Highlights
-
•
A light CNN for efficient detection of COVID-19 from chest CT scans is proposed.
-
•
The accuracy is comparable with that of more complex CNN designs.
-
•
The efficiency is 10 times better than more complex CNNs using pre-processing.
-
•
No GPU acceleration is required and can be executed on middle class computers.
Keywords: Deep Learning, CNN, Pattern Recognition, COVID-19
Abstract
Computer Tomography (CT) imaging of the chest is a valid diagnosis tool to detect COVID-19 promptly and to control the spread of the disease. In this work we propose a light Convolutional Neural Network (CNN) design, based on the model of the SqueezeNet, for the efficient discrimination of COVID-19 CT images with respect to other community-acquired pneumonia and/or healthy CT images. The architecture allows to an accuracy of 85.03% with an improvement of about 3.2% in the first dataset arrangement and of about 2.1% in the second dataset arrangement. The obtained gain, though of low entity, can be really important in medical diagnosis and, in particular, for Covid-19 scenario. Also the average classification time on a high-end workstation, 1.25 s, is very competitive with respect to that of more complex CNN designs, 13.41 s, witch require pre-processing. The proposed CNN can be executed on medium-end laptop without GPU acceleration in 7.81 s: this is impossible for methods requiring GPU acceleration. The performance of the method can be further improved with efficient pre-processing strategies for witch GPU acceleration is not necessary.
1. Introduction
Coronavirus (COVID19) is a world-wide disease that has been declared as a pandemic by the World Health Organization on 11th March 2020. To date, Covid-19 disease counts more than 10 millions of confirmed cases, of which: more than 500 thousands of deaths around the world (mortality rate of 5.3%); more than 5 millions of recovered people. A quick diagnosis is fundamental to control the spread of the disease and increases the effectiveness of medical treatment and, consequently, the chances of survival without the necessity of intensive and sub-intensive care. This is a crucial point because hospitals have limited availability of equipment for intensive care. Viral nucleic acid detection using real-time polymerase chain reaction (RT-PCR) is the accepted standard diagnostic method. However, many countries are unable to provide the sufficient RT-PCR due to the fact that the disease is very contagious. So, only people with evident symptoms are tested. Moreover, it takes several hours to furnish a result. Therefore, faster and reliable screening techniques that could be further confirmed by the PCR test (or replace it) are required.
Computer tomography (CT) imaging is a valid alternative to detect COVID-19 [2] with a higher sensitivity [5] (up to 98% compared with 71% of RT-PCR). CT is likely to become increasingly important for the diagnosis and management of COVID-19 pneumonia, considering the continuous increments in global cases. Early research shows a pathological pathway that might be amenable to early CT detection, particularly if the patient is scanned 2 or more days after developing symptoms [2]. Nevertheless, the main bottleneck that radiologists experience in analysing radiography images is the visual scanning of small details. Moreover, a large number of CT images have to be evaluated in a very short time thus increasing the probability of misclassifications. This justifies the use of intelligent approaches that can automatically classify CT images of the chest.
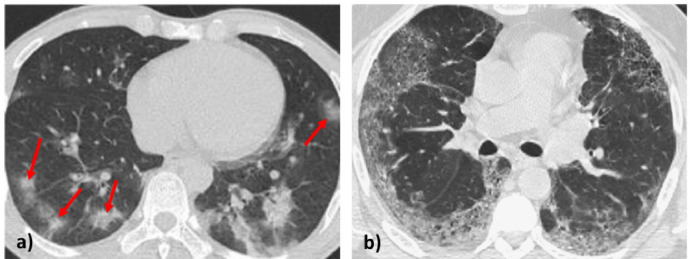
Deep Learning methods have been extensively used in medical imaging. In particular, convolutional neural networks (CNNs) have been used both for classification and segmentation problems, also of CT images [16]. However, CT images of the lungs referred to COVID-19 and not COVID-19 can be easily misclassified especially when damages due to pneumonia referred due to different causes are present at the same time. In fact, the main chest CT findings are pure ground glass opacities (GGO) [6] but also other lesions can be present like consolidations with or without vascular enlargement, interlobular septal thickening, and air bronchogram [11]. As an example, two CT scans of COVID-19 and not COVID-19 are reported in Fig. 1 a and b, respectively. Until now, there are limited datasets for COVID-19 and those available contain a limited number of CT images. For this reason, during the training phase it is necessary to avoid/reduce overfitting (that means the CNN is not learning the discriminant features of COVID-19 CT scans but only memorizing it). Another critical point is that CNN inference requires a lot of computational power. In fact, usually CNNs are executed on particularly expensive GPUs equipped with specific hardware acceleration systems. Anyway, expensive GPUs are still the exception rather than the norm in common computing clusters that usually are CPU based [13]. Even more, this type of machines could not be available in hospitals, especially in emergency situations and/or in developing Countries. At the moment, of the Top 12 Countries with more confirmed cases [12] (Table 1 ), 7 are developing Countries though COVID-19 emergency also is strongly stressing Health Systems of advanced Countries. In this work, we present an automatic method for recognizing COVID-19 and not COVID-19 CT images of lungs. It’s accuracy comparable with complex CNNs supported by massive pre-processing strategies while maintaining a light architecture and high efficiency that makes it executable in low/middle range computers.
Fig. 1.
(a) CT scan of lungs of a patient affected by COVID-19 where some evident GGO are indicated with red arrows (only for illustrative purpose); (b) CT scan of lungs of a patient affected by a not COVID-19 lung disease (diffuse opacity in the outer parts of the lungs). Images extracted from dataset [17].
Table 1.
Top 12 Countries with most confirmed cases of COVID-19 (update to 06/07/2020).
| Country | Confirmed cases | Confirmed deaths |
|---|---|---|
| USA | 2,496,628 | 125,318 |
| Brazil | 1,313,667 | 57,070 |
| Russia | 641,156 | 9,166 |
| India | 548,318 | 16,475 |
| United Kingdom | 311,155 | 43,550 |
| Peru | 275,989 | 9,135 |
| Chile | 271,982 | 5,509 |
| Spain | 248,770 | 28,343 |
| Italy | 240,310 | 34,738 |
| Iran | 222,669 | 10,508 |
| Mexico | 212,802 | 26,381 |
| Pakistan | 206,512 | 4,167 |
We started from the model of the SqueezeNet CNN to discriminate between COVID-19 and community-acquired pneumonia and/or healthy CT images. In fact, SqueezeNet is capable to reach the same accuracy of modern CNNs but with fewer parameters [7]. Moreover, in a recent benchmark [1], SqueezeNet has achieved the best accuracy density (accuracy divided by number of parameters) and the best inference time.
The hyperparameters have been optimized with Bayesian method on two datasets [17], [8]. In addition, Class Activation Mapping (CAM) [18] has been used to understand which parts of the image are relevant for the CNN to classify it and to check that no overfitting occurs.
The paper is structured as follow: in the next section (Materials and Methods) the datasets organization, the used processing equipment and the proposed methodology are presented; Section 3 contains Results and Discussion, including a comparison with recent works on the same argument; finally Section 4 concludes the paper and proposes future improvements.
2. Materials and methods
2.1. Datasets organization
The datasets used are the Zhao et al. dataset [17] and the Italian dataset [8]. Both datasets used in this study comply with Helsinki declaration and guidelines and we also operated in respect to the Helsinki declaration and guidelines. The Zhao et al. dataset is composed by 360 CT scans of COVID-19 subjects and 397 CT scans of other kinds of illnesses and/or healthy subjects. The Italian dataset is composed of 100 CT scans of COVID-19. These datasets are continuously updating and their images is raising at the same time. In this work we used two different arrangements of the datasets, one in which data from both datasets are used separately and the other containing data mixed by both datasets. The first arrangement contains two different test datasets (Test-1 and Test-2). In fact, the Zhao dataset is used alone and divided in train, validation and Test-1. The Italian dataset is integrated into a second test dataset, Test-2 (Table 2 ), while the Zhao dataset is always used in train, validation and Test-2 (in Test-2, the not COVID-19 images of the Zhao dataset are the same of Test-1). The first arrangement is used to check if, even with a small training dataset, it is possible to train a CNN capable to work well also on a completely different and new dataset (the Italian one). In the second arrangement, both datasets are mixed as indicated in Table 3 . In this arrangement the number of images from the italian dataset used to train, validate and Test-1 are 60, 20 and 20, respectively. The second arrangement represents a more realistic case in which both datasets are mixed to increase as possible the training dataset (at the expenses of a Test-2 which, in this case, is absent). In both arrangements, the training dataset has been augmented with the following transformations: a rotation (with a random angle between 0 and 90 degrees), a scale (with a random value between 1.1 and 1.3) and addition of gaussian noise to the original image.
Table 2.
Dataset arrangement 1.
| COVID-19 | Not COVID-19 | Data Augm. | Total | |
|---|---|---|---|---|
| Train | 191 | 191 | × 4 | 1528 |
| Validation | 60 | 58 | No | 118 |
| Test-1 | 98 | 95 | No | 193 |
| Test-2 | 100 | 95 | No | 195 |
Table 3.
Dataset arrangement 2.
| COVID-19 | Not COVID-19 | Data Augm. | Total | |
|---|---|---|---|---|
| Train | 251 | 191 | × 4 | 1768 |
| Validation | 80 | 58 | No | 138 |
| Test-1 | 108 | 95 | No | 203 |
2.2. Computational resources
For the numerical of the proposed CNNs we used two hardware systems: 1) a high level computer with CPU Intel Core i7-67100, RAM 32 GB and GPU Nvidia GeForce GTX 1080 8 GB dedicated memory; 2) a low level laptot with CPU Intel Core i5 processor, RAM 8 GB and no dedicated GPU. The first is used for hyperparameters optimization and to train, validate and test the CNNs; the second is used just for test in order to demonstrate the computational efficiency of the proposed solution.
In both cases we used the development environment Matlab 2020a. Matlab integrates powerful toolboxes for the design of neural networks. Moreover, with Matlab it is possible to export the CNNs in an open source format called ‘ONNX’ useful to share the CNNs with research community. When the high level computer is used, the GPU acceleration is enabled in Matlab environment, based on the technology Nvida Cuda Core provided by the GPU that allows parallel computing. In this way we speed up the prototyping of the CNNs. When final tests are performed on the low level hardware, no GPU acceleration is used.
2.3. CNN design
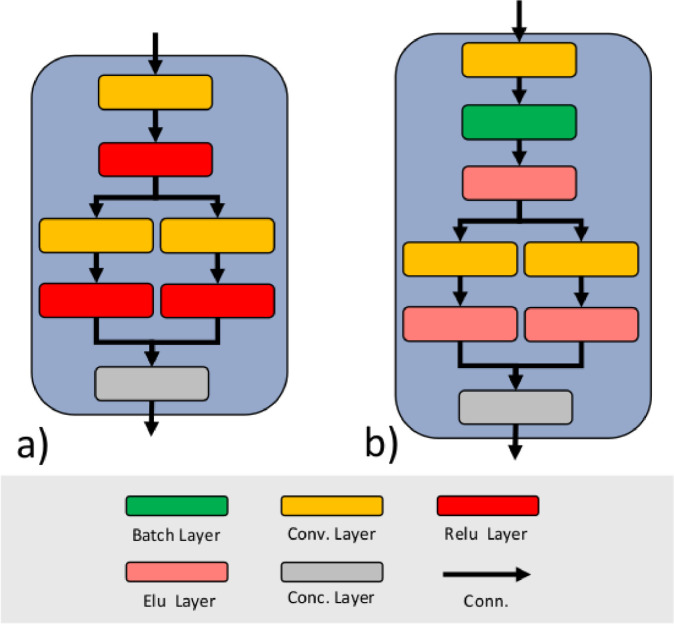
The SqueezeNet is capable of achieving the same level of accuracy of others, more complex, CNN designs which have a huge number of layers and parameters [7]. For example, SqueezeNet can achieve the same accuracy of Alex-Net [9] on the ImageNet dataset [4] with 50X fewer parameters and a model size of less than 0.5MB [7]. The SqueezeNet is composed of blocks called ”Fire Module”. As shown in Fig. 2 a, each block is composed of a squeeze convolution layer (which has 1 × 1 filters) feeding an expanding section of two convolution layers with 1 × 1 and 3 × 3 filters, respectively. Each convolution layer is followed by a ReLU layer. The ReLU layers output of the expanding section are concatenated with a Concatenation layer. To improve the training convergence and to reduce overfitting we added a Batch Normalization layer between the squeeze convolution layer and the ReLU layer (Fig. 2b). Each Batch Normalization layer adds 30% of computation overhead and for this reason we chose to add them only before the expanding section in order to make it more effective while, at the same time, limiting their number. Moreover, we replaced all the ReLU layers with ELU layers because, from literature [3], ELUs networks without Batch Normalization significantly outperform ReLU networks with Batch Normalization.
Fig. 2.
The classic Fire Module of the SqueezeNet (a). Proposed modification to the Fire Module of the SqueezeNet performed to improve convergence and to reduce overfitting (b).
The SqueezeNet has 8 Fire Modules in cascade configuration. Anyway, two more complex architectures exist: one with simple and another with complex bypass. The simple bypass configuration consists in 4 skip connections added between Fire Module 2 and Fire Module 3, Fire Module 4 and Fire Module 5, Fire Module 6 and Fire Module 7 and, finally, between Fire Module 8 and Fire Module 9. The complex bypass added 4 more skip connections (between the same Fire Modules) with a convolutional layer of filter size 1 × 1. From the original paper [7] the better accuracy is achieved by the simpler bypass configuration. For this reason, in this work we test both SqueezeNet without any bypass (to have the most efficient model) and with simple bypass (to have the most accurate model), while complex bypass configuration is not considered.
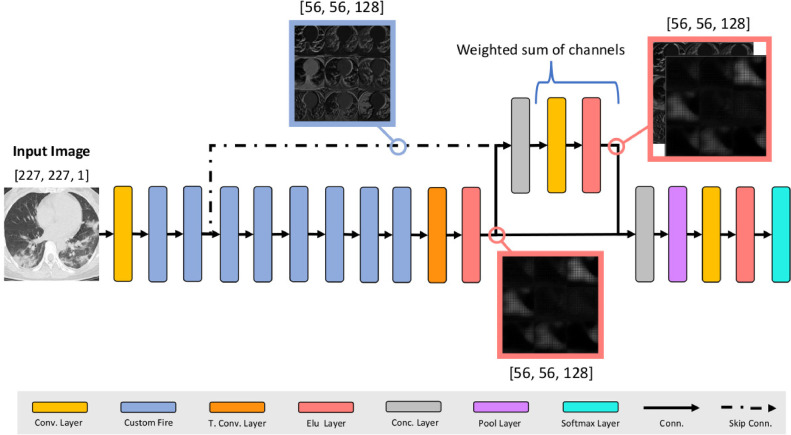
Besides, we propose also a further modify CNN (Fig. 3 ) based on the SqueezeNet without any bypass. Moreover, we added a Transpose Convolutional Layer to the last Custom Fire Module that expands the feature maps 4 times along width and height dimensions. These feature maps are concatenated in depth with the feature maps from the second Custom Fire Module through a skip connection. Weighted sum is performed between them with a Convolution Layer with 128 filters of size 1 × 1. Finally all the feature map are concatenated in depth and averaged with a Global Average Pool Layer. This design allows to combine spatial information (early layers) and features information (last layers) to improve the accuracy.
Fig. 3.
The proposed custom CNN. Spatial information contained in the feature maps from the second Custom Fire Module are weighted with the feature maps of the last Custom Fire Module.
2.4. Hyperparameters tuning
Since we are using a light CNN to classify, the optimization of the training phase is crucial to achieve good results with a limited number of parameters. The training phase of a CNN is highly correlated with settings hyperparameters. Hyperparameters are different from model weights. The former are calculated before the training phase, whereas the latter are optimised during the training phase. Setting of hyperparameters is not trivial and different strategies can be adopted. A first way is to select hyperparameters manually though it would be preferable to avoid it because the number of different configurations is huge. For the same reason, approaches like grid search do not use past evaluations: a lot of time has to be spent for evaluating bad hyperparameters configurations. Instead, Bayesian approaches, by using past evaluation results to build a surrogate probabilistic model mapping hyperparameters to a probability of a score on the objective function, seem to work better.
In this work we used Bayesian optimization for the following hyperparameters:
-
1.
Initial Learning Rate: the rate used for updating weights during the training time;
-
2.
Momentum: this parameter influences the weights update taking into consideration the update value of the previous iteration;
-
3.
L2-Regularization: a regularization term for the weights to the loss function in order to reduce over-fitting.
3. Results and discussion
3.1. Experiments organization and hyperparameters optimization
For each dataset arrangement we organized 4 experiments in which we tested different CNN models, transfer learning and the effectiveness of data augmentation. For each experiment, 30 different attempts (with Bayesian method) have been made with different set of hyperparameters (Initial Learning Rate, Momentum, L2-Regularization). For each attempt, the CNN model has been trained for 20 epochs and evaluated by the accuracy results calculated on the validation dataset. The experiments, all performed on the augumented dataset were:
-
1.
SqueezeNet without bypass and transfer learning;
-
2.
SqueezeNet with simple bypass but without transfer learning;
-
3.
SqueezeNet with simple bypass and transfer learning;
-
4.
the proposed CNN.
Regarding the arrangement 1, the results of the experiments are reported in Table 4 . For a better visualization of the results, we report just the the best accuracy calculated with respect to all the attempts, the accuracy estimated by the objective function at the end of all attempts and the values of the hyperparameters. The best accuracy value is achieved with the experiment #4. Both observed and estimated accuracy are the highest between all the experiments. Regarding the original paper of the SqueezeNet [7], there is not a relevant difference between the model without bypass and with bypass. It is also interesting to note that use transfer learning (experiment #3) from the original weights of the SqueezeNet does not have a relevant effect. Regarding the dataset arrangement 2, the results of the experiments are shown in Table 5 . The experiment #4 is still the best one, though experiment #1 is closer in terms of observed accuracy. By comparing the hyperparameters of the experiment #4 of Tables 4 and 5, a relevant difference in learning rate and L2-Regularization is evident. Regarding the dataset arrangement 1, Table 4 shows that to a decrease of the learning rate corresponds an increment of momentum and vice-versa; the same occurs between the learning rate and L2-Regularization; momentum and L2-regularization have the same behaviour. Regarding the dataset arrangement 2, Table 5 shows that learning rate, L2- Regularization and momentum have concordant trend. This hypothesis is confirmed in all the experiments. The different behaviour between hyperparameters in Tables 4 and 5 suggests that the CNN trained/validated on the dataset arrangement 1 (that we call CNN-1) is different by the CNN trained/validated on dataset arrangement 2 (that we call CNN-2), also confirmed by the evaluation of CAM, presented and discussed in the next sub-section. The results shown in Table 4 and Table 5 confirm that the proposed CNNs (experiment #4) perform better then original SqueezeNet configurations. In particular, CNN-1 design overcomes the original 3 SqueezeNet models in terms of accuracy of 1.6%, 4.0% and 4.0% (3.2% on average), respectively and CNN-2 of 0.7%, 2.2%, and 3.1% (2.1% on average), respectively. Two considerations are necessary: 1) the proposed architecture always overcomes the original ones; 2) an accuracy gain, though of low entity, can be really important in medical diagnosis.
Table 4.
Results on the dataset arrangement 1.
| Exp. | Obs. Acc. | Est. Acc. | Learn. Rate | Mom. | L2-Reg. |
|---|---|---|---|---|---|
| 1 | 88.30% | 82.26% | 0.074516 | 0.58486 | 1.6387e-07 |
| 2 | 85.76% | 82.42% | 0.011358 | 0.97926 | 3.684e-08 |
| 3 | 85.76% | 80.58% | 0.00070093 | 0.96348 | 1.0172e-12 |
| 4 | 89.85% | 87.27% | 0.007132 | 0.87589 | 0.9532e-06 |
Table 5.
Results on the dataset arrangement 2.
| Exp. | Obs. Acc. | Est. Acc. | Learn. Rate | Mom. | L2-Reg. |
|---|---|---|---|---|---|
| 1 | 86,84% | 82.11% | 0.00010091 | 0.70963 | 2.2153e-11 |
| 2 | 85.36% | 81.53% | 0.086175 | 0.59589 | 7.5468e-09 |
| 3 | 84.44% | 80.22% | 0.0016053 | 0.86453 | 1.0048e-10 |
| 4 | 87.56% | 85.87% | 0.089642 | 0.84559 | 0.5895e-07 |
3.2. Training, validation and test
The calculated hyperparameters have been used to train (20 epochs, Learning Rate drop of 0.8 every 5 epochs) both CNN-1 and CNN-2 with a 10-fold cross-validation strategy on both datasets (results are reported in Tables 6 and 7 , respectively).
Table 6.
10-fold cross-validation on dataset arrangement 1.
| Acc. | Sens. | Spec. | Prec. | F1-Score |
|---|---|---|---|---|
| 86.48 | 91.17 | 82.50 | 81.57 | 86.11 |
| 86.48 | 88.23 | 85.00 | 83.33 | 85.71 |
| 82.43 | 79.41 | 85.00 | 81.81 | 80.59 |
| 84.93 | 91.17 | 79.48 | 79.48 | 84.93 |
| 86.48 | 91.17 | 82.50 | 81.57 | 86.11 |
| 87.83 | 94.16 | 82.50 | 82.05 | 87.67 |
| 78.37 | 79.41 | 77.50 | 75.00 | 77.14 |
| 86.48 | 88.23 | 85.00 | 83.33 | 85.71 |
| 85.13 | 94.11 | 77.50 | 78.04 | 85.33 |
| 81.08 | 85.29 | 77.50 | 76.31 | 80.55 |
Table 7.
10-fold cross-validation on dataset arrangement 2.
| Acc. | Sens. | Spec. | Prec. | F1-Score |
|---|---|---|---|---|
| 82.35 | 91.11 | 72.50 | 78.84 | 84.53 |
| 85.88 | 86.66 | 85.00 | 86.66 | 86.66 |
| 81.17 | 86.66 | 75.00 | 79.59 | 82.97 |
| 84.70 | 86.66 | 82.50 | 84.78 | 85.71 |
| 90.58 | 88.88 | 92.50 | 93.02 | 90.90 |
| 83.52 | 88.88 | 77.50 | 81.63 | 85.10 |
| 83.52 | 84.44 | 82.50 | 84.44 | 84.44 |
| 85.88 | 95.55 | 75.00 | 81.13 | 87.75 |
| 82.14 | 84.44 | 79.48 | 82.60 | 83.51 |
| 90.58 | 84.44 | 97.50 | 97.43 | 90.47 |
Each, CNN is evaluated with the following benchmark metrics: Accuracy, Sensitivity, Specificity, Precision and F1-Score.
The average 10-fold cross-validation metrics, summarized in Table 8 , confirm that CNN-1 and CNN-2 behave differently.
Table 8.
CNN-1 and CNN-2 performance.
| CNN | Acc. | Sens. | Spec. | Prec. | F1-Score |
|---|---|---|---|---|---|
| CNN-1 | 84.56 | 88.23 | 81.44 | 80.24 | 83.98 |
| CNN-2 | 85.03 | 87.55 | 81.95 | 85.01 | 86.20 |
Regarding the application of CNN-1 on Test-2, the results are insufficient. In fact, the accuracy reaches just 50.24% because the CNN is capable only to recognize well not COVID-19 images (precision is 80.00%) but has very low performance on COVID-19 images (sensitivity = 19.00%). As affirmed before, the analyses of Test-2 is very hard if we do not use a larger dataset of images.
In order to deeply understand the behaviour of CNN-1 and CNN-2 we used CAM, that gives a visual explanations of the predictions of convolutional neural networks. This is useful to figure out what each CNN has learned and which part of the input of the network is responsible for the classification. It can be useful to identify biases in the training set and to increase model accuracy. With CAM it is also possible to verify if a CNN is overfitting and, in particular, if its predictions are based on relevant image features or on the background. To this aim, we expect that the activations maps are focused on the lungs and especially on those parts affected by COVID-19 (lighter regions with respect to healthy, darker, zones of the lungs).
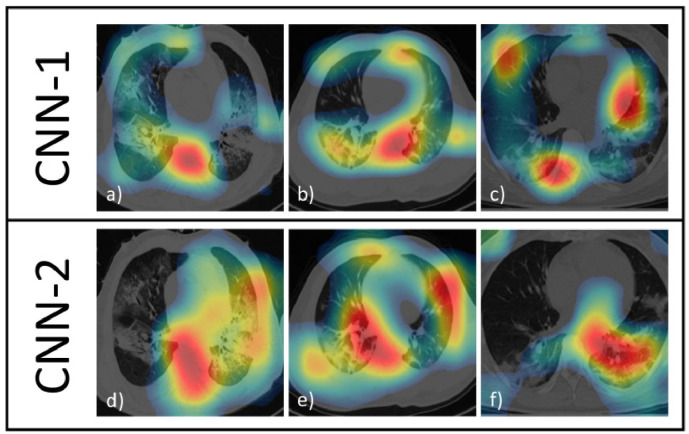
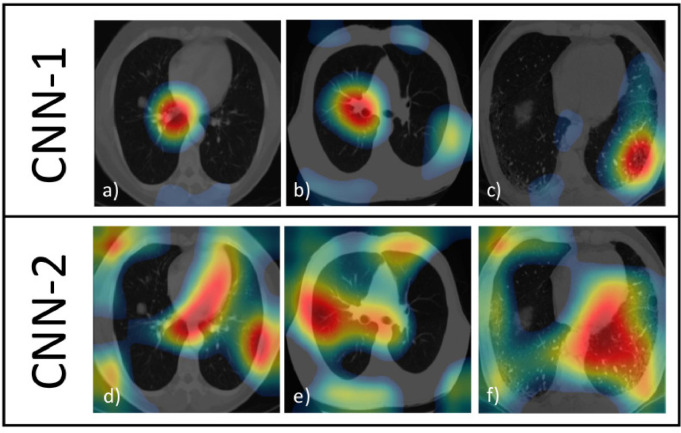
Fig. 4 shows 3 examples of CAMs for each CNNs and, to allow comparisons, we refer them to the same 3 CT images (COVID-19 diagnosed both from radiologists and CNNs) extracted from the training dataset.
Fig. 4.
CAMs of CNN-1 and CNN-2 on 3 COVID-19 CT images. Strongest colors (red) implies greater activations. Colors in CAMs are normalized.
By a visual comparison, for CNN-1 (Fig. 4a–c), the activations are not well localized inside the lungs, though in Fig. 4b the activations are better focused on the lungs than in Fig. 4a and c.
Regarding the CAMs of CNN-2 (Fig. 4d–f), there is an improvement because the activations are more localized on the ill parts of the lungs (this situation is perfectly represented in Fig. 4f). Fig. 5 shows 3 examples of CAMs for each CNNs (as Fig. 4) but with 3 CT images of lungs not affected by COVID-19 and correctly classified by both CNNs. CNN-1 focuses on small isolated zones (Figs. 5a–c): even if these zones are inside the lungs, it is unreasonable to obtain a correct classification with so few information (and without having checked the remaining of the lungs). Instead, in CNN-2, the activations take into consideration the whole region occupied by lungs as demonstrated in Figs. 5d–f.
Fig. 5.
CAMs of CNN-1 and CNN-2 on 3 not COVID-19 CT images. Strongest colors (red) implies greater activations. Colors in CAMs are normalized.
As a conclusion, it is evident that CNN-2 has a better behaviour with respect to CNN-1. Since CNN-1 and CNN-2 have the same model design but different training datatasets, we argue that the training dataset is the responsible of their different behaviour. In fact, the dataset arrangement-2 contains more training images (taken from the Italian dataset) and the CNN-2 seems to be gain by it. Figs. 4 and 5 show that the CNN model, even with a limited number of parameters, is capable to learn the discriminant features of this kind of images. Therefore, the increment of the training dataset should increase also the performance of the CNN.
3.3. Comparison with recent works
We compare the results of the CNN-2 with [10], [14], [15]. Since methods and datasets (training and test) differ and a correct quantitative comparison is arduous, we can have an idea regarding the respective results, summarized in Table 9 .
Table 9.
Comparison with previous works.
| Works | Image Preproc | Accuracy (%) | Sensitivy (%) | Specificity (%) | Precision (%) | F1-Score (%) | #Parameters (Millions) | Sens / #Param |
|---|---|---|---|---|---|---|---|---|
| Wang et al. [14] | No | 73.1 | 67 | 76 | 61 | 63 | 23.9 | 2.8 |
| Xu et al. [15] | Yes | - | 86.7 | - | 81.3 | 83.9 | 11.7 | 7.41 |
| Li et al. [10] | Yes | - | 90 | 96 | - | - | 25.6 | 3.52 |
| The proposed CNN | No | 85.03 | 87.55 | 81.95 | 85.01 | 86.20 | 1.26 | 69.48 |
The method [10] achieves better results than CNN-2. With respect to [14] and [15] our method achieves better results, especially regarding sensitivity.
The average time required by CNN-2 to classify a single CT image is 1.25 s on the previously defined high end workstation. As comparison, the method in [10] requires 4.51 s on a similar high-end workstation (Intel Xeon Processor E5-1620, GPU RAM 16GB, GPU Nvidia Quadro M4000 8GB) when just classification is considered. However, when the time necessary for pre-processing is considered, the method in [10] requires 13.41 s on the same workstation, thus resulting more 10 times slower than CNN-2. The computation time dramatically increases for [10] when considering pre-processing: it includes lungs segmentation through a supplementary CNN (a U-Net), voxel intensity clipping/normalization and, finally, the application of maximum intensity projection. This also makes the method in [10] unpractical for medium-end machines without graphic GPU acceleration. On the contrary, the average classification time for CNN-2 was 7.81 s on a middle class computer.
This represents, for the method proposed therein, the possibility to be used massively on medium-end computers: a dataset of about 4300 images, roughly corresponding to 3300 patients [10], could be classified in about 9.32 h. The improvement in efficiency of the proposed method with respect to the previously compared is demonstrated in Table 9, where the sensitivity value (the only parameter reported by all the compared methods) is rated with respect the number of parameters used to reach it: the resulting ratio confirms that the proposed method greatly overcomes the others in efficiency.
4. Conclusion
In this study, we proposed a CNN design (starting from the model of the SqueezeNet CNN) to discriminate between COVID-19 and other CT images (composed both by community-acquired pneumonia and healthy images). On both dataset arrangements, the proposed CNN-2 outperforms the original SqueezeNet. In particular, CNN-2 achieved 85.03% of accuracy, 87.55% of sensitivity, 81.95% of specificity, 85.01% of precision and 86.20% of F1-Score.
Moreover, CNN-2 is more efficient than other, more complex, CNN designs. In fact, the average classification time is low both on a high-end computer (1.25 s for a single CT image) and on a medium-end laptot (7.81 s for a single CT image). This demonstrates that the proposed CNN is capable to analyze thousands of images per day even with limited hardware resources.
The next step is to further increase the performance of CNN-2 through specific pre-processing strategies. In fact, performant CNN designs [10], [15] mostly use pre-processing with GPU acceleration.
Our future ambitious goal is to obtain specific and efficient pre-processing strategies for middle class computers without GPU acceleration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Bianco S., Cadene R., Celona L., Napoletano P. Benchmark analysis of representative deep neural network architectures. IEEE Access. 2018;6:64270–64277. [Google Scholar]
- 2.Chua F., Armstrong-James D., Desai S.R., Barnett J., Kouranos V., Kon O.M., José R., Vancheeswaran R., Loebinger M.R., Wong J. The role of CT in case ascertainment and management of COVID-19 pneumonia in the UK: insights from high-incidence regions. Lancet Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D.-A. Clevert, T. Unterthiner, S. Hochreiter, Fast and accurate deep network learning by exponential linear units (ELUs), arXiv preprint arXiv:1511.07289 (2015).
- 4.Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE; 2009. ImageNet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 5.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020:200432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Q., Guan H., Sun Z., Huang L., Chen C., Ai T., Pan Y., Xia L. Early CT features and temporal lung changes in COVID-19 pneumonia in Wuhan, China. Eur. J. Radiol. 2020:109017. doi: 10.1016/j.ejrad.2020.109017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.F.N. Iandola, S. Han, M.W. Moskewicz, K. Ashraf, W.J. Dally, K. Keutzer, SqueezeNet: alexnet-level accuracy with 50x fewer parameters and < 0.5 mb model size, arXiv preprint arXiv:1602.07360 (2016).
- 8.S. italiana di Radiologia Medica e Interventistica, Sirm dataset of COVID-19 chest CT scan, (https://www.sirm.org/category/senza-categoria/covid-19/). Accessed: 2020-04-05.
- 9.Krizhevsky A., Sutskever I., Hinton G.E. Advances in Neural Information Processing Systems. 2012. ImageNet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- 10.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020:200905. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. Am. J. Roentgenol. 2020;214(6):1280–1286. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 12.W.H. Organization, World health organization web site, (https://covid19.who.int/?gclid=CjwKCAjw88v3BRBFEiwApwLevSOpc4Ho-5vvQC8vAxoT_VU5VI1x9B9Tzu7LjxJSTF0itWGkawCdnhoCI_MQAvD_BwE). Accessed: 2020-07-01.
- 13.V. Vanhoucke, A. Senior, M.Z. Mao, Improving the speed of neural networks on CPUs (2011).
- 14.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19) medRxiv. 2020 doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.X. Xu, X. Jiang, C. Ma, P. Du, X. Li, S. Lv, L. Yu, Y. Chen, J. Su, G. Lang, et al., Deep learning system to screen coronavirus disease 2019 pneumonia, arXiv preprint arXiv:2002.09334 (2020). [DOI] [PMC free article] [PubMed]
- 16.Xu X., Zhou F., Liu B., Fu D., Bai X. Efficient multiple organ localization in ct image using 3d region proposal network. IEEE Trans. Med. Imaging. 2019;38(8):1885–1898. doi: 10.1109/TMI.2019.2894854. [DOI] [PubMed] [Google Scholar]
- 17.J. Zhao, Y. Zhang, X. He, P. Xie, COVID-CT-dataset: a CT scan dataset about COVID-19, arXiv preprint arXiv:2003.13865 (2020).
- 18.Zhou B., Khosla A., Lapedriza A., Oliva A., Torralba A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Learning deep features for discriminative localization; pp. 2921–2929. [Google Scholar]