchial epithelial brushings from adults with mild to moderate asthma after ICS treatment has been reported.85,136 Corroborating studies also showed that the microbiome of ICS-responsive asthmatics is enriched with Neisseria, Moraxella, and Haemophilus.137 Therefore, ICSs may alter the lung microbiome by promoting the colonization of potentially pathogenic bacterial strains and subsequent ICS unresponsiveness.
In light of these findings, microbial manipulation may represent a novel therapeutic strategy in treating asthma, as microbiota seem to exert a considerable effect on the development of epithelial barriers, local immunological responses, and even the responsiveness to the standard of care for this condition. In this regard, introducing certain commensal strains in the form of probiotic supplementation may reduce allergy and asthma risk. Bacteria used for probiotics mainly belong to lactic acid bacteria (Lactobacillus, Streptococcus, Enterococcus), Bifidobacterium, and non-pathogenic Escherichia coli.138 Specifically, administration of Lactobacillus rhamnosus has been shown to prevent asthma development.139 When asthmatic children were orally administered a combination of Lactobacillus acidophilus, Bifidobacterium bifidum, and Lactobacillus delbrueckii subspecies Bulgaricus, an improvement in lung function and a reduction in asthma exacerbations were observed.140 Lactobacillus paracasei, Lactobacillus fermentum, and Lactobacillus gasseri could also alleviate asthma symptoms and decrease IgE levels in children.141,142 However, two meta-analyses of a total of 31 studies with over 5000 participants separately found no significant changes in the risk of asthma with probiotic supplementation.143,144 These conflicting data on the benefit of probiotics in asthma prevention may be related to individual differences in the ability to incorporate probiotics into the microbiome.145
Supplementing with prebiotics may provide the dietary support required for probiotics. Prebiotics are specific dietary fibers that are used as a nutritional source for commensal gut bacteria. These fermentable fibers are used by gut microbiota to produce SCFAs.146 Prebiotics were found to have a positive influence on microflora diversity, stimulating the growth and activity of beneficial bacteria in the colon.146,147 In one study, infants fed prebiotic formula had an intestinal microflora diversity closer to that of breastfed infants, with breastfeeding known to reduce allergy and asthma risk compared with that associated with regular formulas.148 Bacterial lysates may also be beneficial as asthma therapeutics. In contrast to probiotics, bacterial lysates are not living organisms and, therefore, have only transient effects. For example, OM-85, an alkaline lysate of 21 respiratory tract pathogens, was able to reduce disease exacerbations in asthmatic children.149–151 Similarly, sublingual administration of a bacterial lysate tablet (PMBL) improved asthmatic symptoms.152 These bacterial compounds may exert immunoregulatory mechanisms in the gut, which can act on distant mucosal sites to reduce allergic inflammation and airway hyperreactivity. However, the evidence for the use of probiotics, prebiotics, or other microbial therapeutics for the prevention of asthma is currently too weak for any recommendation to be drawn.144,153 Similarly, in light of the current COVID-19 pandemic, there is no clinical evidence of microbiota modulation for the treatment of this disease. Nevertheless, emerging speculations highlight the role of targeting microbiota as primary or adjuvant therapies.154,155 In this regard, microbiota modulators can be used to alleviate the GI distress and protect the respiratory system from opportunistic bacterial and fungal infections. Additional research is needed before a general recommendation can be provided to harness the therapeutic powers of microbes in asthma and other emerging respiratory diseases.
Conclusions
In conclusion, the microbiota plays an important role in the development of protective barriers and mucous membrane immunity. Correlations exist between immune-microbiome crosstalks, epithelial barrier dysfunction, and asthma development. There is likely to be positive feedback between allergic inflammation-induced dysbiosis that, in turn, further promotes further barrier dysfunction, which leads to increased displacement of allergens/microbes to exacerbate the allergic response. A better appreciation of how these mechanisms interconnect will be crucial for enhancing our understanding of the pathogenesis of asthma and aiding the development of early intervention and therapeutic methods.
Abbreviations
C. difficile, Clostridium difficile; CLR, C-type lectin receptors; DCs, dendritic cells; GIT, gastrointestinal tract; ICSs, inhaled corticosteroids; HMOs, human milk oligosaccharides; PAMPs, pathogen-associated molecular patterns; PRR, pattern-recognition receptors; PAR, protease-activated receptors; PPs, Peyer’s patch; SCFA, short-chain fatty acid; sIgA, secretory immunoglobulin A; sIgE, specific immunoglobulin E; SP, surfactant protein; Th2, T helper 2; TJs, tight junctions; TLR, toll-like receptor; TRIF, TIR-domain-containing adapter-inducing interferon-ß; TSLP, thymic stromal lymphopoietin; ZO, zonula occludens.
Disclosure
The author declares that they have no competing interests for this work.
References
- 1.Stern J, Pier J, Litonjua AA. Asthma epidemiology and risk factors. Semin Immunopathol. 2020;42(1):5–15. doi: 10.1007/s00281-020-00785-1 [DOI] [PubMed] [Google Scholar]
- 2.Kaur R, Chupp G. Phenotypes and endotypes of adult asthma: moving toward precision medicine. J Allergy Clin Immunol. 2019;144(1):1–12. doi: 10.1016/j.jaci.2019.05.031 [DOI] [PubMed] [Google Scholar]
- 3.Kuruvilla ME, Lee FE, Lee GB. Understanding asthma phenotypes, endotypes, and mechanisms of disease. Clin Rev Allergy Immunol. 2019;56(2):219–233. doi: 10.1007/s12016-018-8712-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akdis CA, Arkwright PD, Brüggen MC, et al. Type 2 immunity in the skin and lungs. Allergy. 2020;75(7):1582–1605. doi: 10.1111/all.14318 [DOI] [PubMed] [Google Scholar]
- 5.Barcik W, Boutin RCT, Sokolowska M, Finlay BB. The role of lung and gut microbiota in the pathology of asthma. Immunity. 2020;52(2):241–255. doi: 10.1016/j.immuni.2020.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strachan DP. Hay fever, hygiene, and household size. Br Med J. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wampach L, Heintz-Buschart A, Fritz JV, et al. Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential. Nat Commun. 2018;9(1):5091. doi: 10.1038/s41467-018-07631-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ege MJ. The hygiene hypothesis in the age of the microbiome. Ann Am Thorac Soc. 2017;14(Supplement_5):S348–53. doi: 10.1513/AnnalsATS.201702-139AW [DOI] [PubMed] [Google Scholar]
- 9.Mitre E, Susi A, Kropp LE, Schwartz DJ, Gorman GH, Nylund CM. Association between use of acid-suppressive medications and antibiotics during infancy and allergic diseases in early childhood. JAMA Pediatr. 2018;172(6):e180315. doi: 10.1001/jamapediatrics.2018.0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguilera AC, Dagher IA, Kloepfer KM. Role of the microbiome in allergic disease development. Curr Allergy Asthma Rep. 2020;20(9):44. doi: 10.1007/s11882-020-00944-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukacs NW, Huang YJ. Microbiota-immune interactions in asthma pathogenesis and phenotype. Curr Opin Immunol. 2020;66:22–26. doi: 10.1016/j.coi.2020.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanidad KZ, Zeng MY. Neonatal gut microbiome and immunity. Curr Opin Microbiol. 2020;56:30–37. doi: 10.1016/j.mib.2020.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd-Price J, Mahurkar A, Rahnavard G, et al. Strains, functions and dynamics in the expanded human microbiome project. Nature. 2017;550(7674):61–66. doi: 10.1038/nature23889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sulaiman I, Schuster S, Segal LN. Perspectives in lung microbiome research. Curr Opin Microbiol. 2020;56:24–29. doi: 10.1016/j.mib.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hufnagl K, Pali-Schöll I, Roth-Walter F, Jensen-Jarolim E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin Immunopathol. 2020;42(1):75–93. doi: 10.1007/s00281-019-00775-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugita K, Soyka MB, Wawrzyniak P, et al. Outside-in hypothesis revisited: the role of microbial, epithelial, and immune interactions. Ann Allergy Asthma Immunol. 2020;23:S1081–1206(20)30355–0. [DOI] [PubMed] [Google Scholar]
- 17.Hellings PW, Steelant B. Epithelial barriers in allergy and asthma. J Allergy Clin Immunol. 2020;145(6):1499–1509. doi: 10.1016/j.jaci.2020.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goleva E, Berdyshev E, Leung DY. Epithelial barrier repair and prevention of allergy. J Clin Invest. 2019;129(4):1463–1474. doi: 10.1172/JCI124608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odenwald MA, Turner JR. The intestinal epithelial barrier: a therapeutic target? Nat Rev Gastroenterol Hepatol. 2017;14:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chelakkot C, Ghim J, Ryu SH. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med. 2018;50:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279:70–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knudsen L, Ochs M. The micromechanics of lung alveoli: structure and function of surfactant and tissue components. Histochem Cell Biol. 2018;150:661–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hohlfeld JM. The role of surfactant in asthma. Nutrients. 2018;10:988. [Google Scholar]
- 24.Georas SN, Rezaee F. Epithelial barrier function: at the front line of asthma immunology and allergic airway inflammation. J Allergy Clin Immunol. 2014;134(3):509–520. doi: 10.1016/j.jaci.2014.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gon Y, Hashimoto S. Role of airway epithelial barrier dysfunction in pathogenesis of asthma. Allergol Int. 2018;67(1):12–17. doi: 10.1016/j.alit.2017.08.011 [DOI] [PubMed] [Google Scholar]
- 26.Deckers J, De Bosscher K, Lambrecht BN, Hammad H. Interplay between barrier epithelial cells and dendritic cells in allergic sensitization through the lung and the skin. Immunol Rev. 2017;278:131–144. [DOI] [PubMed] [Google Scholar]
- 27.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6(1):10–21. doi: 10.1016/j.chom.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. 2017;46(4):562–576. doi: 10.1016/j.immuni.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muñoz-Wolf N, Lavelle EC. Innate immune receptors. Methods Mol Biol. 2016;1417(1–43). [DOI] [PubMed] [Google Scholar]
- 30.Nikolakopoulou C, Willment JA, Brown GD. C-type lectin receptors in antifungal immunity. Adv Exp Med Biol. 2020;1204:1–30. [DOI] [PubMed] [Google Scholar]
- 31.Li M, Wang Y, Sun Y, Cui H, Zhu SJ, Qiu HJ. Mucosal vaccines: strategies and challenges. Immunol Lett. 2020;217:116–125. doi: 10.1016/j.imlet.2019.10.013 [DOI] [PubMed] [Google Scholar]
- 32.Vancamelbeke M, Vermeire S. The intestinal barrier: a fundamental role in health and disease. Expert Rev Gastroenterol Hepatol. 2017;11(9):821–834. doi: 10.1080/17474124.2017.1343143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heijink IH, Kuchibhotla VNS, Roffel MP, et al. Epithelial cell dysfunction, a major driver of asthma development. Allergy. 2020;75(8):1898–1913. doi: 10.1111/all.14421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh SS, Wang J, Yannie PJ, Ghosh S. Intestinal barrier dysfunction, LPS translocation, and disease development. J Endocr Soc. 2020;4(2):bvz039. doi: 10.1210/jendso/bvz039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Budden KF, Gellatly SL, Wood DL, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15(1):55–63. doi: 10.1038/nrmicro.2016.142 [DOI] [PubMed] [Google Scholar]
- 36.Smits HH, Lepm VDV, von Mutius E, Hiemstra PS. Childhood allergies and asthma: new insights on environmental exposures and local immunity at the lung barrier. Curr Opin Immunol. 2016;42:41–47. doi: 10.1016/j.coi.2016.05.009 [DOI] [PubMed] [Google Scholar]
- 37.Bartemes KR, Kita H. Innate and adaptive immune responses to fungi in the airway. J Allergy Clin Immunol. 2018;142(2):353–363. doi: 10.1016/j.jaci.2018.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jartti T, Gern JE. Role of viral infections in the development and exacerbation of asthma in children. J Allergy Clin Immunol. 2017;140(4):895–906. doi: 10.1016/j.jaci.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM. Collaborative induction of inflammatory responses by dectin-1 and toll-like receptor 2. J Exp Med. 2003;197(9):1107–1117. doi: 10.1084/jem.20021787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura Y, Chihara K, Honjoh C, et al. Dectin-1-mediated signaling leads to characteristic gene expressions and cytokine secretion via spleen tyrosine kinase (Syk) in rat mast cells. J Biol Chem. 2014;289(45):31565–31575. doi: 10.1074/jbc.M114.581322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oczypok EA, Milutinovic PS, Alcorn JF, et al. Pulmonary receptor for advanced glycation end-products promotes asthma pathogenesis through IL-33 and accumulation of group 2 innate lymphoid cells. J Allergy Clin Immunol. 2015;136(3):747–56 e4. doi: 10.1016/j.jaci.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snelgrove RJ, Gregory LG, Peiro T, et al. Alternaria- derived serine protease activity drives IL-33-mediated asthma exacerbations. J Allergy Clin Immunol. 2014;134(3):583–92 e6. doi: 10.1016/j.jaci.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kouzaki H, O’Grady SM, Lawrence CB, Kita H. Proteases induce production of thymic stromal lymphopoietin by airway epithelial cells through protease-activated receptor-2. J Immunol. 2009;183(2):1427–1434. doi: 10.4049/jimmunol.0900904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenney ES, Kendall MM. Microbiota and pathogen ‘pas de deux’: setting up and breaking down barriers to intestinal infection. Pathog Dis. 2016;74(5):ftw051. doi: 10.1093/femspd/ftw051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z, Quan G, Jiang X, et al. Effects of metabolites derived from gut microbiota and hosts on pathogens. Front Cell Infect Microbiol. 2018;8:314. doi: 10.3389/fcimb.2018.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4(12):953–964. doi: 10.1038/nri1499 [DOI] [PubMed] [Google Scholar]
- 47.Walker RW, Clemente JC, Peter I, Loos RJF. The prenatal gut microbiome: are we colonized with bacteria in utero? Pediatr Obes. 2017;12(Suppl 1):3–17. doi: 10.1111/ijpo.12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Escherich TH. The intestinal bacteria of the neonate and breast-fed infant. Rev Infect Dis. 1988;10(6):1220–1225. doi: 10.1093/clinids/10.6.1220 [DOI] [PubMed] [Google Scholar]
- 49.Burrage S. Bacteria in the supposedly sterile meconium. J Bacteriol. 1927;13:47. [Google Scholar]
- 50.Hall IC. Bacterial flora of first specimens of meconium passed by fifty newborn infants. Arch Pediatr Adolesc Med. 1934;47(6):1279–1285. doi: 10.1001/archpedi.1934.01960130103007 [DOI] [Google Scholar]
- 51.Prevedourakis C, Papadimitriou G, Ioannidou A. Isolation of pathogenic bacteria in the amniotic fluid during pregnancy and labor. Am J Obstet Gynecol. 1970;106(3):400–402. doi: 10.1016/0002-9378(70)90365-0 [DOI] [PubMed] [Google Scholar]
- 52.Lewis JF, Johnson P, Miller P. Evaluation of amniotic fluid for aerobic and anaerobic bacteria. Am J Clin Pathol. 1976;65(1):58–63. doi: 10.1093/ajcp/65.1.58 [DOI] [PubMed] [Google Scholar]
- 53.Perez-Muñoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016;6(1):23129. doi: 10.1038/srep23129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones HE, Harris KA, Azizia M, et al. Differing prevalence and diversity of bacterial species in fetal membranes from very preterm and term labor. PLoS One. 2009;4(12):e8205. doi: 10.1371/journal.pone.0008205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rautava S, Collado MC, Salminen S, Isolauri E. Probiotics modulate host-microbe interaction in the placenta and fetal gut: a randomized, double-blind, placebo-controlled trial. Neonatology. 2012;102(3):178–184. doi: 10.1159/000339182 [DOI] [PubMed] [Google Scholar]
- 57.Makino H, Kushiro A, Ishikawa E, et al. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS One. 2013;8(11):e78331. doi: 10.1371/journal.pone.0078331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madan JC, Hoen AG, Lundgren SN, et al. Association of cesarean delivery and formula supplementation with the intestinal microbiome of 6-week-old infants. JAMA Pediatr. 2016;170(3):212–219. doi: 10.1001/jamapediatrics.2015.3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ardissone AN, de la Cruz DM, Davis-Richardson AG, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One. 2014;9(3):e90784. doi: 10.1371/journal.pone.0090784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6(237):237ra65. doi: 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pannaraj PS, Li F, Cerini C, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171(7):647–654. doi: 10.1001/jamapediatrics.2017.0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dimitriu PA, Iker B, Malik K, Leung H, Mohn WW, Hillebrand GG. New insights into the intrinsic and extrinsic factors that shape the human skin microbiome. mBio. 2019;10(4):e00839–19. doi: 10.1128/mBio.00839-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodríguez JM. The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv Nutr. 2014;5(6):779–784. doi: 10.3945/an.114.007229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martín RO, Langa S, Reviriego C, et al. The commensal microflora of human milk: new perspectives for food bacteriotherapy and probiotics. Trends Food Sci Technol. 2004;15(121–7):121–127. doi: 10.1016/j.tifs.2003.09.010 [DOI] [Google Scholar]
- 65.Gavin A, Ostovar K. Microbiological characterization of human milk. J Food Prot. 1977;40(9):614–616. doi: 10.4315/0362-028X-40.9.614 [DOI] [PubMed] [Google Scholar]
- 66.Hunt KM, Foster JA, Forney LJ, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. 2011;6(6):e21313. doi: 10.1371/journal.pone.0021313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kordy K, Gaufin T, Mwangi M, et al. Contributions to human breast milk microbiome and enteromammary transfer of Bifidobacterium breve. PLoS One. 2020;15(1):e0219633. doi: 10.1371/journal.pone.0219633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ho NT, Li F, Lee-Sarwar KA, et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat Commun. 2018;9(1):4169. doi: 10.1038/s41467-018-06473-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee SA, Lim JY, Kim B-S, et al. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutr Res Pract. 2015;9(3):242–248. doi: 10.4162/nrp.2015.9.3.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lyons KE, Ryan CA, Dempsey EM, Ross RP, Stanton C. Breast milk, a source of beneficial microbes and associated benefits for infant health. Nutrients. 2020;12(4):1039. doi: 10.3390/nu12041039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bidart GN, Rodríguez-Díaz J, Monedero V, Yebra MJ. A unique gene cluster for the utilization of the mucosal and human milk-associated glycans galacto-N-biose and lacto-N-biose in Lactobacillus casei. Mol Microbiol. 2014;93(3):521–538. doi: 10.1111/mmi.12678 [DOI] [PubMed] [Google Scholar]
- 72.Triantis V, Bode L, van Neerven RJJ. Immunological effects of human milk oligosaccharides. Front Pediatr. 2018;6:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weichert S, Koromyslova A, Singh BK, et al. Structural basis for norovirus inhibition by human milk oligosaccharides. J Virol. 2016;90(9):4843–4848. doi: 10.1128/JVI.03223-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang XF, Tan M, Chhabra M, Dai YC, Meller J, Jiang X. Inhibition of histo-blood group antigen binding as a novel strategy to block norovirus infections. PLoS One. 2013;8(7):e69379. doi: 10.1371/journal.pone.0069379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morozov V, Hansman G, Hanisch F-G, Schroten H, Kunz C. Human milk oligosaccharides as promising antivirals. Mol Nutr Food Res. 2018;62(6):1700679. doi: 10.1002/mnfr.201700679 [DOI] [PubMed] [Google Scholar]
- 76.Anand S, Mande SS. Diet, microbiota and gut-lung connection. Front Microbiol. 2018;9:2147. doi: 10.3389/fmicb.2018.02147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rogier EW, Frantz AL, Bruno MEC, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci U S A. 2014;111:3074–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pulvirenti G, Parisi GF, Giallongo A, et al. Lower airway microbiota. Front Pediatr. 2019;7:393. doi: 10.3389/fped.2019.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Biesbroek G, Tsivtsivadze E, Sanders EAM, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190(11):1283–1292. doi: 10.1164/rccm.201407-1240OC [DOI] [PubMed] [Google Scholar]
- 80.Marsland BJ, Trompette A, Gollwitzer ES. The gut-lung axis in respiratory disease. Ann Am Thorac Soc. 2015;12(Suppl 2):S150–6. [DOI] [PubMed] [Google Scholar]
- 81.Madan JC, Koestler DC, Stanton BA, et al. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. mBio. 2012;3(4):e00251–12. doi: 10.1128/mBio.00251-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Segal LN, Alekseyenko AV, Clemente JC, et al. Erratum to: enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2014;2(1):19. doi: 10.1186/2049-2618-2-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Venkataraman A, Bassis CM, Beck JM, et al. Application of a neutral community model to assess structuring of the human lung microbiome. mBio. 2015;6(1):e02284–14. doi: 10.1128/mBio.02284-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marsh RL, Kaestli M, Chang AB, et al. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome. 2016;4:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578. doi: 10.1371/journal.pone.0008578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H, Liu J-S, Peng S-H, et al. Gut-lung crosstalk in pulmonary involvement with inflammatory bowel diseases. World J Gastroenterol. 2013;19(40):6794–6804. doi: 10.3748/wjg.v19.i40.6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Russell SL, Gold MJ, Hartmann M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13(5):440–447. doi: 10.1038/embor.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vital M, Harkema JR, Rizzo M, Tiedje J, Brandenberger C. Alterations of the murine gut microbiome with age and allergic airway disease. J Immunol Res. 2015;2015:892568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang J, Li F, Wei H, Lian ZX, Sun R, Tian Z. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J Exp Med. 2014;211:2397–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berin MC, Sampson HA. Mucosal immunology of food allergy. Curr Biol. 2013;23(9):R389–400. doi: 10.1016/j.cub.2013.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou H, Dai C, Pan J. Pediatric asthma and food allergy. Indian J Pediatr. 2017;84(8):585–590. doi: 10.1007/s12098-017-2326-0 [DOI] [PubMed] [Google Scholar]
- 92.Savage JH, Lee-Sarwar KA, Sordillo J, et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy. 2018;73(1):145–152. doi: 10.1111/all.13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Toscano M, De Grandi R, Grossi E, Drago L. Role of the human breast milk-associated microbiota on the newborns’ immune system: a mini review. Front Microbiol. 2017;8:2100. doi: 10.3389/fmicb.2017.02100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Suzuki K. Diversified IgA-bacteria interaction in gut homeostasis. Adv Exp Med Biol. 2020;1254:105–116. [DOI] [PubMed] [Google Scholar]
- 96.Cebra JJ, Periwal SB, Lee G, Lee F, Shroff KE. Development and maintenance of the gut-associated lymphoid tissue (GALT): the roles of enteric bacteria and viruses. Dev Immunol. 1998;6(1–2):13–18. doi: 10.1155/1998/68382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fadlallah J, El Kafsi H, Sterlin D, et al. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med. 2018;10(439):eaan1217. doi: 10.1126/scitranslmed.aan1217 [DOI] [PubMed] [Google Scholar]
- 98.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303(5664):1662–1665. doi: 10.1126/science.1091334 [DOI] [PubMed] [Google Scholar]
- 99.Yoshida M, Claypool SM, Wagner JS, et al. Human neonatal Fc receptor mediates transport of IgG into luminal secretions for delivery of antigens to mucosal dendritic cells. Immunity. 2004;20(6):769–783. doi: 10.1016/j.immuni.2004.05.007 [DOI] [PubMed] [Google Scholar]
- 100.Janzon A, Goodrich JK, Koren O, Group TS, Waters JL, Ley RE. Interactions between the gut microbiome and mucosal immunoglobulins A, M, and G in the developing infant gut. mSystems. 2019;4(6):e00612–19. doi: 10.1128/mSystems.00612-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.AlKhater SA. Paediatric non-alcoholic fatty liver disease: an overview. Obes Rev. 2015;16(5):393–405. doi: 10.1111/obr.12271 [DOI] [PubMed] [Google Scholar]
- 102.Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107(1):129–134. doi: 10.1067/mai.2001.111237 [DOI] [PubMed] [Google Scholar]
- 103.Penders J, Thijs C, van den Brandt PA, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA birth cohort study. Gut. 2007;56(5):661–667. doi: 10.1136/gut.2006.100164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vael C, Nelen V, Verhulst SL, Goossens H, Desager KN. Early intestinal Bacteroides fragilis colonisation and development of asthma. BMC Pulm Med. 2008;8(1):19. doi: 10.1186/1471-2466-8-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Almqvist C, Cnattingius S, Lichtenstein P, Lundholm C. The impact of birth mode of delivery on childhood asthma and allergic diseases–a sibling study. Clin Exp Allergy. 2012;42(9):1369–1376. doi: 10.1111/j.1365-2222.2012.04021.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Arrieta MC, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. doi: 10.1126/scitranslmed.aab2271 [DOI] [PubMed] [Google Scholar]
- 107.Stiemsma Leah T, Arrieta M-C, Dimitriu Pedro A, et al. Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin Sci. 2016;130(23):2199–2207. doi: 10.1042/CS20160349 [DOI] [PubMed] [Google Scholar]
- 108.Chiu CY, Cheng ML, Chiang MH, et al. Gut microbial‐derived butyrate is inversely associated with IgE responses to allergens in childhood asthma. Pediatr Allergy Immunol. 2019;30(7):689–697. doi: 10.1111/pai.13096 [DOI] [PubMed] [Google Scholar]
- 109.Huang YJ, Lynch SV. The emerging relationship between the airway microbiota and chronic respiratory disease: clinical implications. Expert Rev Respir Med. 2011;5(6):809–821. doi: 10.1586/ers.11.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McCauley K, Durack J, Valladares R, et al. Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J Allergy Clin Immunol. 2019;144(5):1187–1197. doi: 10.1016/j.jaci.2019.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013;131(2):346–52.e1-3. doi: 10.1016/j.jaci.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Korten I, Mika M, Klenja S, et al. Interactions of respiratory viruses and the nasal microbiota during the first year of life in healthy infants. mSphere. 2016;1(6):e00312–16. doi: 10.1128/mSphere.00312-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roth-Walter F, Adcock IM, Benito-Villalvilla C, et al. Comparing biologicals and small molecule drug therapies for chronic respiratory diseases: an EAACI taskforce on immunopharmacology position paper. Allergy. 2019;74(3):432–448. doi: 10.1111/all.13642 [DOI] [PubMed] [Google Scholar]
- 114.Hill DA, Siracusa MC, Abt MC, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18(4):538–546. doi: 10.1038/nm.2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goleva E, Jackson LP, Harris JK, et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. 2013;188(10):1193–1201. doi: 10.1164/rccm.201304-0775OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang YJ, Nelson CE, Brodie EL, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127(2):372–81.e1-3. doi: 10.1016/j.jaci.2010.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pascal M, Perez-Gordo M, Caballero T, et al. Microbiome and allergic diseases. Front Immunol. 2018;9:1584. doi: 10.3389/fimmu.2018.01584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Choy DF, Hart KM, Borthwick LA, et al. TH2 and TH17 inflammatory pathways are reciprocally regulated in asthma. Sci Transl Med. 2015;7(301):301ra129. doi: 10.1126/scitranslmed.aab3142 [DOI] [PubMed] [Google Scholar]
- 119.Bisgaard H, Hermansen MN, Buchvald F, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357(15):1487–1495. doi: 10.1056/NEJMoa052632 [DOI] [PubMed] [Google Scholar]
- 120.Huang YJ, Nariya S, Harris JM, et al. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol. 2015;136(4):874–884. doi: 10.1016/j.jaci.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sverrild A, Kiilerich P, Brejnrod A, et al. Eosinophilic airway inflammation in asthmatic patients is associated with an altered airway microbiome. J Allergy Clin Immunol. 2017;140(2):407–17.e11. doi: 10.1016/j.jaci.2016.10.046 [DOI] [PubMed] [Google Scholar]
- 122.Taylor SL, Leong LEX, Choo JM, et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J Allergy Clin Immunol. 2018;141(1):94–103 e15. doi: 10.1016/j.jaci.2017.03.044 [DOI] [PubMed] [Google Scholar]
- 123.Sharma A, Laxman B, Naureckas ET, et al. Associations between fungal and bacterial microbiota of airways and asthma endotypes. J Allergy Clin Immunol. 2019;144(1214–1227.e7):1214–1227.e7. doi: 10.1016/j.jaci.2019.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schatz M, Rosenwasser L. The allergic asthma phenotype. J Allergy Clin Immunol Pract. 2014;2(6):645–648. doi: 10.1016/j.jaip.2014.09.004 [DOI] [PubMed] [Google Scholar]
- 125.Xu J, Singhera GK, Dorscheid DR. Expression of surfactant protein D in airways of asthmatics and interleukin-13 modulation of surfactant protein D in human models of airway epithelium. Respir Res. 2015;16(1):26. doi: 10.1186/s12931-015-0177-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mishra A, Weaver TE, Beck DC, Rothenberg ME. Interleukin-5-mediated allergic airway inflammation inhibits the human surfactant protein C promoter in transgenic mice. J Biol Chem. 2001;276(11):8453–8459. doi: 10.1074/jbc.M009481200 [DOI] [PubMed] [Google Scholar]
- 127.Jain-Vora S, Wert SE, Temann UA, Rankin JA, Whitsett JA. Interleukin-4 alters epithelial cell differentiation and surfactant homeostasis in the postnatal mouse lung. Am J Respir Cell Mol Biol. 1997;17(5):541–551. doi: 10.1165/ajrcmb.17.5.2883 [DOI] [PubMed] [Google Scholar]
- 128.Ahdieh M, Vandenbos T, Youakim A. Lung epithelial barrier function and wound healing are decreased by IL-4 and IL-13 and enhanced by IFN-gamma. Am J Physiol Cell Physiol. 2001;281(6):C2029–38. doi: 10.1152/ajpcell.2001.281.6.C2029 [DOI] [PubMed] [Google Scholar]
- 129.Hackett T-L, de Bruin HG, Shaheen F, et al. Caveolin-1 controls airway epithelial barrier function. Implications for asthma. Am J Respir Cell Mol Biol. 2013;49(4):662–671. doi: 10.1165/rcmb.2013-0124OC [DOI] [PubMed] [Google Scholar]
- 130.Aktas B, Aslim B. Gut-lung axis and dysbiosis in COVID-19. Turk J Biol. 2020;44(3):265–272. doi: 10.3906/biy-2005-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Khatiwada S, Subedi A. Lung microbiome and coronavirus disease 2019 (COVID-19): possible link and implications. Hum Microb J. 2020;17:100073. doi: 10.1016/j.humic.2020.100073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin Infect Dis. 2020. doi: 10.1093/cid/ciaa709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zuo T, Zhan H, Zhang F, Liu Q, Tso EYK, Lui GCY. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology. 2020;S0016–5085:34852–34856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang L. Genomic diversity of severe acute respiratory syndrome-coronavirus 2 in patients with coronavirus disease 2019. Clin Infect Dis. 2020;71(15):713–720. doi: 10.1093/cid/ciaa203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fan J, Li X, Gao Y, Zhou J, Wang S, Huang B. The lung tissue microbiota features of 20 deceased patients with COVID-19. J Infect. 2020;S0163–4453:30429–30431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang YJ, Boushey HA. The microbiome in asthma. J Allergy Clin Immunol. 2015;135(1):25–30. doi: 10.1016/j.jaci.2014.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Durack J, Lynch SV, Nariya S, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol. 2017;140(1):63–75. doi: 10.1016/j.jaci.2016.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dargahi N, Johnson J, Donkor O, Vasiljevic T, Apostolopoulos V. Immunomodulatory effects of probiotics: can they be used to treat allergies and autoimmune diseases? Maturitas. 2019;119:25–38. doi: 10.1016/j.maturitas.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 139.Du X, Wang L, Wu S, et al. Efficacy of probiotic supplementary therapy for asthma, allergic rhinitis, and wheeze: a meta-analysis of randomized controlled trials. Allergy Asthma Proc. 2019;40(4):250–260. doi: 10.2500/aap.2019.40.4227 [DOI] [PubMed] [Google Scholar]
- 140.Gutkowski P, Madalinski K, Grek M, Dmenska H, Syczewska M, Michalkiewicz J. Effect of orally administered probiotic strains Lactobacillus and Bifidobacterium in children with atopic asthma. Cent Eur J Immunol. 2003;35:233–238. [Google Scholar]
- 141.Huang CF, Chie WC, Wang IJ. Efficacy of Lactobacillus administration in school-age children with asthma: a randomized, placebo-controlled trial. Nutrients. 2018;10(11):E1678. doi: 10.3390/nu10111678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen YS, Jan RL, Lin YL, Chen HH, Wang JY. Randomized placebo-controlled trial of Lactobacillus on asthmatic children with allergic rhinitis. Pediatr Pulmonol. 2010;45(11):1111–1120. doi: 10.1002/ppul.21296 [DOI] [PubMed] [Google Scholar]
- 143.Elazab N, Mendy A, Gasana J, Vieira ER, Quizon A, Forno E. Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics. 2013;132(3):e666–76. doi: 10.1542/peds.2013-0246 [DOI] [PubMed] [Google Scholar]
- 144.Lin J, Zhang Y, He C, Dai J. Probiotics supplementation in children with asthma: a systematic review and meta-analysis. J Paediatr Child Health. 2018;54:953–961. [DOI] [PubMed] [Google Scholar]
- 145.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12(1):5–9. doi: 10.1038/ni0111-5 [DOI] [PubMed] [Google Scholar]
- 146.Trompette A, Gollwitzer ES, Yadava K, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20(2):159–166. doi: 10.1038/nm.3444 [DOI] [PubMed] [Google Scholar]
- 147.Bindels LB, Delzenne NM, Cani PD, Walter J. Opinion: towards a more comprehensive concept for prebiotics. Nat Rev Gastroenterol Hepatol. 2015;12(5):303–310. doi: 10.1038/nrgastro.2015.47 [DOI] [PubMed] [Google Scholar]
- 148.Wopereis H, Sim K, Shaw A, Warner JO, Knol J, Kroll JS. Intestinal microbiota in infants at high risk for allergy: effects of prebiotics and role in eczema development. J Allergy Clin Immunol. 2018;141(1334–42.e5):1334–1342.e5. doi: 10.1016/j.jaci.2017.05.054 [DOI] [PubMed] [Google Scholar]
- 149.Esposito S, Bianchini S, Polinori I, Principi N. Impact of OM-85 given during two consecutive years to children with a history of recurrent respiratory tract infections: a retrospective study. Int J Environ Res Public Health. 2019;16(6):1065. doi: 10.3390/ijerph16061065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lu Y, Li Y, Xu L, Xia M, Cao L. Bacterial lysate increases the percentage of natural killer T cells in peripheral blood and alleviates asthma in children. Pharmacology. 2015;95:139–144. [DOI] [PubMed] [Google Scholar]
- 151.Razi CH, Harmancı K, Abacı A, et al. The immunostimulant OM-85 BV prevents wheezing attacks in preschool children. J Allergy Clin Immunol. 2010;126(4):763–769. doi: 10.1016/j.jaci.2010.07.038 [DOI] [PubMed] [Google Scholar]
- 152.Emeryk A, Bartkowiak-Emeryk M, Raus Z, Braido F, Ferlazzo G, Melioli G. Mechanical bacterial lysate administration prevents exacerbation in allergic asthmatic children-the EOLIA study. Pediatr Allergy Immunol. 2018;29(4):394–401. doi: 10.1111/pai.12894 [DOI] [PubMed] [Google Scholar]
- 153.Osborn DA, Sinn JKH. Prebiotics in infants for prevention of allergy. Cochrane Database Syst Rev. 2013;CD006474. doi: 10.1002/14651858.CD006474.pub3 [DOI] [PubMed] [Google Scholar]
- 154.Antunes AEC, Vinderola G, Xavier-Santos D, Sivieri K. Potential contribution of beneficial microbes to face the COVID-19 pandemic. Food Res Int. 2020;136:109577. doi: 10.1016/j.foodres.2020.109577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.He LH, Ren LF, Li JF, Wu YN, Li X, Zhang L. Intestinal flora as a potential strategy to fight SARS-CoV-2 infection. Front Microbiol. 2020;11:1388. doi: 10.3389/fmicb.2020.01388 [DOI] [PMC free article] [PubMed] [Google Scholar]