Abstract
Chagas disease is considered the most important parasitic disease in Latin America. The protozoan agent, Trypanosoma cruzi, comprises six genetic lineages, TcI-TcVI. Genotyping to link lineage(s) to severity of cardiomyopathy and gastrointestinal pathology is impeded by the sequestration and replication of T. cruzi in host tissues. We describe serology specific for TcI, the predominant lineage north of the Amazon, based on expression of recombinant trypomastigote small surface antigen (gTSSA-I) in the eukaryote Leishmania tarentolae, to allow realistic glycosylation and structure of the antigen. Sera from TcI-endemic regions recognised gTSSA-I (74/146; 50.7%), with no cross reaction with common components of gTSSA-II/V/VI recombinant antigen. Antigenicity was abolished by chemical (periodate) oxidation of gTSSA-I glycosylation but retained after heat-denaturation of conformation. Conversely, non-specific recognition of gTSSA-I by non-endemic malaria sera was abolished by heat-denaturation. TcI-specific serology facilitates investigation between lineage and diverse clinical presentations. Glycosylation cannot be ignored in the search for immunogenic antigens.
Subject terms: ELISA, Diagnostic markers, Parasitic infection
Introduction
American trypanosomiasis (Chagas disease) is a neglected tropical disease caused by the protozoan parasite Trypanosoma cruzi, and is considered the most important human parasitic infection in Latin America. The infection is transmitted by blood sucking triatomine bugs through contamination of human mucous membranes, abraded skin or food, with T. cruzi infected triatomine faeces. Once acquired, unless successfully treated, T. cruzi usually persists as a life-long infection, which can also be transmitted congenitally from mother-to-child, and by transfusion or organ donation. Principal human tissues damaged are the heart and the intestinal tract, and symptoms may develop years after the early infection. An estimated 30% of T. cruzi infected individuals develop chagasic cardiomyopathy and a proportion of those develop chagasic gastrointestinal megasyndromes1. Only two drugs are currently available for treatment, benznidazole and nifurtimox. Due to severe dermatological side effects, treatment of adults frequently fails; a shorter, lower dose schedule for benznidazole and a paediatric formulation have recently been introduced2,3.
Currently WHO estimates 5–6 million cases of T. cruzi infection worldwide4. Despite international control programmes, there is still a high prevalence and incidence in regions such as the Gran Chaco of Bolivia and Argentina. Triatomine vectors are spreading into periurban sites, and Chagas disease is becoming a global health issue among Latin American migrant populations, with an estimated 250,000 infected in the USA, more than 100,000 in Europe and 12,000 in the UK, with risk of global non-vector borne transmission, congenitally and via blood and organ donors5,6.
Trypanosoma cruzi comprises six genetic lineages TcI-TcVI7–9, with TcBat proposed as a seventh lineage related to TcI10. Based on genotyping, TcI is the predominant agent of Chagas disease north of the Amazon, with TcIV a secondary cause in Venezuela11. TcII, TcV and TcVI are prevalent among cases in the Southern Cone countries of South America (Argentina, Bolivia, Brazil, Chile, Paraguay and Uruguay); TcIII is uncommonly found in human infections1.
In 198112 it was proposed that the different geographical distributions of the T. cruzi lineages may contribute to the disparate clinical presentations of Chagas disease in the Southern Cone countries, where megasyndromes are found, compared to northern South America, where they are not reported1. However, it is complex to prove such an association by parasite genotyping, because T. cruzi blood parasitaemia is scanty in chronic Chagas disease, does not necessarily represent lineages sequestered in the internal organs13–16, and growth rate competition occurs between isolates grown in vitro.
One approach to surveillance of clinical, geographical and ecological distributions of the T. cruzi lineages is to develop lineage-specific serology, originally proposed by Di Noia et al.17. Specific epitopes of the T. cruzi trypomastigote small surface antigen (TSSA), a cell surface mucin, have been identified for all six genetic lineages, with the hybrid lineages TcV and TcVI having two epitopes encoded at the heterozygous locus, one of which is shared with TcII, as shown by Bhattacharyya et al.18. Lineage-specific serology with synthetic peptides representing the TcII/V/VI and TcV/VI epitopes enabled surveillance of chagasic patients19, and the discovery of reservoir hosts20,21. Furthermore, TcII/V/VI serology, adaptable to rapid diagnostic test (RDT) format, demonstrated that among Bolivian patients stratified by severity of cardiomyopathy, TcII/V/VI seropositives were five-fold more prevalent in the severe versus no evidence of cardiomyopathy groups22. RDTs also identified TcII/V/VI seropositive sympatric humans and dogs in the Argentine Chaco23.
A long-standing research objective is the validation of a robust and sensitive TcI-specific antigen that would enable the enigma of link between infective lineage and clinical prognosis to be more comprehensively investigated. Furthermore, this would enable systematic low-cost analysis of T. cruzi transmission cycles and evaluation of the risk of emergence of sylvatic lineages into the domestic environment. However, repeated attempts have failed to develop a lineage-specific serological test for the TcI specific epitope, either using an E. coli-expressed recombinant protein or a synthetic peptide19,24–26.
Here, we have expressed the TSSA-I epitope within a related trypanosomatid, Leishmania tarentolae, which enables O-linked and N-linked glycosylation27,28 to determine whether glycosylation and/or structural integrity impart serological recognition of this TcI antigen.
Methods
Ethics
All human sera used here were archived, with consent for research, were anonymised, coded, and did not reveal patient identities. Informed consent was obtained from all subjects or, if subjects are under 18, consent was provided by a parent and/or legal guardian. No samples were collected specifically for this work.
Colombian (Bogotá), Venezuelan and Ecuadorean samples: these were collected as part of routine diagnostic examination, with local institutional ethical approvals Universidad de los Andes, Bogotá, Colombia; (Instituto de Medicina Tropical, Caracas, Venezuela; Pontificia Universidad Católica del Ecuador, Quito, Ecuador) and in accord with EC ethical standards, established as part of the ChagasEpiNet international collaboration (ethical approval from London School of Hygiene and Tropical Medicine, UK). Colombian (Medellín) ethical approval was provided by the Instituto de Biología, Universidad de Antioquia, Medellín, Colombia.
Peruvian and Bolivian samples: the following institutional review boards granted ethical approval: Johns Hopkins Bloomberg School of Public Health, USA; Hospital Universitario Japonés, Bolivia; Universidad Católica Boliviana, Bolivia; Universidad Peruana Cayetano Heredia, Peru; Asociación Benéfica Proyectos en Informatica, Salud, Medicina y Agricultura, Peru; and the Centers for Disease Control and Prevention, USA.
Gambian malaria sera were provided, with consent for further research on diagnostics, from London School of Hygiene and Tropical Medicine archives.
Non-endemic control sera were provided at the London School of Hygiene and Tropical Medicine and used with consent for further research on diagnostics.
Sources of chagasic sera
Chagasic sera were generously provided from the following sources: Colombia (Medellín, n = 55; Bogotá, n = 31); Ecuador (n = 14); Venezuela (n = 4), Peru (n = 42) and Bolivia (n = 10).
Assessing serological recognition of synthetic peptides by ELISA
Colombian (Medellín) sera were assayed by ELISA with synthetic peptides TSSApep-I, -II/V/VI, -III, -IV, -V/VI, according to protocols described previously19. Colombian (Bogotá), Ecuadorean and Venezuelan sera were previously assayed with synthetic peptides TSSApep-I, -II/V/VI, -III, -IV, -V/VI by ELISA; all were negative with TSSApep-I19; Peruvian and Bolivian sera were previously assayed by TSSApep-II/V/VI RDT22.
Prediction of glycosylation sites within gTSSA-I recombinant
In order to determine the presence of O and N glycosylation sites on the TSSA-I epitope, the amino acid sequence was submitted to NetOGlyc 4.0 (www.cbs.dtu.dk/services/NetOGlyc/) and NetNGlyc 1.0 (www.cbs.dtu.dk/services/NetNGlyc/) online servers. Prediction included coverage of the SUMO component of gTSSA-I.
L. tarentolae production of recombinant antigens gTSSA-I and gTSSA-II/V/VI
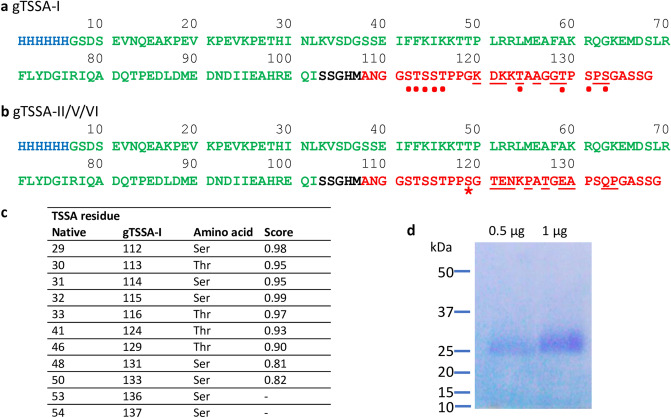
The TSSA-specific sequences were cloned into separate expression plasmids pLEXSY_I-blecherry3 with an upstream His tag and SUMO fusion partner to aid solubility of the resulting recombinant proteins, hereafter called gTSSA-I or gTSSA-II/V/VI, expressed in the L. tarentolae system (Jena Biosciences, Germany). Figure 1a,b depict the sequences of these recombinant antigens, with N-terminal histidine tag (blue), SUMO sequence (green) and TSSA-I sequence (red).
Figure 1.
T. cruzi TSSA recombinant proteins produced in L. tarentolae expression system. (a) gTSSA-I protein sequence (based on GenBank GU059925) and (b) gTSSA-II/V/VI protein sequence (based on GenBank GU075675): histidine tag (blue), SUMO sequence (green), linker sequences (black) and TSSA sequences (red). Dots: Residues predicted to be O-glycosylated. Starred: Ser residue present in native TSSA-II/V/VI but absent in native TSSA-I. Underlined: lineage-specific polymorphic residues. (c) Predicted likelihood scores for O-glycosylation of TSSA-I sequence range between 0 (least)—1 (highest); scores below 0.5 not listed. (d) Coomassie blue stained gel of purified gTSSA-I.
Production of recombinant protein, based on the methodology of Rooney et al.29 was carried out in 1 L baffled Erlenmeyer flasks in BHI medium (supplemented with antibiotics and hemin) and the medium was harvested when the OD600 reached 4 (approx. 70 h post inoculation, 108 cells/ml). All media components were from Jena Bioscience. Clarified medium was concentrated 20- fold on a Pellicon XL 50 Ultrafiltration cassette (10 kDa MWCO) and diluted four times in binding buffer (20 mM Phosphate, 500 mM NaCl, 10 mM Imidazole) before addition to an equilibrated His Trap (GE Healthcare) FPLC column. Bound proteins were eluted using an increasing Imidazole gradient (20 mM Phosphate, 500 mM NaCl, 500 mM Imidazole). Peak protein containing fractions, as determined by A280 nm measurement, were combined, desalted and concentrated by centrifugation in Amicon Ultra-15 device (5 kDa MWCO). Purified recombinant gTSSA-I was run on Coomassie blue-stained pre-cast 4–12% BisTris gradient SDS-PAGE gel (Novex) using the MOPS running system, along with Precision Plus Protein Unstained Standards (BioRad). The final protein was stored at 1 mg/ml in solution in PBS, 15% glycerol at – 20 °C.
Glycoproteomics
For glycoproteomics 20 µg of the glycoprotein with the gTSSA-I epitope were denaturated in 10 µL 8 M guanidine hydrochloride solution and reduced with 1 µL of 10 mM DTT (Thermo Fisher Scientific) in 50 mM ammonium bicarbonate buffer (pH 8.4) for 30 min at 56 °C. 1 µL of 55 mM iodoacetamide (Thermo Fisher Scientific) was added and incubated at room temperature for 30 min in the dark. Finally, the digest was diluted with 30 µl of 50 mM ammonium bicarbonate buffer prior to adding 1 µL of trypsin (1:20 w/w enzyme:protein ratio).
The online ESI-LC–MS data were recorded in MSE mode for 60 min using a Waters Synapt G2 mass spectrometer (Waters, Milford, MA). Separations were achieved on C18 Acquity UPLC M-Class column (HSS T3 1.8 µm 75 µm × 150 mm) equilibrated at 40 °C. The mobile phases were A: 0.1% (v/v) formic acid (Biosolve Chemicals) in LC–MS grade water (Greyhound) and B: 0.1% (v/v) formic acid (Biosolve Chemicals) in acetonitrile (Greyhound). Lockmass was set to m/z: 785.80 Glu-1-Fibrinopeptide B (200 nmol/µl, Waters). The Synapt G2-S mass range was operated between 50–2000 m/z.
Initial peptide mapping was carried out using BiopharmaLynx 1.3.3 (Waters, Milford, MA) to locate the epitope region of the gTSSA-I construct in the chromatogram. Interpretations of glycopeptide MS data were performed manually by previously reported methods based on amino acid and sugar masses together with the known fragmentation of glycopeptides30, with the assistance of MassLynxV4.1. (Waters, Milford, MA).
Assessing serological efficacy of the recombinant antigen gTSSA-I
Separate wells of a 96-well flat bottomed ELISA plates (735–0465: Immulon 4HBX, VWR) were coated with 50 μl/well of 1 µg/ml L. tarentolae-expressed gTSSA-I, and with 100 μl/well of 2 µg/ml lysate of T. cruzi TcII strains IINF/PY/00/Chaco23 or MHOM/BR/00/Y in coating buffer (15 mM Na2CO3, 34 mM NaHCO3, pH 9.6). Lysates were prepared as described previousy19. After overnight incubation at 4 °C, plates were washed three times with PBS/0.05% (v/v) Tween 20 (P7949: Sigma Aldrich, UK) (PBST), then blocked with 200 μl of PBS/2% skimmed milk powder (PBSM) at 37 °C for 2 h. Following three washes, 100 μl of 1:200 dilutions of serum in PBSM with 0.05% Tween 20 (PBSMT) was applied and incubated at 37 °C for 1 h; after six further washes, 100 μl of donkey anti-human IgG (H + L)-HRP (709–035–149: Jackson ImmunoResearch, USA) diluted 1:2000 (Peruvian and Bogotá sera) or 1:4000 (Medellín, Ecuadorean, Venezuelan, Bolivian sera) in PBSMT was added and incubated at 37 °C for 1 h. Following six washes, wells were developed with 100 µl/well of 50 mM phosphate/citrate buffer (pH 5.0) containing 2 mM o-phenylenediamine HCl (P1526: Sigma Aldrich) and 0.005% (vol/vol) H2O2 (216763: Sigma Aldrich); the plates were incubated in the dark at room temperature for approximately 10 min. Reactions were stopped by 50 µl/well of 2 M H2SO4, and absorbance values read at a wavelength of 490 nm. For Gambian malaria samples, sera were used at 1:100 and anti-human IgG 1:2000.
Excluding cross reaction with the SUMO component of recombinant antigens
A subset of 34 Colombian (Medellín) samples were assayed for serological cross reactivity with the SUMO component of the recombinant antigens. This was done by parallel performance of ELISAs, as described above, with the recombinant antigen gTSSA-II/V/VI coated in separate wells from the gTSSA-I.
Assessing the contribution of glycosylation and secondary structure to antigenicity of gTSSA-I
Contribution of glycosylation to antigenicity was assessed using an assay based on the protocol of Woodward et al.31, and employed subsequently32,33, which described the oxidative cleavage of carbohydrate vicinal –OH groups and subsequent reduction of generated aldehyde groups to prevent non-specific antibody binding. After the blocking and washing steps of the ELISA, described above, all wells were rinsed in periodate buffer (50 mM sodium acetate buffer, pH 4.5), and the wells that had been coated with gTSSA-I received 5 mM freshly-made sodium (meta)periodate (71859: Sigma Aldrich) in periodate buffer; the remaining wells received periodate buffer only. Plates were incubated in the dark at room temperature for 1 h, followed by rinsing of all wells with periodate buffer. The periodate-treated wells were then reduced with freshly-made 50 mM sodium borohydride (71320: Sigma Aldrich) in PBS for 30 min; the remaining wells received PBS only. Following this step, wells were washed three times with PBST before addition of sera and subsequent processing, as described above.
To investigate the contribution of secondary structure to the antigenicity of gTSSA-I, an aliquot of gTSSA-I in coating buffer was heated > 95 °C for up to 10 min, prior to coating onto the plate and performance of the serological efficacy was assessed as described above.
Statistical analysis
Replica ELISA plates were run in duplicate simultaneously. Cut-off values were determined by first subtracting the background absorbance values (i.e., mean of wells with coating buffer only, no antigen) from the mean reading for each sample; samples that were then greater than three standard deviations above the mean of seronegative non-endemic controls were considered positive. P values were determined by performing two-sample T test either unpaired (gTSSA-I against gTSSA-II/V/VI) or paired (unmodified gTSSA-I against oxidised or denatured) with GraphPad Prism (GraphPad Software, San Diego, USA).
Results
Synthetic peptides were not recognised by TcI endemic chagasic sera
All Colombian (Medellín and Bogotá), Ecuadorean, Venezuelan, and northern Peruvian sera samples (n = 146) were from regions considered to be endemic for TcI principally11, and were predominantly from chronic cases of Chagas disease in rural locations1,34. All sera were seropositive with T. cruzi lysate; (TcII and TcI lysate antigens do not discriminative between lineage infections). Apart from Colombia (Medellín) and Peruvian samples, all had been previously assayed with synthetic peptide TSSApep-I, and no reaction had been identified19. Here, there was no TSSApep-I recognition by the Colombian sera (Medellín) and no indication of the presence of infection with any of the other synthetic peptides.
Glycosylation prediction
Figure 1c shows the bioinformatic analysis predicting the likelihood score for O-linked glycosylation of the TSSA-I sequence used in gTSSA-I. No N-linked glycosylation was predicted by NetNGlyc 1.0, nor was any glycosylation predicted on the SUMO component of the recombinant proteins.
Recombinant antigens produced in L. tarentolae: gTSSA-I and gTSSA-II/V/VI
Recombinant proteins were produced by the L. tarentolae LEXSY system and purified by NiNTA from the culture media. The predicted mass is 15 kDa but a broad smear between 25 and 35 kDa is typical for a glycosylated protein (Fig. 1d, and supplementary Figure S1).
Mass spectrometry analysis proves glycosylation of recombinant gTSSA-I
Confirmation of glycosylation was demonstrated by mass spectrometry based glycoproteomics. An O-glycosylated glycopeptide was identified, consisting of the sequence TAAGGTPSPSGASSG substituted with a single N-acetylhexosamine (HexNAc) residue (Supplementary Figure S2). Such O-glycosylation with a single HexNAc residue is consistent with previous reports of L. tarentolae recombinant glycoprotein glycosylation, confirming the prediction of the NetOGlyc 4.0 programme28. As predicted by NetNGlyc 1.0, no N-linked glycosylation was found.
Recognition of gTSSA-I
Overall (74/146; 50.7%) of chagasic sera from TcI endemic regions of northern South America were seropositive with gTSSA-I (Table 1).
Table 1.
Recognition of gTSSA-I by chagasic sera from northern South America.
| Source | gTSSA-I reactive (%) |
|---|---|
| Colombia (Medellín) | 47/55 (85.5%) |
| Colombia (Bogotá) | 7/31 (22%) |
| Ecuador | 6/14 (42.9%) |
| Venezuela | 2/4 (50%) |
| Peru | 12/42 (28.6%) |
Absence of SUMO cross-reactivity between gTSSA-I and gTSSA-II/V/VI
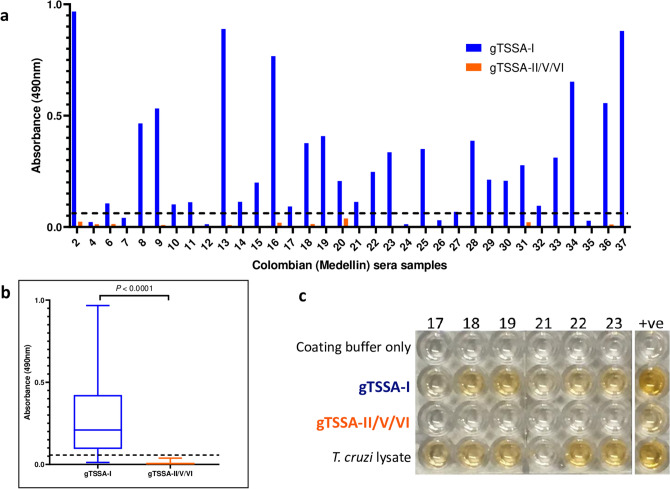
A subset of Colombian (Medellín) sera positive for gTSSA-I (n = 34) were assayed by ELISA against gTSSA-II/V/VI, to assess levels of potential cross-reactivity to the SUMO component of gTSSA-I. No recognition of gTSSA-II/V/VI was observed with these sera (Fig. 2a). Grouping these data, there was a significant difference in the absorbance values between these two recombinant antigens (P < 0.0001) (Fig. 2b). Thus, there was no cross reactivity between these two recombinant antigens attributable to antibody recognition of the SUMO component. Bolivian sera shown previously to be reactive with synthetic peptide TSSApep-II/V/VI (n = 5) reacted with gTSSA-II/V/VI; two sera also recognising gTSSA-I (Fig. 2c), suggesting TcII/V/VI and TcI co-infection.
Figure 2.
Antigenicity of gTSSA-I is due to TSSA-I sequence. (a) Absorbance values with Colombian (Medellín) sera recognising gTSSA-I (blue bars) and gTSSA-II/V/VI (orange bars). (b) Overall absorbance values for Colombian (Medellín) sera against gTSSA-I (blue) and gTSSA-II/V/VI (orange); gTSSA-I is recognised, whereas gTSSA-II/V/VI is not (P < 0.0001). (c) ELISA plate illustrating the recognition of gTSSA-I but not of gTSSA-II/V/VI; all samples were seropositive with lysate, and coating buffer controls were negative. Sample numbers correspond with (a). Positive control: serum from a Bolivian patient previously shown to be reactive with synthetic peptide TSSApep-II/V/VI and also seropositive with gTSSA-I, indicating co-infection (see text).
gTSSA-I antigenicity is principally dependent on glycosylation, not structure
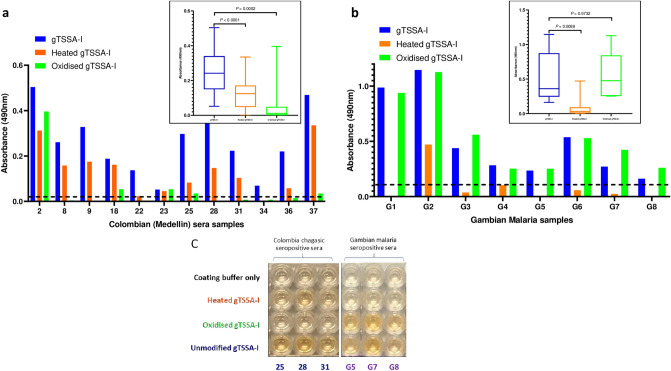
A subset of Colombian (Medellín) samples that recognised gTSSA-I (n = 12) were assayed against periodate-treated, heat denatured and unmodified gTSSA-I on the same ELISA plates. Recognition of gTSSA-I decreased substantially after periodate treatment, compared to the heat denatured antigen, for all samples except sample 2 (Fig. 3a). The absorbance values were significantly lower (P = 0.0002) after periodate treatment, with only a single sample retaining the same level of recognition as with the unmodified antigen. After heat denaturation the absorbance values of all 12 samples were reduced to a much lesser extent, albeit with a significant difference (P < 0.0001) (Fig. 3a).
Figure 3.
gTSSA-I antigenicity is dependent on glycosylation, whereas recognition by non-endemic malaria sera is dependent on structure. (a) Absorbance values of Colombian Medellín sera recognising unmodified gTSSA-I (blue bars), heat denatured gTSSA-I (orange bars) and oxidised gTSSA-I (green bars); oxidation decreases absorbance values substantially more than heat denaturing for all samples except 2. (b) Absorbance values of Gambian malaria (G) sera recognising unmodified gTSSA-I (blue bars), heat denatured gTSSA-I (orange bars) and oxidised gTSSA-I (green bars); heat denaturing decreases absorbance values substantially, whereas oxidation does not. For each of the data sets represented in (a) and (b) individual samples are presented on the main graph and composite absorbance values in the inset box-and-whisker plots. (c) ELISA plate illustrating the effects of modifications: coating with of unmodified gTSSA-I (fourth row); heat denatured gTSSA-I (second row), with slight signal reduction for Colombian sera but ablation for Gambian malaria sera; oxidised gTSSA-I (third row), with considerable reduction for Colombian sera, yet no reduction with Gambian malaria sera; coating buffer only (no antigen) control (first row). Sample numbers correspond with (a) and (b).
gTSSA-I recognition by some Gambian malaria sera is dependent on structure
Unexpectedly, recognition of gTSSA-I was observed with 13/24 Gambian malaria sera samples. Eight of these reactive samples were assayed against periodate-treated (oxidised) and heat denatured gTSSA-I. Oxidation had little effect on recognition of gTSSA-I, with no significant difference between absorbance values (P = 0.5732) and recognition of gTSSA-I was retained by all 8 samples. However, this was abolished by heat denaturation of this antigen (P = 0.0089). One sample remained above the cut-off, though the absorbance value had been greatly reduced (Fig. 3b,c). Any recognition of gTSSA-I by non-endemic healthy controls (NEHC) was also abrogated by heat-denaturation.
Discussion
Lineage-specific serology for T. cruzi infections provides a powerful tool for understanding the epidemiology and ecology of Chagas disease. Here, we have investigated lineage-specific epitopes to determine the importance of glycosylation in antigen recognition, particularly in relation to TcI.
The L. tarentolae expression system has been used previously to produce Trypanosoma brucei gambiense surface antigens, as a low cost alternative to harvesting diagnostic antigens from T. b. gambiense grown in vitro29. Expression in L. tarentolae has also been applied in attempts to improve diagnostic antigens for the trypanosomatids Leishmania braziliensis35 and Leishmania donovani36.
Here, we applied L. tarentolae expression to produce recombinant T. cruzi gTSSA-I, and to determine whether consequent glycosylation or more bona fide structural conformation of the protein conferred serological recognition upon the antigen.
All Colombian, Venezuelan, and Ecuadorean sera that were positive here with gTSSA-I were previously seronegative with the synthetic peptide TSSApep-I. The Medellín samples showed by far the highest proportion that recognised gTSSA-I; 85.5%. Reasons for this are unclear, but may be related to lower levels of anti-T. cruzi IgG in the other sera; in some localities there are low IgG antibody levels in T. cruzi infections37. Furthermore, the gTSSA-I positive samples were not seropositive with gTSSA-II/V/VI, demonstrating that there was no serological cross reaction with the SUMO component of the recombinant antigens. Thus, we have demonstrated clear, robust TcI lineage-specific serology with sera originating from countries where TcI has been identified by genotyping as the principal cause of Chagas disease.
In comparison with synthetic peptides or production of recombinants in the bacterium E. coli, heterologous expression in the eukaryote L. tarentolae enables glycosylation of the trypanosomatid proteins and adoption of a more natural conformation. O- and N-linked glycosylation of recombinant proteins in L. tarentolae has been described, and the wild type pattern in T. cruzi also shown to be O-linked (GlcNAc)28,38. Assays with periodate-treated antigen showed that the protein glycosylation (predicted by the online algorithms and demonstrated by glycoproteomics), and not the protein conformation, was important for serological recognition of gTSSA-I. Thus, expression in L. tarentolae revealed antigenic properties of TSSA-I that are not evident in synthetic peptides or E. coli-expressed recombinant proteins.
The specificity of gTSSA-I was questioned, due to some cross-reactivity with non-endemic (Gambian) malaria control sera. The sera from malaria were included due to breadth of immune response associated with antigenic variation, and recent research interest in whether antibodies to Plasmodium may recognise glycosylated antigens39. Two of these controls were also seropositive with the lateral flow diagnostic test specifically to detect exposure to T. b. gambiense infection (data not shown), suggesting an explanation for their cross reactivity. However, heating gTSSA-I abolished recognition by all Gambian malaria sera, demonstrating that this was due to analogous protein structure of gTSSA-I, not glycosylation. Since this is contrary to gTSSA-I recognition by chagasic sera, heating of gTSSA-I prior to performance of diagnostic ELISA or to incorporation into RDTs provides TcI-specific diagnosis via recognition of glycosylation antigenicity.
We have previously demonstrated a link between serological recognition of TSSApep-II/V/VI and severity of chagasic cardiac symptoms19,22. Availability of gTSSA-I serology enables parallel investigation of clinical status associated with TcI infection, and serological detection of sporadic TcII/V/VI and TcI co-infections, which occur in some Bolivian and Brazilian endemic foci40,41. As with application of TSSA-II/V/VI serology to sylvatic mammals20,21,23,42,43, gTSSA-I can be deployed for resolution of TcI domestic and sylvatic transmission cycles, and for discovery of reservoir hosts. Thus, expression of L. tarentolae recombinant antigens representing epitopes specific to each of the T. cruzi lineages, may facilitate comprehensive epidemiological investigations. Our results encourage L. tarentolae expression to improve efficacy of candidate diagnostic antigens.
Proteins have been preferentially pursued for decades as highly sensitive and specific diagnostic antigens, and as vaccine candidates. Perhaps most importantly, as recognised in recent research on malaria39 and bacteria44 we have indicated here that glycosylation cannot be ignored in the search for improved diagnostics or efficacious vaccines.
Supplementary information
Acknowledgements
We thank Juan-David Ramirez (Universidad del Rosario, Bogotá, Colombia), Felipe Guhl (Universidad de los Andes, Bogotá, Colombia), Hernan Carrasco (Universidad Central de Venezuela Instituto de Medicina Tropical, Caracas, Venezuela), Mario Grijalva (Pontificia Universidad Católica del Ecuador, Quito, Ecuador), Caryn Bern (School of Medicine, University of California, San Francisco, USA) and Louisa Messenger (LSHTM, UK) for provision of chagasic serum samples. NM was funded by the Sir Halley Stewart Trust; BR was funded by BBSRC (BB/N012496/1) and the Sir Halley Stewart Trust; AK and SHM were funded by (BB/P02789X/1); the views expressed within this article are those of the authors and not necessarily those of the Trust.
Author contributions
Produced the recombinant antigens: B.R., C.M.S. Provided sera: O.T.C., R.H.G., K.T., C.D. Performed glycoproteomics: A.K., S.M.H. Performed ELISAs: N.M., T.B., V.O.R., M.P., J.T. Analysed the data: N.M., B.R., T.B., J.B-S., M.A.M. Wrote the manuscript: N.M., T.B., B.R., M.A.M. Supervised the project: M.A.M.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Niamh Murphy and Barrie Rooney.
Supplementary information
is available for this paper at 10.1038/s41598-020-73390-9.
References
- 1.Messenger LA, Miles MA, Bern C. Between a bug and a hard place: Trypanosoma cruzi genetic diversity and the clinical outcomes of Chagas disease. Expert Rev. Antiinfect. Ther. 2015;13:995–1029. doi: 10.1586/14787210.2015.1056158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altcheh J, et al. Population pharmacokinetic study of benznidazole in pediatric Chagas disease suggests efficacy despite lower plasma concentrations than in adults. PLoS Negl. Trop. Dis. 2014;8:e2907. doi: 10.1371/journal.pntd.0002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez ML, et al. Pharmacokinetic and pharmacodynamic responses in adult patients with Chagas disease treated with a new formulation of benznidazole. Mem. Inst. Oswaldo Cruz. 2016;111:218–221. doi: 10.1590/0074-02760150401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization Chagas disease in Latin America: an epidemiological update based on 2010 estimates. Wkly. Epidemiol. Rec. 2015;90:33–43. [PubMed] [Google Scholar]
- 5.Lidani KCF, et al. Chagas disease: from discovery to a worldwide health problem. Front Public Health. 2019;7:166. doi: 10.3389/fpubh.2019.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manne-Goehler J, Umeh CA, Montgomery SP, Wirtz VJ. Estimating the burden of chagas disease in the United States. PLoS Negl. Trop. Dis. 2016;10:e0005033. doi: 10.1371/journal.pntd.0005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zingales B, et al. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI–TcVI. Mem. Inst. Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/S0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- 8.Zingales B. Trypanosoma cruzi genetic diversity: something new for something known about Chagas disease manifestations, serodiagnosis and drug sensitivity. Acta Trop. 2018;184:38–52. doi: 10.1016/j.actatropica.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Breniere SF, Waleckx E, Barnabe C. Over six thousand Trypanosoma cruzi strains classified into discrete typing units (DTUs): attempt at an inventory. PLoS Negl. Trop. Dis. 2016;10:e0004792. doi: 10.1371/journal.pntd.0004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lima L, et al. Genetic diversity of Trypanosoma cruzi in bats, and multilocus phylogenetic and phylogeographical analyses supporting Tcbat as an independent DTU (discrete typing unit) Acta Trop. 2015;151:166–177. doi: 10.1016/j.actatropica.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Carrasco HJ, et al. Geographical distribution of Trypanosoma cruzi genotypes in Venezuela. PLoS Negl. Trop. Dis. 2012;6:e1707. doi: 10.1371/journal.pntd.0001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miles MA, et al. Do radically dissimilar Trypanosoma cruzi strains (zymodemes) cause Venezuelan and Brazilian forms of Chagas' disease? Lancet. 1981;1:1338–1340. doi: 10.1016/S0140-6736(81)92518-6. [DOI] [PubMed] [Google Scholar]
- 13.Vago AR, et al. Genetic characterization of Trypanosoma cruzi directly from tissues of patients with chronic Chagas disease: differential distribution of genetic types into diverse organs. Am. J. Pathol. 2000;156:1805–1809. doi: 10.1016/S0002-9440(10)65052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgos JM, et al. Molecular identification of Trypanosoma cruzi discrete typing units in end-stage chronic Chagas heart disease and reactivation after heart transplantation. Clin. Infect. Dis. 2010;51:485–495. doi: 10.1086/655680. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez HO, et al. Trypanosomacruzi strains cause different myocarditis patterns in infected mice. Acta Trop. 2014;139:57–66. doi: 10.1016/j.actatropica.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Lewis MD, Francisco AF, Taylor MC, Jayawardhana S, Kelly JM. Host and parasite genetics shape a link between Trypanosoma cruzi infection dynamics and chronic cardiomyopathy. Cell Microbiol. 2016;18:1429–1443. doi: 10.1111/cmi.12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Noia JM, Buscaglia CA, De Marchi CR, Almeida IC, Frasch AC. A Trypanosoma cruzi small surface molecule provides the first immunological evidence that Chagas' disease is due to a single parasite lineage. J. Exp. Med. 2002;195:401–413. doi: 10.1084/jem.20011433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharyya T, et al. Analysis of molecular diversity of the Trypanosoma cruzi trypomastigote small surface antigen reveals novel epitopes, evidence of positive selection and potential implications for lineage-specific serology. Int. J. Parasitol. 2010;40:921–928. doi: 10.1016/j.ijpara.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharyya T, et al. Development of peptide-based lineage-specific serology for chronic Chagas disease: geographical and clinical distribution of epitope recognition. PLoS Negl. Trop. Dis. 2014;8:e2892. doi: 10.1371/journal.pntd.0002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerr CL, et al. Lineage-specific serology confirms Brazilian Atlantic forest lion tamarins, Leontopithecus chrysomelas and Leontopithecus rosalia, as reservoir hosts of Trypanosoma cruzi II (TcII) Parasit. Vectors. 2016;9:584. doi: 10.1186/s13071-016-1873-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClean MCW, et al. A lineage-specific rapid diagnostic test (ChagasSero K-SeT) identifies Brazilian Trypanosoma cruzi II/V/VI reservoir hosts among diverse mammalian orders. PLoS ONE. 2020;15:e0227828. doi: 10.1371/journal.pone.0227828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharyya T, et al. Severity of Chagasic cardiomyopathy is associated with response to a novel rapid diagnostic test for Trypanosoma cruziTcII/V/VI. Clin. Infect. Dis. 2018;67:519–524. doi: 10.1093/cid/ciy121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy N, et al. Lineage-specific rapid diagnostic tests can resolve Trypanosoma cruzi TcII/V/VI ecological and epidemiological associations in the Argentine Chaco. Parasit. Vectors. 2019;12:424. doi: 10.1186/s13071-019-3681-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bisio M, et al. Urbanization of congenital transmission of Trypanosoma cruzi: prospective polymerase chain reaction study in pregnancy. Trans. R. Soc. Trop. Med. Hyg. 2011;105:543–549. doi: 10.1016/j.trstmh.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Canepa GE, Degese MS, Budu A, Garcia CR, Buscaglia CA. Involvement of TSSA (trypomastigote small surface antigen) in Trypanosoma cruzi invasion of mammalian cells. Biochem. J. 2012;444:211–218. doi: 10.1042/BJ20120074. [DOI] [PubMed] [Google Scholar]
- 26.Longhi, S. A. et al. Cytokine production but lack of proliferation in peripheral blood mononuclear cells from chronic Chagas' disease cardiomyopathy patients in response to T. cruzi ribosomal P proteins. PLoS Negl. Trop. Dis.8, e2906 (2014). [DOI] [PMC free article] [PubMed]
- 27.Breitling R, et al. Non-pathogenic trypanosomatid protozoa as a platform for protein research and production. Protein Expr. Purif. 2002;25:209–218. doi: 10.1016/S1046-5928(02)00001-3. [DOI] [PubMed] [Google Scholar]
- 28.Klatt S, et al. Production of glycosylated soluble amyloid precursor protein alpha (sAPPalpha) in Leishmania tarentolae. J. Proteome Res. 2013;12:396–403. doi: 10.1021/pr300693f. [DOI] [PubMed] [Google Scholar]
- 29.Rooney, B., Piening, T., Buscher, P., Roge, S. & Smales, C. M. Expression of Trypanosoma brucei gambiense Antigens in Leishmania tarentolae. Potential for use in rapid serodiagnostic tests (RDTs). PLoS Negl. Trop. Dis.9, e0004271 (2015). [DOI] [PMC free article] [PubMed]
- 30.Panico M, et al. Mapping the complete glycoproteome of virion-derived HIV-1 gp120 provides insights into broadly neutralizing antibody binding. Sci. Rep. 2016;6:32956. doi: 10.1038/srep32956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodward MP, Young WW, Jr, Bloodgood RA. Detection of monoclonal antibodies specific for carbohydrate epitopes using periodate oxidation. J. Immunol. Methods. 1985;78:143–153. doi: 10.1016/0022-1759(85)90337-0. [DOI] [PubMed] [Google Scholar]
- 32.Gonzaga HT, et al. Metaperiodatedeglycosylation of Strongyloides venezuelensis larvae: immunochemical characterization and antigen production for human strongyloidiasis diagnosis. Acta Trop. 2018;182:27–33. doi: 10.1016/j.actatropica.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Choi JW, et al. Use of CA153 for screening breast cancer: an antibodylectin sandwich assay for detecting glycosylation of CA153 in sera. Oncol. Rep. 2018;40:145–154. doi: 10.3892/or.2018.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomez-Palacio A, et al. Multilocus analysis indicates that Trypanosoma cruzi I genetic substructure associated with sylvatic and domestic cycles is not an attribute conserved throughout Colombia. Infect. Genet. Evol. 2016;38:35–43. doi: 10.1016/j.meegid.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 35.de Souza LMB, et al. Production of a kinesin-related recombinant protein (Lbk39) from Leishmania braziliensis by Leishmania tarentolaepromastigotes and its application in the serodiagnosis of leishmaniasis. One Health. 2019;8:100111. doi: 10.1016/j.onehlt.2019.100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rezaei Z, et al. Expression of a rK39 homologue from an Iranian Leishmania infantum isolate in Leishmania tarentolae for serodiagnosis of visceral leishmaniasis. Parasit Vectors. 2019;12:593. doi: 10.1186/s13071-019-3839-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin DL, et al. Regional variation in the correlation of antibody and T-cell responses to Trypanosoma cruzi. Am. J. Trop. Med. Hyg. 2014;90:1074–1081. doi: 10.4269/ajtmh.13-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acosta-Serrano A, Almeida IC, Freitas-Junior LH, Yoshida N, Schenkman S. The mucin-like glycoprotein super-family of Trypanosoma cruzi: structure and biological roles. Mol. Biochem. Parasitol. 2001;114:143–150. doi: 10.1016/S0166-6851(01)00245-6. [DOI] [PubMed] [Google Scholar]
- 39.Goddard-Borger ED, Boddey JA. Implications of plasmodium glycosylation on vaccine efficacy and design. Future Microbiol. 2018;13:609–612. doi: 10.2217/fmb-2017-0284. [DOI] [PubMed] [Google Scholar]
- 40.Llewellyn MS, et al. Deep sequencing of the Trypanosoma cruzi GP63 surface proteases reveals diversity and diversifying selection among chronic and congenital Chagas disease patients. PLoS Negl. Trop. Dis. 2015;9:e0003458. doi: 10.1371/journal.pntd.0003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Perez A, et al. Prevalence of Trypanosoma cruzi discrete typing units in a cohort of Latin American migrants in Spain. Acta Trop. 2016;157:145–150. doi: 10.1016/j.actatropica.2016.01.032. [DOI] [PubMed] [Google Scholar]
- 42.Cimino RO, et al. Immuno-enzymatic evaluation of the recombinant TSSA-II protein of Trypanosoma cruzi in dogs and human sera: a tool for epidemiological studies. Parasitology. 2011;138:995–1002. doi: 10.1017/S0031182011000540. [DOI] [PubMed] [Google Scholar]
- 43.Floridia-Yapur N, et al. Evaluation of recombinant antigens of Trypanosoma cruzi to diagnose infection in naturally infected dogs from Chaco region Argentina. Parasite Immunol. 2014;36(694):699. doi: 10.1111/pim.12144. [DOI] [PubMed] [Google Scholar]
- 44.Kay E, Cuccui J, Wren BW. Recent advances in the production of recombinant glycoconjugate vaccines. NPJ Vac. 2019;4:16. doi: 10.1038/s41541-019-0110-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.