Abstract
Vibrio parahaemolyticus is a major foodborne pathogen worldwide. The increasing number of cases of V. parahaemolyticus infections in China indicates an urgent need to evaluate the prevalence and genetic diversity of this pathogenic bacterium. In this paper, we introduce the Foodborne Vibrio parahaemolyticus genome database (FVPGD), the first scientific database of foodborne V. parahaemolyticus distribution and genomic data in China, based on our previous investigations of V. parahaemolyticus contamination in different kinds of food samples across China from 2011 to 2016. The dataset includes records of 2,499 food samples and 643 V. parahaemolyticus strains from supermarkets and marketplaces distributed over 39 cities in China; 268 whole-genome sequences have been deposited in this database. A spatial view on the risk situations of V. parahaemolyticus contamination in different food types is provided. Additionally, the database provides a functional interface of sequence BLAST, core genome multilocus sequence typing, and phylogenetic analysis. The database will become a powerful tool for risk assessment and outbreak investigations of foodborne pathogens in China.
Subject terms: Pathogens, Infection, Microbial ecology
| Measurement(s) | Microbial Contamination • sequence_assembly • DNA |
| Technology Type(s) | the most probable number method • sequence assembly process • DNA sequencing |
| Sample Characteristic - Organism | Vibrio parahaemolyticus |
| Sample Characteristic - Location | China |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.12629891
Background & Summary
Vibrio parahaemolyticus is a halophilic, gram-negative bacterium that is commonly found in estuarine and marine environments worldwide. This microorganism is recognized as one of the most prevalent foodborne pathogens and typically causes acute gastroenteritis in humans1. This bacterium preferentially grows in warm and low-salinity marine water and sometimes colonizes aquatic hosts such as mollusks, shrimp, and fish2. Most people are infected by eating raw or undercooked seafood3,4. V. parahaemolyticus can also cause necrotizing fasciitis through wound infection5. V. parahaemolyticus outbreaks have been reported in many countries such as Bangladesh6, Italy7, Japan8, Brazil9, and the USA10.
China, a vast country with the largest population in the world, has a high rate of seafood consumption. As a result of this, V. parahaemolyticus has been the leading cause of foodborne outbreaks and cases of infectious diarrhea in China, especially in its coastal regions11,12. In the city of Sanya, China, alone, there were 29 outbreaks caused by V. parahaemolyticus resulting in 499 cases from 2010 to 2016, accounting for about half of all foodborne microbial infections13. To characterize the prevalence and genetic diversity of V. parahaemolyticus in foods in China, we collected food samples from all over China from July 2011 to July 2016 and tested for contamination by V. parahaemolyticus14–17. Remarkably, V. parahaemolyticus contamination was not only found with a high rate in aquatic products but also in ready-to-eat (RTE) foods in most cities in China17. As all previous studies were published independently and focused on a coarse food type-level, there is a need to integrate this information, with more complete records that include the genetic background of foodborne V. parahaemolyticus strains at the whole-genome level.
Here, we have constructed the Foodborne Vibrio parahaemolyticus Genome Database (FVPGD), a database comprising 2,499 records of aquatic products and RTE foods collected from 39 cites in China, from which, a total of 643 V. parahaemolyticus strains were isolated. The whole-genome sequences of most strains were obtained using a next-generation sequencing strategy and deposited in this database. A core genome multilocus sequence typing (cgMLST) scheme was provided to analyze the epidemiology of V. parahaemolyticus in China according to the genomic information. This database demonstrates the risk level of V. parahaemolyticus contamination in different kinds of food samples collected from all over China and provides a series of high-quality genomes for investigating the genetic relationships of foodborne V. parahaemolyticus strains from multiple temporal-spatial niches. Moreover, this database will facilitate the risk assessment of foodborne V. parahaemolyticus in China and contamination tracing of V. parahaemolyticus infections in the future.
Methods
Data collection
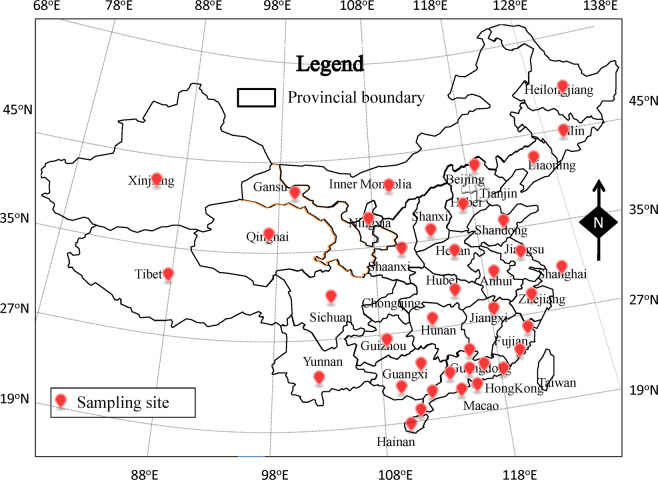
From July 2011 to July 2016, a total of 2,499 food samples, including 1,640 aquatic products and 859 RTE foods, were collected from supermarkets and marketplaces from most provincial capitals in China (Fig. 1). A total of 643 V. parahaemolyticus strains were isolated from 574 positive samples according to the GB 4789.7-2013 food microbiological examination of V. parahaemolyticus (National Food Safety Standards of China) and the most probable number method14–17. They were further identified by the analysis of oxidase activity, Gram staining, 3.5% NaCl triple sugar iron tests, halophilism tests, and API 20E diagnostic strips (Biomerieux Company, France). For epidemiological analysis, we also sampled 16 clinical V. parahaemolyticus strains isolated from patients in Guangdong, China, from 2011 to 2018.
Fig. 1.
Food sampling sites in China for this study.
Genome sequencing
V. parahaemolyticus strains were grown overnight at 37 °C in tryptic soytone broth medium (HuanKai Microbial, Guangzhou, China). Genomic DNA was extracted from V. parahaemolyticus strains using a genomic DNA extraction kit (Magen Biotech, Guangzhou, China) according to the manufacturer’s instructions. DNA samples were fragmented into 400-bp fragments by a Covaris M200 sonicator and used to generate sequencing libraries. Whole genomes were sequenced on the Life Ion S5 platform or Illumina Miseq/Nextseq 550 platform with an average coverage of 100× as previously described18. Raw reads were subjected to adapter clipping and quality filtering, and the obtained clean reads were assembled de novo by SPAdes v3.6.2 software19. All sequence data were checked for contamination using the Contamination Screen module of the submission system of NCBI, and then the sequences that needed to be trimmed and/or excluded were removed from the assemblies accordingly.
Definition of cgMLST scheme
For the cgMLST scheme construction, the complete genome sequences of six reference V. parahaemolyticus strains [RIMD2210633 (GenBank accession no. GCA_000196095.1), ATCC17802 (GenBank accession no. GCA_001558495.1), BB22OP (GenBank accession no. GCA_000328405.1), CDC_K4557 (GenBank accession no. GCA_000430425.1), FDA_R31 (GenBank accession no. GCA_000430405.1), and CHN25 (GenBank accession no. GCA_001700835.1)] were downloaded from NCBI and added to our database. The protein-coding genes of all genome sequences were annotated using Prokka v1.1120. The output of Prokka was used to create the pan-genome of V. parahaemolyticus with Roary v3.11.221. The core genes were then determined for each isolate with a BLASTN identity cutoff of 85% and with a cutoff of their presence in more than 99% of the strains, as previously described18. To ensure the computing speed of further analysis, core genes that lacked functional annotation or were annotated as hypothetical proteins were excluded. Genes that were present in more than one copy in any of the reference genomes were also removed. Finally, 672 core genes were selected, and their reference sequences (the same as that of strain RIMD2210633) were used for the cgMLST scheme. A genome-wide gene-by-gene cgMLST comparison was performed with every genome queried against the reference sequences, with the BLASTN threshold of identity >85% and coverage >90%. Alleles for each gene were assigned automatically by a local script. The combination of all alleles in each strain formed an allelic profile and the missing core genes were excluded from this profile.
Phylogenetic analysis of V. parahaemolyticus strains
A phylogenetic analysis of V. parahaemolyticus strains was performed using the concatenated alignment of 672 core genes. Missing core genes in each genome were ignored for the estimation of phylogenetic relationships as previously reported22. Each nucleotide sequence was aligned with MAFFT v7.31023, and the concatenated alignment was used to infer an approximate maximum likelihood phylogeny by FastTree v2.1.10 with default parameters (FastTree -gtr -nt alignment_file > tree_file)24.
BLAST search
The NCBI BLAST + v2.7.1 software was integrated as a web-service in our database. The genome sequences, protein sequences, and protein-coding sequences from each V. parahaemolyticus strain were used as the BLAST database.
Data Records
The dataset consists of two groups of data. The first group has the full information of all collected food samples, including their spatial and temporal distributions, sample types (e.g., fish, shrimp, and squid), detail of collection (supermarket or marketplace), and condition of contamination. The second group describes the background of all isolated V. parahaemolyticus strains, including their spatial and temporal distributions, isolated sources, serotypes, and sequencing information. These two data groups are connected through a unique ID for the food samples and have been uploaded to the figshare repository25. Additionally, genome sequences and genomic annotations were linked to V. parahaemolyticus strains through a unique ID for each strain and the corresponding files have been deposited in figshare26. All sequencing reads have been deposited in the NCBI Sequence Read Archive database under the SRA study accession SRP25345827.
The dataset is managed locally in a shared database and is accessible publicly at http://210.77.86.67/VP.html. The online dataset is updated when new genomes of V. parahaemolyticus strains are sequenced and when there are new collections of food samples.
Technical Validation
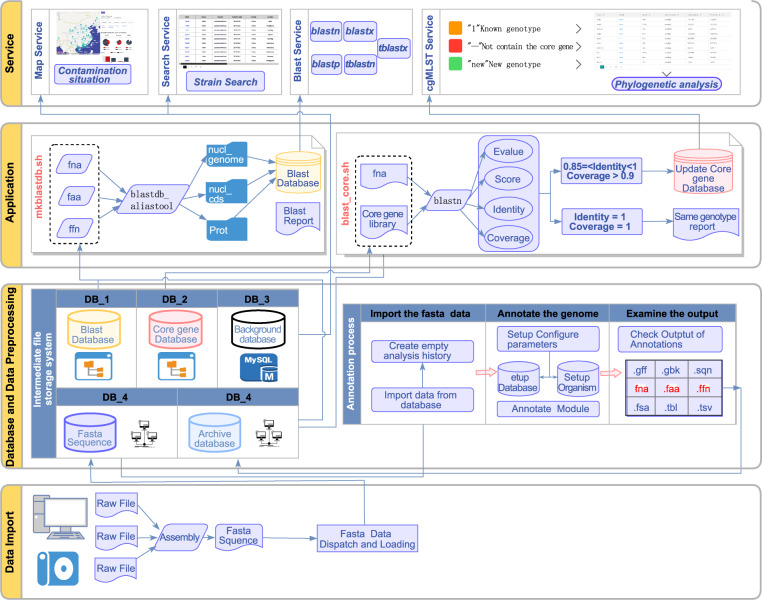
Information on the food samples and V. parahaemolyticus strains has been deposited as primary data in the FVPGD and is managed locally to ensure its validity and timeliness. The genome sequencing data of V. parahaemolyticus strains were processed following the schematic overview shown in Fig. 2. All genome sequences were checked for their GC content (within 44%~46%), contig number (≤300), and contig N50 (>50 kb) (Supplementary Table 1) before importing to the database. The processed data were transferred to the web service for browsing and comparison.
Fig. 2.
Schematic overview of the structure and data schedule of the FVPGD.
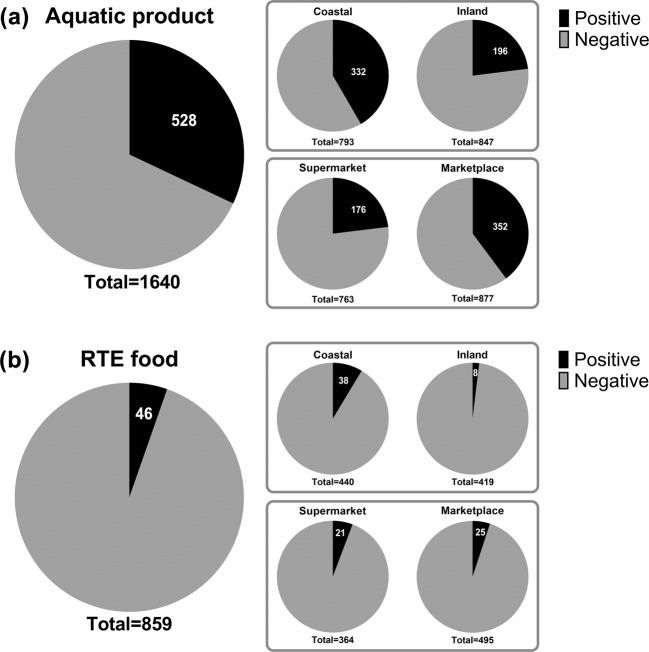
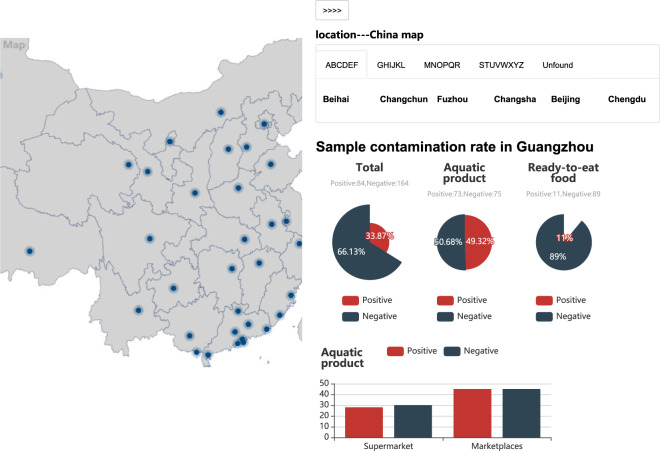
For aquatic products, the total rate of V. parahaemolyticus contamination in China was 32.20% (Fig. 3a). However, there was a great difference between coastal provinces and inland provinces. The contamination rate in coastal provinces (41.87%) was higher than that in inland provinces (23.14%), which was in accordance with the high level of foodborne V. parahaemolyticus infections in the coastal cities in China11,28. Additionally, aquatic products collected from marketplaces showed a much higher contamination rate of V. parahaemolyticus (40.14%) than those collected from supermarkets (23.07%), reflecting that the standardized procurement conditions in supermarkets might significantly reduce the risk of V. parahaemolyticus infection. The total rate of V. parahaemolyticus contamination in RTE foods from China was 5.36% (Fig. 3b). Although lower than that of aquatic products, its risk should not be ignored because RTE foods do not require heat treatment or other forms of curing before eating17. The V. parahaemolyticus contamination rate of RTE foods was much higher in coastal provinces (8.64%) than in inland provinces (1.91%), which is similar to that observed in aquatic products. However, there was no difference between RTE foods collected from supermarkets (5.77%) and marketplaces (5.05%). This might be an indication of V. parahaemolyticus persistence in RTE foods18.
Fig. 3.
Statistics of V. parahaemolyticus contamination situations in China. (a) Contamination rate of V. parahaemolyticus in aquatic products. (b) Contamination rate of V. parahaemolyticus in RTE foods.
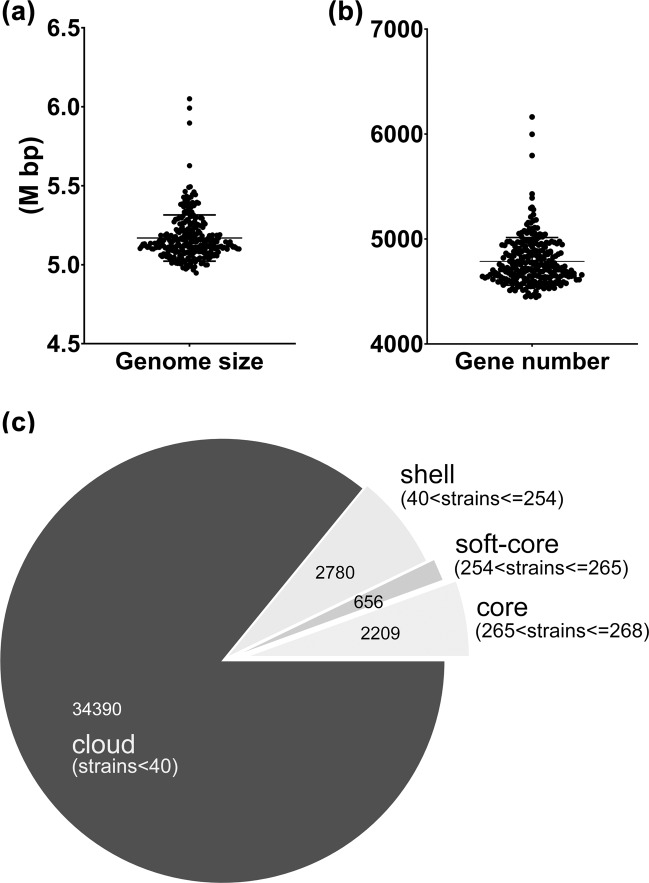
Up to now, the FVPGD has recorded the genome sequences of 268 V. parahaemolyticus strains, including 6 reference strains, 16 clinical strains, and 246 food-sourced strains. Among them, the sequences of 39 food-sourced strains have been previously reported18. The genome sizes of these strains ranged from 4.95 to 6.05 M bp (Fig. 4a). These genomes contained an average protein-coding gene number of 4787 (Fig. 4b). Additionally, the pan-genome of these strains consisted of 40,035 protein-coding genes, of which, 2,209 core genes were identified (Fig. 4c). It has been reported that V. parahaemolyticus harbors an open pan-genome, the gene content of which will increase as more strains are analyzed29,30. The pan-genome size obtained in this analysis was rather large. In contrast, the content of core genes remains stable and is very close to the results of previous analyses29,31. Further analysis revealed that 672 core genes were annotated with definitive functions, and these genes were used for the cgMLST scheme of this database according to a previous report31. Although they differed in topological structure, the phylogenetic trees generated from this gene set and full core genes were identical in the clustering of homologous strains (Supplementary Fig. 1), indicating that the selected gene set is sufficient to distinguish related and unrelated strains in a genome-wide resolution.
Fig. 4.
Overview of the genomic data deposited in the FVPGD. (a) Genome sizes of V. parahaemolyticus strains. (b) Protein-coding number of V. parahaemolyticus strains. (c) Pan-genome distribution of V. parahaemolyticus strains.
Usage Notes
V. parahaemolyticus strains included in the FVPGD can be directly retrieved according to their characteristics, such as location, source, and gene name. Furthermore, the information on V. parahaemolyticus strains can also be linked through a geographical map showing the risk situations of V. parahaemolyticus contamination in different cities in China (Fig. 5).
Fig. 5.
Spatial view on the risk situations of V. parahaemolyticus contamination in the FVPGD.
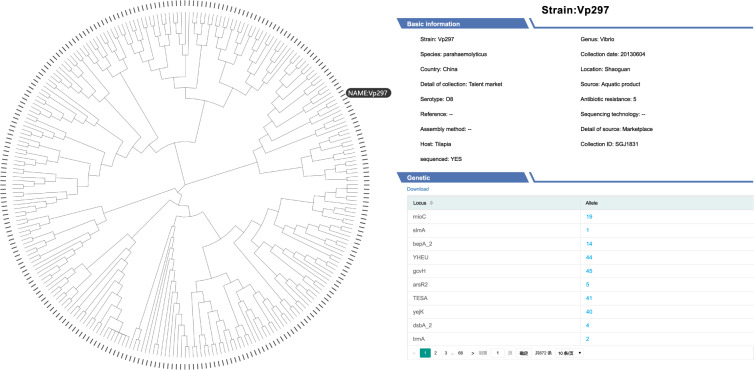
In the functional interface of cgMLST, users can upload the genome sequence of their V. parahaemolyticus strains. The sequence will be automatically BLASTed to the reference sequences of all core genes. If a core gene of a query sequence is identical to one in the sequences stored in the FVPGD, then the corresponding genotype number will be displayed; otherwise, the query core gene will be annotated as a new genotype. The users can compare their strains to all the strains in this database through a phylogenetic analysis based on the aligned sequences of all core genes. From the output phylogenetic tree, the input V. parahaemolyticus strain can be traced to the most homologous strain to determine its potential source (Fig. 6).
Fig. 6.
Phylogenetic tracing of V. parahaemolyticus strains in the FVPGD.
Supplementary information
Acknowledgements
This work was supported by the Key Project of National Natural Science Foundation of China (Grant No. 31730070), the Natural Science Foundation of Guangdong Province, China (Grant No. 2019A1515010810), and GDAS’s Special Project of Science and Technology Development (2019GDASYL-0201001). We thank Dunjin Chen, Zilun Pu, Qianli Zeng, Li Zhang, Yingpeng Cao, and Xinghao Bi from MingLead Gene Technology Co., Ltd. (Guangzhou, China) for software support.
Author contributions
Q.W. coordinated the project. R.P. and Q.W. designed the study and drafted the manuscript. M.C., H.Z., T.L., Y.D., J.W., S.W., Q.Y. and JM.Z. conducted sample collection and isolation. Y.L. performed genome sequencing. R.P. and Y.L. analyzed the sequencing data and compiled the data records. R.P. and JH.Z. contributed to geographic information positioning and data visualization.
Code availability
The versions of any software used and any specific variables or parameters used to process the current dataset have been detailed in the Methods section. The custom code produced during the validation of this dataset and the source code that underlies the interactive web interface (http://210.77.86.67/VP.html) have been shared on GitHub (https://github.com/pr839ok/FVPGD).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41597-020-00671-3.
References
- 1.Letchumanan V, Chan KG, Lee LH. Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 2014;5:705. doi: 10.3389/fmicb.2014.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DePaola A, Hopkins LH, Peeler JT, Wentz B, McPhearson RM. Incidence of Vibrio parahaemolyticus in U.S. coastal waters and oysters. Appl. Environ. Microbiol. 1990;56:2299–2302. doi: 10.1128/AEM.56.8.2299-2302.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniels NA, et al. Emergence of a new Vibrio parahaemolyticus serotype in raw oysters: a prevention quandary. JAMA. 2000;284:1541–1545. doi: 10.1001/jama.284.12.1541. [DOI] [PubMed] [Google Scholar]
- 4.McLaughlin JB, et al. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N. Engl. J. Med. 2005;353:1463–1470. doi: 10.1056/NEJMoa051594. [DOI] [PubMed] [Google Scholar]
- 5.Ralph A, Currie BJ. Vibrio vulnificus and V. parahaemolyticus necrotising fasciitis in fishermen visiting an estuarine tropical northern Australian location. J. Infect. 2007;54:e111–114. doi: 10.1016/j.jinf.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Akther F, et al. Major tdh(+)Vibrio parahaemolyticus serotype changes temporally in the Bay of Bengal estuary of Bangladesh. Infect. Genet. Evol. 2016;41:153–159. doi: 10.1016/j.meegid.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Caburlotto G, et al. Occurrence and molecular characterisation of Vibrio parahaemolyticus in crustaceans commercialised in Venice area, Italy. Int. J. Food Microbiol. 2016;220:39–49. doi: 10.1016/j.ijfoodmicro.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Arakawa E, et al. Emergence and prevalence of a novel Vibrio parahaemolyticus O3:K6 clone in Japan. Jpn. J. Infect. Dis. 1999;52:246–247. [PubMed] [Google Scholar]
- 9.Leal NC, et al. Vibrio parahaemolyticus serovar O3:K6 gastroenteritis in northeast Brazil. J. Appl. Microbiol. 2008;105:691–697. doi: 10.1111/j.1365-2672.2008.03782.x. [DOI] [PubMed] [Google Scholar]
- 10.Shaw KS, Sapkota AR, Jacobs JM, He X, Crump BC. Recreational swimmers’ exposure to Vibrio vulnificus and Vibrio parahaemolyticus in the Chesapeake Bay, Maryland, USA. Environ. Int. 2015;74:99–105. doi: 10.1016/j.envint.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, et al. Trends of foodborne diseases in China: lessons from laboratory-based surveillance since 2011. Front. Med. 2018;12:48–57. doi: 10.1007/s11684-017-0608-6. [DOI] [PubMed] [Google Scholar]
- 12.Paudyal N, et al. A meta-analysis of major foodborne pathogens in Chinese food commodities between 2006 and 2016. Foodborne Pathog. Dis. 2018;15:187–197. doi: 10.1089/fpd.2017.2417. [DOI] [PubMed] [Google Scholar]
- 13.Deng C, Deng Y, Yi J. Analysis of microbial food poisoning from 2010 to 2016 in Sanya city. Hainan Med. J. 2017;28:2723–2725. [Google Scholar]
- 14.Xu X, et al. Prevalence, pathogenicity, and serotypes of Vibrio parahaemolyticus in shrimp from Chinese retail markets. Food Control. 2014;46:81–85. doi: 10.1016/j.foodcont.2014.04.042. [DOI] [Google Scholar]
- 15.Xie, T., Wu, Q., Xu, X., Zhang, J. & Guo, W. Prevalence and population analysis of Vibrio parahaemolyticus in aquatic products from South China markets. FEMS Microbiol. Lett. 362, 10.1093/femsle/fnv178 (2015). [DOI] [PubMed]
- 16.Xu X, Cheng J, Wu Q, Zhang J, Xie T. Prevalence, characterization, and antibiotic susceptibility of Vibrio parahaemolyticus isolated from retail aquatic products in North China. BMC Microbiol. 2016;16:32. doi: 10.1186/s12866-016-0650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie T, Xu X, Wu Q, Zhang J, Cheng J. Prevalence, molecular characterization, and antibiotic susceptibility of Vibrio parahaemolyticus from Ready-to-Eat foods in China. Front. Microbiol. 2016;7:549. doi: 10.3389/fmicb.2016.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pang R, et al. Comparative genomic analysis reveals the potential risk of Vibrio parahaemolyticus isolated from Ready-To-Eat foods in China. Front. Microbiol. 2019;10:186. doi: 10.3389/fmicb.2019.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 21.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nabil-Fareed A, Zhemin Z, Sergeant MJ, Achtman M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018;14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura T, Yamada KD, Tomii K, Katoh K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 2018;34:2490–2492. doi: 10.1093/bioinformatics/bty121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pang R, Wu Q. 2020. A database for risk assessment and comparative genomic analysis of foodborne Vibrio parahaemolyticus in China. figshare. [DOI] [PMC free article] [PubMed]
- 26.Pang R, Wu Q. 2020. Genome assemblies and annotations of food-borne Vibrio parahaemolyticus strains. figshare. [DOI]
- 27.Pang R, 2020. Comparative genomic analysis of foodborne Vibrio parahaemolyticus in China. NCBI Sequence Read Archive. SRP253458 [DOI] [PMC free article] [PubMed]
- 28.Chen Y, et al. Foodborne disease outbreaks in 2006 report of the National Foodborne Disease Surveillance Network, China. Wei Sheng Yan Jiu. 2010;39:331–334. [PubMed] [Google Scholar]
- 29.Li L, et al. Comparative genomic analysis of clinical and environmental strains provides insight into the pathogenicity and evolution of Vibrio parahaemolyticus. BMC Genomics. 2014;15:1135. doi: 10.1186/1471-2164-15-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McInerney JO, McNally A, O’Connell MJ. Why prokaryotes have pangenomes. Nat. Microbiol. 2017;2:17040. doi: 10.1038/nmicrobiol.2017.40. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez-Escalona N, Jolley KA, Reed E, Martinez-Urtaza J. Defining a core genome multilocus sequence typing scheme for the global epidemiology of Vibrio parahaemolyticus. J. Clin. Microbiol. 2017;55:1682–1697. doi: 10.1128/JCM.00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Pang R, Wu Q. 2020. A database for risk assessment and comparative genomic analysis of foodborne Vibrio parahaemolyticus in China. figshare. [DOI] [PMC free article] [PubMed]
- Pang R, Wu Q. 2020. Genome assemblies and annotations of food-borne Vibrio parahaemolyticus strains. figshare. [DOI]
- Pang R, 2020. Comparative genomic analysis of foodborne Vibrio parahaemolyticus in China. NCBI Sequence Read Archive. SRP253458 [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The versions of any software used and any specific variables or parameters used to process the current dataset have been detailed in the Methods section. The custom code produced during the validation of this dataset and the source code that underlies the interactive web interface (http://210.77.86.67/VP.html) have been shared on GitHub (https://github.com/pr839ok/FVPGD).