Abstract
Clonal propagation and genetic engineering of plants requires regeneration, but many species are recalcitrant and there is large variability in explant responses. Here, we perform a genome-wide association study using 190 natural Arabidopsis accessions to dissect the genetics of shoot regeneration from root explants and several related in vitro traits. Strong variation is found in the recorded phenotypes and association mapping pinpoints a myriad of quantitative trait genes, including prior candidates and potential novel regeneration determinants. As most of these genes are trait- and protocol-specific, we propose a model wherein shoot regeneration is governed by many conditional fine-tuning factors and a few universal master regulators such as WUSCHEL, whose transcript levels correlate with natural variation in regenerated shoot numbers. Potentially novel genes in this last category are AT3G09925, SUP, EDA40 and DOF4.4. We urge future research in the field to consider multiple conditions and genetic backgrounds.
Subject terms: Plant regeneration, Natural variation in plants, Gene expression, Plant hormones, Genome-wide association studies
Robin Lardon et al. report a genome-wide association study of shoot regeneration in Arabidopsis under 2 different in vitro incubation conditions. They find wide variation in regeneration phenotypes, attributable to allelic variants in key developmental genes, and show that genetic association patterns differ depending on environmental factors.
Introduction
Plant cells exhibit remarkable developmental plasticity, enabling them to reconstruct tissues and organs upon wounding during post-embryonic development1,2. This feature also allows for regeneration of entire plants from tissue explants cultured in vitro, which is widely exploited for clonal propagation of elite varieties, virus sanitization and the creation of transgenic crops3,4. As the success of such practices is highly variable among species and cultivars, many studies have focused on the model organism Arabidopsis thaliana to obtain insight into the molecular framework of de novo organ formation3. Here, shoots can be regenerated from root explants by a two-step protocol in which root segments are placed on auxin-rich callus-inducing medium (CIM), before being transferred to cytokinin (CK)-rich shoot induction medium (SIM)5.
During CIM preincubation, auxin signals transmitted by the SCF–TIR1/AFB receptor complex, Aux/IAA repressors and auxin response factors (ARFs) activate division of designated pericycle cells to form a mass of organogenic callus6,7. This tissue resembles lateral root primordia and expresses root meristem genes such as PLETHORA (PLT) 1 & 2, WUSCHEL-RELATED HOMEOBOX (WOX) 5, SHORT ROOT (SHR), and SCARECROW (SCR)8,9. The convergence of hormone signals (e.g., auxin-induced PLT3, 5 & 7/CUP-SHAPED COTYLEDON (CUC) 1 & 2 and WOX11/LATERAL ORGAN BOUNDARIES DOMAIN (LBD) 16 modules10,11) with stress and wounding responses (e.g., mediated by WOUND-INDUCED DEDIFFERENTIATION (WIND) 112) on CIM also underlies the acquisition of competence to regenerate shoots later on13, by reactivating the cell cycle and installing progressive epigenetic changes such as DNA demethylation and histone modifications (e.g., H3K4me2 and H3K27me3)14–17. High cytokinin levels in the SIM then trigger a phosphorelay consisting of Arabidopsis histidine kinases (AHK2–4), phosphotransfer proteins (AHP1–5) and response regulators (ARRs) to repress root markers and induce coordinated expression of shoot apical meristem (SAM) genes such as WUSCHEL (WUS), SHOOT MERISTEMLESS (STM), ENHANCED SHOOT REGENERATION (ESR) 1 & 2, and LIGHT-SENSITIVE HYPOCOTYLS (LSH) 3 & 47,18. This results in the “transdifferentiation” of root-like protuberances to shoot primordia, which is limited to a narrow developmental window6,19,20.
The homeobox transcription factor (TF) WUS has been put forward as a master regulator of de novo SAM establishment21. Under steady-state meristem growth, it forms a feedback loop with CLAVATA3 (CLV3) to maintain the size and position of the stem cell niche in the SAM22. On SIM, WUS is induced following a two-step model that involves the dilution of repressive epigenetic marks by CK-induced cell division prior to direct activation by B-type ARRs21,23–25. The latter interact with HD-ZIP III TFs PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV) to spatially confine WUS expression and they limit auxin responses by blocking YUC transcription, further contributing to the elevated WUS levels21,23. WUS in turn reinforces CK responses by inhibition of A-type ARRs. PHB, PHV and REV also promote expression of the shoot determinants STM and RAP2.6L and WIND1 contributes to the events on SIM by directly activating ESR113,26.
Besides the type and physiological status of explants, incubation conditions such as hormone concentrations, temperature, and light affect regeneration and despite the conservation of cytokinin signals and WUS in the control of the SAM, natural Arabidopsis accessions show strong variation in their capacity to regenerate2,27. Previously, linkage mapping uncovered five quantitative trait loci (QTLs) responsible for the difference in regenerative capacity between accessions Nok-3 and Ga-0 and local association analyses resolved RECEPTOR-LIKE PROTEIN KINASE (RPK) 1 as a potential candidate gene28. Here, we further exploit natural variation in de novo shoot formation by conducting a genome-wide association study (GWAS), which is combined with the analysis of chromosome substitution lines (CSLs), T-DNA insertion mutants and gene expression levels. Known shoot meristem regulators and potential novel mediators of organogenesis are identified, confirming that regeneration is a complex trait controlled by multiple loci. We find that most rate-limiting factors are specific for the applied protocol and a few genes act as master regulators, including WUS, AT3G09925, SUP, EDA40, and DOF4.4.
Results
Shoot regeneration is subject to natural variation
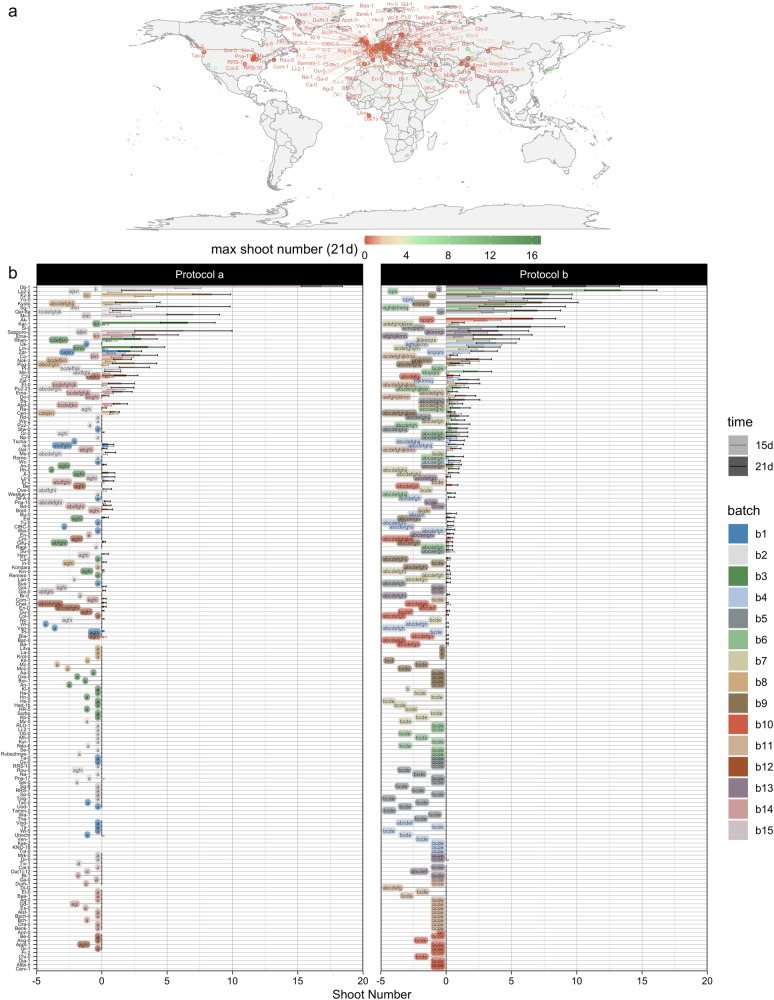
We have subjected 190 natural Arabidopsis accessions to the two-step protocol for shoot regeneration from root explants and scored various phenotypes that reflect the organogenic potential, including the number of shoots, shoot primordia and roots (details of the recorded traits are presented in the “Methods” section and Supplementary Fig. 1; Supplementary Fig. 2 provides corresponding bar charts). To incorporate the environmental effects, we have tested two protocol variants designated a and b, differing in explant age, light quality, and cytokinin concentration in the SIM. Irrespective of the protocol, the number of regenerated shoots follows an exponential trend, in which around half of the tested accessions do not form shoots at all (Fig. 1b). This similarity between the results of both protocols not only shows that the data are robust, but also indicates an important contribution by the genotype. A compact letter display based on nonparametric statistics further reveals that accessions can be divided over 14 and 19 different classes according to regenerated shoot numbers with protocol a and b, respectively. This multitude of levels in the phenotype can only be explained by numerous small allelic contributions, which suggests that de novo shoot organogenesis is a multigenic trait, a notion that agrees with the state of the art13.
Fig. 1. Variation in shoot regeneration among natural Arabidopsis thaliana accessions.
a Geographic distribution of accessions ranked according to the maximum number of regenerated shoots after 21 days with protocol a or b. b Bar chart of shoot numbers after 15 and 21 days with protocol a and b. Colors indicate the batches in which accessions were analysed and labels reflect a compact letter display of two-sided pairwise comparisons (on count data after 21 days) using Dunn’s nonparametric test at a false discovery rate (FDR) of 5% with n = 12 independent biological replicates. Global Kruskal–Wallis tests yielded p-values < 2e−16 for both protocols.
Plotting the maximum of regenerated shoot numbers under either protocol on a world map using accession coordinates reveals no obvious pattern (Fig. 1a), implying that if regeneration yields an evolutionary advantage, it is not coupled to geographic parameters. Besides, strong differences were recorded among explants of the same genotype (reflected by the error bars in Fig. 1b), indicating that there is an important residual effect likely contributed by the environment and the physiological state of explants. Hence, our data illustrate that higher order variation in de novo shoot organogenesis in Arabidopsis is explained by the genotype and variability within accessions is likely due to environmental fluctuations and epigenetic effects. Apart from variation in the extent of shoot regeneration, there is also a lot of diversity in the morphology of regenerated structures, going from hairy root-like outgrowths to trichome-covered shoots, leaf-like structures, and flower buds. Finally, the equal distribution of accessions analysed in different batches (represented by different colors in Fig. 1b) across the graph shows that there was no substantial blocking effect.
Association patterns differ with environmental conditions
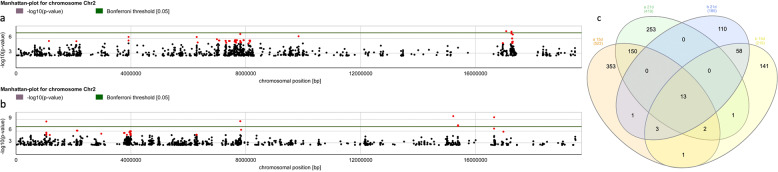
To uncover the genetic framework of the observed regenerative variation, we computed trait correlations for single nucleotide polymorphisms (SNPs) across the genome (see “Methods” section), displayed in Manhattan plots (Fig. 2a, b and Supplementary Fig. 3). Multiple significant associations were found for regenerated shoot numbers (grouped in 5–10 distinct regions per chromosome), with an increased density of highly significant SNPs on chromosomes 3 and 2 for data from protocol a and b, respectively. Evaluating all genes within 10 kb of significant SNPs as potential quantitative trait genes (QTGs) pinpoints URH1 (or SAP4), MIR393A (or MYB25), AT3G09925 (or IPS1), SAUR23, WUS, QUL2 (or EFM), ARF20, and MSL3 (or RLP9) as important players (Supplementary Data 1; refer to the “Discussion” for an in-depth review of candidate genes). This myriad of QTLs is in line with the genetic complexity expected from the phenotypic trend (Fig. 1b). Although there is partial overlap in association patterns under protocol a and b, the ranking of the SNPs is altered, suggesting that different factors are rate-limiting under different conditions. This is also observed in a Venn diagram of SNPs with p ≤ 1e−5 and MAF (minor allele frequency) > 5% for shoot numbers after 15 and 21 days following protocol a and b, showing that relatively few SNPs are highly significant for both procedures (Fig. 2c). The fraction of SNPs linked to regenerated shoots after 15 and 21 days under either protocol is higher, indicating a common regulatory network for de novo SAM establishment and subsequent shoot development.
Fig. 2. SNP associations for shoot regeneration.
a, b Manhattan plots of chromosome 2, showing the significance of SNP associations with regenerated shoot numbers after 21 days under protocol a (a) or b (b). The green line marks a Bonferroni threshold of 5%, deduced from an efficient mixed-model association expedited (EMMAX) test with n = 129 and n = 149 independent samples for protocol a and b, respectively. SNPs considered significant in subsequent analyses (raw p-value ≤ 1e−5) are highlighted in red. c Venn diagram showing overlap in genome-wide SNPs that were significantly linked to shoot numbers under protocol a (orange and green parts) or b (blue and yellow parts) after 15d (orange and yellow parts) or 21d (green and blue parts).
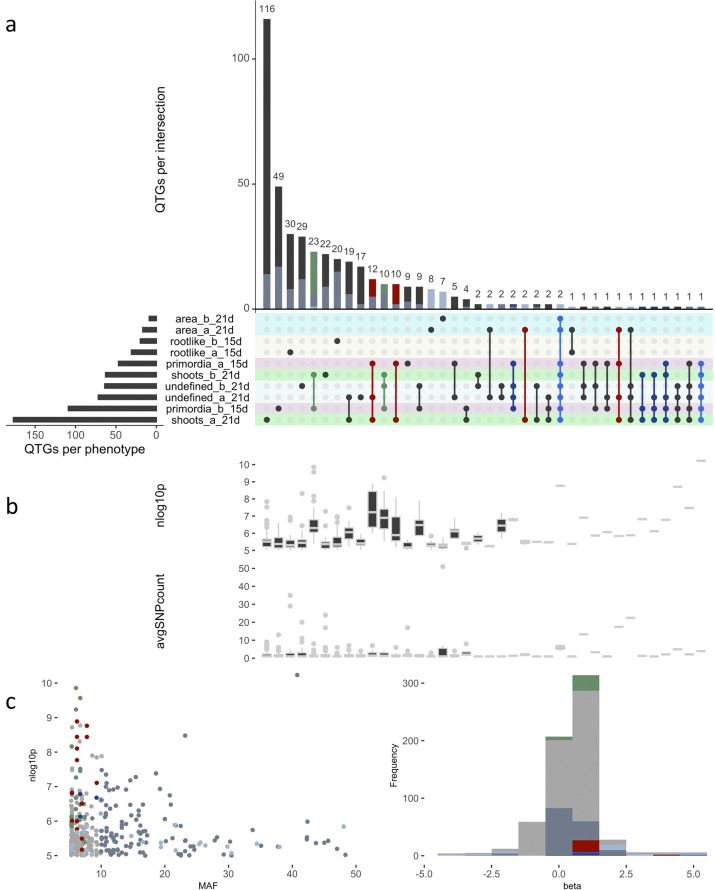
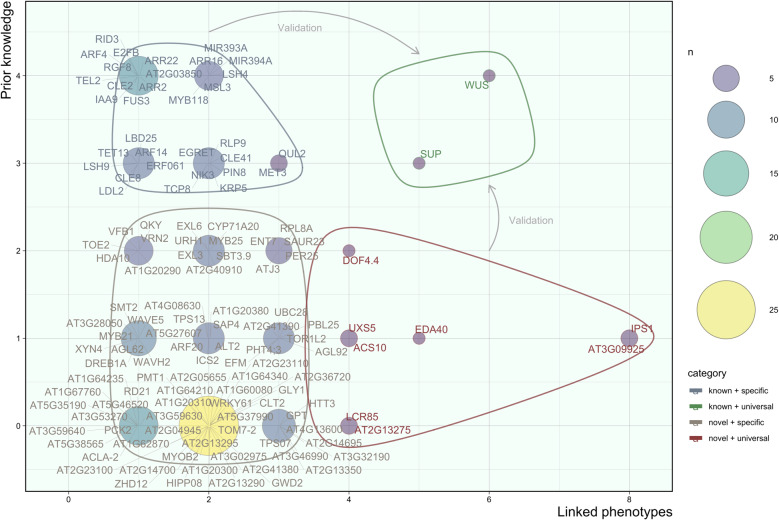
Given the above, we explored the overlap in genes harboring allelic variation important for different phenotypes. Hereto, the genes closest to SNPs significantly associated (p ≤ 1e−5) with ten traits of interest were selected and a comparison of the subsets was made using the UpSetR package in R (Fig. 3a)29. Attribute plots were constructed to assess the significance, allele frequency, impact and number of SNPs underlying the genes in each category (Fig. 3b, c). Because few genes are found in higher-order intersections, Fig. 3a reveals that most candidates only play a role for specific traits under specific conditions. Moreover, intersections corresponding to different characteristics under the same protocol contain more genes than intersections spanning the two procedures, suggesting that critical determinants are shared between various regeneration pathways under similar conditions. However, candidates linked to root-like structures form a distinct category. Intriguingly, around ten factors are linked to many phenotypes across protocols and genes in highlighted intersections are supported by larger SNP clusters with low p-values (Fig. 3b). Moreover, their positive alleles are rare (low MAF) and often correspond to beta values at the edge of the distribution, meaning they contribute substantially to regenerative variation (Fig. 3c). The two genes found in most sets are AT3G09925 (encoding a pollen ole e 1 and extensin family protein) and IPS1—likely corresponding to a single peak, followed by WUS, SUP/FLO10, EDA40, DOF4.4, LCR85, AT2G13275, ACS10, and EMB2296. Notably, many of these genes act in embryo or flower development (see “Discussion” section).
Fig. 3. Overlap in candidate genes for various regeneration traits.
a Upset diagram showing overlap in QTGs (i.e., genes closest to SNPs with p ≤ 1e−5) between phenotypes, protocols and time points. Bright blue, dark blue, red, and green colors respectively indicate the most common sets (degree ≥ 6), selected intersections spanning both protocols and the highest order groups within protocol a or b. The same color codes are used for individual SNPs and genes, which are further highlighted in light or dark steel blue if beta (i.e., regression coefficient) ≥ 1.0 or MAF ≥ 10%. b Box plots showing −log10(p-value) (nlog10p) and number of SNPs (avgSNPcount) supporting the genes in each intersection (hinges reflect quartiles). c Scatter plot of SNP significance vs. minor allele frequency (MAF) and histogram of beta values.
GWAS highlights the importance of WUS and uncovers novel candidate genes
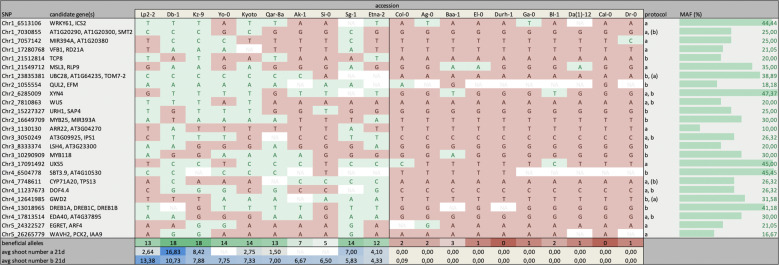
Another strategy we followed to assign priorities to QTLs existed in ranking the allelic divergence of significant SNPs (p ≤ 1e−5 for shoot numbers after 21 days under protocol a or b) in the ten best and ten of the worst regenerating accessions (Fig. 4). The most interesting candidates according to this criterium are UBC28, QUL2, DREB1A, MIR393A, GWD2, and EDA40 (for which at least 6 out of 10 strong regenerators, but at most 1 out of 10 poor regenerators carries the beneficial allele). Moreover, of the 25 loci with the most pronounced allele distinction between good and bad performers, five are linked to at least four different phenotypes (AT3G09925, WUS, EDA40, DOF4.4, and UXS5; Supplementary Data 1). Looking at the allele distribution in Fig. 4 reveals that a variety of beneficial SNP combinations can lead to strong regeneration, whereas poorly regenerating accessions tend to have all negative alleles. In other words, the proposed candidates appear to provide more positive than negative selection, suggesting that they act as stimulators of regeneration rather than suppressors. Also, different sets of favorable SNPs are required for optimal performance under different protocols (i.e., there is a slight reorganization among top accessions under different conditions), implying a change in rate-limiting factors and epistasis effects depending on the environment.
Fig. 4. Allelic variation in the top candidate genes.
Distribution of selected SNPs among the ten best and ten of the worst regenerating accessions. Green and red boxes respectively indicate superior and inferior alleles and the three bottom rows depict gradients based on the accumulation of beneficial SNPs and regenerated shoot numbers after 21 days under protocol a or b. The two leftmost columns specify the SNP position along with nearby genes and the two rightmost columns show whether the SNP was significantly linked (p ≤ 1e−5) to data from protocol a and/or b (with brackets reflecting insignificant associations) and the percentage of accessions carrying the minor and usually superior allele in the population (MAF). Blank fields contain missing data.
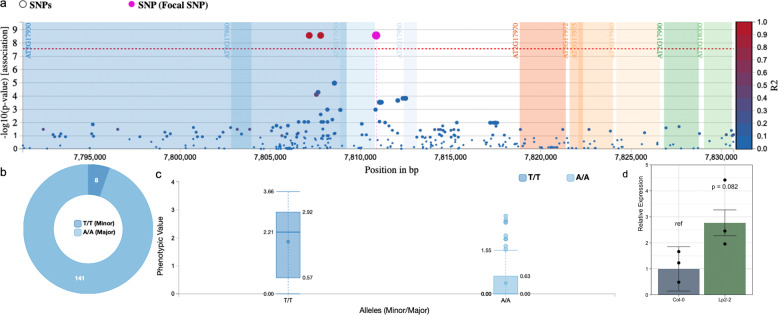
Among the top candidates is WUSCHEL and a close-up of the association with shoot numbers under protocol b unveils three significant SNPs surrounding its open reading frame (ORF), 2 of which are located downstream and 1 is in the promoter sequence (67 bp from the transcription start site or TSS; Fig. 5a). They are highly significant, exhibit strong allelic distinction and exert a large phenotypic effect (Fig. 5a–c), independent of the conditions. As there are no nucleotide changes in the gene body and hence no potential effects on protein conformation, we investigated possible transcriptional regulation. RT-qPCR revealed three-fold higher mRNA levels in Lp2-2 than in Col-0 after 3 days on SIM (Fig. 5d) and as these are respectively good and bad regenerators, changes in WUS expression might indeed be responsible for natural variation in shoot regeneration. Nonetheless, our GWAS shows that various other factors contribute to the observed variability, which is likely a result of differential regulation at various stages of de novo shoot organogenesis, including founder cell specification, pluripotency acquisition, and SAM patterning.
Fig. 5. Details of the SNPs near WUS and their effect on shoot regeneration.
a SNP associations and linkage (R2) in the region around WUS, showing one upstream SNP (67 bp from the TSS; in pink) and two linked downstream SNPs (in red). b Allele frequencies of the pink SNP. c Box plot of the square root of regenerated shoot numbers after 21 days under protocol b in accessions with either variant of the highlighted SNP (hinges depict quartiles and whiskers extend to 1.5 times the interquartile range). d WUS expression relative to UBC9 and TIP41L in Col-0 and Lp2-2 (respectively a poorly and a strongly regenerating accession) after 3 days on SIM following protocol b. Error bars reflect standard errors and the p-value (corresponding to an estimated effect size of −1.47 within a 95% confidence interval from −3.24 to 0.30) is deduced from a linear model using a two-sided ANOVA with n = 3 independent biological replicates (represented as black dots).
Chromosome substitution lines refine the GWAS
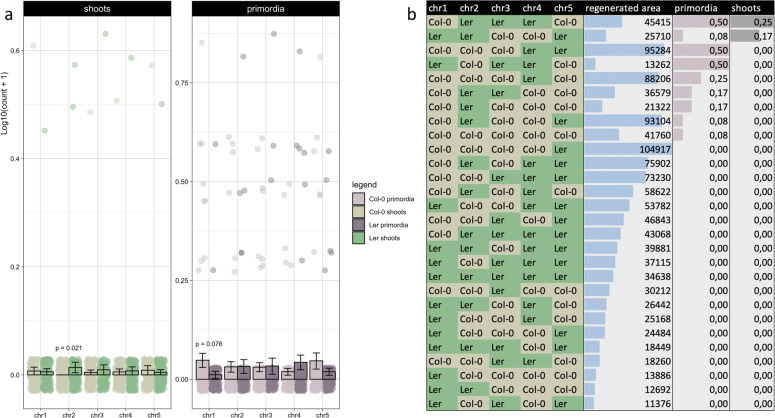
Considering the genetic complexity of de novo shoot organogenesis and the possibility that epistasis contributes to variation between accessions, we phenotyped a full set of Col-0 × Ler CSLs30 using protocol b. Although the potential for mapping is restricted because of limited differences in the regenerative potential of Col-0 and Ler, there is also limited genetic variation between them at the loci of interest and overlap with association data thus allows to refine certain QTLs. According to the results, lines carrying a Ler chromosome 2 regenerate significantly better than the ones with a corresponding Col-0 variant (irrespective of the rest of the genome; Fig. 6a). Of the GWAS candidates found on this chromosome, only WUS and QUL2 carry different alleles in Col-0 and Ler. As the positive allele of QUL2 is found in the Col-0 variant, however, WUS is the most likely cause of variation between these accessions. Intriguingly, a nucleotide substitution at 341 bp upstream of the TSS (GAGT to GATT) creates an additional ARR binding motif in the WUS promoter on Ler chromosome 2, which agrees with the notion that transcriptional regulation of WUSCHEL could be underlying regenerative differences between accessions. Chromosome 1 appears to be important as well, because lines with a Col-0 variant form more shoot primordia than those with the Ler version (Fig. 6a) and significant interactions with shoot regeneration were found between chr3:chr4 and chr3:chr5 (with respective FDRs of 0.03 and 0.01, effect sizes of 42.60 and −61.82 and deviances of 6.2 and 11.1). Together with the small regenerative difference between Col-0 and Ler, this suggests that WUS is not the only factor at play and variation between these accessions is orchestrated by a combination of positive and negative inputs. Ranking the CSLs reveals a complex pattern, wherein no particular interaction is highlighted (Fig. 6b).
Fig. 6. Shoot regeneration in chromosome substitution lines.
a Bar plot of log-transformed shoot and shoot primordium numbers in CSLs between Col-0 and Ler after 21 days on SIM following protocol b. Per chromosome, the averages of all lines with the Col-0 variant and the Ler variant are compared (irrespective of the other chromosomes) and individual data points are overlaid in jittered strip charts. Error bars reflect standard errors and FDR-adjusted p-values are deduced from negative binomial generalized linear models using two-sided likelihood ratio tests with n = 12 independent biological replicates per CSL and 321 or 318 residual degrees of freedom for shoots or primordia. The two significant terms had estimated sizes of 39.27 (effect of chr2 on shoots; deviance = 7.55) and −2.52 (effect of chr1 on primordia; deviance = 5.90). b Chromosome patterns of the CSLs (with Col-0 alleles in khaki and Ler variants in green), ranked by regenerated shoots, primordia and area.
Novel candidate regeneration genes are highly context-dependent
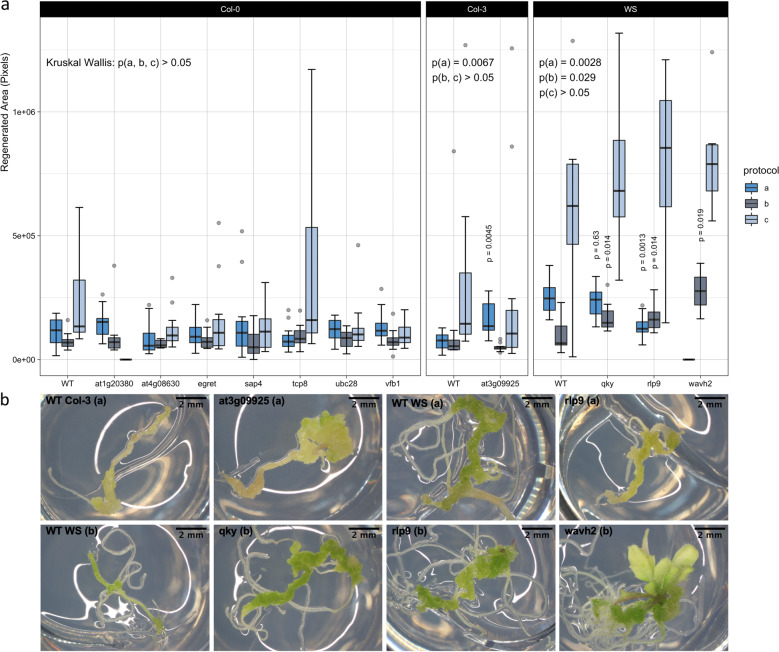
To validate the importance of novel candidates put forward by association mapping, we ordered T-DNA insertion lines for 25 genes, selected by significance, literature, specificity, allelic distribution and commercial availability (Supplementary Data 1). We obtained homozygous mutants for 11 of these genes (Supplementary Fig. 4) and analysed their phenotype using protocols a and b. In addition, a third protocol c was applied (similar to protocol b, but with 6 days of CIM incubation and continuous light exposure during SIM) to improve comparison with wild type Col-0, which regenerates poorly using protocols a and b. Disruption of AT3G09925, QKY, RLP9, or WAVH2 causes significant changes in de novo shoot formation (Fig. 7a), but none of the lines completely lost their regenerative capacity, potentially because of gene redundancy or incomplete loss-of-function. For comparison, we tested nine unrelated T-DNA mutants and five multiple gene knockouts linked to light signaling and phosphorylation, revealing that while the insertions in Fig. 7 are not as detrimental as higher order mutations, they yield more severe defects than random single gene disruptions (Supplementary Fig. 5). Visual inspection of the explants suggests that whereas QKY only affects callus growth, AT3G09925 and RLP9 are active at the stage of primordium formation and WAVH2 determines both shoot initiation and development (Fig. 7b). However, rlp9 shows decreased regeneration under protocol a, while an improvement is detected under protocol b and the opposite holds for at3g09925. The effects of individual T-DNA insertions are also small compared to variation between protocols and although this could again be attributed to redundancy or weak null alleles, it suggests that single gene contributions are subordinate to environmental changes and that a combination of multiple alleles accounts for differential regeneration among accessions. Notably, protocol c yields much better regeneration rates in different wild type backgrounds and in many mutants. These results illustrate that many determinants of regeneration are highly context-dependent and thus we propose that only a small set of master regulators such as WUS are critical under various conditions.
Fig. 7. Shoot regeneration in mutants for GWAS candidate genes.
a Box plot of the regenerated area in 11 T-DNA insertion lines compared to wild type (WT) plants after 21 days under protocol a, b, or c (respectively shown in bright blue, dark steel blue, and light steel blue). Hinges mark the first and third quartile, horizontal lines reflect the median and whiskers extend to the furthest data point within 1.5 times the interquartile range of the nearest hinge. FDR-adjusted p-values are deduced from a Kruskal–Wallis test and Dunn’s many-to-one tests for two-sided pairwise comparisons to a single control on n = 12 independent biological replicates (quantile estimates are 3.18, −0.47, 2.59, −3.60, 2.59, and 2.42 for significant terms from left to right, i.e., at3g09925 (a), qky (a, b), rlp9 (a, b), and wavh2 (b)). No data was available for at1g20380 under protocol c and wavh2 under protocol a. b Representative images for lines that differ significantly from their wild type counterparts under the respective conditions (protocol a, b or c).
Discussion
In line with previous reports, our extended and genome-wide association analysis found substantial variation in the regenerative potential of 190 Arabidopsis thaliana accessions under two different protocols28,31. This agrees with the finding that root hormone levels differ among accessions32, because hormone responsivity is a key determinant of regeneration2,33. In planta developmental traits such as rosette morphology, leaf expansion, and flowering time are subject to natural variation as well34–36, implying that care must be taken when extrapolating observations regarding plant development made in a particular ecotype, but also highlighting the potential of GWAS. Association mapping revealed de novo shoot organogenesis to be a complex trait, similar to what was reported for in vivo shoot development34. It is controlled by several QTGs, including ARFs and ARRs, MYB and AP2/ERF2 family TFs, miRNAs, receptor-like kinases, F-box proteins, chromatin remodelers and various biosynthetic and cell wall modifying enzymes. Based on literature, we recognized 18 a priori candidates, 17 homologs of prior candidates, 19 genes involved in similar processes, and 75 unrelated or unknown genes. Plotting the number of phenotypes these factors support against a score for prior links with organogenesis revealed four categories (Fig. 8), which are elaborated below. Combined with the analysis of T-DNA mutants, Fig. 8 reveals a major group of fine-tuning factors (~95%) whose role depends on the protocol and a minor group of master regulators (~5%), that are critical for multiple tested procedures and traits. This shift in rate-limiting factors depending on the conditions could explain why several studies attempting to map natural variation in shoot regeneration have pinpointed different QTGs28,31. Accordingly, a GWAS on embryonic callus regeneration in maize showed that only 15 out of 63 QTNs were retained in multiple environments and highlighted WUSCHEL-RELATED HOMEOBOX 2 (WOX2), although other candidates are distinct from ours37. Most QTGs we identified are also new compared to association studies on adventitious shoot regeneration in roses38, callus formation in poplar39 and rice40 and in vitro regeneration of cucumber41 and tomato42. However, these studies do report similar functional classes of candidates (e.g., embryogenesis and meristem genes, reprogramming factors, hormone-related proteins, receptor-like kinases, and TFs from the LBD, ERF, MYB, and WOX families)37–40 and in cases where multiple traits, protocols or techniques are evaluated, overlap between them is limited37,38, suggesting that the difference in experimental systems could be part of the cause. Notably, several established SAM genes, epigenetic factors and cell cycle regulators (e.g., STM, CUCs, ESRs, PLTs, WIND1, MET1, and CYCD313) were not detected in our assay, which might be due to a lack of functional sequence variation at these loci in the tested population43. In turn, this could be the result of stringent selection against harmful mutations in genes that are vital to embryonic development, wound repair, and rooting. Possibly, epigenetic, transcriptional or post-translational regulation is favored for key survival genes to allow for better fine-tuning. Investigating the role of these mechanisms in natural regenerative variability by means of eQTL mapping and methylome-wide associations is a promising future prospect44.
Fig. 8. Visual representation of the four categories of candidate regeneration genes.
The number of associated phenotypes, i.e., shoots, shoot primordia, green area, root-like structures, and undefined structures under protocol a or b (x-axis) was plotted against prior links with shoot regeneration based on literature, where 0 = no link, 1 = possible link, 2 = plausible link, 3 = indirect link, 4 = direct link (y-axis). The size and color of circles reflects the number of genes in each set (n). Blue, green, gray and red outlines respectively define known conditional factors, known master regulators, unknown conditional factors, and unknown master regulators.
A strong correlation was found between regenerated shoot numbers and allelic variation in the promoter of WUSCHEL, and we propose that this is due to differential transcription. WUS is essential for formation and maintenance of the SAM and knock-out of this gene severely impairs regeneration18,22. Moreover, its expression marks shoot progenitor cells on SIM and overexpression induces somatic embryogenesis and shoot regeneration21,45. Hence, this association demonstrates the robustness of the GWAS. CSL analyses suggest that WUS could also contribute to regenerative differences between Col-0 and Ler, and sequence comparison uncovered a SNP that introduces an additional ARR binding motif in the promoter of the beneficial Ler allele (GAGT to GATT; 341 bp upstream of the TSS). This SNP is also present in Lp2-2, a strong regenerator with 3-fold higher WUS mRNA levels on SIM than poorly regenerating Col-0 plants, which is in line with recent reports showing direct WUS induction by B-type ARRs21,23. However, at present we cannot distinguish whether improved shoot formation in Lp2-2 is due to an increased number of WUS-expressing foci or elevated WUS levels in individual foci. Lastly, the SNP upstream of WUS overlaps with one of four unresolved QTLs obtained by linkage mapping28.
Two other prime candidates are LSH4 and CLE2, whose linked SNPs show high trait specificity and are downstream of the ORF. LSH4 is induced by CUC1 in the boundary cells of shoot organs to coordinate differentiation and overexpression leads to ectopic development of WUS-expressing meristems46. It has also been used successfully as a marker for shoot regeneration47. CLE2 is a homolog of CLV3, known for its role in the regulatory feedback loop with WUS that controls the size of the SAM in vivo48. It is induced by ESR1 during the early stages of regeneration49 and upregulated in ick1/2/5/6/7 mutants showing increased regenerative potential50. E2FB, another TF we picked up, is also regulated by ICK/KRP genes51 and acts in the translation of environmental stimuli and auxin signals to cell cycle progression in root and shoot apexes52,53. In addition, we found associations near two microRNAs: MIR393A (overexpression of which impedes de novo SAM formation by repressing TIR154) and MIR394A (known to spread from the L1 layer of the SAM to the L3 layer, where it represses LCR to potentiate WUS activity and therefore maintain stem cell pluripotency55). Both miRNAs are differentially expressed between totipotent and non-totipotent calli56. We also found two auxin-related and three cytokinin-related prior candidates: IAA9 is upregulated during CIM57 preincubation and implicated in lateral root formation58 and somatic embryogenesis59, while ARF4 acts redundantly with ARF3 to control organ polarity60 and lateral root initiation61. Recently, it was discovered that these ARFs undergo differential methylation by MET1 during shoot regeneration62, and they promote organogenesis by repressing STM through histone deacetylation63. On the other hand, ARR2 is reported to associate with HD-ZIP III TFs to control WUS expression during regeneration and impair the process when knocked-out21, whereas ARR16 and ARR22 are A-type response regulators. The former is downregulated during CIM preincubation and induced by ARR2 on SIM9,64, and the latter suppresses B-type ARRs65. Seven lesser-known prior candidates mainly involved in lateral rooting, callus formation or somatic embryogenesis are MSL366, MYB11867, RGF868, RID369, FUS370, TEL271, and AT2G038509.
Among indirect QTGs (i.e., homologs of prior candidates) is the pectin methyltransferase QUL2/PMT5, a paralog of QUA2/TSD2 (expressed in meristems and mutation of which causes “shooty” callus formation in vitro, paralleled by enlarged expression domains of KNAT1&2 and elevated transcription of STM72). It is near six highly significant SNPs, but corresponding T-DNA lines were not viable. Another indirect candidate, TCP8, harbors a missense variant with large phenotypic effect and low p-value. Although TCP8 (a class I TCP) has only been linked to immunity, auxin homeostasis and leaf development73–75, repression of TCP3 (a class II TCP) yields ectopic shoot formation, because it suppresses CUC genes and KNOX1 factors through interaction with AS276–78. Moreover, strong redundancy is observed within and between TCP clades and class I TCPs can also modulate KNOX1 and cell cycle gene expression to control lateral organ growth. Some even affect cytokinin sensitivity or bind to AHPs75,77,79. Accordingly, a T-DNA mutant showed increased regeneration with protocol c. Next, ERF061 belongs to the DREB A-6 subfamily of ERF/AP2 TFs also containing WIND1 (able to bypass wounding and auxin pre-incubation and enhance callus formation and shoot regeneration by upregulating ESR1)26,80. This functionality is conserved in WIND2-480, but ERF061 is only supported by one downstream SNP. Two further indirect QTGs compete with a direct candidate: RLP9 is a homolog of CLV2/RLP10 (promoting stem cell differentiation by repressing WUS in intact SAMs81) that lies upstream of six SNPs in the gene body of MSL3 (mutation of which induces callus production at the shoot apex, coupled to an altered cytokinin to auxin ratio66). Nonetheless, disruption of RLP9, respectively decreased and increased the number of regenerated shoots under protocol a and b, so both genes might contribute to the association. Likewise, EGRET/IDD13 is a C2HC zinc finger protein from the same family as JKD and MGP (known for their roles in root patterning82), but regeneration was not affected in egret mutants, suggesting that nearby ARF4 is causing the correlation. Lastly, LDL2 is homologous to LDL3, recently found to eliminate H3K4me2 during callus formation and thus facilitate the induction of shoot markers on SIM17.
Some candidates are plausible QTGs because they act in processes related to shoot regeneration. For example, QKY is a component of SUB signaling required for tissue morphogenesis and organ development, mutation of which alters the morphology of the L2 SAM layer83,84 and T-DNA insertion significantly increased regeneration under protocol b. Another such QTG is VFB1, encoding an F-box protein from the same family as TIR1 and AFB1-5. Loss of all four VFB genes compromises lateral root formation and DR5:GUS expression, but VFB2 cannot functionally substitute TIR185, which agrees with our finding that disruption of VFB1 had no significant effect. SUP/FLO10 (a TF that defines the boundary between stamens and carpels in the floral meristem by regulating cell proliferation and floral homeotic genes such as AP3 and PI86), DOF4.4 (related to shoot branching87) and EMB2296 are plausible QTGs that underlie at least three investigated phenotypes. Other candidates in this category worth mentioning are URH1 (potentially involved in CK metabolism88), ENT7 (a possible CK transporter89), AT1G20290 (a SWI-SNF-related chromatin-binding protein), VRN2 (a Polycomb protein), and HDA10 (a histone deacetylase).
Finally, a possible link was found for a few poorly annotated candidates: AT3G09925 is a Pollen Ole e 1 (POE1) allergen and extensin family gene and recent reports suggest that these genes could act in various aspects of plant development, as they exhibit specific transcription patterns and some members are even regulated by H3K27me3 (which also restricts WUS during SIM incubation)16,90. According to our data, this gene is the second most important regulator of regeneration (after WUS), because it harbors nine highly significant SNPs (in the ORF and downstream region) that contribute strongly to variation in eight out of ten recorded traits and show pronounced allelic differences between poor and strong regenerators. Curiously, T-DNA insertion had a slightly negative effect under protocols b and c, but it increased the regenerated area using protocol a. Another less obvious candidate is WAVH2, T-DNA disruption of which significantly increased regeneration under protocol b. Along with homologs WAV3 and WAVH1, it has been attributed a role in root gravitropism and phototropism (triple wav3 wavh1 wavh2 mutants showing abnormal auxin signals in the root)91. Intriguingly, another member of this gene family named EDA40 underlies five of the recorded phenotypes in our GWAS and shows strong allelic distinction between good and bad accessions. Hence, the WAVY GROWTH E3 ligases likely play a role in de novo organogenesis. The E2 ubiquitin-conjugating enzyme UBC28 is interesting because transcriptomic comparison of accessions with the beneficial allele against those with the weak variant revealed downregulation of this gene in the good accessions (Supplementary Data 2), but no significant changes were found in the T-DNA line. Other possible candidates are DREB1A/CBF3 (involved in cold stress92) and ACS10 (an aminotransferase without ACC synthase activity93).
Our take-home message is that natural variation in tissue regeneration is associated with allelic differences in master meristem regulators and conserved genes that play a role in embryogenesis and flower development, but only a minority of these genes have broad functionality. Most regeneration determinants are context-dependent, which also holds for novel candidates put forward by GWAS, whose precise role in de novo SAM formation remains to be elucidated. Because in vitro responses are highly variable among accessions and rate-limiting molecular factors depend on the applied protocol, we urge future research in the field of regeneration to consider multiple conditions and validate results in different genetic backgrounds.
Methods
Plant materials and growth conditions
The GWAS included 190 natural Arabidopsis thaliana (L.) Heynh. accessions sequenced by the 1001 Genomes Consortium94 (N76636; NASC). Col-0 × Ler chromosome substitution lines were kindly provided by Cris L. Wijnen, Erik Wijnker, and Joost Keurentjes (Wageningen University)30. T-DNA insertion mutants were retrieved from NASC (SALK and SAIL lines) and INRA (FLAG lines): SALK_121407C (N654861; at1g20380), SALK_012673C (N670321; at4g08630), SALK_044769C (N674790; egret), SALK_152010C (N681875; sap4), SAIL_656_F11 (N862668; tcp8), SALK_040325C (N653158; ubc28), SALK_128933C (N667298; vfb1), SAIL_208_E09C1 (N867008; at3g09925), FLAG_124D07 (DSH18; qky), FLAG_056E09 (DLL1; rlp9), FLAG_109A12 (DYB211; wavh2). T-DNA inserts were checked by PCR (Supplementary Fig. 4) using the primers in Supplementary Table 1.
Seeds were sterilized by exposure to chlorine gas for 4 h and sown on Gamborg B5 medium (3.1 g B5 salts including vitamins per liter of medium, with 0.05% 2-(4-morpholino)-ethane sulfonic acid (MES), 2% (w/v) glucose and 0.7% agar at pH 5.8) and vernalized for 4 days at 5 °C. Three procedures were used for shoot regeneration, respectively designated as protocol a, b and c (adapted from the work of Valvekens et al.5). In protocol a, seedlings were grown under cool white fluorescent tungsten tubes (70 µM m−2 s−1) at 21 °C following a 14/10 h light/dark regime. After 7 days, 7 mm long root segments including the tip were excised and placed on CIM (B5 supplemented with 2.2 µM 2,4-dichlorophenoxy acetic acid (2,4-D) and 0.2 µM kinetin) for 4 days. Next, the explants were transferred to SIM (B5 supplemented with 25 µM 2-isopentenyl adenine (2-IP) and 0.86 µM 3-indole acetic acid (IAA)) and incubated for 3 weeks. Protocol b differs from protocol a in three ways: warm white light was used (70 µM m−2 s−1), explants were excised after 10 days and only 5 µM 2-IP was used in the SIM. Protocol c is a slight modification of protocol b, whereby explants were kept on CIM for 6 days and they were placed in continuous light during SIM incubation. Pictures were taken under binoculars (6.3×) after 15 days and after 21 days.
Genome-wide association study
Accessions were randomly divided over 15 batches (15–40 accessions per batch) and within batches the regenerative capacity was assessed following either protocol a or b, in such a way that all accessions were subjected to both protocols. Pictures were taken from every well after 15 days and 21 days and used to manually count shoots, primordia, root-like structures and undefined structures (representative images showing how these features were distinguished are provided in Supplementary Fig. 1) and assign a score from 0 to 5 for callus formation and greening. Raw data were processed in R (version 3.5.2; Supplementary Code) and multiple secondary phenotypes were calculated, including the regeneration index (defined as the percentage of explants forming at least 1 shoot), shoot variability (measured as the standard deviation of shoot numbers divided by the regeneration index) and shoot formation rate (the number of shoots formed per day between 15 days and 21 days of SIM incubation). Images were also analysed with ImageJ (version 2.0.0-rc-69/1.52j; Supplementary Code) to determine the area of green structures as a proxy for the regenerative potential. Finally, trait averages per accession were calculated and data were curated to handle missing sequence data, incomplete germination or contamination (requiring at least six explants per accession) and a distributional error at the stock center95, leaving 129 and 149 observations for the association analysis with protocol a and b, respectively (for Zal-1, Tol-0, KNO-18, Kas-2, Ven-1, Yo-0, Bu-0, Bs-1, Wa-1, Tamm-2, Su-0, Si-0, Tha-1, Ak-1, Cerv-1, Ba-1, Altai-5, Dja-1, Baz-0, Chi-0, Fr-2, Anz-0, and Di-G, no data was available under protocol a and for Tiv-1, Hey-1 and Mr-0, no data was available under protocol b). Note that for each phenotype, accessions showing excessive variability were also eliminated. Corresponding bar plots are provided in Supplementary Fig. 2.
Bioinformatics
For every phenotype described above, accession averages were uploaded to easyGWAS for correlation analyses96. A square root transformation was applied in the case of count values to approximate a normal distribution and three principle components were included in the model to account for higher order population effects. The minor allele frequency (MAF) was cut off at 5% and the EMMAX algorithm was used for computation. Resulting Manhattan plots and a list of candidate genes selected using custom python code (version 2.7.15; Supplementary Code) can be found in Supplementary Fig. 3 and Supplementary Data 1, respectively. Individual transcriptome analyses were performed in R for each of 297 manually selected candidate SNPs by considering accessions with identical alleles in that position as biological replicates and contrasting the two variant groups using limma (after converting counts to FPKM values)97 to get differentially expressed genes (Supplementary Data 2; Supplementary Code). Three principle components were incorporated in the design matrix to correct for population structure.
RT-qPCR
Three biological replicates of Arabidopsis thaliana Col-0 and Lp2-2 root segments were sampled after 3 days on SIM following protocol b (~100 mg fresh weight/sample). RNA extraction was done with the Qiagen RNeasy Plant Mini Kit and cDNA synthesis was done with the Promega GoScriptTM system using random hexamer primers. Yield and purity were assessed by nanodrop. WUS expression was normalized to UBC9 and TIP41L levels and a sample maximization strategy was applied for the plate layout (assays were spread over three runs, including two technical replicates, no-RT controls, and triplicate no-template controls). RT-qPCR was set-up with the GoTaq® master mix using primers described in Supplementary Table 1 and run on a Stratagene Mx3005P cycler. Results were analysed according to the ∆∆Ct method using the R package pcr98 (Supplementary Code).
Statistics and reproducibility
For phenotyping, 12 explants were analysed per combination of line (i.e., accession, CSL, or T-DNA insertion mutant) and protocol, divided over two sets of six that were incubated on multi-well plates according to a completely randomized block design. This number was determined by prior experience, feasibility and the use of 24-well plates (with two lines per plate). Several accessions were phenotyped repeatedly to confirm that objective phenotypic values were obtained. Association mapping was performed using a mixed-linear model on square-root-transformed phenotypes to correct for population stratification and data distribution. Count and area data were analysed using two-sided likelihood ratio tests on negative binomial generalized linear models or consecutive global and pairwise nonparametric tests (i.e., two-sided Kruskal–Wallis and Dunn’s tests). Triplicate RT-qPCR data were analysed according to the ∆∆Ct method and ANOVA on a linear model. Details of the statistics for each figure are provided in the corresponding caption.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Hoang Khai Trinh for helping with experiments requested by the referees, Tim De Meyer and Wim Van Criekinge for advice on the computational analyses and Monica Höfte for sharing equipment. This work was supported by the Research Foundation Flanders (Fonds Wetenschappelijk Onderzoek; FWO), project numbers 1S48517N and G094619N.
Author contributions
R.L. designed and performed experiments, analysed and visualized data and wrote the manuscript. D.G. conceptualized research, supervised the project and edited the manuscript. E.W. and J.K. provided resources.
Data availability
The data generated and/or analysed in the current study are either included in this article as Supplementary Data or submitted to public repositories. Raw phenotype data (Fig. 1 and Supplementary Fig. 2) are available from AraPheno at 10.21958/study:8099 and full GWAS data (Figs. 2, 5a–c and Supplementary Fig. 3) are available in easyGWAS at https://easygwas.ethz.ch/gwas/myhistory/public/24/ (accession codes for phenotypic averages underlying these results are AT1P23868, AT1P24025, AT1P24028, AT1P24031, AT1P24043, AT1P24048, AT1P24065, AT1P24068, AT1P24071, AT1P24088, AT1P25078, AT1P26148). Associations for raw phenotypes submitted to AraPheno were also recomputed using a permutation-based pipeline and published in the AraGWAS Catalog (10.21958/gwas:1290, 10.21958/gwas:1283, 10.21958/gwas:1276, 10.21958/gwas:1269, 10.21958/gwas:1288, 10.21958/gwas:1281, 10.21958/gwas:1274, 10.21958/gwas:1267, 10.21958/gwas:1289, 10.21958/gwas:1282, 10.21958/gwas:1275, 10.21958/gwas:1268, 10.21958/gwas:1291, 10.21958/gwas:1284, 10.21958/gwas:1277, 10.21958/gwas:1270, 10.21958/gwas:1294, 10.21958/gwas:1287, 10.21958/gwas:1280, 10.21958/gwas:1273, 10.21958/gwas:1292, 10.21958/gwas:1285, 10.21958/gwas:1278, 10.21958/gwas:1271, 10.21958/gwas:1293, 10.21958/gwas:1286, 10.21958/gwas:1279, 10.21958/gwas:1272). For questions on data availability, contact robin.lardon@ugent.be.
Code availability
Custom code for image processing (ImageJ), handling phenotypic data (R), selection of candidate genes based on association data (python), transcriptome and RT-qPCR analysis (R) is available in the Supplementary Code (plain text).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s42003-020-01274-9.
References
- 1.Perianez-Rodriguez J, Manzano C, Moreno-Risueno MA. Post-embryonic organogenesis and plant regeneration from tissues: two sides of the same coin? Front. Plant Sci. 2014;5:219. doi: 10.3389/fpls.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikeuchi M, Ogawa Y, Iwase A, Sugimoto K. Plant regeneration: cellular origins and molecular mechanisms. Development. 2016;143:1442–1451. doi: 10.1242/dev.134668. [DOI] [PubMed] [Google Scholar]
- 3.Motte H, Vereecke D, Geelen D, Werbrouck S. The molecular path to in vitro shoot regeneration. Biotechnol. Adv. 2014;32:107–121. doi: 10.1016/j.biotechadv.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Pulianmackal AJ, Kareem AVK, Durgaprasad K, Trivedi ZB, Prasad K. Competence and regulatory interactions during regeneration in plants. Front. Plant Sci. 2014;5:142. doi: 10.3389/fpls.2014.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl Acad. Sci. USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atta R, et al. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 2009;57:626–644. doi: 10.1111/j.1365-313X.2008.03715.x. [DOI] [PubMed] [Google Scholar]
- 7.Schaller GE, Bishopp A, Kieber JJ. The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell. 2015;27:44–63. doi: 10.1105/tpc.114.133595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugimoto K, Jiao Y, Meyerowitz EM. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell. 2010;18:463–471. doi: 10.1016/j.devcel.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Che P, Lall S, Nettleton D, Howell SH. Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 2006;141:620–637. doi: 10.1104/pp.106.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kareem A, et al. PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 2015;25:1017–1030. doi: 10.1016/j.cub.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, et al. The WOX11 - LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant Cell Physiol. 2018;59:734–743. doi: 10.1093/pcp/pcy010. [DOI] [PubMed] [Google Scholar]
- 12.Iwase A, et al. WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J. Plant Res. 2015;128:389–397. doi: 10.1007/s10265-015-0714-y. [DOI] [PubMed] [Google Scholar]
- 13.Ikeuchi M, et al. Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 2019;70:377–406. doi: 10.1146/annurev-arplant-050718-100434. [DOI] [PubMed] [Google Scholar]
- 14.Che P, Lall S, Howell SH. Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta. 2007;226:1183–1194. doi: 10.1007/s00425-007-0565-4. [DOI] [PubMed] [Google Scholar]
- 15.Shemer O, Landau U, Candela H, Zemach A, Eshed Williams L. Competency for shoot regeneration from Arabidopsis root explants is regulated by DNA methylation. Plant Sci. 2015;238:251–261. doi: 10.1016/j.plantsci.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 16.He C, Chen X, Huang H, Xu L. Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 2012;8:1–13. doi: 10.1371/journal.pgen.1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishihara H, et al. Primed histone demethylation regulates shoot regenerative competency. Nat. Commun. 2019;10:1786. doi: 10.1038/s41467-019-09386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon SP, et al. Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development. 2007;134:3539–3548. doi: 10.1242/dev.010298. [DOI] [PubMed] [Google Scholar]
- 19.Rosspopoff O, et al. Direct conversion of root primordium into shoot meristem relies on timing of stem cell niche development. Development. 2017;144:1187–1200. doi: 10.1242/dev.142570. [DOI] [PubMed] [Google Scholar]
- 20.Sugimoto K, Gordon SP, Meyerowitz EM. Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 2011;21:212–218. doi: 10.1016/j.tcb.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Zhang T-Q, et al. A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell Online. 2017;29:1073–1087. doi: 10.1105/tpc.16.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoof H, et al. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- 23.Meng WJ, et al. Type-B Arabidopsis ressponse regulators specify the shoot stem cell niche by dual regulation of Wuschel. Plant Cell. 2017;29:1357–1372. doi: 10.1105/tpc.16.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai X, et al. ARR12 promotes de novo shoot regeneration in Arabidopsis thaliana via activation of Wuschel expression. J. Integr. Plant Biol. 2017;59:747–758. doi: 10.1111/jipb.12567. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Zhang H, Dong YX, Hao YJ, Zhang XS. DNA methyltransferase1-mediated shoot regeneration is regulated by cytokinin-induced cell cycle in Arabidopsis. New Phytol. 2018;217:219–232. doi: 10.1111/nph.14814. [DOI] [PubMed] [Google Scholar]
- 26.Iwase A, et al. WIND1 promotes shoot regeneration through transcriptional activation of enhancer of shoot regeneration1 in Arabidopsis. Plant Cell. 2017;29:54–69. doi: 10.1105/tpc.16.00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nameth B, et al. The shoot regeneration capacity of excised Arabidopsis cotyledons is established during the initial hours after injury and is modulated by a complex genetic network of light signalling. Plant. Cell Environ. 2013;36:68–86. doi: 10.1111/j.1365-3040.2012.02554.x. [DOI] [PubMed] [Google Scholar]
- 28.Motte H, et al. Combining linkage and association mapping identifies Receptor-like protein kinase1 as an essential Arabidopsis shoot regeneration gene. Proc. Natl Acad. Sci. USA. 2014;111:8305–8310. doi: 10.1073/pnas.1404978111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wijnen, C. L. et al. A complete chromosome substitution mapping panel reveals genome-wide epistasis in Arabidopsis. bioRxiv10.1101/436154 (2018). [DOI] [PMC free article] [PubMed]
- 31.Zhang H, et al. Thioredoxin-mediated ROS homeostasis explains natural variation in plant regeneration. Plant Physiol. 2018;176:2231–2250. doi: 10.1104/pp.17.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S, I Sergeeva L, Vreugdenhil D. Natural variation of hormone levels in Arabidopsis roots and correlations with complex root architecture. J. Integr. Plant Biol. 2018;60:292–309. doi: 10.1111/jipb.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cary A, Uttamchandani SJ, Smets R, Van Onckelen HA, Howell SH. Arabidopsis mutants with increased organ regeneration in tissue culture are more competent to respond to hormonal signals. Planta. 2001;213:700–707. doi: 10.1007/s004250100565. [DOI] [PubMed] [Google Scholar]
- 34.Marchadier E, et al. The complex genetic architecture of shoot growth natural variation in Arabidopsis thaliana. PLoS Genet. 2019;15:1–27. doi: 10.1371/journal.pgen.1007954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Hause RJ, Borevitz JO, Borevitz JO. Natural genetic variation for growth and development revealed by high-throughput phenotyping in Arabidopsis thaliana. G3. 2012;2:29–34. doi: 10.1534/g3.111.001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma L, et al. Genetic dissection of maize embryonic callus regenerative capacity using multi-locus Genome-Wide Association Studies. Front. Plant Sci. 2018;9:561. doi: 10.3389/fpls.2018.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen THN, et al. Genetic dissection of adventitious shoot regeneration in roses by employing genome-wide association studies. Plant Cell Rep. 2017;36:1493–1505. doi: 10.1007/s00299-017-2170-8. [DOI] [PubMed] [Google Scholar]
- 39.Tuskan GA, et al. Defining the genetic components of callus formation: a GWAS approach. PLoS ONE. 2018;13:e0202519. doi: 10.1371/journal.pone.0202519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, et al. Genome‐wide association study of callus induction variation to explore the callus formation mechanism of rice. J. Integr. Plant Biol. 2019;61:1134–1150. doi: 10.1111/jipb.12759. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, et al. Genetic analysis and identification of a candidate gene associated with in vitro regeneration ability of cucumber. Theor. Appl. Genet. 2018;131:2663–2675. doi: 10.1007/s00122-018-3182-7. [DOI] [PubMed] [Google Scholar]
- 42.Trujillo-Moya C, Gisbert C, Vilanova S, Nuez F. Localization of QTLs for in vitro plant regeneration in tomato. BMC Plant Biol. 2011;11:140. doi: 10.1186/1471-2229-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korte A, Farlow A. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods. 2013;9:29. doi: 10.1186/1746-4811-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawakatsu T, et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell. 2016;166:492–505. doi: 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuo J, Niu Q-W, Frugis G, Chua N-H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002;30:349–359. doi: 10.1046/j.1365-313X.2002.01289.x. [DOI] [PubMed] [Google Scholar]
- 46.Takeda S, et al. Cup-shaped cotyledon1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. 2011;66:1066–1077. doi: 10.1111/j.1365-313X.2011.04571.x. [DOI] [PubMed] [Google Scholar]
- 47.Motte H, et al. Phenyl-Adenine, identified in a light-dependent short HYPOCOTYLS4-assisted chemical screen, is a potent compound for shoot regeneration through the inhibition of cytokinin oxidase/dehydrogenase activity. Plant Physiol. 2013;161:1229–1241. doi: 10.1104/pp.112.210716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strabala TJ, et al. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 2006;140:1331–1344. doi: 10.1104/pp.105.075515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuo N, Mase H, Makino M, Takahashi H, Banno H. Identification of enhancer of shoot regeneration 1-upregulated genes during in vitro shoot regeneration. Plant Biotechnol. 2009;26:385–393. doi: 10.5511/plantbiotechnology.26.385. [DOI] [Google Scholar]
- 50.Cheng Y, et al. Down-regulation of multiple CDK inhibitor ICK/KRP genes promotes cell proliferation, callus induction and plant regeneration in Arabidopsis. Front. Plant Sci. 2015;6:825. doi: 10.3389/fpls.2015.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng Y, et al. Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. Plant J. 2013;75:642–655. doi: 10.1111/tpj.12228. [DOI] [PubMed] [Google Scholar]
- 52.Li X, et al. Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc. Natl Acad. Sci. USA. 2017;114:2765–2770. doi: 10.1073/pnas.1618782114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Magyar Z, et al. The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell. 2005;17:2527–2541. doi: 10.1105/tpc.105.033761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Liu Z, Qiao M, Xiang F. miR393 inhibits in vitro shoot regeneration in Arabidopsis thaliana via repressing TIR1. Plant Sci. 2018;266:1–8. doi: 10.1016/j.plantsci.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 55.Knauer S, et al. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell. 2013;24:125–132. doi: 10.1016/j.devcel.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 56.Qiao M, et al. Proper regeneration from in vitro cultured Arabidopsis thaliana requires the microRNA-directed action of an auxin response factor. Plant J. 2012;71:14–22. doi: 10.1111/j.1365-313X.2012.04944.x. [DOI] [PubMed] [Google Scholar]
- 57.Che P, Gingerich DJ, Lall S, Howell SH. Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell. 2002;14:2771–2785. doi: 10.1105/tpc.006668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ivanchenko MG, et al. The cyclophilin A DIAGEOTROPICA gene affects auxin transport in both root and shoot to control lateral root formation. Development. 2015;142:712–721. doi: 10.1242/dev.113225. [DOI] [PubMed] [Google Scholar]
- 59.Gliwicka M, Nowak K, Balazadeh S, Mueller-Roeber B, Gaj MD. Extensive modulation of the transcription factor transcriptome during somatic embryogenesis in Arabidopsis thaliana. PLoS ONE. 2013;8:e69261. doi: 10.1371/journal.pone.0069261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marin E, et al. miR390, Arabidopsis TAS3 tasiRNAs, and their Auxin response factor targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell. 2010;22:1104–1117. doi: 10.1105/tpc.109.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li W, et al. DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet. 2011;7:e1002243. doi: 10.1371/journal.pgen.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chung Y, et al. Auxin response factors promote organogenesis by chromatin-mediated repression of the pluripotency gene shootmeristemless. Nat. Commun. 2019;10:886. doi: 10.1038/s41467-019-08861-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Che P, Lall S, Howell SH. Acquiring competence for shoot development in Arabidopsis: ARR2 directly targets A-type ARR genes that are differentially activated by CIM preincubation. Plant Signal. Behav. 2008;3:99–101. doi: 10.4161/psb.3.2.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wallmeroth N, et al. ARR22 overexpression can suppress plant two-component regulatory systems. PLoS ONE. 2019;14:e0212056. doi: 10.1371/journal.pone.0212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilson ME, Mixdorf M, Berg RH, Haswell ES. Plastid osmotic stress influences cell differentiation at the plant shoot apex. Development. 2016;143:3382–3393. doi: 10.1242/dev.136234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X, et al. Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 2009;19:224–235. doi: 10.1038/cr.2008.276. [DOI] [PubMed] [Google Scholar]
- 68.Fernandez A, et al. The GLV6/RGF8/CLEL2 peptide regulates early pericycle divisions during lateral root initiation. J. Exp. Bot. 2015;66:5245–5256. doi: 10.1093/jxb/erv329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tamaki H, et al. Identification of novel meristem factors involved in shoot regeneration through the analysis of temperature-sensitive mutants of Arabidopsis. Plant J. 2009;57:1027–1039. doi: 10.1111/j.1365-313X.2008.03750.x. [DOI] [PubMed] [Google Scholar]
- 70.Horstman A, et al. The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 2017;175:848–857. doi: 10.1104/pp.17.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anderson GH, et al. Diversification of genes encoding Mei2-like RNA binding proteins in plants. Plant Mol. Biol. 2004;54:653–670. doi: 10.1023/B:PLAN.0000040819.33383.b6. [DOI] [PubMed] [Google Scholar]
- 72.Frank M, et al. Tumorous shoot development (TSD) genes are required for co-ordinated plant shoot development. Plant J. 2002;29:73–85. doi: 10.1046/j.1365-313x.2002.01197.x. [DOI] [PubMed] [Google Scholar]
- 73.Wang X, et al. TCP transcription factors are critical for the coordinated regulation of Isochorismate synthase 1 expression in Arabidopsis thaliana. Plant J. 2015;82:151–162. doi: 10.1111/tpj.12803. [DOI] [PubMed] [Google Scholar]
- 74.Kim SH, et al. The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 2014;78:978–989. doi: 10.1111/tpj.12527. [DOI] [PubMed] [Google Scholar]
- 75.Aguilar-Martínez JA, Sinha N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 2013;4:406. doi: 10.3389/fpls.2013.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Z, Li B, Shen W-H, Huang H, Dong A. TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 2012;71:99–107. doi: 10.1111/j.1365-313X.2012.04973.x. [DOI] [PubMed] [Google Scholar]
- 77.Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell Online. 2007;19:473–484. doi: 10.1105/tpc.106.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. TCP transcription factors regulate the activities of asymmetric leaves1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell. 2010;22:3574–3588. doi: 10.1105/tpc.110.075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suzuki T, Sakurai K, Ueguchi C, Mizuno T. Two types of putative nuclear factors that physically interact with histidine-containing phosphotransfer (Hpt) domains, signaling mediators in His-to-Asp phosphorelay, in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:37–45. doi: 10.1093/pcp/pce011. [DOI] [PubMed] [Google Scholar]
- 80.Iwase A, et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in arabidopsis. Curr. Biol. 2011;21:508–514. doi: 10.1016/j.cub.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 81.Green KA, Prigge MJ, Katzman RB, Clark SE. CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell Online. 2005;17:691–704. doi: 10.1105/tpc.104.026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Welch D, et al. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 2007;21:2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vaddepalli P, et al. The C2-domain protein QUIRKY and the receptor-like kinase STRUBBELIG localize to plasmodesmata and mediate tissue morphogenesis in Arabidopsis thaliana. Development. 2014;141:4139–4148. doi: 10.1242/dev.113878. [DOI] [PubMed] [Google Scholar]
- 84.Fulton L, et al. DETORQUEO, QUIRKY, and ZERZAUST represent novel components involved in organ development mediated by the receptor-like kinase STRUBBELIG in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000355. doi: 10.1371/journal.pgen.1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwager KM, et al. Characterization of the VIER F-BOX PROTEINE genes from Arabidopsis reveals their importance for plant growth and development. Plant Cell. 2007;19:1163–1178. doi: 10.1105/tpc.105.040675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Prunet N, Yang W, Das P, Meyerowitz EM, Jack TP. SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proc. Natl Acad. Sci. 2017;114:7166–7171. doi: 10.1073/pnas.1705977114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zou H-F, et al. The transcription factor AtDOF4.2 regulates shoot branching and seed coat formation in Arabidopsis. Biochem. J. 2013;449:373–388. doi: 10.1042/BJ20110060. [DOI] [PubMed] [Google Scholar]
- 88.Jung B, et al. Uridine-ribohydrolase is a key regulator in the uridine degradation pathway of Arabidopsis. Plant Cell. 2009;21:876–891. doi: 10.1105/tpc.108.062612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.SUN J, et al. Arabidopsis SOI33/AtENT8 gene encodes a putative equilibrative nucleoside transporter that is involved in cytokinin transport in planta. J. Integr. Plant Biol. 2005;47:588–603. doi: 10.1111/j.1744-7909.2005.00104.x. [DOI] [Google Scholar]
- 90.Hu B, et al. Epigenetic control of Pollen Ole e 1 allergen and extensin family gene expression in Arabidopsis thaliana. Acta Physiol. Plant. 2014;36:2203–2209. doi: 10.1007/s11738-014-1597-6. [DOI] [Google Scholar]
- 91.Sakai T, et al. The WAVY GROWTH 3 E3 ligase family controls the gravitropic response in Arabidopsis roots. Plant J. 2012;70:303–314. doi: 10.1111/j.1365-313X.2011.04870.x. [DOI] [PubMed] [Google Scholar]
- 92.Zhou M, Chen H, Wei D, Ma H, Lin J. Arabidopsis CBF3 and DELLAs positively regulate each other in response to low temperature. Sci. Rep. 2017;7:39819. doi: 10.1038/srep39819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamagami T, et al. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 2003;278:49102–49112. doi: 10.1074/jbc.M308297200. [DOI] [PubMed] [Google Scholar]
- 94.‘The 1001 Genomes Consortium’. 1135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016;166:481–491. doi: 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pisupati R, et al. Verification of Arabidopsis stock collections using SNPmatch, a tool for genotyping high-plexed samples. Sci. Data. 2017;4:1–9. doi: 10.1038/sdata.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grimm DG, et al. easyGWAS: a cloud-based platform for comparing the results of genome-wide association studies. Plant Cell. 2017;29:5–19. doi: 10.1105/tpc.16.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahmed M, Kim DR. pcr: an R package for quality assessment, analysis and testing of qPCR data. PeerJ. 2018;6:e4473. doi: 10.7717/peerj.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lardon, R. Genetic dissection of shoot regeneration from root explants in Arabidopsis. 10.21958/study:80 (2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data generated and/or analysed in the current study are either included in this article as Supplementary Data or submitted to public repositories. Raw phenotype data (Fig. 1 and Supplementary Fig. 2) are available from AraPheno at 10.21958/study:8099 and full GWAS data (Figs. 2, 5a–c and Supplementary Fig. 3) are available in easyGWAS at https://easygwas.ethz.ch/gwas/myhistory/public/24/ (accession codes for phenotypic averages underlying these results are AT1P23868, AT1P24025, AT1P24028, AT1P24031, AT1P24043, AT1P24048, AT1P24065, AT1P24068, AT1P24071, AT1P24088, AT1P25078, AT1P26148). Associations for raw phenotypes submitted to AraPheno were also recomputed using a permutation-based pipeline and published in the AraGWAS Catalog (10.21958/gwas:1290, 10.21958/gwas:1283, 10.21958/gwas:1276, 10.21958/gwas:1269, 10.21958/gwas:1288, 10.21958/gwas:1281, 10.21958/gwas:1274, 10.21958/gwas:1267, 10.21958/gwas:1289, 10.21958/gwas:1282, 10.21958/gwas:1275, 10.21958/gwas:1268, 10.21958/gwas:1291, 10.21958/gwas:1284, 10.21958/gwas:1277, 10.21958/gwas:1270, 10.21958/gwas:1294, 10.21958/gwas:1287, 10.21958/gwas:1280, 10.21958/gwas:1273, 10.21958/gwas:1292, 10.21958/gwas:1285, 10.21958/gwas:1278, 10.21958/gwas:1271, 10.21958/gwas:1293, 10.21958/gwas:1286, 10.21958/gwas:1279, 10.21958/gwas:1272). For questions on data availability, contact robin.lardon@ugent.be.
Custom code for image processing (ImageJ), handling phenotypic data (R), selection of candidate genes based on association data (python), transcriptome and RT-qPCR analysis (R) is available in the Supplementary Code (plain text).