Abstract
ShuFeng JieDu capsule (SFJDC), a traditional Chinese medicine, has been recommended for the treatment of COVID-19 infections. However, the pharmacological mechanism of SFJDC still remains vague to date. The active ingredients and their target genes of SFJDC were collected from TCMSP. COVID-19 is a type of Novel Coronavirus Pneumonia (NCP). NCP-related target genes were collected from GeneCards database. The ingredients-targets network of SFJDC and PPI networks were constructed. The candidate genes were screened by Venn diagram package for enrichment analysis. The gene-pathway network was structured to obtain key target genes. In total, 124 active ingredients, 120 target genes of SFJDC and 251 NCP-related target genes were collected. The functional annotations cluster 1 of 23 candidate genes (CGs) were related to lung and Virus infection. RELA, MAPK1, MAPK14, CASP3, CASP8 and IL6 were the key target genes. The results suggested that SFJDC cloud be treated COVID-19 by multi-compounds and multi-pathways, and this study showed that the mechanism of traditional Chinese medicine (TCM) in the treatment of disease from the overall perspective.
Keywords: ShuFeng JieDu capsule; Novel Coronavirus Pneumonia; network pharmacology, mechanism, pathway; candidate genes
Introduction
Since December 2019, a novel coronavirus pneumonia (NCP) caused by new coronavirus (SARS-COV-2) has been prevalent in China and other countries, such as United States and Korea 1-3. WHO named this novel coronavirus pneumonia COVID-19 on February 11, 2020 4 and there was a total of 20 million reported cases of COVID-19 globally and 750,000 deaths as of August 10, 2020 5.
Its transmission route is mainly through respiratory droplets, but also through contact transmission, which has the characteristics of rapid spread, strong infectivity and general susceptibility of various groups of people. COVID-19 mild patients present with fever, fatigue, dry cough and other symptoms, whereas severe patients can appear with dyspnea, acute respiratory distress syndrome (ARDS) or septic shock and other symptoms. There is no special drug at present 6,7.
The treatment of COVID-19 mainly consisted of bed rest; intensive supportive treatment; oxygen therapy; antiviral therapy; antimicrobial therapy and Chinese medicine treatment. Critical cases need respiratory support (high flow nasal oxygen therapy, non-invasive ventilator or invasive mechanical ventilator); circulatory support for critically ill patients; plasma treatment from recovered patients and immunotherapy 8,9. Most of the infectious diseases caused by virus belong to the category of "plague" in ancient Chinese traditional medicine, which is caused by many evil spirits invading the body 10. The traditional medicine, including traditional Chinese medicine (TCM), has a good therapeutic effect on it 11,12. The Health and Health Commission of China and the State Administration of traditional Chinese Medicine in the "circular on the issuance of a new type of coronavirus infection pneumonia diagnosis and treatment program (version 5)" requested to strengthen the integration of Chinese and western medicine, and recommended a number of proprietary Chinese medicine in the process of diagnosis and treatment 13. On the basis of the national plan and in accordance with the principle of "three conditions and conditions", local prevention and control projects have also been successively issued according to local conditions 14. Recommended Chinese medicines include MaXing ShiGan Tang, QingFei PaiDu Tang, HuoXiang ZhengQi Capsules, JinHua QingGan Granules, LianHua QingWen Capsules or ShuFeng JieDu capsule 8. One clinical study showed that LianHua QingWen could improve the symptoms of COVID-19 patients and shorten the course of disease 15. A retrospective analysis study showed that the time of disappearance of clinical symptoms, recovery of body temperature, average length of stay in the integrated Chinese and western medicine treatment group (34) was significantly lower than that of the western medicine group (18) among the 52 COVID-19 patients 16.With QingFei PaiDu Tang combined with western medicine to treat the COVID-19 could significantly improve the patient's symptoms and achieved better results 17.
ShuFeng JieDu capsule (SFJDC) is a traditional Chinese medicine used to treat influenza in China 18. SFJDC is composed of Polygoni Cuspidati Rhizoma Et Radix (PCRR), Forsythiae Fructus (FF), Isatidis Radix (IR), Herba Patriniae (HP), Phragmitis Rhizoma (PR), Verbenae Herb (VH), licorice (I), Radix Bupleuri (RB) (Table 1). SFJDC has antiviral, anti-inflammatory, antipyretic and immune regulatory effects 19. SFJDC was commonly used for upper respiratory tract infection, pulmonary infection, AECOPD and other disease 20.This drug now is also recommended for the treatment of COVID-19 infections in the latest Diagnosis and Treatment of Pneumonia Caused by COVID-19 (version 5) 13,21. Currently, SFJDC is recommended in the Diagnosis and Treatment of Pneumonia Caused by COVID-19 in 5 provinces and cities 22.
Table 1.
Herb composition of Shu Feng Jie Du Capsule (SFJDC)
| English translation | Latin name | Chinese name |
|---|---|---|
| Hu-Zhang | Polygoni Cuspidati Rhizoma Et Radix | 虎杖 |
| Lian-Qiao | Forsythiae Fructus | 连翘 |
| Ban-Lan-Gen | Isatidis Radix | 板蓝根 |
| Chai-Hu | Herba Patriniae | 柴胡 |
| Bai-Jiang-Cao | Phragmitis Rhizoma | 败酱草 |
| Ma-Bian-Cao | Verbenae Herb | 马鞭草 |
| Lu-Gen | licorice | 芦根 |
| Gan-Cao | Radix Bupleuri | 甘草 |
Network pharmacology is a new discipline based on the theory of system biology, which analyzes the biological systems and selects specific signal nodes for multi-target drug molecular design. Network pharmacology emphasizes the multi-pathway regulation of signaling pathways and the regulation of multi-component, multi-target, multi-pathway, linking active components in traditional chinese medicine with target genes from molecular and biological aspects 23. Network pharmacology will help to understand the relationship among ingredients, genes and diseases and is suitable for the study of complex TCM or TCM compounds. The potential mechanism of preventing COVID-19 by HuoXiang ZhengQi oral solution was realized by network pharmacology and molecular docking 24. The research group Jing Zhao elucidated the mechanism of QingFei PaiDu Tang in the treatment of COVID-19 using network pharmacology 25. SFJDC could be efficacious for COVID-19, but active incredients, target genes and putative mechanism are not known.
In the present study, the network pharmacological was used to investigate the possible mechanism and target of SFJDC in the treatment of COVID-19. COVID-19 is a type of Novel Coronavirus Pneumonia (NCP). The active ingredients and their target genes of SFJDC were collected from TCMSP. NCP-related target genes were collected from GeneCards database. The putative mechanism of SFJDC against NCP were analyzed by GO and KEGG pathway. The flowchart of network pharmacology was shown in Figure 1. The study provided possible theoretical reference for SFJDC in the prevention and treatment of COVID-19.
Figure 1.
The flowchart of the whole manuscript base on network pharmacology.
Materials and Methods
Screening of active Ingredients in SFJDC
We identified the active ingredients of SFJDC from Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP http://tcmspw.com/tcmsp.php) 26. TCMSP is a unique herbal pharmacology platform that captures the relationship between drugs, target genes and diseases. The database includes the detection of natural compounds such as chemical, target and drug target networks. ADME is pharmacokinetics, which refers to the absorption, distribution, metabolism and excretion of exogenous chemicals by myosome. The four key parameters of ADME were blood-brain barrier (BBB), oral bioavailability (OB), Caco-2 permeability (Caco-2) and drug-likeness (DL) 27. In this study select candidate compounds which has OB≥30%, DL≥ 0.18, Caco-2≥-0.4, BBB≥-0.3.Then we sorted out each active ingredient for identification of targets.
Identification of SFJDC putative target genes
This study used the TCMSP platform to obtain the putative target genes of active ingredients of SFJDC. The Uniprot (https://www.uniprot.org/) 28 database provides a comprehensive, high quality and freely available source of protein sequence and function information. The putative target information corresponding to the active ingredients were input into UniProt database to obtain the standard name of the action target genes.
Screening of NCP related targets
COVID-19 is a type of Novel Coronavirus Pneumonia (NCP). So We collected NCP related targets from GeneCards (https://www.genecards.org/), which is a searchable, integrative database that provides comprehensive, user-friendly information on all annotated and predicted human genes 29. The key word “Novel Coronavirus Pneumonia” was used in the GeenCards database.
PPI (Protein-Protein Interaction) network construction of SFJDC putative and NCP related target genes
The PPI network of SFJDC putative and NCP related targets would be obtained from STRING (https://string-db.org/ ver11.0, update Jan 2019) 30. Active interaction sources were set as follows: Textmining, Co-expression, Neighborhood, Experiments, Databases, Gene Fusion and Co-occurrence. The required minimum interaction score was set at 0.4 in PPI network of SFJDC related targets, PPI network of NCP was set at 0.9. The barplot were generated by the R software (https://www.r-project.org/ver 3.6.2) based on counts value.
Construction of SFJDC ingredient-target network
Perl (https://www.perl.org/get.html) is a programming language suitable for writing simple scripts as well as complex applications. We used Strawberry Perl 5.30.1.1 to prepare the ingredient-target network. Cytoscape is a universal open source software for large-scale integrated development of molecular interaction networks working data. Then the ingredients-targets network of SFJDC was constructed using Cytoscape 3.7.2 software 31.
PPI network construction of SFJDC against NCP
In order to reveal the mechanism of SFJDC against NCP, a PPI network was constructed by the BisoGenet client which is a Cytoscape plugin was used to visualize. In this plugin, Protein-protein interactions information is taken from the DIP, BIOGRID, HPRD, INTACT, MINT, BIND 32. CytoNCA is a Cytoscape plugin integrating calculation, evaluation and visualization analysis for multiple centrality measure measures including Betweenness Centrality (BC), Degree Centrality (DC), Colseness Centrality (CC), Local average connectivity-based method (LAC), Eigenvector Centrality (EC) and Network Centrality (NC) 33.
Identification of candidate genes (CGs) and enrichment analysis of CGs
The CGs were filtered with R software using the Venn Diagram package (https://cran.r-project.org/web/packages/VennDiagram/index.html). The CGs would be used for Gene Ontology (GO) analysis (including biological processes (BP), molecular functions (MF), and cellular components (CC)) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. GO and KEGG pathway analyses results were processed by the “enrichplot” (http://www.bioconductor.org/packages/release/bioc/html/enrichplot.html) “clusterProfiler” (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) and “ggplot2” packages by R software. A P value of less than 0.05 was used regarded as statistically significant. At the same time, we input CGs into DAVID (https://david.ncifcrf.gov/) for functional enrichment analysis to obtain disease clustering.
Construction of gene-pathway network
KEGG pathways that had significant changes of P<0.05 were further analyzed. The genes that significantly regulated pathways for gene-pathway network construction. The key target genes of SFJDC against NCP were screened by gene-pathway network.
Results
The active ingredients of each herb contained in SFJDC
One hundred and thirty-seven active ingredients were screened out of TCMSP based on ADME, 4 in PCRR, 17 in FF, 25 in IR, 9 in HP, 7 in PR, 7 in VH, 1 in I, 67 in RB and 13 of which were repeated. Finally, 124 candidate active components of each herb contained in SFJDC were screened for further analysis after removing duplation (Table 2).
Table 2.
The active ingredients of each herb contained in SFJDC
| Mol ID | Molecule Name | OB (%) | Caco-2 | BBB | DL | Source |
|---|---|---|---|---|---|---|
| MOL000173 | wogonin | 30.68 | 0.79 | 0.04 | 0.23 | FF |
| MOL000211 | Mairin | 55.38 | 0.73 | 0.22 | 0.78 | FF; RB |
| MOL000239 | Jaranol | 50.83 | 0.61 | -0.22 | 0.29 | RB |
| MOL000358 | beta-sitosterol | 36.91 | 1.32 | 0.99 | 0.75 | PR; PCRR; IR; FF; VH |
| MOL000359 | sitosterol | 36.91 | 1.32 | 0.87 | 0.75 | PR; IR; RB |
| MOL000392 | formononetin | 69.67 | 0.78 | 0.02 | 0.21 | RB |
| MOL000449 | Stigmasterol | 43.83 | 1.44 | 1 | 0.76 | PR; IR; HP; I; VH |
| MOL000497 | licochalcone a | 40.79 | 0.82 | -0.21 | 0.29 | RB |
| MOL000791 | bicuculline | 69.67 | 0.72 | 0.02 | 0.88 | FF |
| MOL000953 | CLR | 37.87 | 1.43 | 1.13 | 0.68 | IR |
| MOL001484 | Inermine | 75.18 | 0.89 | 0.4 | 0.54 | RB |
| MOL001645 | Linoleyl acetate | 42.1 | 1.36 | 1.08 | 0.2 | HP |
| MOL001663 | (4aS,6aR,6aS,6bR,8aR,10R,12aR,14bS)-10-hydroxy-2,2,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,6a,7,8,8a,10,11,12,13,14b-tetradecahydropicene-4a-carboxylic acid | 32.03 | 0.61 | 0.39 | 0.76 | VH |
| MOL001676 | Vilmorrianine C | 33.96 | 0.59 | 0.14 | 0.22 | PR |
| MOL001677 | asperglaucide | 58.02 | 0.28 | -0.22 | 0.52 | PR |
| MOL001689 | acacetin | 34.97 | 0.67 | -0.05 | 0.24 | PR; IR |
| MOL001697 | Sinoacutine | 63.39 | 0.72 | 0.36 | 0.53 | PR |
| MOL001749 | ZINC03860434 | 43.59 | 1.04 | 0.6 | 0.35 | IR |
| MOL001755 | 24-Ethylcholest-4-en-3-one | 36.08 | 1.46 | 1.22 | 0.76 | IR |
| MOL001756 | quindoline | 33.17 | 1.5 | 0.99 | 0.22 | IR |
| MOL001769 | beta-sitosterol dodecantate | 34.57 | 1.28 | 0.57 | 0.57 | IR |
| MOL001771 | poriferast-5-en-3beta-ol | 36.91 | 1.45 | 1.14 | 0.75 | IR |
| MOL001774 | Ineketone | 37.14 | 0.39 | 0.1 | 0.3 | IR |
| MOL001779 | Sinoacutine | 49.11 | 0.7 | 0.39 | 0.46 | IR |
| MOL001781 | Indigo | 38.2 | 0.83 | 0.02 | 0.26 | IR |
| MOL001782 | (2Z)-2-(2-oxoindolin-3-ylidene)indolin-3-one | 48.4 | 0.85 | -0.06 | 0.26 | IR |
| MOL001783 | 2-(9-((3-methyl-2-oxopent-3-en-1-yl)oxy)-2-oxo-1,2,8,9-tetrahydrofuro[2,3-h]quinolin-8-yl)propan-2-yl acetate | 64 | 0.39 | -0.09 | 0.57 | IR |
| MOL001792 | DFV | 32.76 | 0.51 | -0.29 | 0.18 | IR; RB |
| MOL001793 | (E)-2-[(3-indole)cyanomethylene-]-3-indolinone | 54.59 | 1.06 | 0.22 | 0.32 | IR |
| MOL001800 | rosasterol | 35.87 | 1.28 | 0.89 | 0.75 | IR |
| MOL001803 | Sinensetin | 50.56 | 1.12 | 0.04 | 0.45 | IR |
| MOL001804 | Stigmasta-5,22-diene-3beta,7alpha-diol | 43.04 | 1.35 | 0.84 | 0.82 | IR |
| MOL001806 | Stigmasta-5,22-diene-3beta,7beta-diol | 42.56 | 1.37 | 0.81 | 0.83 | IR |
| MOL001810 | 6-(3-oxoindolin-2-ylidene)indolo[2,1-b]quinazolin-12-one | 45.28 | 1.19 | 0.48 | 0.89 | IR |
| MOL001814 | (E)-3-(3,5-dimethoxy-4-hydroxy-benzylidene)-2-indolinone | 57.18 | 0.69 | 0.16 | 0.25 | IR |
| MOL001820 | (E)-3-(3,5-dimethoxy-4-hydroxyb-enzylidene)-2-indolinone | 65.17 | 0.28 | -0.17 | 0.25 | IR |
| MOL001828 | 3-[(3,5-dimethoxy-4-oxo-1-cyclohexa-2,5-dienylidene)methyl]-2,4-dihydro-1H-pyrrolo[2,1-b]quinazolin-9-one | 51.84 | 0.81 | 0.03 | 0.56 | IR |
| MOL002311 | Glycyrol | 90.78 | 0.71 | -0.2 | 0.67 | RB |
| MOL002565 | Medicarpin | 49.22 | 1 | 0.53 | 0.34 | RB |
| MOL002773 | beta-carotene | 37.18 | 2.25 | 1.52 | 0.58 | VH |
| MOL003281 | 20(S)-dammar-24-ene-3β,20-diol-3-acetate | 40.23 | 0.93 | 0.28 | 0.82 | FF |
| MOL003290 | (3R,4R)-3,4-bis[(3,4-dimethoxyphenyl)methyl]oxolan-2-one | 52.3 | 0.78 | 0.17 | 0.48 | FF |
| MOL003295 | (+)-pinoresinol monomethyl ether | 53.08 | 0.69 | 0 | 0.57 | FF |
| MOL003306 | ACon1_001697 | 85.12 | 0.76 | 0 | 0.57 | FF |
| MOL003308 | (+)-pinoresinol monomethyl ether-4-D-beta-glucoside_qt | 61.2 | 0.7 | 0.12 | 0.57 | FF |
| MOL003315 | 3beta-Acetyl-20,25-epoxydammarane-24alpha-ol | 33.07 | 0.75 | 0.24 | 0.79 | FF |
| MOL003322 | FORSYTHINOL | 81.25 | 0.59 | -0.08 | 0.57 | FF |
| MOL003330 | (-)-Phillygenin | 95.04 | 0.75 | 0.07 | 0.57 | FF |
| MOL003344 | β-amyrin acetate | 42.06 | 1.36 | 1.1 | 0.74 | FF |
| MOL003347 | hyperforin | 44.03 | 0.87 | 0.4 | 0.6 | FF |
| MOL003348 | adhyperforin | 44.03 | 0.93 | 0.58 | 0.61 | FF |
| MOL003365 | Lactucasterol | 40.99 | 0.88 | 0.5 | 0.85 | FF |
| MOL003370 | Onjixanthone I | 79.16 | 0.84 | 0.04 | 0.3 | FF |
| MOL003656 | Lupiwighteone | 51.64 | 0.68 | -0.23 | 0.37 | RB |
| MOL003896 | 7-Methoxy-2-methyl isoflavone | 42.56 | 1.16 | 0.56 | 0.2 | RB |
| MOL004598 | 3,5,6,7-tetramethoxy-2-(3,4,5-trimethoxyphenyl)chromone | 31.97 | 0.75 | 0.08 | 0.59 | HP |
| MOL004609 | Areapillin | 48.96 | 0.6 | -0.29 | 0.41 | HP |
| MOL004624 | Longikaurin A | 47.72 | 0.08 | 0.09 | 0.53 | HP |
| MOL004628 | Octalupine | 47.82 | 0.48 | 0.3 | 0.28 | HP |
| MOL004644 | Sainfuran | 79.91 | 0.9 | 0.23 | 0.23 | HP |
| MOL004653 | (+)-Anomalin | 46.06 | 0.46 | 0 | 0.66 | HP |
| MOL004718 | α-spinasterol | 42.98 | 1.28 | 0.79 | 0.76 | HP |
| MOL004805 | (2S)-2-[4-hydroxy-3-(3-methylbut-2-enyl)phenyl]-8,8-dimethyl-2,3-dihydropyrano[2,3-f]chromen-4-one | 31.79 | 1 | 0.25 | 0.72 | RB |
| MOL004806 | euchrenone | 30.29 | 1.09 | 0.39 | 0.57 | RB |
| MOL004808 | glyasperin B | 65.22 | 0.47 | -0.09 | 0.44 | RB |
| MOL004810 | glyasperin F | 75.84 | 0.43 | -0.15 | 0.54 | RB |
| MOL004811 | Glyasperin C | 45.56 | 0.71 | 0.07 | 0.4 | RB |
| MOL004814 | Isotrifoliol | 31.94 | 0.53 | -0.25 | 0.42 | RB |
| MOL004815 | (E)-1-(2,4-dihydroxyphenyl)-3-(2,2-dimethylchromen-6-yl)prop-2-en-1-one | 39.62 | 0.66 | -0.12 | 0.35 | RB |
| MOL004820 | kanzonols W | 50.48 | 0.63 | 0.04 | 0.52 | RB |
| MOL004828 | Glepidotin A | 44.72 | 0.79 | 0.06 | 0.35 | RB |
| MOL004829 | Glepidotin B | 64.46 | 0.46 | -0.09 | 0.34 | RB |
| MOL004833 | Phaseolinisoflavan | 32.01 | 1.01 | 0.46 | 0.45 | RB |
| MOL004835 | Glypallichalcone | 61.6 | 0.76 | 0.23 | 0.19 | RB |
| MOL004838 | 8-(6-hydroxy-2-benzofuranyl)-2,2-dimethyl-5-chromenol | 58.44 | 1 | 0.34 | 0.38 | RB |
| MOL004848 | licochalcone G | 49.25 | 0.64 | -0.04 | 0.32 | RB |
| MOL004849 | 3-(2,4-dihydroxyphenyl)-8-(1,1-dimethylprop-2-enyl)-7-hydroxy-5-methoxy-coumarin | 59.62 | 0.4 | -0.23 | 0.43 | RB |
| MOL004855 | Licoricone | 63.58 | 0.53 | -0.14 | 0.47 | RB |
| MOL004856 | RBnin A | 51.08 | 0.8 | 0.13 | 0.4 | RB |
| MOL004857 | RBnin B | 48.79 | 0.58 | -0.1 | 0.45 | RB |
| MOL004863 | 3-(3,4-dihydroxyphenyl)-5,7-dihydroxy-8-(3-methylbut-2-enyl)chromone | 66.37 | 0.52 | -0.13 | 0.41 | RB |
| MOL004866 | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-6-(3-methylbut-2-enyl)chromone | 44.15 | 0.48 | -0.28 | 0.41 | RB |
| MOL004879 | Glycyrin | 52.61 | 0.59 | -0.13 | 0.47 | RB |
| MOL004882 | Licocoumarone | 33.21 | 0.84 | 0.06 | 0.36 | RB |
| MOL004883 | Licoisoflavone | 41.61 | 0.37 | -0.27 | 0.42 | RB |
| MOL004884 | Licoisoflavone B | 38.93 | 0.46 | -0.18 | 0.55 | RB |
| MOL004885 | licoisoflavanone | 52.47 | 0.39 | -0.22 | 0.54 | RB |
| MOL004891 | shinpterocarpin | 80.3 | 1.1 | 0.68 | 0.73 | RB |
| MOL004907 | Glyzaglabrin | 61.07 | 0.34 | -0.2 | 0.35 | RB |
| MOL004908 | Glabridin | 53.25 | 0.97 | 0.36 | 0.47 | RB |
| MOL004910 | Glabranin | 52.9 | 0.97 | 0.31 | 0.31 | RB |
| MOL004911 | Glabrene | 46.27 | 0.99 | 0.04 | 0.44 | RB |
| MOL004912 | Glabrone | 52.51 | 0.59 | -0.11 | 0.5 | RB |
| MOL004913 | 1,3-dihydroxy-9-methoxy-6-benzofurano[3,2-c]chromenone | 48.14 | 0.48 | -0.19 | 0.43 | RB |
| MOL004915 | Eurycarpin A | 43.28 | 0.43 | -0.06 | 0.37 | RB |
| MOL004941 | (2R)-7-hydroxy-2-(4-hydroxyphenyl)chroman-4-one | 71.12 | 0.41 | -0.25 | 0.18 | RB |
| MOL004945 | (2S)-7-hydroxy-2-(4-hydroxyphenyl)-8-(3-methylbut-2-enyl)chroman-4-one | 36.57 | 0.72 | -0.04 | 0.32 | RB |
| MOL004948 | Isoglycyrol | 44.7 | 0.91 | 0.05 | 0.84 | RB |
| MOL004957 | HMO | 38.37 | 0.79 | 0.25 | 0.21 | RB |
| MOL004959 | 1-Methoxyphaseollidin | 69.98 | 1.01 | 0.48 | 0.64 | RB |
| MOL004966 | 3'-Hydroxy-4'-O-Methylglabridin | 43.71 | 1 | 0.73 | 0.57 | RB |
| MOL004974 | 3'-Methoxyglabridin | 46.16 | 0.94 | 0.47 | 0.57 | RB |
| MOL004978 | 2-[(3R)-8,8-dimethyl-3,4-dihydro-2H-pyrano[6,5-f]chromen-3-yl]-5-methoxyphenol | 36.21 | 1.12 | 0.61 | 0.52 | RB |
| MOL004980 | Inflacoumarin A | 39.71 | 0.73 | -0.24 | 0.33 | RB |
| MOL004985 | icos-5-enoic acid | 30.7 | 1.22 | 1.09 | 0.2 | RB |
| MOL004988 | Kanzonol F | 32.47 | 1.18 | 0.56 | 0.89 | RB |
| MOL004989 | 6-prenylated eriodictyol | 39.22 | 0.4 | -0.29 | 0.41 | RB |
| MOL004991 | 7-Acetoxy-2-methylisoflavone | 38.92 | 0.74 | 0.16 | 0.26 | RB |
| MOL004996 | gadelaidic acid | 30.7 | 1.2 | 0.94 | 0.2 | RB |
| MOL005000 | RBnin G | 60.44 | 0.78 | 0.23 | 0.39 | RB |
| MOL005001 | RBnin H | 50.1 | 0.6 | -0.14 | 0.78 | RB |
| MOL005003 | Licoagrocarpin | 58.81 | 1.23 | 0.61 | 0.58 | RB |
| MOL005007 | Glyasperins M | 72.67 | 0.49 | -0.04 | 0.59 | RB |
| MOL005012 | Licoagroisoflavone | 57.28 | 0.71 | 0.09 | 0.49 | RB |
| MOL005016 | Odoratin | 49.95 | 0.42 | -0.24 | 0.3 | RB |
| MOL005017 | Phaseol | 78.77 | 0.76 | -0.06 | 0.58 | RB |
| MOL005018 | Xambioona | 54.85 | 1.09 | 0.52 | 0.87 | RB |
| MOL005020 | dehydroglyasperins C | 53.82 | 0.68 | -0.12 | 0.37 | RB |
| MOL005229 | Artemetin | 49.55 | 0.81 | -0.09 | 0.48 | VH |
| MOL005503 | Cornudentanone | 39.66 | 0.47 | 0.09 | 0.33 | VH |
| MOL008752 | Dihydroverticillatine | 42.69 | 0.56 | 0.11 | 0.84 | VH |
| MOL013281 | 6,8-Dihydroxy-7-methoxyxanthone | 35.83 | 0.68 | 0.1 | 0.21 | PCRR |
| MOL013287 | Physovenine | 106.21 | 0.51 | 0.2 | 0.19 | PCRR |
| MOL013288 | Picralinal | 58.01 | 0.23 | -0.21 | 0.75 | PCRR |
Putative target genes of each herb in SFJDC and NCP related target genes
The 124 candidate active components were imported into TCMSP database and Uniport database to identify the Putative target genes of each herb in SFJDC. One hundred and ten components were finally selected after removing 14 ingredients which did not link to any target genes. The target genes of 110 compounds were collected. 1705 genes were identified, 103 in PR, 209 in IR, 65 in HP, 1052 in RB, 75 in PCRR, 173 in FF and 27 in I. There were 1585 genes of the eight herbs overlapped, which was suggestive of potential interaction between the compounds of SFJDCA in the course of treatment. A total of 120 genes were identified after removing duplation (Table 3). And 251 NCP related target genes were identified from Gene Cards database (Table 4).
Table 3.
Putative target genes of each herb in SFJDC
| Herb | Mol ID | Molname | Target genes |
|---|---|---|---|
| FF | MOL000173 | wogonin | ADRB2 AHSA1 AKT1 AR BAX BBC3 BCL2 CALM1 CASP3 CASP9 CCL2 CCND1 CDK2 CDKN1A CHEK1 DPP4 EIF6 ESR1 FSD1 GABRA1 GSK3B HSP90AA1 IL6 IL8RA JUN KDR MAPK14 MCL1 MMP1 NOS2 PPARG PRKCD PRSS1 PTGER3 PTGS1 PTGS2 RELA RXRA SCN5A TEP1 TNFSF15 TP63 |
| RB FF | MOL000211 | Mairin | PGR |
| RB | MOL000239 | Jaranol | AR CALM1 CDK2 CHEK1 DPP4 ESR2 HSP90AA1 NCOA2 NOS2 PRSS1 PTGS1 PTGS2 SCN5A |
| PR IR; PCRR; FF | MOL000358 | beta-sitosterol | ADRA1A ADRA1B ADRB2 BAX BCL2 CASP3 CASP8 CASP9 CHRM1 CHRM2 CHRM3 CHRM4 CHRNA2 DRD1 GABRA1 HSP90AA1 JUN KCNH2 MAP2 NCOA2 OPRM1 PGR PON1 PRKCA PTGS1 PTGS2 SCN5A SLC6A4 |
| PR IR RB | MOL000359 | sitosterol | NCOA2 NR3C2 PGR |
| RB | MOL000392 | formononetin | ACHE ADRA1A ADRB2 AR ATP5F1B CALM1 CCNA2 CDK2 CHEK1 CHRM1 DPP4 ESR1 ESR2 GSK3B HSD3B1 HSD3B2 HSP90AA1 HTR IL4 JUN MAOB MAPK14 ND6 NOS2 PKIA PPARG PPARG PRSS1 PTGS1 PTGS2 RXRA SLC6A3 SLC6A4 |
| PR IR HP I | MOL000449 | Stigmasterol | ADH1C ADRA1A ADRA1B ADRA2A ADRB1 ADRB2 AKR1B1 CHRM1 CHRM2 CHRM3 CTRB1 GABRA1 IGHG1 LTA4H MAOA MAOB NCOA1 NCOA2 NR3C2 PGR PLAU PTGS1 PTGS2 RXRA SCN5A SLC6A2 SLC6A3 |
| RB | MOL000497 | licochalcone a | ADRA1B ADRB2 AR BCL2 CA2 CALM1 CCNA2 CCND1 CDK2 CDK4 CHEK1 CHRM1 EIF6 ESR1 ESR2 FOSL2 GSK3B HSP90AA1 MAPK1 MAPK14 NCOA2 NOS2 PPARG PTGS1 PTGS2 RB1 RELA SCN5A SLC6A3 STAT3 |
| FF | MOL000791 | bicuculline | ACHE ALDH3A1 AR BMPR2 CRH FOS GABBR1 GJA1 GJB1 GNRH1 GNRHR GRIN2D GRM1 GRM5 HSP90AA1 HTR KCNH2 KDR PTGS1 PTGS2 SCN5A SLC6A2 VCP |
| IR | MOL000953 | CLR | NCOA2 NR3C2 PGR |
| RB | MOL001484 | Inermine | ADRA1B ADRA1D ADRB2 CALM1 CHRM1 CHRM3 HSP90AA1 HTR3A IGHG1 OPRM1 PRSS1 PTGS1 PTGS2 RXRA SCN5A |
| HP | MOL001645 | Linoleyl acetate | NCOA2 PTGS1 PTGS2 RXRA |
| PR | MOL001677 | asperglaucide | HTR KCNH2 PRSS1 PTGS2 |
| PR; IR | MOL001689 | acacetin | ADRB2 AR BAX BCL2 CALM1 CASP3 CASP8 CDK2 CDKN1A CHEK1 CYP19A1 DPP4 FASLG FASN HSP90AA1 NCOA1 NCOA2 NOS2 PRSS1 PTGS1 PTGS2 RELA TP63 |
| PR | MOL001697 | Sinoacutine | ACHE ADRA1A ADRA1B AR CHRM1 CHRM2 CHRM3 CHRM4 CHRM5 ESR1 ESR2 GABRA1 HTR OPRD1 OPRM1 PTGS1 PTGS2 SCN5A |
| IR | MOL001749 | ZINC03860434 | ADRB2 CHRM1 CHRM3 SCN5A |
| IR | MOL001755 | 24-Ethylcholest-4-en-3-one | NR3C2 PGR |
| IR | MOL001756 | quindoline | MAOB NCOA2 PKIA PTGS1 PTGS2 |
| IR | MOL001771 | poriferast-5-en-3beta-ol | NCOA2 PGR |
| IR | MOL001774 | Ineketone | NR3C2 |
| IR | MOL001779 | Sinoacutine | ACHE ADRA1B AR CALM1 CHRM1 CHRM3 CHRM5 DPP4 ESR1 ESR2 HSP90AA1 HTR NOS2 OPRD1 OPRM1 PTGS1 PTGS2 RXRA SCN5A |
| IR | MOL001781 | Indigo | CCNA2 CDK2 PTGS1 PTGS2 RXRA |
| IR | MOL001782 | (2Z)-2-(2-oxoindolin-3-ylidene)indolin-3-one | AR CCNA2 CDK2 CHEK1 ESR1 GABRA1 GSK3B HSP90AA1 MAPK14 NOS2 PTGS1 PTGS2 RXRA |
| IR | MOL001783 | 2-(9-((3-methyl-2-oxopent-3-en-1-yl)oxy)-2-oxo-1,2,8,9-tetrahydrofuro[2,3-h]quinolin-8-yl)propan-2-yl acetate | HSP90AA1 KCNH2 NCONA2 PRSS1 PTGS2 |
| IR RB | MOL001792 | DFV | ADRB2 ESR1 HSP90AA1 MAOB PKIA PTGS1 PTGS2 RXRA SLC6A4 |
| IR | MOL001793 | (E)-2-[(3-indole)cyanomethylene-]-3-indolinone | AR CCNA2 CDK2 CHEK1 ESR1 GSK3B HSP90AA1 MAPK14 NOS2 PTGS1 PTGS2 RXRA |
| IR | MOL001800 | rosasterol | PGR |
| IR | MOL001803 | Sinensetin | ACHE ADRA1B ADRB2 AR CALM1 CHEK1 DPP4 ESR2 F7 HSP90AA1 HTR KCNH2 NCOA1 NCOA2 NOS2 PRSS1 PTGS1 PTGS2 SCN5A |
| IR | MOL001804 | Stigmasta-5,22-diene-3beta,7alpha-diol | NCOA2 PGR |
| IR | MOL001810 | 6-(3-oxoindolin-2-ylidene)indolo[2,1-b]quinazolin-12-one | ESR1 KDR PRSS1 PTGS1 PTGS2 |
| IR | MOL001814 | (E)-3-(3,5-dimethoxy-4-hydroxy-benzylidene)-2-indolinone | GABRA1 HSP90AA1 PTGS1 PTGS2 RXRA SCN5A |
| IR | MOL001820 | (E)-3-(3,5-dimethoxy-4-hydroxyb-enzylidene)-2-indolinone | ADRB2 CHRM1 GABRA1 HSP90AA1 PTGS1 PTGS2 RXRA SCN5A |
| IR | MOL001828 | 3-[(3,5-dimethoxy-4-oxo-1-cyclohexa-2,5-dienylidene)methyl]-2,4-dihydro-1H-pyrrolo[2,1-b]quinazolin-9-one | F7 HSP90AA1 KCNH2 PRSS1 PTGS1 PTGS2 SCN5A |
| RB | MOL002311 | Glycyrol | CCNA2 CHEK1 ESR1 GSK3B HTR KDR MAPK14 NOS2 PPARG PTGS2 |
| RB | MOL002565 | Medicarpin | ADRA1A ADRA1B ADRA1D ADRB2 CALM1 CCNA2 CDK2 CHRM1 CHRM2 CHRM3 CHRM4 CHRM5 DPP4 DRD1 ESR1 ESR2 HSP90AA1 MAPK10 NOS2 OPRD1 OPRM1 PRSS1 PTGS1 PTGS2 RXRA SCN5A SLC6A3 SLC6A4 |
| FF | MOL003290 | (3R,4R)-3,4-bis[(3,4-dimethoxyphenyl)methyl]oxolan-2-one | ADRA1B ADRA1D ADRB2 CALM1 CHRM3 ESR1 F7 HSP90AA1 KCNH2 NCOA2 PTGS2 SCN5A SLC6A3 |
| FF | MOL003295 | (+)-pinoresinol monomethyl ether | ADRA1B ADRB2 CALM1 HSP90AA1 KCNH2 NCOA1 NCOA2 PTGS1 PTGS2 RXRA RXRB SCN5A |
| FF | MOL003306 | ACon1_001697 | ADRA1B ADRB2 CALM1 HSP90AA1 KCNH2 NCOA1 NCOA2 PTGS1 PTGS2 SCN5A |
| FF | MOL003308 | (+)-pinoresinol monomethyl ether-4-D-beta-glucoside_qt | ADRB2 CALM1 HSP90AA1 KCNH2 NCOA1 NCOA2 PTGS2 SCN5A |
| FF | MOL003315 | 3beta-Acetyl-20,25-epoxydammarane-24alpha-ol | NR3C1 |
| FF | MOL003322 | FORSYTHINOL | ADRA1B ADRB2 CALM1 HSP90AA1 KCNH2 NCOA1 NCOA2 PTGS2 SCN5A |
| FF | MOL003330 | (-)-Phillygenin | ADRA1B ADRB2 CALM1 CHRM1 CHRM3 CHRM5 HSP90AA1 IGHG1 KCNH2 NCOA2 PTGS2 SCN5A |
| FF | MOL003347 | hyperforin | CYP3A4 ICAM1 IL8RA NR1I2 |
| FF | MOL003370 | Onjixanthone I | CALM1 CHEK1 DPP4 ESR2 HSP90AA1 NOS2 PTGS1 PTGS2 RXRA SCN5A |
| RB | MOL003656 | Lupiwighteone | AR CALM1 CCNA2 CDK2 CHEK1 DPP4 ESR1 ESR2 GSK3B HSP90AA1 HTR MAPK14 NCOA2 NOS2 PPARG PRSS1 PTGS2 SCN5A |
| RB | MOL003896 | 7-Methoxy-2-methyl isoflavone | ACHE ADRA1B ADRA1D ADRB1 ADRB2 AR CALM1 CCNA2 CDK2 CHEK1 CHRM1 CHRM3 CHRM5 DPP4 DRD1 ESR1 ESR2 GABRA1 GSK3B HSP90AA1 HTR IGHG1 LTA4H MAOB MAPK14 NCOA1 NCOA2 NOS2 OPRM1 PKIA PPARG PRSS1 PTGS1 PTGS2 RXRA SCN5A SLC6A3 SLC6A4 |
| HP | MOL004598 | 3,5,6,7-tetramethoxy-2-(3,4,5-trimethoxyphenyl)chromone | ACHE AR CALM1 ESR1 ESR2 F7 HTR NCOA2 PRSS1 PTGS2 |
| HP | MOL004609 | Areapillin | AR CALM1 DPP4 ESR2 F7 HSP90AA1 HTR IGHG1 NCOA1 NCOA2 NOS2 PRSS1 PTGS2 SCN5A |
| HP | MOL004624 | Longikaurin A | CHRM1 CHRM2 PRSS1 |
| HP | MOL004653 | (+)-Anomalin | DPP4 HTR KCNH2 PTGS2 |
| HP | MOL004718 | α-spinasterol | NCOA2 NR3C2 PGR |
| RB | MOL004805 | (2S)-2-[4-hydroxy-3-(3-methylbut-2-enyl)phenyl]-8,8-dimethyl-2,3-dihydropyrano[2,3-f]chromen-4-one | AR CALM1 ESR1 ESR2 GSK3B KCNH2 MAPK14 NOS2 PPARG PTGS2 |
| RB | MOL004806 | euchrenone | BACE1 CALM1 ESR1 ESR2 KCNH2 NOS2 PTGS2 SCN5A |
| RB | MOL004808 | glyasperin B | ACHE AR CALM1 CCNA2 CDK2 DPP4 ESR1 ESR2 F7 GSK3B HSP90AA1 HTR KDR NCOA2 NOS2 PPARG PRSS1 PTGS2 |
| RB | MOL004810 | glyasperin F | AR CALM1 CCNA2 CDK2 ESR1 ESR2 GSK3B HSP90AA1 MAPK14 NOS2 PPARG PRSS1 PTGS1 PTGS2 SCN5A |
| RB | MOL004811 | Glyasperin C | ACHE AR CALM1 CCNA2 CDK2 CHEK1 DPP4 ESR1 ESR2 GSK3B HSP90AA1 HTR KCNH2 MAPK14 NCOA2 NOS2 PPARG PRSS1 PTGS2 RXRA SCN5A |
| RB | MOL004814 | Isotrifoliol | AR CCNA2 CDK2 CHEK1 ESR1 ESR2 GSK3B HSP90AA1 MAPK14 NOS2 PTGS2 |
| RB | MOL004815 | (E)-1-(2,4-dihydroxyphenyl)-3-(2,2-dimethylchromen-6-yl)prop-2-en-1-one | ADRA1B AR CA2 CALM1 CCNA2 CDK2 CHEK1 ESR1 ESR2 GSK3B MAPK14 NCOA2 NOS2 PPARG PTGS1 PTGS2 RXRA SCN5A |
| RB | MOL004820 | kanzonols W | AR CALM1 CCNA2 CDK2 CHEK1 ESR1 ESR2 GSK3B MAPK14 NCOA1 NCOA2 NOS2 PPARG PRSS1 PTGS1 PTGS2 RXRA SCN5A |
| RB | MOL004828 | Glepidotin A | AR CALM1 CCNA2 CDK2 CHEK1 DPP4 ESR1 F7 GSK3B HSP90AA1 HTR IGHG1 KDR MAPK14 NOS2 PPARG PRSS1 PTGS1 PTGS2 RXRA SCN5A |
| RB | MOL004829 | Glepidotin B | ADRA1B CALM1 ESR1 F7 HSP90AA1 IGHG1 NCOA1 PTGS1 PTGS2 RXRA SCN5A |
| RB | MOL004833 | Phaseolinisoflavan | ACHE ADRA1B ADRB2 AR CALM1 CCNA2 CDK2 CHEK1 CHRM1 ESR1 ESR2 GSK3B MAPK14 NCOA1 NOS2 PPARG PRSS1 PTGS2 RXRA SCN5A |
| RB | MOL004835 | Glypallichalcone | ADRA1B ADRB2 AR CA2 CALM1 CCNA2 CDK2 CHEK1 CHRM1 ESR1 ESR2 GSK3B HSP90AA1 LTA4H MAOB MAPK14 NCOA1 NOS2 PKIA PPARG PTGS1 PTGS2 SCN5A SLC6A3 SLC6A4 |
| RB | MOL004838 | 8-(6-hydroxy-2-benzofuranyl)-2,2-dimethyl-5-chromenol | ESR1 HSP90AA1 NOS2 PTGS2 RXRA |
| RB | MOL004848 | licochalcone G | AR CALM1 CCNA2 CDK2 ESR1 ESR2 GSK3B HSP90AA1 IGHG1 KDR MAPK14 NCOA2 NOS2 PPARG PTGS2 |
| RB | MOL004849 | 3-(2,4-dihydroxyphenyl)-8-(1,1-dimethylprop-2-enyl)-7-hydroxy-5-methoxy-coumarin | AR CALM1 CDK2 CHEK1 DPP4 ESR1 ESR2 F7 GSK3B HSP90AA1 HTR KCNH2 KDR MAPK14 NCOA1 NCOA2 NOS2 PPARG PRSS1 PTGS2 |
| RB | MOL004855 | Licoricone | AR CALM1 CHEK1 ESR1 HTR KCNH2 KDR NCOA2 NOS2 PPARG PRSS1 PTGS2 |
| RB | MOL004856 | RBnin A | ACHE AR CALM1 CCNA2 CHEK1 DPP4 ESR1 ESR2 GSK3B HSP90AA1 HTR NCOA2 NOS2 PPARG PRSS1 PTGS2 SCN5A |
| RB | MOL004857 | RBnin B | ADRA1B ADRB2 AR CALM1 CCNA2 CHEK1 DPP4 ESR1 ESR2 F7 GSK3B HSP90AA1 HTR KDR NCOA2 NOS2 PPARG PRSS1 PTGS2 |
| RB | MOL004863 | 3-(3,4-dihydroxyphenyl)-5,7-dihydroxy-8-(3-methylbut-2-enyl)chromone | AR CALM1 CCNA2 CDK2 CHEK1 ESR1 GSK3B HSP90AA1 HTR MAPK14 NCOA2 NOS2 PPARG PRSS1 PTGS2 |
| RB | MOL004866 | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-6-(3-methylbut-2-enyl)chromone | ADRB2 AR CALM1 CCNA2 CDK2 CHEK1 DPP4 F7 HSP90AA1 HTR PPARG PRSS1 PTGS2 SCN5A |
| RB | MOL004879 | Glycyrin | AR CALM1 CHEK1 DPP4 ESR1 ESR2 HTR KCNH2 KDR NCOA2 NOS2 PPARG PRSS1 PTGS2 |
| RB | MOL004882 | Licocoumarone | AR CCNA2 CDK2 ESR1 ESR2 GSK3B HSP90AA1 |
| RB | MOL004883 | Licoisoflavone | AR CALM1 CCNA2 CDK2 CHEK1 DPP4 ESR1 HSP90AA1 HTR KDR MAPK14 NCOA2 NOS2 PPARG PRSS1 PTGS2 |
| RB | MOL004884 | Licoisoflavone B | ACHE AR CALM1 CCNA2 CDK2 CHEK1 ESR1 ESR2 GSK3B HTR NOS2 PPARG PRSS1 PTGS2 |
| RB | MOL004885 | licoisoflavanone | ACHE AR CALM1 CCNA2 CDK2 ESR1 ESR2 F7 GSK3B HSP90AA1 NCOA1 NOS2 PPARG PRSS1 PTGS1 PTGS2 SCN5A |
| RB | MOL004891 | shinpterocarpin | ADRA1B ADRA1D ADRB2 AR CALM1 CCNA2 CDK2 CHRM1 CHRM3 ESR1 ESR2 GSK3B HTR3A KCNH2 MAPK14 NCOA1 NOS2 OPRD1 OPRM1 PPARG PRSS1 PTGS1 PTGS2 RXRA RXRB SCN5A |
| RB | MOL004907 | Glyzaglabrin | AR CCNA2 CDK2 CHEK1 DPP4 ESR1 ESR2 GSK3B HSP90AA1 MAPK14 NOS2 PPARG PRSS1 PTGS1 PTGS2 |
| RB | MOL004908 | Glabridin | ACHE ADRA1B ADRB2 AR CALM1 CCNA2 CDK2 CHEK1 CHRM1 ESR1 ESR2 GSK3B IGHG1 MAPK14 NCOA1 NCOA2 NOS2 PPARG PRSS1 PTGS2 RXRA RXRB SCN5A |
| RB | MOL004910 | Glabranin | CALM1 ESR1 HSP90AA1 NOS2 PTGS1 PTGS2 SCN5A |
| RB | MOL004911 | Glabrene | ADRB2 AR CALM1 CDK2 ESR1 ESR2 GSK3B HSP90AA1 MAPK14 NCOA2 NOS2 PPARG PRSS1 PTGS1 PTGS2 RXRA SCN5A |
| RB | MOL004912 | Glabrone | ACHE AR CALM1 CCNA2 CDK2 CHEK1 DPP4 ESR1 ESR2 GSK3B HTR MAPK14 NOS2 PPARG PRSS1 PTGS1 PTGS2 RXRA SCN5A |
| RB | MOL004913 | 1,3-dihydroxy-9-methoxy-6-benzofurano[3,2-c]chromenone | CCNA2 CDK2 CHEK1 ESR1 ESR2 GSK3B HSP90AA1 MAPK14 PPARG |
| RB | MOL004915 | Eurycarpin A | AR CALM1 CCNA2 CDK2 CHEK1 DPP4 ESR1 ESR2 GSK3B HSP90AA1 HTR MAPK14 NOS2 PPARG PRSS1 PTGS2 SCN5A |
| RB | MOL004941 | (2R)-7-hydroxy-2-(4-hydroxyphenyl)chroman-4-one | PTGS1 ESR1 PTGS2 RXRA ADRB2 HSP90AA1 MAOB PKIA CALM1 GABRA1 SLC6A4 |
| RB | MOL004945 | (2S)-7-hydroxy-2-(4-hydroxyphenyl)-8-(3-methylbut-2-enyl)chroman-4-one | NOS2 |
| RB | MOL004948 | Isoglycyrol | AR DPP4 ESR1 GSK3B NOS2 PTGS2 |
| RB | MOL004957 | HMO | ADRB2 AR CALM1 CCNA2 CDK2 CHEK1 CHRM1 DPP4 ESR1 ESR2 GSK3B IGHG1 MAOB MAPK14 NOS2 PKIA PPARG PRSS1 PTGS1 PTGS2 RXRA SCN5A SLC6A3 SLC6A4 |
| RB | MOL004959 | 1-Methoxyphaseollidin | ADRA1B ADRA1D ADRB2 AR CALM1 CCNA2 CDK2 ESR1 ESR2 GSK3B HSP90AA1 HTR KCNH2 KDR MAPK14 NCOA1 NCOA2 NOS2 PPARG PRSS1 PTGS1 PTGS2 RXRA SCN5A |
| RB | MOL004966 | 3'-Hydroxy-4'-O-Methylglabridin | ADRA1B ADRB2 AR CALM1 CCNA2 CDK2 CHEK1 ESR1 ESR2 F7 GSK3B HSP90AA1 KCNH2 KDR MAPK14 NCOA1 NCOA2 NOS2 PPARG PRSS1 PTGS1 PTGS2 SCN5A |
| RB | MOL004974 | 3'-Methoxyglabridin | ACHE ADRA1B ADRB2 AR CALM1 CCNA2 CDK2 CHEK1 ESR1 ESR2 F7 GSK3B HSP90AA1 KCNH2 MAPK14 NCOA1 NCOA2 NOS2 PPARG PRSS1 PTGS1 PTGS2 RXRA SCN5A |
| RB | MOL004978 | 2-[(3R)-8,8-dimethyl-3,4-dihydro-2H-pyrano[6,5-f]chromen-3-yl]-5-methoxyphenol | ACHE ADRA1B ADRB2 AR CALM1 CCNA2 CDK2 CHEK1 CHRM1 CHRM3 ESR1 ESR2 GSK3B KCNH2 MAPK14 NCOA1 NCOA2 NOS2 PPARG PRSS1 PTGS1 PTGS2 RXRA RXRB SCN5A SLC6A3 |
| RB | MOL004980 | Inflacoumarin A | ADRB2 AR CALM1 DPP4 ESR1 HSP90AA1 HTR NCOA2 PPARG PRSS1 PTGS1 PTGS2 SCN5A |
| RB | MOL004985 | icos-5-enoic acid | NCOA2 |
| RB | MOL004988 | Kanzonol F | AR CALM1 ESR1 ESR2 NCOA2 PTGS2 |
| RB | MOL004989 | 6-prenylated eriodictyol | CALM1 ESR1 F7 HSP90AA1 NOS2 PTGS2 SCN5A |
| RB | MOL004991 | 7-Acetoxy-2-methylisoflavone | ACHE ADRA1B ADRA1D ADRB2 AR CALM1 CDK2 CHEK1 DPP4 ESR1 GABRA1 GSK3B HSP90AA1 HTR MAPK14 NCOA2 NOS2 PPARG PRSS1 PTGS1 PTGS2 RXRA SCN5A |
| RB | MOL004996 | gadelaidic acid | NCOA2 |
| RB | MOL005000 | RBnin G | AR CALM1 CCNA2 CHEK1 DPP4 ESR1 ESR2 GSK3B HSP90AA1 HTR MAPK14 NCOA2 NOS2 PPARG PRSS1 PTGS2 |
| RB | MOL005001 | RBnin H | AR CALM1 CCNA2 ESR1 HSP90AA1 KDR NCOA2 PRSS1 PTGS2 |
| RB | MOL005003 | Licoagrocarpin | ACHE ADRA1B ADRB2 AR CALM1 CCNA2 CDK2 CHRM1 CHRM3 CHRM5 ESR1 ESR2 GSK3B HSP90AA1 HTR KCNH2 MAPK14 NCOA2 NOS2 PPARG PRSS1 PTGS1 PTGS2 RXRA RXRB SCN5A |
| RB | MOL005007 | Glyasperins M | ACHE AR CALM1 CCNA2 CDK2 ESR1 ESR2 F7 GSK3B HSP90AA1 KCNH2 KDR NCOA1 NCOA2 NOS2 PPARD PPARG PRSS1 PTGS1 PTGS2 SCN5A |
| RB | MOL005012 | Licoagroisoflavone | AR CALM1 CCNA2 CDK2 CHEK1 DPP4 ESR1 ESR2 GSK3B HTR MAPK14 NOS2 PPARG PRSS1 PTGS2 SCN5A |
| RB | MOL005016 | Odoratin | AR CALM1 CCNA2 CDK2 CHEK1 DPP4 ESR1 ESR2 GSK3B HSP90AA1 MAPK14 NCOA2 NOS2 PPARG PRSS1 PTGS1 PTGS2 RXRA SCN5A |
| RB | MOL005017 | Phaseol | AR CCNA2 CDK2 CHEK1 ESR1 GSK3B HSP90AA1 HTR KDR MAPK14 PPARG PTGS2 |
| RB | MOL005018 | Xambioona | CALM1 ESR1 ESR2 NCOA2 NOS2 PTGS2 |
| RB | MOL005020 | dehydroglyasperins C | ADRB2 AR CALM1 CCNA2 CDK2 CHEK1 ESR1 ESR2 HSP90AA1 MAPK14 NCOA2 NOS2 PPARG PRSS1 PTGS2 SCN5A |
| PCRR | MOL013281 | 6,8-Dihydroxy-7-methoxyxanthone | ADRB2 CA2 CDK2 CHEK1 DPP4 GSK3B HSP90AA1 MAPK14 PKIA PTGS1 PTGS2 |
| PCRR | MOL013287 | Physovenine | ACHE ADRA1A ADRA1B ADRA2B ADRB2 AR CA2 CCNA2 CDK2 CHRM1 CHRM2 CHRM3 CHRNA2 DRD1 ESR1 ESR2 GABRA1 GRIA2 GSK3B HSP90AA1 HTR NOS2 OPRD1 OPRM1 PRSS1 PTGS1 PTGS2 RXRA SCN5A SLC6A2 SLC6A3 SLC6A4 |
| PCRR | MOL013288 | Picralinal | AR OPRD1 OPRM1 SCN5A |
Table 4.
Known therapeutic target genes for COVID-19
| Gene | GC Id | Score | Gene | GC Id | Score |
|---|---|---|---|---|---|
| TNF | GC06P033397 | 33.08 | ITGAL | GC16P030472 | 4.07 |
| IL6 | GC07P022765 | 31.28 | STAT6 | GC12M057095 | 4.04 |
| CXCL8 | GC04P073740 | 31.05 | BAK1 | GC06M033572 | 4.03 |
| CD40LG | GC0XP136649 | 30.56 | PIK3CG | GC07P106865 | 4.02 |
| IL10 | GC01M206767 | 30.33 | FOS | GC14P075278 | 4.01 |
| IFNG | GC12M068064 | 27.48 | HELLS | GC10P094501 | 4 |
| CRP | GC01M159715 | 25.76 | CP | GC03M149162 | 3.96 |
| STAT1 | GC02M190964 | 22.73 | APOA1 | GC11M116835 | 3.95 |
| MBL2 | GC10M052760 | 22.1 | RPS27A | GC02P055231 | 3.91 |
| TP53 | GC17M007661 | 19 | CREBBP | GC16M003726 | 3.87 |
| CCL2 | GC17P034255 | 18.13 | TFRC | GC03M196027 | 3.83 |
| IL2 | GC04M122451 | 17.68 | LMAN1 | GC18M059327 | 3.82 |
| CCL5 | GC17M035871 | 16.71 | PLA2G4A | GC01P186798 | 3.81 |
| IFNA1 | GC09P021478 | 16.65 | CEACAM5 | GC19P041709 | 3.65 |
| EGFR | GC07P055019 | 16.29 | PRKCA | GC17P066302 | 3.65 |
| CXCL10 | GC04M076021 | 15.3 | EIF2S1 | GC14P067359 | 3.65 |
| TGFB1 | GC19M041301 | 14.98 | CLEC12A | GC12P009951 | 3.61 |
| IL1B | GC02M112829 | 13.78 | SUMO1 | GC02M202206 | 3.59 |
| ACE2 | GC0XM015494 | 12.32 | CCR3 | GC03P046227 | 3.56 |
| CSF2 | GC05P132073 | 11.95 | UBB | GC17P016380 | 3.53 |
| PPARG | GC03P012287 | 11.93 | MAPKAPK2 | GC01P206684 | 3.48 |
| CCR5 | GC03P046384 | 11.37 | CD3D | GC11M118338 | 3.47 |
| CXCL9 | GC04M076001 | 11.3 | CHKB | GC22M050578 | 3.43 |
| GPT | GC08P144502 | 11.12 | PPIA | GC07P044808 | 3.43 |
| MAPK1 | GC22M021754 | 11.09 | RUNX1 | GC21M034787 | 3.42 |
| CASP3 | GC04M184627 | 10.88 | BCL2L1 | GC20M031664 | 3.4 |
| IFNB1 | GC09M021077 | 10.77 | GZMA | GC05P055102 | 3.38 |
| ALB | GC04P073397 | 10.68 | IRF1 | GC05M132481 | 3.35 |
| FGF2 | GC04P122826 | 10.53 | CD81 | GC11P002377 | 3.35 |
| SFTPD | GC10M079937 | 10.47 | CST3 | GC20M023608 | 3.29 |
| CXCR3 | GC0XM071615 | 10.18 | PTGS1 | GC09P122370 | 3.24 |
| IL4 | GC05P132673 | 10.12 | F10 | GC13P113122 | 3.22 |
| HLA-B | GC06M031289 | 9.84 | CBL | GC11P119206 | 3.18 |
| CD79A | GC19P041877 | 9.73 | CXCL11 | GC04M076033 | 3.13 |
| CXCL2 | GC04M074097 | 9.61 | MAVS | GC20P003827 | 3.12 |
| ACE | GC17P063477 | 9.6 | KPNB1 | GC17P047649 | 3.1 |
| TMPRSS2 | GC21M041464 | 9.59 | SLC17A5 | GC06M073593 | 3.07 |
| IRF3 | GC19M049659 | 9.51 | ITGA5 | GC12M054396 | 3 |
| MAPK3 | GC16M030117 | 9.37 | ARF1 | GC01P228082 | 2.99 |
| IL17A | GC06P052186 | 9.29 | IFNL1 | GC19P039296 | 2.97 |
| IL5 | GC05M132541 | 9.27 | GRB2 | GC17M075318 | 2.86 |
| ICAM1 | GC19P010270 | 9.22 | CD3E | GC11P118304 | 2.84 |
| CCL3 | GC17M036088 | 9.2 | ATF2 | GC02M175072 | 2.78 |
| IL13 | GC05P132656 | 9.19 | CEACAM3 | GC19P041796 | 2.72 |
| MAPK8 | GC10P048306 | 9.08 | HAVCR2 | GC05M157063 | 2.7 |
| TTR | GC18P031557 | 9.04 | JAK1 | GC01M064833 | 2.69 |
| IL18 | GC11M112143 | 8.58 | NPM1 | GC05P171387 | 2.67 |
| ANPEP | GC15M089784 | 8.58 | TBK1 | GC12P064451 | 2.64 |
| PIK3R1 | GC05P068215 | 8.57 | F11 | GC04P186265 | 2.63 |
| CTSL | GC09P087725 | 8.52 | VHL | GC03P010205 | 2.63 |
| CD209 | GC19M007739 | 8.45 | IL16 | GC15P081159 | 2.59 |
| DDX58 | GC09M032455 | 8.2 | KPNA2 | GC17P068035 | 2.57 |
| FURIN | GC15P090868 | 8.08 | RELB | GC19P045002 | 2.57 |
| ADA | GC20M044620 | 7.97 | FCER2 | GC19M007689 | 2.56 |
| APOE | GC19P044906 | 7.97 | PIK3CB | GC03M138652 | 2.55 |
| MAPK14 | GC06P046047 | 7.77 | PRSS2 | GC07P144731 | 2.54 |
| DPP4 | GC02M161992 | 7.64 | RAPGEF3 | GC12M047736 | 2.52 |
| NFKB1 | GC04P102501 | 7.61 | BECN1 | GC17M042810 | 2.51 |
| HLA-A | GC06P033211 | 7.44 | HAVCR1 | GC05M157007 | 2.48 |
| SERPINE1 | GC07P101127 | 7.43 | ISG15 | GC01P001001 | 2.41 |
| PIK3CA | GC03P179148 | 7.27 | PML | GC15P073994 | 2.41 |
| PTGS2 | GC01M186640 | 7.24 | PRKCE | GC02P045651 | 2.39 |
| CD14 | GC05M140631 | 7.16 | CEACAM1 | GC19M042507 | 2.38 |
| MX1 | GC21P041420 | 7.07 | PIK3CD | GC01P009629 | 2.37 |
| IFIH1 | GC02M162267 | 6.99 | ERN1 | GC17M064039 | 2.37 |
| BCL2 | GC18M063123 | 6.96 | IFITM1 | GC11P000313 | 2.36 |
| FCGR2A | GC01P161505 | 6.67 | IRAK3 | GC12P066188 | 2.35 |
| CDK4 | GC12M057743 | 6.64 | NPTX1 | GC17M080466 | 2.35 |
| HSPA5 | GC09M125234 | 6.59 | HFE | GC06P026087 | 2.34 |
| BAX | GC19P048954 | 6.53 | TLR10 | GC04M038773 | 2.33 |
| CCL11 | GC17P034285 | 6.47 | SLC40A1 | GC02M189560 | 2.3 |
| CAT | GC11P034460 | 6.43 | LCK | GC01P032251 | 2.29 |
| HMOX1 | GC22P035380 | 6.28 | EIF2AK3 | GC02M088637 | 2.27 |
| SOD1 | GC21P031659 | 6.25 | POU5F1 | GC06M031177 | 2.25 |
| G6PD | GC0XM154531 | 6.06 | VAPA | GC18P009904 | 2.15 |
| CD4 | GC12P006786 | 6.01 | CARD9 | GC09M136361 | 2.15 |
| TF | GC03P133666 | 5.96 | TRIM25 | GC17M056836 | 2.13 |
| CTRL | GC16M067927 | 5.95 | HNRNPA1 | GC12P054280 | 2.05 |
| IL1A | GC02M112773 | 5.93 | CCND3 | GC06M041934 | 1.99 |
| PIK3C2A | GC11M017165 | 5.92 | MYOM2 | GC08P002045 | 1.97 |
| PARP1 | GC01M226360 | 5.91 | PRKRA | GC02M178431 | 1.96 |
| RELA | GC11M065653 | 5.89 | SOCS3 | GC17M078356 | 1.95 |
| NOS2 | GC17M027756 | 5.85 | LCN1 | GC09P135521 | 1.91 |
| EIF2AK2 | GC02M037099 | 5.83 | EIF4E | GC04M098871 | 1.91 |
| GAPDH | GC12P006630 | 5.81 | ICAM2 | GC17M064002 | 1.89 |
| NOS3 | GC07P150990 | 5.77 | BST2 | GC19M017403 | 1.88 |
| CTSB | GC08M011842 | 5.72 | IFITM2 | GC11P000300 | 1.87 |
| CCL4 | GC17P036103 | 5.69 | KPNA4 | GC03M160494 | 1.83 |
| CASP8 | GC02P201233 | 5.65 | DROSHA | GC05M031401 | 1.78 |
| ANXA5 | GC04M121667 | 5.59 | USP7 | GC16M008892 | 1.78 |
| F8 | GC0XM154835 | 5.58 | CD46 | GC01P207752 | 1.74 |
| CREB1 | GC02P207529 | 5.55 | AHSG | GC03P186612 | 1.73 |
| SH2D3A | GC19M006752 | 5.54 | BAG3 | GC10P119651 | 1.72 |
| HLA-DRB1 | GC06M032578 | 5.48 | TMPRSS11A | GC04M067909 | 1.69 |
| TMPRSS11D | GC04M067820 | 5.38 | APOD | GC03M195568 | 1.66 |
| BMP6 | GC06P007726 | 5.32 | PRKCB | GC16P023872 | 1.64 |
| SMAD3 | GC15P067063 | 5.2 | RHOB | GC02P020447 | 1.64 |
| MASP2 | GC01M011026 | 5.13 | ITGA6 | GC02P172427 | 1.63 |
| IFITM3 | GC11M000319 | 5.11 | STAT2 | GC12M056341 | 1.62 |
| HLA-C | GC06M031272 | 5.11 | CALM1 | GC14P090396 | 1.61 |
| BAD | GC11M064273 | 5.04 | OAS1 | GC12P112906 | 1.6 |
| CANX | GC05P179678 | 4.97 | BCL2L2 | GC14P025033 | 1.6 |
| MCL1 | GC01M150673 | 4.77 | IFI27 | GC14P094104 | 1.6 |
| CCL7 | GC17P034270 | 4.71 | PSMC6 | GC14P052707 | 1.55 |
| CASP6 | GC04M109688 | 4.7 | TFR2 | GC07M100620 | 1.5 |
| EGR1 | GC05P138465 | 4.66 | SPI1 | GC11M059694 | 1.45 |
| ITGB1 | GC10M032900 | 4.64 | IGKC | GC02M089081 | 1.44 |
| RNASE3 | GC14P020891 | 4.63 | PHB2 | GC12M006965 | 1.44 |
| STING1 | GC05M139476 | 4.5 | CD151 | GC11P000883 | 1.42 |
| CD34 | GC01M207880 | 4.48 | ITGA1 | GC05P052788 | 1.42 |
| DUSP1 | GC05M172768 | 4.42 | FAH | GC15P080152 | 1.4 |
| RB1 | GC13P048303 | 4.41 | NUDT2 | GC09P034329 | 1.36 |
| ADAM17 | GC02M009488 | 4.4 | AQP1 | GC07P030911 | 1.35 |
| HSPB1 | GC07P076302 | 4.33 | TMPRSS13 | GC11M117900 | 1.32 |
| EEF1A1 | GC06M073515 | 4.33 | CD3G | GC11P118344 | 1.28 |
| TOLLIP | GC11M001274 | 4.31 | PCSK5 | GC09P075890 | 1.23 |
| CCR1 | GC03M046218 | 4.3 | CBLB | GC03M105655 | 1.21 |
| EZR | GC06M158765 | 4.27 | TMEM233 | GC12P119594 | 1.18 |
| LCN2 | GC09P128149 | 4.26 | ANXA11 | GC10M080150 | 1.13 |
| TRAF3 | GC14P104312 | 4.23 | CLEC4D | GC12P008509 | 1.1 |
| SMAD7 | GC18M048919 | 4.18 | NMRAL1 | GC16M004461 | 1.07 |
| TXN | GC09M110243 | 4.17 | HPGDS | GC04M094298 | 0.84 |
| ICAM3 | GC19M010335 | 4.15 | SLC39A14 | GC08P022367 | 0.83 |
| VCP | GC09M035056 | 4.15 | OR8U9 | GC11Pi00193 | 0.35 |
| NLRP12 | GC19M053793 | 4.14 | C8G | GC09P136944 | 0.31 |
| ANXA2 | GC15M060347 | 4.12 |
PPI network of SFJDC putative and NCP related target genes
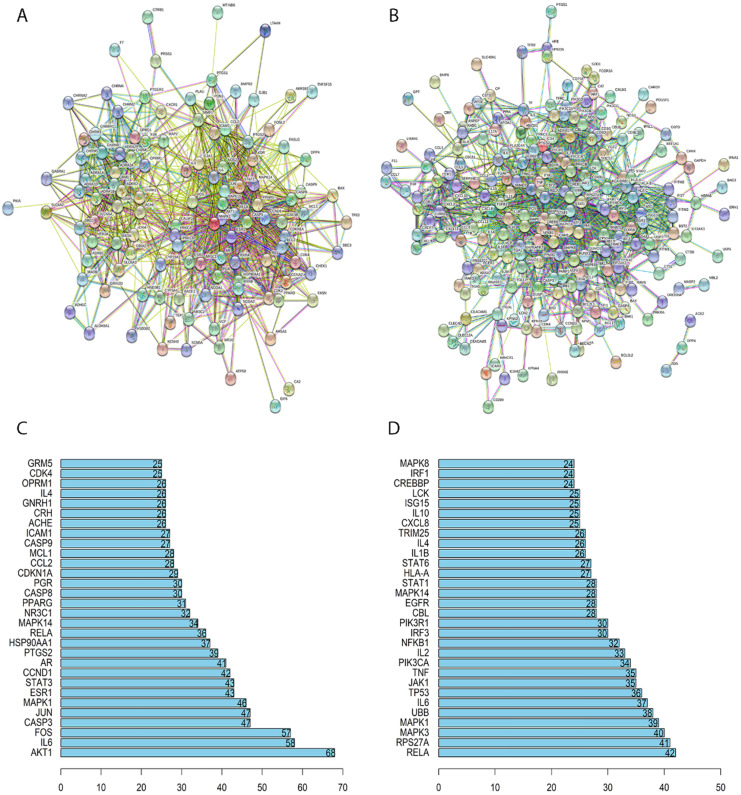
In this study, we constructed the PPI network of SFJDC putative and NCP related target genes separately. The network of SFJDC putative target genes which minimum interaction score was set at 0.4 contained 119 nodes and 1108 edges which indicated the target genes interactions after removing the discrete points (Figure 2A). According the PPI network, the top thirty genes were listed in Figure 2B. After hiding the discrete points, NCP-related target genes PPI network contained 248 nodes and 1235 edges (Figure 2C). Similarly, the first 30 related genes were shown in Figure 2D.
Figure 2.
PPI network of SFJDC putative and NCP related target genes and the Barplot of PPI. (A) PPI network of SFJDC putative target genes. (B) PPI network of NCP related target genes. (C) Barplot showing the significant genes in PPI network of SFJDC. (D) Barplot showing the significant genes in PPI network of NCP. PPI, protein-protein interaction; SFJDC: ShuFeng JieDu capsule; NCP: Novel Coronavirus Pneumonia.
SFJDC ingredient-target network analysis
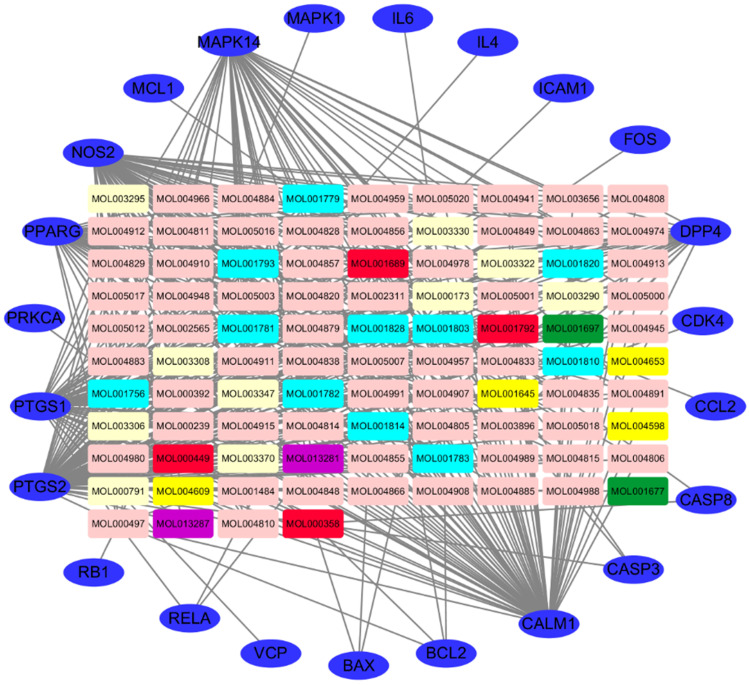
The ingredient-target network of SFJDC was constructed using the screened ingredients and their targets as shown in Figure 3. The network contained 117 nodes and 419 edges which indicated the compound-target genes interaction. A median of 110 candidate compouds was 5 degrees which indicating that most compounds of SFJDC were affected by multiple target genes. The top three effective ingredient according were Wogonin, licochalcone a and acacetin. Wogonin, licochalcone a and acacetin have 42, 30 and 23 target genes, respectively. And the OB of Wogonin, licochalcone a and acacetin were 30.68, 40.79 and 34.97%, respectively. Hence, they might be the crucial effective compounds of SFJDC according the network.
Figure 3.
Ingredient-target network of SFJDC. The blue ovals represent target genes; the green, light blue, yellow, pink, purple and light yellow rectangulars represent the ingredients from PR, IR, HP, RB, PCRR, FF; the red rectangulars represent the ingredients from mlti-herb. PR: Phragmitis Rhizoma; IR: Isatidis Radix; HP: Herba Patriniae; RB:Radix Bupleuri; PCRR: Polygoni Cuspidati Rhizoma Et Radix; FF: Forsythiae Fructus.
PPI network analysis of SFJDC against NCP
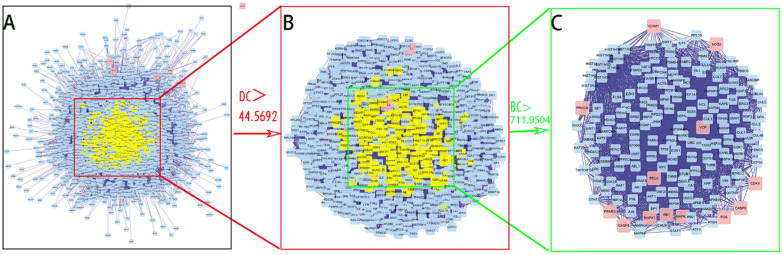
PPI network of SFJDC against NCP were visualized using Cytoscape software. The network contained 2407 nodes and 53639 edges was shown in Figure 4A. The average degree of all nodes was 44.5692 and we selected the nodes with more than 44.5692 degrees as significant genes. A subnetwork of significant genes for SFJDC against NCP was constructed which consisted of 766 nodes and 28872 edges (Figure 4B). The average value of BC was 711.9504. The significant genes were further screened and a new network was constructed with 169 nodes and 4238 edges (Figure 4C). 169 genes were eventually identified for SFJDC against NCP including 156 other human genes and 13 target genes.
Figure 4.
PPI network of SFJDC against NCP. (A)The whole network of SFJDC against NCP contained 2,407 nodes and 53,639 edges. (B) A subnetwork of significant genes from A consisted of 766 nodes and 28872 edges. (C) PPI network of more significant genes from B with 169 nodes and 4238 edges. BC: Betweenness Centrality; DC: Degree Centrality.
Identification of candidate genes (CGs) and Enrichment analysis of CGs
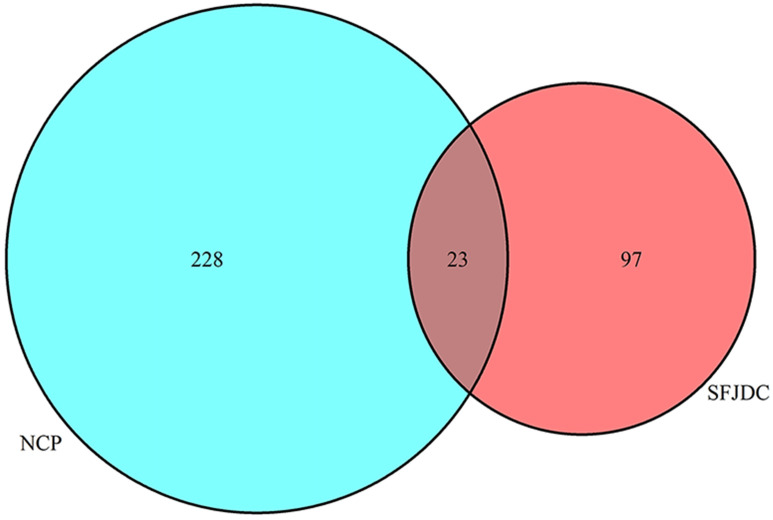
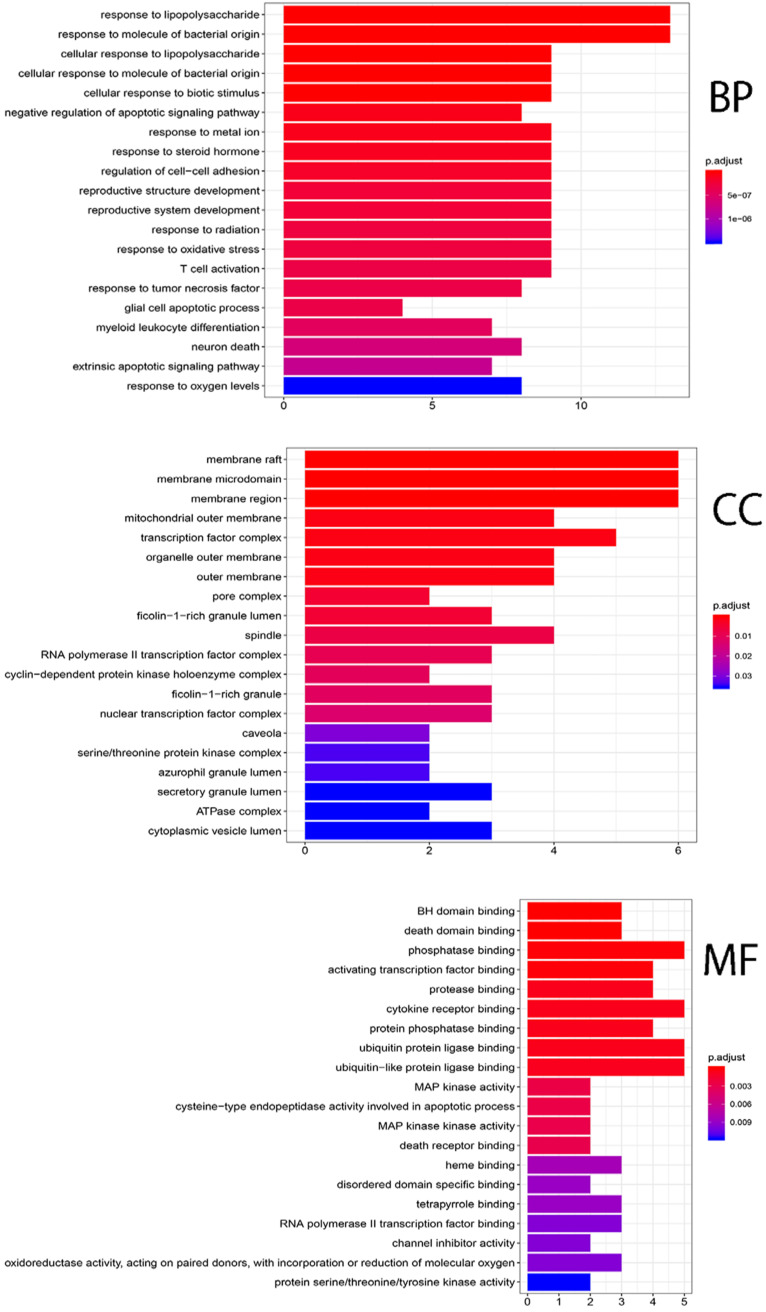
Twenty-three candidate genes (CGs) were identified by using the VennDiagram package (Figure 5). Then R software was used to perform GO and KEGG pathway analysis of the CGs. GO of CGs was analyzed based on BP, CC, MF. 1215 GO terms were significantly enriched (P<0.05), 1148 in BP, 28 in CC, 39 in MF. Top 20 terms were shown in Figure 6. The data of top 20 GO analysis were listed in Table 5. Based on these GO terms data, we found that most significantly terms were response to lipopolysaccharide, response to molecule of bacterial origin, membrane raft, membrane microdomain, BH domain binding and death domain binding, suggested that SFJDC could treat NCP with multiple synergies.
Figure 5.
Twenty-three overlapping genes between SFJDC and NCP.
Figure 6.
Gene ontology terms of CGs. The top 20 GO functional terms were selected (P<0.05). BP: biological processes; CC: cellular components; MF: molecular functions.
Table 5.
The data of top twenty GO terms including BP, CC, MF
| GO category | ID | Description | P-value | P.adjust | Count |
|---|---|---|---|---|---|
| BP | GO:0032496 | response to lipopolysaccharide | 1.27E-17 | 2.31E-14 | 13 |
| BP | GO:0002237 | response to molecule of bacterial origin | 2.10E-17 | 2.31E-14 | 13 |
| BP | GO:0071222 | cellular response to lipopolysaccharide | 1.39E-12 | 1.02E-09 | 9 |
| BP | GO:0071219 | cellular response to molecule of bacterial origin | 1.89E-12 | 1.04E-09 | 9 |
| BP | GO:0071216 | cellular response to biotic stimulus | 4.96E-12 | 2.18E-09 | 9 |
| BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 1.96E-10 | 7.20E-08 | 8 |
| BP | GO:0010038 | response to metal ion | 2.37E-10 | 7.45E-08 | 9 |
| BP | GO:0048545 | response to steroid hormone | 3.89E-10 | 1.07E-07 | 9 |
| BP | GO:0022407 | regulation of cell-cell adhesion | 5.82E-10 | 1.42E-07 | 9 |
| BP | GO:0048608 | reproductive structure development | 1.05E-09 | 2.23E-07 | 9 |
| BP | GO:0061458 | reproductive system development | 1.12E-09 | 2.23E-07 | 9 |
| BP | GO:0009314 | response to radiation | 1.48E-09 | 2.65E-07 | 9 |
| BP | GO:0006979 | response to oxidative stress | 1.57E-09 | 2.65E-07 | 9 |
| BP | GO:0042110 | T cell activation | 2.01E-09 | 3.16E-07 | 9 |
| BP | GO:0034612 | response to tumor necrosis factor | 2.19E-09 | 3.22E-07 | 8 |
| BP | GO:0034349 | glial cell apoptotic process | 2.37E-09 | 3.25E-07 | 4 |
| BP | GO:0002573 | myeloid leukocyte differentiation | 3.55E-09 | 4.59E-07 | 7 |
| BP | GO:0070997 | neuron death | 5.17E-09 | 6.31E-07 | 8 |
| BP | GO:0097191 | extrinsic apoptotic signaling pathway | 6.78E-09 | 7.86E-07 | 7 |
| BP | GO:0070482 | response to oxygen levels | 1.36E-08 | 1.50E-06 | 8 |
| CC | GO:0045121 | membrane raft | 1.27E-06 | 6.17E-05 | 6 |
| CC | GO:0098857 | membrane microdomain | 1.30E-06 | 6.17E-05 | 6 |
| CC | GO:0098589 | membrane region | 1.61E-06 | 6.17E-05 | 6 |
| CC | GO:0005741 | mitochondrial outer membrane | 4.97E-05 | 0.001243834 | 4 |
| CC | GO:0005667 | transcription factor complex | 5.41E-05 | 0.001243834 | 5 |
| CC | GO:0031968 | organelle outer membrane | 7.97E-05 | 0.001361141 | 4 |
| CC | GO:0019867 | outer membrane | 8.29E-05 | 0.001361141 | 4 |
| CC | GO:0046930 | pore complex | 0.000324441 | 0.004663842 | 2 |
| CC | GO:1904813 | ficolin-1-rich granule lumen | 0.000392171 | 0.005011074 | 3 |
| CC | GO:0005819 | spindle | 0.000640704 | 0.007368092 | 4 |
| CC | GO:0090575 | RNA polymerase II transcription factor complex | 0.000869852 | 0.009093909 | 3 |
| CC | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex | 0.001089346 | 0.010439568 | 2 |
| CC | GO:0101002 | ficolin-1-rich granule | 0.001253428 | 0.011088015 | 3 |
| CC | GO:0044798 | nuclear transcription factor complex | 0.001590231 | 0.013062616 | 3 |
| CC | GO:0005901 | caveola | 0.003891901 | 0.029837911 | 2 |
| CC | GO:1902554 | serine/threonine protein kinase complex | 0.004688015 | 0.03369511 | 2 |
| CC | GO:0035578 | azurophil granule lumen | 0.005004373 | 0.03385311 | 2 |
| CC | GO:0034774 | secretory granule lumen | 0.005946804 | 0.035510845 | 3 |
| CC | GO:1904949 | ATPase complex | 0.006127975 | 0.035510845 | 2 |
| CC | GO:0060205 | cytoplasmic vesicle lumen | 0.006856895 | 0.035510845 | 3 |
| MF | GO:0051400 | BH domain binding | 2.29E-07 | 1.91E-05 | 3 |
| MF | GO:0070513 | death domain binding | 2.29E-07 | 1.91E-05 | 3 |
| MF | GO:0019902 | phosphatase binding | 3.42E-06 | 0.000170898 | 5 |
| MF | GO:0033613 | activating transcription factor binding | 4.09E-06 | 0.000170898 | 4 |
| MF | GO:0002020 | protease binding | 2.08E-05 | 0.000629425 | 4 |
| MF | GO:0005126 | cytokine receptor binding | 2.82E-05 | 0.000629425 | 5 |
| MF | GO:0019903 | protein phosphatase binding | 2.96E-05 | 0.000629425 | 4 |
| MF | GO:0031625 | ubiquitin protein ligase binding | 3.02E-05 | 0.000629425 | 5 |
| MF | GO:0044389 | ubiquitin-like protein ligase binding | 4.02E-05 | 0.000746042 | 5 |
| MF | GO:0004707 | MAP kinase activity | 0.000145644 | 0.00243226 | 2 |
| MF | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process | 0.000167918 | 0.002549304 | 2 |
| MF | GO:0004708 | MAP kinase kinase activity | 0.000191755 | 0.002668588 | 2 |
| MF | GO:0005123 | death receptor binding | 0.00021715 | 0.002789547 | 2 |
| MF | GO:0020037 | heme binding | 0.000687548 | 0.008201468 | 3 |
| MF | GO:0097718 | disordered domain specific binding | 0.000832459 | 0.00883078 | 2 |
| MF | GO:0046906 | tetrapyrrole binding | 0.000846063 | 0.00883078 | 3 |
| MF | GO:0001085 | RNA polymerase II transcription factor binding | 0.001026133 | 0.009707469 | 3 |
| MF | GO:0016248 | channel inhibitor activity | 0.001103999 | 0.009707469 | 2 |
| MF | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | 0.001104443 | 0.009707469 | 3 |
| MF | GO:0004712 | protein serine/threonine/tyrosine kinase activity | 0.001412492 | 0.011727316 | 2 |
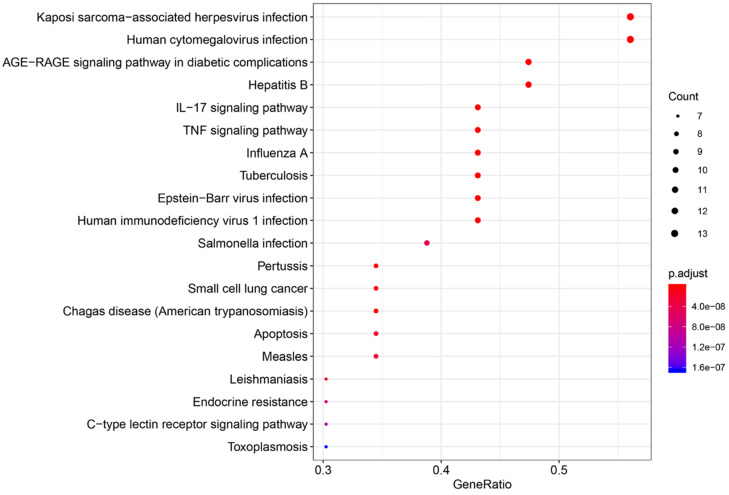
The pathways that were significantly affected by SFJDC in the process of treating NCP were identified by KEGG pathway. 110 KEGG pathways were significantly enriched (P<0.05). Top twenty pathways were shown in Figure 7, color represented P value and size of the spot represented count of genes. Based on the analysis of KEGG pathway data (Table 6), the top five pathways such as Kaposi sarcoma-associated herpesvirus infection, AGE-RAGE signaling pathway in diabetic complications, Human cytomegalovirus infection, IL-17 signaling pathway and Hepatitis B, might be the core pharmacological mechanism of SFJDC for NCP.
Figure 7.
KEGG pathway enrichment of CGs. The top 20 pathways were identified. Color represented P value and size of the spot represented count of genes.
Table 6.
The data of top twenty KEGG pathway
| ID | Description | P-value | P.adjust | Count |
|---|---|---|---|---|
| hsa05167 | Kaposi sarcoma-associated herpesvirus infection | 5.39E-16 | 8.46E-14 | 13 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 1.13E-15 | 8.85E-14 | 11 |
| hsa05163 | Human cytomegalovirus infection | 6.58E-15 | 3.45E-13 | 13 |
| hsa04657 | IL-17 signaling pathway | 4.27E-14 | 1.68E-12 | 10 |
| hsa05161 | Hepatitis B | 2.60E-13 | 6.81E-12 | 11 |
| hsa04668 | TNF signaling pathway | 2.60E-13 | 6.81E-12 | 10 |
| hsa05164 | Influenza A | 1.78E-11 | 4.00E-10 | 10 |
| hsa05133 | Pertussis | 2.57E-11 | 5.04E-10 | 8 |
| hsa05152 | Tuberculosis | 3.16E-11 | 5.50E-10 | 10 |
| hsa05169 | Epstein-Barr virus infection | 9.46E-11 | 1.48E-09 | 10 |
| hsa05170 | Human immunodeficiency virus 1 infection | 1.60E-10 | 2.29E-09 | 10 |
| hsa05142 | Chagas disease (American trypanosomiasis) | 2.86E-10 | 3.74E-09 | 8 |
| hsa05140 | Leishmaniasis | 1.57E-09 | 1.90E-08 | 7 |
| hsa04210 | Apoptosis | 2.89E-09 | 3.24E-08 | 8 |
| hsa05162 | Measles | 3.24E-09 | 3.40E-08 | 8 |
| hsa05132 | Salmonella infection | 4.64E-09 | 4.55E-08 | 9 |
| hsa01522 | Endocrine resistance | 8.69E-09 | 8.03E-08 | 7 |
| hsa04625 | C-type lectin receptor signaling pathway | 1.32E-08 | 1.15E-07 | 7 |
| hsa05145 | Toxoplasmosis | 2.22E-08 | 1.83E-07 | 7 |
| hsa05130 | Pathogenic Escherichia coli infection | 6.53E-08 | 5.13E-07 | 8 |
In this study, we chose the functional annotation clustering and set the classification stringency as high in DAVID. A total of 20 functional annotation clusters were obtained (Table 7). Annotation Cluster 1 (enrichment score 6.04) contains three categories: Asthma, Bronchiolitis Viral, Respiratory Syncytial Virus Infections, respiratory syncytial virus bronchiolitis, and all of them were lung related diseases and Virus infection disease.
Table 7.
The functional annotation clustering of CGs
| Annotation Cluster | Term | Count | P-value |
|---|---|---|---|
| Annotation Cluster 1 (Score:6.04) | Asthma|Bronchiolitis, Viral|Respiratory Syncytial Virus Infections | 7 | 8.50E-07 |
| respiratory syncytial virus bronchiolitis | 7 | 8.50E-07 | |
| Bronchiolitis, Viral|Respiratory Syncytial Virus Infections | 7 | 1.04E-06 | |
| Annotation Cluster 2 (Score:4.91) | Coronary Artery Disease|Inflammation | 5 | 5.45E-07 |
| non-Hodgkin lymphoma | 4 | 1.90E-06 | |
| Recurrence|Venous Thromboembolism | 5 | 2.48E-06 | |
| Arthritis | 5 | 2.66E-06 | |
| Brain Ischemia|Hypertension|Osteoporosis|Stroke | 5 | 3.69E-06 | |
| diabetes, type 1 | 6 | 1.38E-05 | |
| melanoma | 5 | 2.10E-05 | |
| Inflammation|Venous Thromboembolism | 4 | 2.24E-05 | |
| Chlamydia Infections|Inflammation|Trachoma | 4 | 2.24E-05 | |
| Brain Ischemia|Inflammation|Stroke | 4 | 2.24E-05 | |
| Pre-Eclampsia | 4 | 3.32E-04 | |
| Migraine Disorders | 4 | 4.52E-04 | |
| Annotation Cluster 3 (Score:4.89) | Chorioamnionitis|Fetal Membranes, Premature Rupture|Infection of amniotic sac and membranes | 7 | 4.94E-07 |
| Chorioamnionitis|Fetal Membranes, Premature Rupture|Infection of amniotic sac and membranes|Obstetric Labor, Premature|Pre-Eclampsia|Premature Birth | 7 | 5.10E-07 | |
| Coronary Artery Disease | 7 | 1.56E-04 | |
| Alzheimer's disease | 8 | 7.10E-04 | |
| Annotation Cluster 4 (Score:4.44) | Hodgkin Disease|Inflammation | 4 | 3.40E-06 |
| Sarcoidosis | 5 | 4.99E-06 | |
| Adenocarcinoma|Stomach Neoplasms | 4 | 5.74E-05 | |
| kidney failure, chronic | 5 | 2.02E-04 | |
| esophageal cancer | 4 | 3.20E-04 | |
| Annotation Cluster 5 (Score:4.26) |
Lymphoma, Non-Hodgkin|Lymphoma, Non-Hodgkin's | 5 | 3.30E-05 |
| Leukemia, Myelogenous, Chronic, BCR-ABL Positive|Neovascularization, Pathologic | 4 | 4.02E-05 | |
| Leukemia, Myelogenous, Chronic, BCR-ABL Positive | 4 | 1.29E-04 | |
| Annotation Cluster 6 (Score:4.09) | Tuberculosis, Pulmonary | 5 | 2.31E-06 |
| systemic lupus erythematosus | 5 | 8.88E-05 | |
| hepatitis C, chronic | 4 | 3.56E-04 | |
| Tuberculosis | 4 | 6.16E-04 | |
| Annotation Cluster 7 (Score:4.04) | Helicobacter Infections|Inflammation|Precancerous Conditions|Stomach Neoplasms | 4 | 9.23E-07 |
| Stomach Neoplasms | 5 | 3.54E-05 | |
| patent ductus arteriosus | 5 | 6.13E-05 | |
| Cystic Fibrosis | 4 | 1.29E-04 | |
| stomach cancer | 4 | 5.46E-04 | |
| rheumatoid arthritis | 4 | 0.003880586 | |
| Annotation Cluster 8 (Score:3.94) | Infection|Inflammation|Premature Birth | 5 | 5.26E-05 |
| Inflammation|Premature Birth | 5 | 5.77E-05 | |
| Connective Tissue Diseases|Fetal Diseases|Inflammation|Musculoskeletal Diseases|Pregnancy Complications, Hematologic|Premature Birth|Skin Diseases | 5 | 5.77E-05 | |
| Asthma | 4 | 9.96E-04 | |
| Annotation Cluster 9 (Score:3.84) | Atherosclerosis | 7 | 2.07E-05 |
| Myocardial Infarction | 7 | 2.05E-04 | |
| Alzheimer's disease | 8 | 7.10E-04 | |
| Annotation Cluster 10 (Score:3.78) | Brain Ischemia|Stroke | 5 | 1.86E-05 |
| Peripheral Vascular Diseases | 4 | 4.02E-05 | |
| Cardiovascular Diseases | 5 | 2.60E-04 | |
| Hypercholesterolemia|LDLC levels | 4 | 0.004054968 | |
| Annotation Cluster 11 (Score:3.55) | Restenosis | 4 | 1.81E-04 |
| Arthritis, Rheumatoid|Rheumatoid Arthritis | 5 | 1.98E-04 | |
| Endometriosis | 4 | 6.16E-04 | |
| Annotation Cluster 12 (Score:3.28) | Alcoholism|Liver Cirrhosis, Alcoholic | 3 | 2.12E-04 |
| Esophageal Neoplasms|Hyperglycemia|Oesophageal neoplasm | 3 | 2.12E-04 | |
| Biliary Tract Neoplasms|Inflammation | 3 | 5.66E-04 | |
| Arthritis, Psoriatic|Psoriatic arthropathy | 3 | 6.21E-04 | |
| cardiovascular | 3 | 0.002488197 | |
| Annotation Cluster 13 (Score:3.18) | Otitis Media|Recurrence | 3 | 2.47E-04 |
| Brucellosis | 3 | 5.12E-04 | |
| Graft vs Host Disease|Hematologic Neoplasms|Neoplasm Recurrence, Local | 3 | 5.12E-04 | |
| Kawasaki disease | 3 | 6.21E-04 | |
| Atopy | 3 | 0.003077333 | |
| Annotation Cluster 14 (Score:2.82) | Atherosclerosis|Inflammation|Retinal Vein Occlusion | 3 | 1.23E-04 |
| Dermatitis, Atopic|Eczema allergic | 3 | 7.41E-04 | |
| juvenile arthritis | 3 | 0.001084437 | |
| graft-versus-host disease | 3 | 0.002834545 | |
| Graft vs Host Disease | 3 | 0.005354403 | |
| hepatitis C | 3 | 0.008600238 | |
| Annotation Cluster 15 (Score:2.80) | Uveitis, Anterior | 3 | 1.23E-04 |
| Pancreatitis, Chronic | 3 | 8.05E-04 | |
| stroke, ischemic | 3 | 0.004142278 | |
| Glomerulonephritis, IGA | 3 | 0.016033469 | |
| Annotation Cluster 16 (Score:2.76) | giant cell arteritis | 3 | 5.12E-04 |
| Malaria, Falciparum | 3 | 0.002163435 | |
| Malaria | 3 | 0.004882851 | |
| Annotation Cluster 17 (Score:2.72) | Cardiovascular Diseases|Inflammation | 3 | 1.50E-04 |
| skin cancer, non-melanoma | 3 | 0.001084437 | |
| Adenoma|Colorectal Neoplasms | 3 | 0.002954755 | |
| Depression | 3 | 0.028362664 | |
| Annotation Cluster 18 (Score:2.68) | Endometriosis|Uterine Diseases | 3 | 1.50E-04 |
| Hepatitis B, Chronic | 3 | 0.006357795 | |
| Pulmonary Disease, Chronic Obstructive | 3 | 0.009415863 | |
| Annotation Cluster 19 (Score:2.63) | respiratory syncytial virus | 3 | 3.68E-04 |
| Q fever | 3 | 4.61E-04 | |
| Graves' disease|Graves' disease | 3 | 0.001490775 | |
| Graves' disease | 3 | 0.001579503 | |
| Diabetes Mellitus, Insulin-Dependent|Diabetes Mellitus, Type 1 | 3 | 0.006532757 | |
| Premature Birth | 3 | 0.01182929 | |
| Kidney Diseases | 3 | 0.012294446 | |
| Annotation Cluster 20 (Score:2.47) | Carcinoma, Squamous Cell|Mouth Neoplasms | 3 | 0.001084437 |
| Helicobacter Infections|Stomach Neoplasms | 3 | 0.002377534 | |
| Precursor Cell Lymphoblastic Leukemia-Lymphoma | 3 | 0.015509841 |
Gene-pathway network analysis
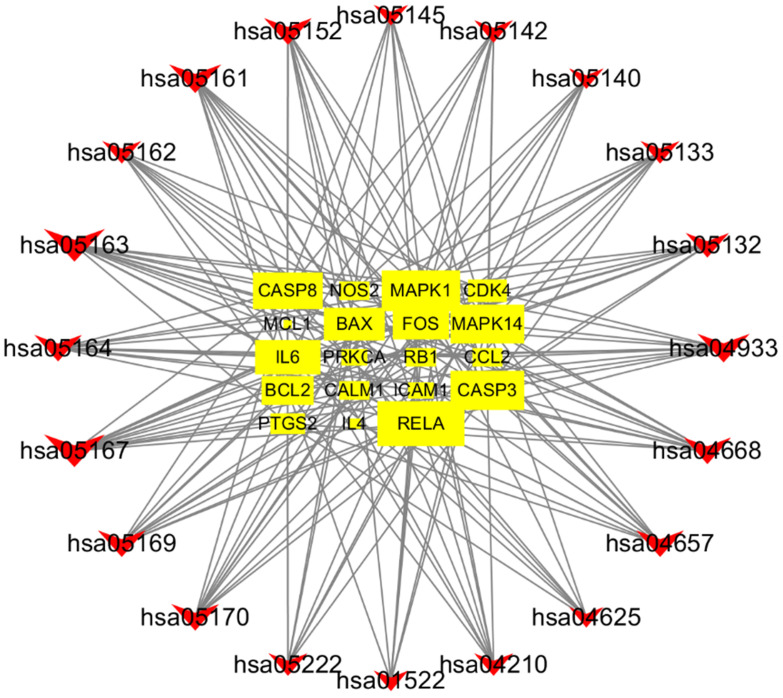
The construction of gene-pathway network is based on significant enrichment pathway and regulated these ways, which was shown in Figure 8. The V shapes represented pathway and the squares represent target genes in the network. The network showed that RELA was the core target gene which had largest degree. Other five genes also had larger degree such as MAPK1, MAPK14, CASP3, CASP8 and IL6. They might be the key target genes using SFJDC in the process of treating NCP. All of the above analysis could reveal a new strategy for drug development on NCP.
Figure 8.
Gene-pathway network of SFJDC against NCP. The V shapes represented pathway and the squares represent target genes in the network.
Discussion
The theory of TCM has been formed and developed for thousands of years in China. In China, TCM has a good therapeutic effect on COVID-19, which has been written into the diagnosis and treatment guidelines. The guideline points out that the combination of traditional Chinese and western medicine should be strengthened in the treatment process 34. SFJDC is a traditional Chinese medicine, mainly used to treat upper respiratory tract infections, such as influenza, sore throat, mumps, streptococcus, etc. 21. Now, SFJDC has become an effective drug for the treatment of COVID-19 35. In recent years, the research on Chinese medicine prescriptions has developed to the level of effective parts, components, components. Network pharmacology can better understand and demonstrate the interaction between multi-component multi-target and disease 36. This study aims to analyze the active components and potential mechanism of SFJDC in the treatment of COVID-19 through network pharmacology.
In the present study, the ingredients-targets network of SFJDC was constructed using 110 ingredients and 120 targets. The network contained 117 nodes and 419 edges which indicated the compound-target genes interaction. The results showed that most compounds of SFJDC were affected by multiple target genes, such as Wogonin, licochalcone a and acacetin acted on 42, 30 and 23 target genes, respectively. Various compounds of SFJDC may have the same targets to achieve synergy. Wogonin, a naturally occurring flavonoid, has been shown to multi-activity, such as anti-inflammatory, anti-fibrosis, anti-cancer and chondroprotective properties 37. Study showed that wogonin had an anti-infulenza activity by modulation of AMPK pathway 38. Licochalcone a, a flavonoid extracted from licorice toot, was known for its anti-inflammatory, anti-cancer, anti-oxidative and anti-bacterial bioactivity 39. Acacetin, a flavone compound, played an important role in anti-inflammatory and anti-peroxidative 40.
In addition, they have high OB and acacetin from 2 herbs (PR, IR) of SFJDC. The three main ingredients were anti-inflammatory and COVID-19 caused by a series of inflammatory storms. Hence, they might be the crucial effective compounds of SFJDC according the network.
PPI network of SFJDC against NCP were visualized using Cytoscape software to obtain the candidate target genes. In order to obtain the more accurate genes, two parameters including DC and BC were used to screen nodes and structure a new network. 169 genes were eventually identified for SFJDC against NCP including 156 other human genes and 13 target genes.
Twenty-three candidate genes (CGs) were identified by using the VennDiagram package. CGs were enriched in BP, CC, MF by GO enrichment analysis. Based on GO terms data, we found that some terms were response to lipopolysaccharide or bacterial origin, membrane raft, membrane microdomain, BH domain binding and cytokine receptor binding. COVID-19 infections leaded to a strong immune response and inflammatory storm in which a large number of cytokines were activated, so SFJDC might regulate COVID-19 through the above biological processes.
SFJDC, as a TCM formula, has multi-component, multi-target-gene, multi-pathway. In the present study, 110 KEGG pathways were significantly enriched. Seven of the top 20 pathways were associated with viral infection including Kaposi sarcoma-associated herpesvirus infection, Human cytomegalovirus infection, Hepatitis B, Influenza A, Epstein-Barr virus infection, Human immunodeficiency virus 1 infection and Measles, and three were associated with lung disease contained tuberculosis, pertussis and small cell lung cancer. Multiple targets of SFJDC may also inhibit the activation of cytokines and reduce inflammation by regulating cytokine pathways, such as IL-17 signaling pathway and TNF signaling pathway. In this study, we obtained 20 functional annotation clusters through DAVID. Annotation Cluster1 including Asthma, Bronchiolitis Viral, Respiratory Syncytial Virus Infections, respiratory syncytial virus bronchiolitis were lung related diseases and Virus infection disease.
Gene-pathway network was constructed to the core and key target genes. The network showed that RELA had largest degree, was the core target gene. Other top five genes such as MAPK1, MAPK14, CASP3, CASP8 and IL6 might be the key target genes. The pathophysiological process of Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-COV-2) infection is similar to that of SARS-CoV infection, with a strong inflammatory response. The SARS-COV-2 virus mainly targets respiratory epithelial cells, alveolar epithelial cells, vascular endothelial cells and pulmonary macrophages, all of which express Angiotensin converting enzyme 2 (ACE2) receptor, triggering the generation of pro- inflammatory cytokines and chemokines (including IL-6, TNF, IL-10 and MCP1) 41. The NF-kB family member RELA is a widely expressed and effective transcriptional activator that activates the expression of many inflammatory through exposure to pathogens and inflammatory cytokines 42. RELA may play an important role in the infection of COVID-19. MAPK1 and MAPK14 are members of the MAPK family, which can regulate multiple cellular processes, such as response to oxidative stress, anti-inflammatory, immune response, apoptosis and cell proliferation 43. Joseph et al showed SASR-CoV-2 could induce severe inflammation by directly activating p38 MAPK pathway and many p38 MAPK inhibitors are in the clinical stage and should be considered for clinical trial for severe COVID-19 infection 44. CASP3 and CASP8, a family of cysteine-dependent proteases, play an important role in these events through activation of other apoptotic proteins mediated by proteolysis and cleavage of nuclear proteins 45. In Krahling's study, infection of 293/ACE2 cells with SARS-CoV activated apoptosis-associated events, such as caspase3, caspase 846. Therefore, we conclude that CASP3 and CASP8 may be activated and play an important role in the pathophysiological process of COVID-19. Higher plasma level of IL-6 was found in ICU patients with COVID-1947. Tocilizumab, a recombinant humanized anti-human IL-6 receptor monoclonal antibody, improved the clinical outcome in 20 severe and critical COVID-19 patients and is an effective treatment to reduce mortality 48.
It has been clinically confirmed that SFJDC is effective in the treatment of COVID-19. Wang et al shown that conventional treatment combined with SFJDC treatment for 4 cases of COVID-19 patients could significantly improve symptoms and promote viral negative conversion 49. Another study including 70 COVID-19 patients found that SFJDC combined with Arbidol for COVID-19 compared with single using Arbidol could significantly shorten the time of clinical symptoms improvement and COVID-19 negative conversion 50.
To summarise, the compound and targets of SFJDC were systematically studied by applying network pharmacology. Wogonin, licochalcone a and acacetin regulated the most targets associated with NCP. RELA, MAPK1, MAPK14, CASP3, CASP8 and IL6 were the core and key genes in the gene-network of SFJDC for the treatment of NCP. SFJDC regulated novel coronavirus pneumonia by multi-compound and multi-target, which provided theoretical support for SFJDC against COVID-19. More mechanism and roles require further clinical validation.
Acknowledgments
Author contributions
YQQ, XC designed the study; YHY and MYZ performed the data collection; JYL and RL analyzed the data; XC drafted the manuscript; YQQ revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the fund from New Coronavirus Pneumonia emergency research project of Shandong University (2020XGA02).
Data Accessibility
Publicly available databases were analyzed in our study. The active ingredients and putative target genes of SFJDC from TCMSP can be found in http://tcmspw.com/tcmsp.php. NCP-related target genes were from GeneCards (https://www.genecards.org/).
Abbreviations
- ACE2
Angiotensin converting enzyme 2
- ARDS
acute respiratory distress syndrome
- BBB
blood-brain barrier
- BC
Betweenness Centrality
- BP
biological processes
- Caco-2
Caco-2 permeability
- CC
cellular components
- CC
Colseness Centrality
- CG
candidate genes
- DC
Degree Centrality
- DL
drug-likeness (DL)
- EC
Eigenvector Centrality
- FF
Forsythiae Fructus
- GO
Gene Ontology
- HP
Herba Patriniae
- I
licorice
- IR
Isatidis Radix
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LAC
Local average connectivity-based method
- LHQWG
LianHua QingWen granules
- MF
molecular functions
- NC
Network Centrality
- NCP
Novel Coronavirus Pneumonia
- OB
oral bioavailability
- PCRR
Polygoni Cuspidati Rhizoma Et Radix
- PPI
protein-protein interaction
- PR
Phragmitis Rhizoma
- RB
Radix Bupleuri
- SFJDC
ShuFeng JieDu capsule
- SARS-COV-2
Severe Acute Respiratory Syndrome-Coronavirus-2
- TCM
Traditional Chinese Medicine
- TCMSP
Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform
- VH
Verbenae Herb
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J. et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim JY, Choe PG, Oh Y, Oh KJ, Kim J, Park SJ. et al. The First Case of 2019 Novel Coronavirus Pneumonia Imported into Korea from Wuhan, China: Implication for Infection Prevention and Control Measures. J Korean Med Sci. 2020;35:e61. doi: 10.3346/jkms.2020.35.e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H. et al. First Case of 2019 Novel Coronavirus in the United States. New England Journal of Medicine. 2020;382:929–36. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. WHO Director-General's remarks at the media briefing on 2019-nCoV on 11 February 2020. Available online: https:// www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020.
- 5.World Health Organization. WHO Director-General's opening remarks at the media briefing on COVID-19 - 10 August 2020. Available online: https:// www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19--10-august-2020.
- 6.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y. et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020;395:507–13. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin YH, Cai L, Cheng ZS, Cheng H, Deng T, Fan YP. et al. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil Med Res. 2020;7:4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu F, Du L, Ojcius DM, Pan C, Jiang S. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020. [DOI] [PMC free article] [PubMed]
- 10.Li S, Feng X, Bi L, Liu X. Selection Analysis and Pharmacological Research Progress of Chinese Patent Medicines in Diagnosis and Treatment of Novel Coronavirus Pneumonia. Journal of Chinese Medicinal Materials. 2020.
- 11.Wang ZF, Wang YP, Zhang HM, Fan YP, Lu C, Wang YY. Thinking on Clinical rational use of TCM injection in the treatment of novel coronavirus pneumonia (COVID-19) Zhonghua Yi Xue Za Zhi. 2020;100:E016. doi: 10.3760/cma.j.issn.cn112137-20200221-00388. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Dh, Wu Kl, Zhang X, Deng S-q, Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. Journal of Integrative Medicine. 2020. [DOI] [PMC free article] [PubMed]
- 13.Lin L, Li T.Interpretation of "Guidelines for the Diagnosis, Treatment of Novel Coronavirus (2019-nCoV) Infection by the National Health Commission (Trial Version 5)". Zhonghua Yi Xue Za Zhi. 2020. [DOI] [PubMed]
- 14.Li T. Diagnosis and clinical management of 2019 novel coronavirus infection: an operational recommendation of Peking Union Medical College Hospital (V2.0) Emerg Microbes Infect. 2020;9(1):582–585. doi: 10.1080/22221751.2020.1735265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Chang L, Wei C, Jia Z. Theoretical and research basis and Clinical efficacy of lianhua qingwen in Treating Coronavirus disease (COVID-19) World Chinese Medicine. 2020.
- 16.Xia W, An C, Zheng C, Zhang J, Huang M, Wang Y, Clinical study on 34 cases of 2019-ncov pneumonia treated with combination of traditional Chinese and Western medicine. Journal of Traditional Chinese Medicine. 2020.
- 17.Tian Z, Qin K, Xiang J, Li Y, Chen X, Ge J, Novel coronavirus pneumonia treated by Qingfei detoxification Decoction: theoretical analysis and clinical practice. World Chinese Medicine. 2020.
- 18.Tao Z. Complementary and alternative medicine is expected to make greater contribution in controlling the prevalence of influenza. BioScience Trends. 2013;7(5):253–256. [PubMed] [Google Scholar]
- 19.Yuan Y, Liao Q, Xue M, Shi Y, Rong L, Song Z. et al. Shufeng Jiedu Capsules Alleviate Lipopolysaccharide-Induced Acute Lung Inflammatory Injury via Activation of GPR18 by Verbenalin. Cell Physiol Biochem. 2018;50:629–39. doi: 10.1159/000494184. [DOI] [PubMed] [Google Scholar]
- 20.Ji S, Bai Q, Wu X, Zhang DW, Wang S, Shen JL. et al. Unique synergistic antiviral effects of Shufeng Jiedu Capsule and oseltamivir in influenza A viral-induced acute exacerbation of chronic obstructive pulmonary disease. Biomed Pharmacother. 2020;121:109652. doi: 10.1016/j.biopha.2019.109652. [DOI] [PubMed] [Google Scholar]
- 21.Xiong W, Ran J, Xie X, Xia Y, Lan B, Wang M, Pharmacological Effect and Clinical Application of Chinese Patent Medicine in Novel Coronavirus Pneumonia 2019. Herald of Medicine. 2020.
- 22.Zheng W, Zhang J, Yang F, Wang Y, Liu Q, Zhang B. Comprehensive analysis of diagnosis and treatment of new type of coronavirus and viral pneumonia with traditional Chinese medicine. Journal of Traditional Chinese Medicine. 2020.
- 23.Boezio B, Audouze K, Ducrot P, Taboureau O. Network-based Approaches in Pharmacology. Mol Inform. 2017. 36. [DOI] [PubMed]
- 24.Deng Y, Liu B, Study on active compounds from Huoxiang Zhengqi Oral Liquid for prevention of novel coronavirus pneumonia (COVID-19) based on network pharmacology and molecular docking. Chinese Traditional and Herbal Drugs. 2020.
- 25.Zhao J, Tian S, Yang J, Liu J, Zhang W. Investigating the mechanism of Qing-Fei-Pai-Du-Tang for the treatment of Novel Coronavirus Pneumonia by network pharmacology. Chinese Traditional and Herbal Drugs. 2020.
- 26.Ru. J, Li. P, Wang. J, Zhou. W, Li. B, Huang. C, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014. 6. [DOI] [PMC free article] [PubMed]
- 27.Shen L, Xu Z, Wang Y, Yun N. Study on anti-tumor mechanism of Salvia miltiorrhiza based on network pharmacology. Tianjin Pharmacy. 2019. 31.
- 28.UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D15. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S. et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016;54:1. doi: 10.1002/cpbi.5. 30 1-1 3. [DOI] [PubMed] [Google Scholar]
- 30.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Martin A, Ochagavia ME, Rabasa LC, Miranda J, Fernandez-de-Cossio J, Bringas R. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinformatics. 2010;11:91. doi: 10.1186/1471-2105-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Xie L, Hao X, Luo J. Research progress on diagnosis and treatment of 2019-ncov pneumonia with traditional Chinese medicine. World Chinese Medicine. 2020.
- 35.Li H, Wang Y, Xu J, Cao B. Potential antiviral therapeutics for 2019 Novel Coronavirus. Zhonghua Jie He He Hu Xi Za Zhi. 2020. [DOI] [PubMed]
- 36.Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chinese Journal of Natural Medicines. 2013;11:110–20. doi: 10.1016/S1875-5364(13)60037-0. [DOI] [PubMed] [Google Scholar]
- 37.Khan NM, Haseeb A, Ansari MY, Devarapalli P, Haynie S, Haqqi TM. Wogonin, a plant derived small molecule, exerts potent anti-inflammatory and chondroprotective effects through the activation of ROS/ERK/Nrf2 signaling pathways in human Osteoarthritis chondrocytes. Free Radical Biology and Medicine. 2017;106:288–301. doi: 10.1016/j.freeradbiomed.2017.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seong RK, Kim JA, Shin OS. Wogonin, a flavonoid isolated from Scutellaria baicalensis, has anti-viral activities against influenza infection via modulation of AMPK pathways. Acta virologica. 2018;62:78–85. doi: 10.4149/av_2018_109. [DOI] [PubMed] [Google Scholar]
- 39.Chu X, Ci X, Wei M, Yang X, Cao Q, Guan M. et al. Licochalcone a inhibits lipopolysaccharide-induced inflammatory response in vitro and in vivo. J Agric Food Chem. 2012;60:3947–54. doi: 10.1021/jf2051587. [DOI] [PubMed] [Google Scholar]
- 40.Sun LC, Zhang HB, Gu CD, Guo SD, Li G, Lian R. et al. Protective effect of acacetin on sepsis-induced acute lung injury via its anti-inflammatory and antioxidative activity. Arch Pharm Res. 2018;41:1199–210. doi: 10.1007/s12272-017-0991-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ngo KA, Kishimoto K, Davis-Turak J, Pimplaskar A, Cheng Z, Spreafico R. et al. Dissecting the Regulatory Strategies of NF-kappaB RelA Target Genes in the Inflammatory Response Reveals Differential Transactivation Logics. Cell Rep. 2020;30:2758–75. doi: 10.1016/j.celrep.2020.01.108. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Meng T, Hao N, Tao H, Zou S, Li M. et al. Immune regulation mechanism of Astragaloside IV on RAW264.7 cells through activating the NF-kappaB/MAPK signaling pathway. Int Immunopharmacol. 2017;49:38–49. doi: 10.1016/j.intimp.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Grimes JM, Grimes KV. p38 MAPK inhibition: A promising therapeutic approach for COVID-19 published online ahead of print, 2020 May 16. J Mol Cell Cardiol. 2020;144:63–65. doi: 10.1016/j.yjmcc.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prokhorova EA, Kopeina GS, Lavrik IN, Zhivotovsky B. Apoptosis regulation by subcellular relocation of caspases. Scientific Reports. 2018. 8. [DOI] [PMC free article] [PubMed]
- 46.Krähling V, Stein DA, Spiegel M, Weber F, Mühlberger E. Severe acute respiratory syndrome coronavirus triggers apoptosis via protein kinase R but is resistant to its antiviral activity. J Virol. 2009;83(5):2298–2309. doi: 10.1128/JVI.01245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodríguez Y, Novelli L, Rojas M, Autoinflammatory and autoimmune conditions at the crossroad of COVID-19 [published online ahead of print, 2020 Jun 16. J Autoimmun. 2020. 102506. [DOI] [PMC free article] [PubMed]
- 48.Xu X, Han M, Li T. et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci U S A. 2020;117(20):10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Chen X, Lu Y, Chen F, Zhang W. Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Biosci Trends. 2020. [DOI] [PubMed]
- 50.Qu X, Hao S, Ma J, Wei G, Song K, Tang C, Observation on the clinical effect of Shufeng Jiedu Capsule combined with Arbidol Hydrochloride Capsules in the treatment of COVID-19. Chinese Traditional and Herbal Drugs. 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available databases were analyzed in our study. The active ingredients and putative target genes of SFJDC from TCMSP can be found in http://tcmspw.com/tcmsp.php. NCP-related target genes were from GeneCards (https://www.genecards.org/).