Abstract
STUDY QUESTION
What are the trends and developments in preimplantation genetic testing (PGT) in 2013–2015 as compared to previous years?
SUMMARY ANSWER
The main trends observed in the retrospective data collections 2013–2015, representing valuable data on PGT activity in (mainly) Europe, are the increased application of trophectoderm biopsy at the cost of cleavage stage biopsy and the continuing expansion of comprehensive testing technology in PGT for chromosomal structural rearrangements and for aneuploidies (PGT-SR and PGT-A).
WHAT IS KNOWN ALREADY
Since it was established in 1997, the ESHRE PGT Consortium has been collecting data from international PGT centres. To date, 15 data sets and an overview of the first 10 years of data collections have been published.
STUDY DESIGN, SIZE, DURATION
Collection of (mainly) European data by the PGT Consortium for ESHRE. The data for PGT cycles performed between 1 January 2013 and 31 December 2015 were provided by participating centres on a voluntary basis. For the collection of cycle, pregnancy and baby data, separate, pre-designed MS Excel tables were used.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Data were submitted by 59, 60 and 59 centres respectively for 2013, 2014 and 2015 (full PGT Consortium members). Records with incomplete or inconsistent data were excluded from the calculations. Corrections, calculations, figures and tables were made by expert co-authors.
MAIN RESULTS AND THE ROLE OF CHANCE
For data collection XVI/XVII/XVIII, 59/60/59 centres reported data on 8164/9769/11 120 cycles with oocyte retrieval: 5020/6278/7155 cycles for PGT-A, 2026/2243/2661 cycles for PGT for monogenic/single gene defects, 1039/1189/1231 cycles for PGT-SR and 79/59/73 cycles for sexing for X-linked diseases. From 2013 until 2015, the uptake of biopsy at the blastocyst stage was mainly observed in cycles for PGT-A (from 23% to 36%) and PGT-SR (from 22% to 36%), alongside the increased application of comprehensive testing technology (from 66% to 75% in PGT-A and from 36% to 58% in PGT-SR).
LIMITATIONS, REASONS FOR CAUTION
The findings apply to the 59/60/59 participating centres and may not represent worldwide trends in PGT. Data were collected retrospectively and no details of the follow-up on PGT pregnancies and babies born were provided.
WIDER IMPLICATIONS OF THE FINDINGS
Being the largest data collection on PGT worldwide, detailed information about ongoing developments in the field is provided.
STUDY FUNDING/COMPETING INTEREST(S)
The study has no external funding and all costs are covered by ESHRE. There are no competing interests declared.
TRIAL REGISTRATION NUMBER
N/A.
Keywords: PGT, PGD, structural rearrangements, monogenic disorders, aneuploidy, human embryo, registry, data collection, preimplantation genetic testing
Introduction
The ESHRE PGT Consortium was established in 1997: its main objectives are to collect prospective and retrospective data, produce consensus guidelines for preimplantation genetic testing (PGT) laboratories and to promote best practice.
Recently, the PGT consortium in collaboration with the ESHRE Special Interest Group Embryology issued four consensus papers on Recommendations for good practice in Preimplantation Genetic Testing (ESHRE PGT Consortium Steering Committee, 2020; ESHRE PGT Consortium and SIG-Embryology Biopsy Working Group, 2020; ESHRE PGT-M Working Group, 2020; ESHRE PGT-SR/PGT-A Working Group, 2020).
To date, 15 data collections have been published, covering all aspects of PGT (Geraedts et al., 1999, 2000; ESHRE PGD Consortium Steering Committee, 2002; Sermon et al., 2005, 2007; Harper et al., 2006, 2008, 2010a,b; Goossens et al., 2008, 2009, 2012; Moutou et al., 2014; De Rycke et al., 2015, 2017). A 10-year overview of PGT was published in 2012 (Harper et al., 2012). No other data of this magnitude exists in the literature.
As of 2017, the International Committee for Monitoring Assisted Reproductive Technologies (ICMART) revised terms and definitions were adopted. They comprise PGT for monogenic/single gene defects (PGT-M) for monogenic diseases, PGT for chromosomal structural rearrangements (PGT-SR) for chromosomal structural rearrangements and PGT for aneuploidies (PGT-A) for aneuploidies (Zegers-Hochschild et al., 2017). Previous Consortium data collections included a separate file for PGT cycles performed for sexing for X-linked diseases and it was decided to keep this format. Traditionally, PGT cycles for chromosomal numerical aberrations of high genetic risk have been included in what currently is the PGT-SR category, and this structure was maintained as well.
This article will present summary data from PGT cycles performed between 1 January 2013 and 31 December 2015 in terms of overall trends, highlighting how the practice of PGT has evolved over the collection period. As such, these data collections have a different format compared to that of previous years. Due to a huge increase in the number of PGT cycles reported to the ESHRE PGT Consortium each year, alongside the ever growing complexity of the PGT treatments, it has become extremely difficult to mine the data and produce accurate tables. The FileMakerPro database that has been used up until data collection XV, has proven to be no longer fit for purpose and efforts have been undertaken to create a new database that will meet current needs and allows centres to register and analyse their PGT data in real time. However, the data collection rejuvenation project has faced significant setbacks causing serious delay for the data collections of the following years. At the same time, new techniques in both the IVF and genetic laboratories were being developed and implemented, changing the PGT landscape and the way PGT cycles were being managed. In an effort to record this transition period, members of the ESHRE PGT Consortium were asked to submit summary data on PGT cycles performed in the years 2013–2015. Concurrently, a questionnaire was sent to ESHRE PGT Consortium members to try and make an inventory of the uptake of innovations in ART and genetic diagnostic technologies in PGT practice.
The present manuscript is based on these data.
Materials and methods
The report includes PGT treatments started between 1 January 2013 and 31 December 2015, and covers data on PGT indication, biopsy method, diagnostic technology, efficiency of the different procedures and clinical PGT outcome in terms of positive hCG and positive heartbeat.
Summary data on PGT cycles for 2013, 2014 and 2015 were provided by 59/60/59 Consortium members, respectively, the majority of which are based in European countries. Data cover the following treatment modalities: PGT-A, PGT-M, PGT-SR and PGT for sexing for X-linked diseases.
For the collection of the data, representatives of the participating PGT centres were asked to anonymize their data, import them in pre-designed MS Excel tables and send these to the ESHRE Central Office Science Manager. Initial curation of the submitted data allowed the identification of omissions and any ambivalent data entries. If data were incomplete or inconsistencies were detected, the ESHRE Science Manager contacted the centres’ representative for clarification. Any remaining records with incomplete or inconsistent data were excluded from further processing. Data from all participating centres were subsequently collated and processed by expert members of the ESHRE PGT Consortium Steering Committee for in-depth analysis.
The terminology used in this report was based on the glossary of ICMART (Zegers-Hochschild et al., 2017). A clinical pregnancy was defined as a pregnancy diagnosed by ultrasonographic visualization of one or more gestational sacs or definitive clinical signs of pregnancy.
To study additional trends in the management and workflow of PGT cycles, two questionnaires were sent to ESHRE PGT Consortium members. The first at the end of 2013 addressing cycles carried out between 1 January and 1 October 2013 and the second 2 years later, addressing a comparable period.
Results
This report includes data from 59 centres, presented in Tables I–IV and Figs 1–6. The nature of the summary data provided by the ESHRE PGT Consortium members for 2013–2015 does not allow the calculation of cumulative data for collections I–XVIII.
Table I.
Overview of overall PGT treatment cycles in 2013–2015.
| PGT-overall |
PGT-A |
PGT-M |
PGT-SR |
PGT for sexing for X-linked diseases |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2013 | 2014 | 2015 | 2013 | 2014 | 2015 | 2013 | 2014 | 2015 | 2013 | 2014 | 2015 | 2013 | 2014 | 2015 | |
| Cycles to OR | 8164 | 9769 | 11 120 | 5020 | 6278 | 7155 | 2026 | 2243 | 2661 | 1039 | 1189 | 1231 | 79 | 59 | 73 |
| ART method | 8164 | 9758 | 11 114 | 5020 | 6277 | 7155 | 2026 | 2234 | 2655 | 1039 | 1189 | 1231 | 79 | 58 | 73 |
| IVF | 280 (3) | 198 (2) | 143 (1) | 166 (3) | 117 (2) | 95 (1) | 0 | 0 | 0 | 94 (9) | 72 (6) | 39 (3) | 20 (25) | 9 (16) | 9 (12) |
| ICSI | 7213 (88) | 9133 (94) | 10 449 (94) | 4515 (90) | 6079 (97) | 6948 (97) | 1802 (89) | 1943 (87) | 2356 (89) | 856 (82) | 1068 (90) | 1093 (89) | 40 (51) | 43 (74) | 52 (71) |
| IVF + ICSI | 246 (3) | 335 (3) | 371 (3) | 0 | 0 | 0 | 206 (10) | 291 (13) | 299 (11) | 29 (3) | 38 (3) | 60 (5) | 11 (14) | 6 (10) | 12 (16) |
| Unknown | 425 (5) | 92 (1) | 151 (1) | 339 (7) | 81 (1) | 112 (2) | 18 (1) | 0 | 0 | 60 (6) | 11 (1) | 39 (3) | 8 (10) | 0 | 0 |
| Cycles to PGT | 7800 | 9159 | 10 292 | 4780 | 5819 | 6512 | 1952 | 2143 | 2524 | 989 | 1141 | 1187 | 79 | 56 | 69 |
| FISH only | 2087 (27) | 1696 (19) | 1380 (13) | 1400 (29) | 1088 (19) | 872 (13) | 0 | 1 (<1) | 3 (<1) | 627 (63) | 568 (50) | 464 (39) | 60 (76) | 39 (70) | 41 (59) |
| PCR only | 1814 (23) | 1885 (21) | 2215 (22) | 16 (<1) | 0 | 4 (<1) | 1775 (91) | 1872 (87) | 2194 (87) | 7 (1) | 4 (<1) | 10 (1) | 16 (20) | 9 (16) | 7 (10) |
| qPCR | 222 (3) | 584 (6) | 783 (8) | 219 (5) | 463 (8) | 728 (11) | 0 | 111 (5) | 25 (1) | 3 (<1) | 10 (1) | 30 (3) | 0 | 0 | 0 |
| WGA | |||||||||||||||
| + PCR | 123 (2) | 89 (1) | 106 (1) | 0 | 8 | 10 (<1) | 123 (6) | 80 (4) | 96 (4) | 0 | 1 (<1) | 0 | 0 | 0 | 0 |
| + qPCR | 0 | 0 | 3 (<1) | 0 | 0 | 0 | 0 | 0 | 3 (<1) | 0 | 0 | 0 | 0 | 0 | 0 |
| + SNP Array | 2 (<1) | 32 (<1) | 121 (1) | 2 (<1) | 2 (<1) | 0 | 0 | 16 (1) | 96 (4) | 0 | 14 (1) | 25 (2) | 0 | 0 | 0 |
| + CGH Array | 3513 (45) | 4718 (52) | 5061 (49) | 3104 (65) | 4127 (71) | 4322 (66) | 54 (3) | 63 (3) | 100 (4) | 352 (36) | 520 (46) | 618 (52) | 3 (4) | 8 (14) | 21 (30) |
| + NGS | 39 (1) | 155 (2) | 623 (6) | 39 (1) | 131 (2) | 576 (9) | 0 | 0 | 7 | 0 | 24 (2) | 40 (3) | 0 | 0 | 0 |
| Biopsy method | 7800 | 9141 | 10 292 | 4780 | 5819 | 6512 | 1952 | 2143 | 2522 | 989 | 1123 | 1187 | 79 | 56 | 71 |
| Polar body | 771 (10) | 657 (7) | 626 (6) | 687 (14) | 593 (10) | 547 (8) | 63 (3) | 53 (2) | 64 (3) | 21 (2) | 11 (1) | 15 (1) | 0 | 0 | 0 |
| Cleavage stage | 5423 (70) | 5935 (65) | 6007 (58) | 2865 (60) | 3229 (55) | 3057 (47) | 1739 (89) | 1834 (86) | 2149 (85) | 745 (75) | 818 (73) | 743 (63) | 74 (94) | 54 (96) | 58 (82) |
| Morula day4 | 154 (2) | 184 (20 | 591 (6) | 145 (3) | 184 (3) | 591 (9) | 0 | 0 | 0 | 9 (1) | 0 | 0 | 0 | 0 | 0 |
| Blastocyst | 1452 (19) | 2365 (26) | 3068 (30) | 1083 (23) | 1813 (31) | 2317 (36) | 150 (8) | 256 (12) | 309 (12) | 214 (22) | 294 (26) | 429 (36) | 5 (6) | 2 (4) | 13 (18) |
| Embryology | |||||||||||||||
| Successfully biopsied | 39 492 | 46 534 | 49 505 | 22 741 | 27 612 | 29 217 | 11 588 | 12 651 | 14 592 | 4748 | 5998 | 5310 | 415 | 273 | 386 |
| Diagnosed | 36 922 (93) | 43 037 (92) | 43 562 (88) | 21 760 (96) | 26 212 (95) | 25 198 (86) | 10 327 (89) | 11 129 (88) | 13 053 (90) | 4480 (94) | 5450 (91) | 4953 (93) | 355 (86) | 246 (90) | 358 (93) |
| Transferable | 12 118 (33) | 14 153 (33) | 15 352 (35) | 6152 (28) | 7799 (30) | 7934 (31) | 4594 (44) | 4832 (43) | 5916 (45) | 1203 (27) | 1410 (26) | 1378 (28) | 169 (48) | 112 (46) | 124 (35) |
| Transferred | 6997 (58) | 7568 (53) | 6969 (45) | 4171 (68) | 4706 (60) | 3875 (489) | 1966 (42) | 1974 (41) | 2303 (39) | 786 (65) | 834 (59) | 742 (54) | 74 (44) | 54 (48) | 49 (40) |
| Clinical outcome | |||||||||||||||
| Cycles to ET | 5052 | 5617 | 5641 | 2892 | 3392 | 3295 | 1456 | 1560 | 1718 | 652 | 632 | 591 | 52 | 33 | 37 |
| hCG positive | 2340 | 2750 | 2841 | 1440 | 1819 | 1757 | 594 | 654 | 823 | 280 | 265 | 252 | 26 | 12 | 9 |
| Positive heart beat | 2104 | 2418 | 2219 | 1354 | 1680 | 1443 | 496 | 514 | 576 | 234 | 215 | 193 | 20 | 9 | 7 |
Data are presented as number (%). % are calculated per total of ART method, cycles to PGT or biopsy method. In the embryology section, % are calculated per total of the row above.
CGH, comparative genomic hybridization; ET, embryo transfer; NGS, next-generation sequencing; OR, oocyte retrieval; PGT, preimplantation genetic testing; PGT-A, PGT for aneuploidies; PGT-M, PGT for monogenic/single gene defects; PGT-SR, PGT for chromosomal structural rearrangements; qPCR, quantitative PCR; SNP, single-nucleotide polymorphism; WGA, whole genome amplification.
Table IV.
Overview of indications for PGT-SR in 2013–2015.
| 2013 | 2014 | 2015 | |
|---|---|---|---|
| Robertsonian translocation, male carrier | 178 (17) | 225 (19) | 166 (13) |
| Robertsonian translocation, female carrier | 74 (7) | 108 (9) | 127 (10) |
| Reciprocal translocation, male carrier | 350 (34) | 352 (30) | 371 (30) |
| Reciprocal translocation, female carrier | 290 (28) | 332 (28) | 393 (32) |
| Deletion | 38 (4) | 29 (2) | 21 (2) |
| Inversion | 60 (6) | 74 (6) | 82 (7) |
| Other | 30 (3) | 54 (5) | 71 (6) |
| Unknown | 19 (2) | 15 (1) | 0 |
| Total | 1039 | 1189 | 1231 |
Data are given as number (%).
Figure 1.
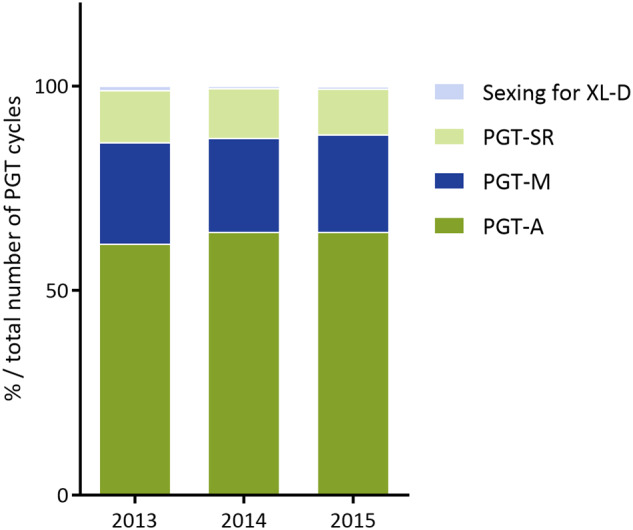
Distribution of PGT indications in 2013–2015. In 2013, preimplantation genetic testing for aneuploidies (PGT-A), monogenic/single gene defects (PGT-M), chromosomal structural rearrangements (PGT-SR) and PGT for X-linked diseases (XL-D) accounted for 61%, 25%, 13% and 1% of total number of PGT cycles, respectively. In 2014, PGT-A, PGT-M, PGT-SR and PGT for XL-D accounted for 64%, 23%, 12% and <1% of total number of PGT cycles, respectively. In 2015, PGT-A, PGT-M, PGT-SR and PGT for XL-D accounted for 64%, 24%, 11% and <1% of total number of PGT cycles, respectively.
Figure 2.
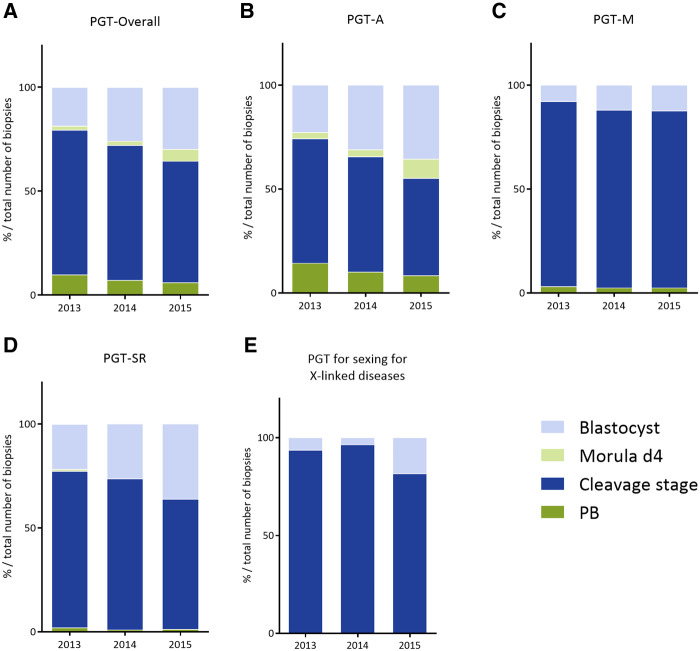
Distribution of biopsy stage in 2013–2015. (A) Overall, (B) PGT-A, (C) PGT-M, (D) PGT-SR, (E) PGT for sexing for X-linked diseases. PB, polar body, d4, day 4. Exact numbers and percentages can be found in Table I.
Figure 3.
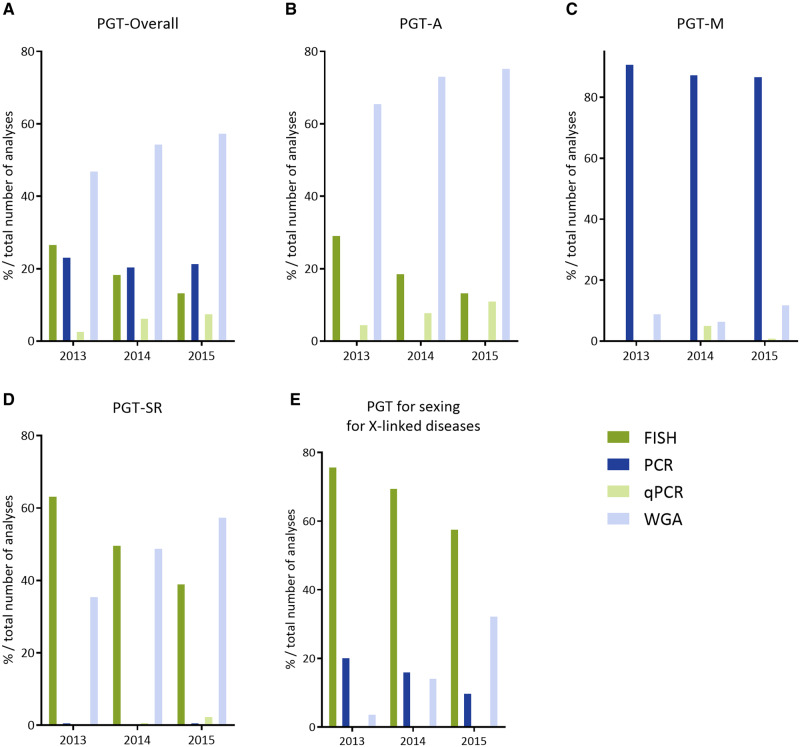
Method of PGT analysis in 2013–2015. (A) Overall, (B) PGT-A, (C) PGT-M, (D) PGT-SR, (E) PGT for sexing for X-linked diseases. qPCR, quantitative PCR; WGA, whole genome amplification. Exact numbers and percentages can be found in Table I.
Figure 4.
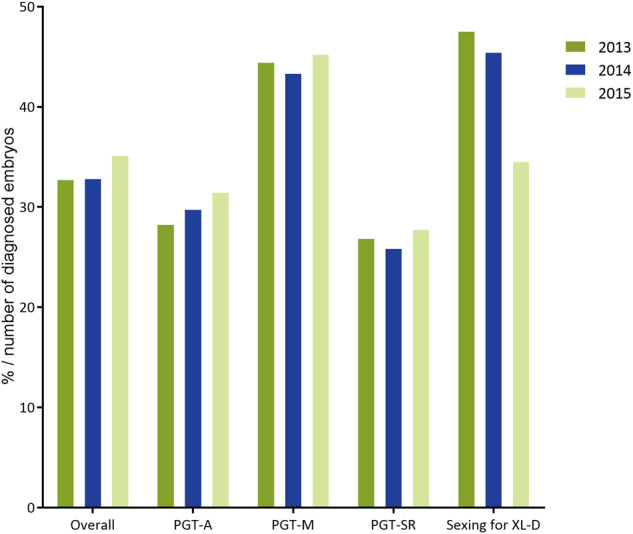
Percentage of genetically transferable embryos per number of embryos diagnosed in 2013–2015. Exact numbers and percentages can be found in Table I.
Figure 5.
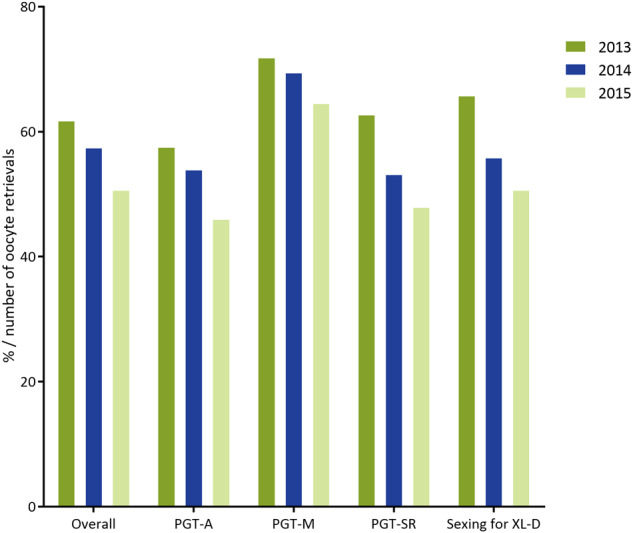
Percentage of cycles to embryo transfer per number of oocyte retrievals in 2013–2015. Exact numbers can be found in Table I.
Figure 6.
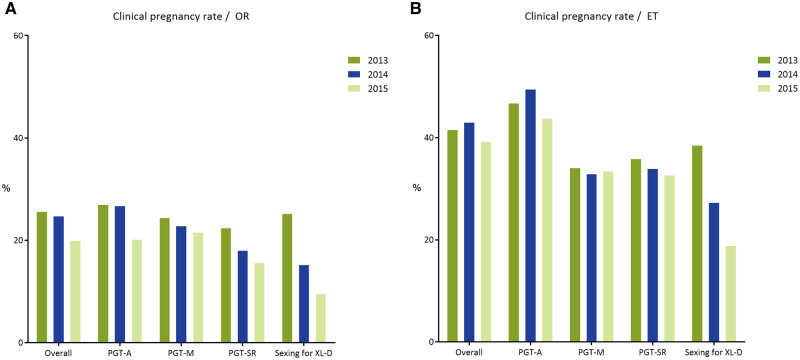
Clinical pregnancy rate per oocyte retrieval (A) and per embryo transfer (B) in 2013–2015. ET, embryo transfer. Exact numbers can be found in Table I.
Table I provides an overview of the current data collection and covers a total of 8164 cycles in 2013, 9769 cycles in 2014 and 11 120 cycles in 2015, indicating a continuing expansion of treatment numbers in the participating centres. As seen in Fig. 1, PGT-A cycles account for the majority of PGT treatments and distribution among the various PGT indication categories has remained stable over the data collection period.
For all PGT indications, ICSI was the method most often used for fertilization. It was used almost exclusively in PGT-M (either for all available oocytes or a proportion thereof). It was used the least in PGT for sexing for X-linked diseases (Table I). These figures do not differ significantly from the ones reported in the previous data collection (80% in PGT-SR, 99% in PGT-M, 80% in PGT-A and 66% in X-linked disorders).
For the total number of cycles to biopsy in 2013, 2014 and 2015, cleavage-stage biopsy was still the most common method applied (70%, 65% and 58%, respectively). However, as shown in Fig. 2 and Table I, remarkable differences were noted for the individual PGT indication categories. In PGT-M and PGT for sexing for X-linked diseases, cleavage stage biopsy was performed for the vast majority of cases during the whole collection period, whereas in PGT-SR and PGT-A, the percentage of cycles performed with blastocyst biopsy steadily increased over the years and amounted to 36% in the data collection of 2015.
PGT-A
Supplementary Table SI summarizes the 5020, 6278 and 7155 PGT-A cycles to oocyte retrieval (OR) per indication in 2013, 2014 and 2015, respectively. Table II shows the different indications for the PGT-A cycles performed. Advanced maternal age (AMA) is by far the most common indication for PGT-A (44%, 47% and 51% in 2013, 2014 and 2015, respectively), followed by repeated implantation failure (RIF) in combination with AMA and recurrent miscarriage (RM) in combination with AMA. Other indications were RIF and RM as a single indication, severe male factor, previous abnormal pregnancies and carrier status of abnormal karyotype. A small number of PGT-A cycles was performed as an integrated part of an oocyte donation treatment and a growing number (compared to previous data collections) of PGT-A cycles was performed without a reported medical indication.
Table II.
Overview of indications for PGT-A in 2013–2015.
| 2013 | 2014 | 2015 | |
|---|---|---|---|
| AMA | 2189 (43.6) | 2936 (46.8) | 3636 (50.8) |
| AMA + RM | 561 (11.2) | 503 (8.0) | 563 (7.9) |
| AMA + RIF | 550 (11.0) | 673 (10.7) | 702 (9.8) |
| RM | 346 (6.9) | 394 (6.3) | 376 (5.3) |
| RIF | 361 (7.2) | 358 (5.7) | 416 (5.8) |
| SMF | 298 (5.9) | 436 (6.9) | 395 (5.5) |
| Previous abnormal pregnancies | 91 (1.8) | 122 (1.9) | 102 (1.4) |
| Carrier status of abnormal karyotype | 98 (2.0) | 90 (1.4) | 65 (0.9) |
| Ovum donation | 85 (1.7) | 101 (1.6) | 131 (1.8) |
| No indication | 327 (6.5) | 562 (9.0) | 372 (5.2) |
| Other | 114 (2.3) | 103 (1.6) | 397 (5.5) |
| Total | 5020 (100.0) | 6278 (100.0) | 7155 (100.0) |
Data are given as number (%).
AMA, advanced maternal age; RIF, repeated implantation failure; RM, recurrent miscarriage; SMF, severe male factor.
In 90–97% of all cycles to OR, ICSI was used for fertilization (Table I), a figure that is similar to the one from previous data sets. The vast majority of cycles to OR (≥91%) had embryos available for biopsy. The majority of biopsies were still performed at cleavage stage, with blastocyst biopsy advancing and accounting for 36% of all PGT-A cycles in 2015 (Fig. 2B).
FISH as the genetic analysis was greatly outnumbered by quantitative PCR and comprehensive, whole genome amplification (WGA)-based methodologies, the latter of which amounted to 75% of cases in 2015 (Fig. 3B).
Of the embryos that were successfully biopsied, 96% in 2013 and 95% in 2014 gave a diagnosis. For unknown reasons, this figure was remarkably lower in 2015 (86%) (Table I). In general, about 30% of diagnosed embryos were genetically transferable (Fig. 4) and between 50% and 68% of these were actually transferred. As the percentage of transferred embryos was lower compared to previous data collections and decreasing over the years, this parameter might be confounded by the fact that some participants only recorded fresh embryo transfers whereas trophectoderm (TE) biopsy in combination with comprehensive diagnosis usually requires cryopreservation of embryos and transfer in a subsequent frozen cycle.
From the 3-year total of 18 453 cycles to OR, only 52% resulted in an embryo transfer procedure (Fig. 5). This is lower than the mean percentage reported in previous data sets (62%), which again might be explained by the missing frozen embryo transfers.
A positive hCG was obtained in 5016 cycles (3-year total), with a positive heartbeat in 4477 cycles (24% per OR and 47% per embryo transfer) (Fig. 6). Historically, the clinical pregnancy rate is stable at around 20% per OR and 30% per embryo transfer procedure. Not surprisingly, the poorest outcome was obtained for AMA + RIF, with a 20% clinical pregnancy rate per OR and 41% per embryo transfer (Supplementary Table SI).
No parameters further downstream (i.e. implantation rate, delivery rate, miscarriage rate) could be calculated because of too many missing data.
PGT-M
Supplementary Table SII summarizes the 2026, 2243 and 2661 PGT-M cycles to OR per indication in 2013, 2014 and 2015, respectively. Half of all the PGT-M cycles was performed for an autosomal dominant disease, followed by an autosomal recessive indication, which accounted for 26% of PGT-M cycles (Table III).
Table III.
Overview of indications for PGT-M in 2013–2015.
| 2013 | 2014 | 2015 | |
|---|---|---|---|
| Autosomal recessive | 552 (27) | 564 (25) | 659 (25) |
| Autosomal dominant | 981 (48) | 1189 (53) | 1339 (50) |
| X-linked (no sexing) | 319 (16) | 330 (15) | 374 (14) |
| HLA only | 16 (1) | 7 (0) | 15 (1) |
| HLA + monogenic disorder | 99 (5) | 101 (5) | 70 (3) |
| Other | 59 (3) | 38 (2) | 204 (8) |
| Other PGD + PGS | 0 (0) | 14 (1) | 0 (0) |
| Total | 2026 | 2243 | 2661 |
Data are given as number (%).
PGS, preimplantation genetic screening.
In all cycles to OR, ICSI was used for fertilization (for all or part of the oocytes), a figure that is similar to the one from previous data sets. Nearly all cycles to OR (96%) had embryos available for biopsy, which was most commonly applied to cleavage-stage embryos (86%) (Fig. 2C) and PCR still being the most widely used first-line method of DNA amplification (88%) (Fig. 3C). The uptake of blastocyst biopsy remained low compared to other indications (8% in 2013 and 12% in 2015), as did the use of comprehensive diagnostic technology (9% in 2013 and 12% in 2015).
Of the embryos successfully biopsied, 88–90% gave a diagnostic result, of which on average 44% was genetically transferable (Fig. 4). As was to be expected, the lowest chance of a transferable embryo was found in the HLA and HLA plus monogenic disorder group (Supplementary Table SII). Of all transferable embryos, between 39% and 42% were actually transferred, either in a fresh or a frozen cycle.
From the 3-year total of 6930 cycles to OR, 68% resulted in an embryo transfer procedure, the highest proportion among the different PGT categories. This is slightly higher than the mean percentage reported in previous data sets (64%).
A positive hCG was obtained in 2071 cycles (3-year total), with a positive heartbeat in 1586 cycles (23% per OR and 34% per embryo transfer) (Fig. 6). Historically, the clinical pregnancy rate is stable at around 24% per OR and 30% per embryo transfer procedure.
No parameters further downstream (i.e. implantation rate, delivery rate, miscarriage rate) could be calculated because of too many missing data.
PGT-SR
Supplementary Table SIII summarizes the 1039, 1189 and 1231 PGT-SR cycles to OR per indication in 2013, 2014 and 2015, respectively.
As for previous years, data from 2013 to 2015 showed that PGT for reciprocal translocations was performed more often than for any other type of structural rearrangement (62%, 58% and 62%, respectively, Table IV). For reciprocal translocations, the number of cycles performed for female carriers was very similar to that for male carriers, whereas for Robertsonian translocations (24%, 28% and 23% of total), the number of cycles performed for male carriers was roughly 2-fold that of female carriers.
In 82–90% of all cycles to OR, ICSI was used for fertilization, a figure that is similar to the one from previous data sets. Nearly all cycles to OR (≥95%) had embryos available for biopsy, which was most commonly applied to cleavage-stage embryos (75% in 2013, 73% in 2014 and 63% in 2015). Biopsy at the blastocyst stage gained ground and increased from 22% in 2013 to 36% in 2015 (Fig. 2D). The downward trend in the use of FISH as diagnostic methodology continued and accounted for only 39% of cycles to analysis in data collection 2015. Comprehensive, WGA-based technology steadily increased its share from 36% to 58% between 2013 and 2015 (Fig. 3D).
Of the embryos successfully biopsied, 91–94% gave a diagnostic result, of which only one out of four was genetically transferable (Fig. 4). This figure has remained stable over the years. As was to be expected, the lowest percentage of transferable embryos was found in the reciprocal translocation group (on average 23% for male and 20% for female carriers). Of all transferable embryos, between 54% and 65% were actually transferred, either in a fresh or a frozen cycle.
From the 3-year total of 3459 cycles to OR, only 54% resulted in an embryo transfer procedure (Fig. 5). This is lower than the mean percentage reported in previous data sets (64%).
A positive hCG was obtained in 797 cycles (3-year total), with a positive heartbeat in 642 cycles (19% per OR and 34% per embryo transfer) (Fig. 6). Historically, the clinical pregnancy rate is stable at around 18% per OR and 28% per embryo transfer procedure.
The poorest outcome, 13% positive heartbeat per OR, was found in the group of deletion carriers. This is linked with the lowest percentage of transferable embryos available for this group.
No parameters further downstream (i.e. implantation rate, delivery rate, miscarriage rate) could be calculated because of too many missing data.
PGT cycles for sexing for X-linked diseases
Supplementary Table SIV summarizes the 79, 59 and 73 PGT-sexing for X-linked diseases cycles to OR in 2013, 2014 and 2015, respectively.
As holds true for the other PGT categories, the majority of cycles for X-linked diseases was performed with ICSI (64%) (Table I). Biopsy was primarily performed at cleavage-stage (82% in 2015, Fig. 2E) and FISH, although declining, was still the most frequently used method (59% in 2015, Fig. 3E); PCR was applied in 10% and WGA/array comparative genomic hybridization (CGH) showed a steep increase in 2015 and was applied in 30% of cycles.
Of the embryos successfully biopsied, 89% gave a diagnostic result, of which 42% were transferable. Half of the cycles to OR resulted in an embryo transfer. A positive hCG was obtained in 47 cycles, with a positive heartbeat in 36 cycles, yielding a clinical pregnancy rate of 17% per OR and 30% per embryo transfer, comparable to previous data sets (20% per OR and 30% per embryo transfer).
No parameters further downstream (i.e. implantation rate, delivery rate, miscarriage rate) could be calculated because of too many missing data.
Pregnancies and babies
The pre-designed Excel tables included parameters on pregnancies and babies, but as there was too many missing data, results no longer reflected reality. From previous data collections it was known that many centres have difficulties with collecting these data and they are often missing or inconsistent. However, follow-up of the children born after PGT remains of great importance.
Misdiagnoses
No misdiagnoses were reported in the current dataset.
Trends in the management and workflow of PGT cycles
Forty PGT centres participated in both questionnaires, reporting on practices and technologies applied in PGT. Apart from the previously described trends that also emerged from the data collections, the following parameters were noteworthy: numerous intra- and inter-departmental set-ups for the collaborative provision of ART and genetic diagnosis required for a PGT service were identified. Many centres had achieved accreditation by 2015: 71% of IVF units and 68% of diagnostic units. The use of time-lapse technology in PGT increased from 21% of PGT cycles in 2013 to 25% in 2015. Oocyte and embryo vitrification before biopsy showed a decline from 7% to 2% and from 12% to 3% of PGT cycles, respectively. By 2015, the use of Acidic Tyrode’s solution for zona breaching was almost fully replaced by laser-assisted zona drilling, the latter practice covering 98% of all PGT cycles. A switch in embryo transfer strategy from classic ‘fresh embryo transfer’ towards a ‘biopsy and freeze all’ strategy was observed concurrent with the increased uptake of TE biopsy.
Discussion
This data report of the ESHRE PGT Consortium involves summary data from three consecutive calendar years, 2013–2015. The most time-consuming step in data processing involves data ‘cleaning’: inconsistent and missing data need to be followed up by (multiple rounds of) contact between the relevant centre and ESHRE’s Science Manager. Increasing volumes of more complex data prompted the Consortium to sort out a new database structure. As data analysis had fallen behind due to serious delay during this procedure it was decided to collect summary PGT data from multiple years in order to record the major changes that had occurred during the period of technical transition in both the IVF and genetics labs.
Nevertheless, fewer participants than usual contributed to the current data set. We are confident, however, that the data presented is robust and reliably reflects the major trends in PGT at the time. Data submission is a time-consuming activity and the steering committee greatly acknowledges the effort of all contributing centres.
With a proportion of 61% of all reported PGT cycles in 2013 and 64% in 2014 and 2015, the relevant contribution of PGT-A to the ESHRE PGT Consortium data set has increased, following years of stability at 52% (De Rycke et al., 2017).
The majority of centres contributing PGT-A data to the current data set (75%) have moved away from targeted FISH analysis on cleavage stage embryos to applying comprehensive, WGA-based technology on either cleavage stage embryos (64% in 2015) or blastocysts (36% in 2015). Such was suggested by the ESHRE PGT Consortium in the consensus paper that was published in 2010 (Harper et al., 2010a,b). The major indication for PGT-A remains AMA, although an increasing proportion of cycles is being performed without a recorded medical indication.
The uptake of new technologies is much slower in PGT-M. The proportion of cycles with blastocyst biopsy comprised 12% of the total number performed in 2015 and this is reflected by the 12% cycles performed with comprehensive diagnostic technology.
Targeted (short tandem repeat marker) analysis works well with a single cell (Day 3) biopsy while single-cell WGA may suffer from allele dropout and be less suitable as input for comprehensive testing strategies. Apparently, the need to change the PGT procedure is less urgent for the detection of monogenic disease than it is for other PGT indications.
Widespread application of new technologies is visible in PGT-SR, albeit less strong than in PGT-A, and the uptake of blastocyst biopsy (7% in 2012, 22% in 2013 and 36% in 2015) goes hand in hand with that of comprehensive diagnostics (13% in 2012, 36% in 2013 and 58% in 2015). Comprehensive diagnostics provide a generic platform, circumventing the need to develop locus-and family-specific tests, but consumables and equipment are relatively expensive.
Turning from cleavage stage to blastocyst biopsy places high demands on the embryo culture system and because of time restrains is usually combined with embryo vitrification and embryo transfer in a deferred cycle.
The number of genetically transferable embryos has not decreased, despite the fact that comprehensive analysis involves copy number calling of the non-indication chromosomes alongside the analysis of the indication chromosomes.
As PGT for sex selection for X-linked diseases does not fit any of the ICMART 2017 PGT categories it is evaluated as a separate entity. An increasing proportion of cycles for X-linked diseases is being performed with mutation detection rather than sex selection. For those cycles that still are being performed with sex selection, new technologies are on the rise. Where comprehensive technology (i.e. array CGH) used to be applied in a small minority of cases (4% in 2013), in 2015 it is used in 30% of cycles performed.
The retrospective data collections 2013–2015 represent valuable data on PGT activity in (mainly) Europe. The main trends observed in this ESHRE PGT Consortium report are the increased application of TE biopsy followed by a ‘vitrify all’ strategy and a deferred frozen embryo transfer, at the cost of cleavage stage biopsy combined with a fresh embryo transfer and the continuing expansion of comprehensive testing technology in all PGT categories, but in particular in PGT-SR and PGT-A.
Supplementary data
Supplementary data are available at Human Reproduction Open online.
Supplementary Material
Acknowledgements
A big thank you goes out to the participating centres that contributed to the creation of this data set.
List of the centres participating in one or more data collections discussed in this report: Argentina: Fecunditas, Department of Genetics and IVF, Buenos Aires; Belgium: Brussels Free University, Centre of Medical Genetics, Brussels; Ghent University Hospital, Infertility Centre, Ghent; Leuven University Fertility Center, Campus Gasthuisberg, Leuven; Brazil: Fertility—Centro de Fertilzação Assis, Sao Paoloa; Conceber-Centro de Medicina Reprodutiva, Curitiba; Canada: Mount Sinai Hospital, Ppathology and Laboratory Medicine, Toronto; Czech Republic: Institute Pronatal, Genetics, Praha; Denmark: Centre for preimplantation genetic diagnosis, Fertility Clinic, Aalborg University Hospital; Egypt: The Egyptian IVF-ET Center; France: C.M.C.O.-SIHCUS, CECOS Alsace, Unité de Diagnostic pré-implantatoire, Service de Biologie de la Reproduction, Strasbourg; Institut Universitaire de Recherche Clinique, Laboratoire de Génétique Moléculaire, Montpellier; Germany: Zentrum Für Humangenetik, Humangenetisches Labor, Regensburg; MVZ Fertility Center, Hamburg; Kinderwunsch centrum München, IVF Labor, Munich; Gyn-Gen-Lehel, Munich; Landes-Frauen und Kinderklinik Linz, Human Genetics; Kinderwunschzentrum an der Gedächteniskirche; University of Heidelberg, Molecular Genetics Unit of Department of Gynaecological Endocrinology and Reproductive Medicine, Heidelberg; CERF, Dr Thiemann, Dr Wetzka, Dr Wolk, Dr Hanjalic-Beck, Dr Friebel, Freiburg; Greece: University of Athens, St. Sophia’s Children’s Hospital, Department of Medical Genetics, Athens; EMBRYOGENESIS, Genetics, Thessaloniki; PGD laboratory, Genesis Athens Clinic, Centre for Human Reproduction, Athens; Embryolab, Thessaloniki; India: Krishna IVF Clinic, Visakhapatnam, Andhrapradesh; Ireland: Cork Fertility centre, Fernhurst House, Cork; Israel: Lis Maternity Hospital, Dept. of IVF, Tel Aviv; Shaare Zedek Medical Centre, Zohar PGD Unit, IVF and Medical Genetics Institute, Jerusalem; Italy: S.I.S.M.E.R. s.r.l. Bologna; European Hospital, Medicina della Riproduzione, Rome; Genoma, Molecular Genetics laboratories, Rome; Clinica Valle Giulia, Rome; Igenomix, Marostica; Fondazione IRCCS, Ca’ Granda Ospedale, Maggiore Policlinico, Milano; Japan: St. Mother Hospital, Kitakyushu; Takeuchi Ladies Clinic/Infertility Center, Aira-shi, Kagoshima; Poland: INVICTA, Gdansk; Portugal: University of Porto Faculty of Medicine, Dept. Genetics, Faculty of Medicine, Porto; Russia: Center for Reproductive Medicine MAMA; Scotland: Glasgow Royal Infirmary, Assisted conception services, Glasgow; Royal Edingburgh Infirmary, Edingburgh Fertility and Reproductive Endocrine Centre, Edingburgh; Singapore: Singapore General Hospital, Centre for Assisted Reproduction (CARE), Singapore; Spain: Institut Universitari Dexeus, Obstetrícia, Ginecologia i Reproducció, Barcelona; Instituto Valenciano de Infertilidad (IVI); Instituto Marques, Infertility department, Barcelona; Sistemas Genomicos SL, Reproductive Medicine, Valencia; Instituto de Reproduccion CEFER, Barcelona; U.R.H. Garcia del real; Hospital Quiron Madrid, Laboratorio de Reproduccion Asistida, Madris; Fundacion Puigvert, Seminologia i Reproduccio, Barcelona; Sweden: Karolinska University Hospital, Department of Clinical Genetics; Sahlgrenska University Hospital, Department of Ob/Gyn; Switzerland: Synlab Genetics, Bioggio; Taiwan: Chang Gung Memorial Hospital, Obstetries and Gynecology, Taoyuan County; The Netherlands: Maastricht University Medical Center, Obstetrics and Gynecology, Maastricht; Center for Reproductive Medicine, Amsterdam Medical Center, Amsterdam; University Medical Center Groningen, IVF/fertility laboratory, Groningen; University Medical Center Utrecht, Utrecht; Turkey: Istanbul Memorial Hospital, Reproductive endocrinology & ART centre, Istanbul; UK: Hammersmith Hospital, Institute of Ob/Gyn-RPMS, London; Ukrain: Clinic of Reproductive Medicine ‘Nadiya’, ART Department, Kyiv.
Authors’ roles
E.C. drafted the manuscript, contributed to the tables, designed the figures and was responsible for final editing of the manuscript. A.v.M. contributed to the tables, designed and drafted the figures and edited the manuscript. F.C., G.K., C.M., C.R. and M.D.R. edited the manuscript. V.G. was responsible for raw data curation, contributed to the tables and parts of the manuscript and edited the manuscript. All authors revised and approved the final manuscript.
Funding
The study has no external funding and all costs are covered by the European Society of Human Reproduction and Embryology.
Conflict of interest
There are no competing interests declared.
References
- De Rycke M, Belva F, Goossens V, Moutou C, SenGupta SB, Traeger-Synodinos J, Coonen E. ESHRE PGD Consortium data collection XIII: cycles from January to December 2010 with pregnancy follow-up to October 2011. Hum Reprod 2015;30:1763–1789. [DOI] [PubMed] [Google Scholar]
- De Rycke M, Goossens V, Kokkali G, Coonen E, Moutou C. ESHRE PGD Consortium data collection XIV-XV: cycles from January 2011 to December 2012 with pregnancy follow-up to October 2013. Hum Reprod 2017;32:1974–1994. [DOI] [PubMed] [Google Scholar]
- ESHRE PGD Consortium Steering Committee. ESHRE Preimplantation Genetic Diagnosis Consortium: data collection III (May 2001). Hum Reprod 2002;17:233–246. [DOI] [PubMed] [Google Scholar]
- ESHRE PGT Consortium and SIG-Embryology Biopsy Working Group, Kokkali G, Coticchio G, Bronet F, Celebi C, Cimadomo D, Goossens V, Liss J, Nunes S, Sfontouris I, Vermeulen Net al. ESHRE PGT Consortium and SIG Embryology Good practice recommendations for polar body and embryo biopsy for PGT. Hum Reprod Open 2020;2020:hoaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESHRE PGT Consortium Steering Committee, Carvalho F, Coonen E, Goossens V, Kokkali G, Rubio C, Meijer-Hoogeveen M, Moutou C, Vermeulen N,, De Rycke M.ESHRE PGT Consortium good practice recommendations for the organisation of PGT. Hum Reprod Open 2020;2020:hoaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESHRE PGT-M Working Group, Carvalho F, Moutou C, Dimitriadou E, Dreesen J, Giménez C, Goossens V, Kakourou G, Vermeulen N, Zuccarello D, De Rycke M.ESHRE PGT Consortium good practice recommendations for the detection of monogenic disorders. Hum Reprod Open 2020;2020:hoaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESHRE PGT-SR/PGT-A Working Group, Coonen E, Rubio C, Christopikou D, Dimitriadou E, Gontar J, Goossens V, Maurer M, Spinella F, Vermeulen N, De Rycke M.ESHRE PGT Consortium Good practice recommendations for the detection of structural and numerical chromosomal aberrations. Hum Reprod Open 2020;2020:hoaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraedts J, Handyside A, Harper J, Liebaers I, Sermon K, Staessen C, Thornhill A, Vanderfaeillie A, Viville S. ESHRE Preimplantation Genetic Diagnosis (PGD) Consortium: preliminary assessment of data from January 1997 to September 1998. ESHRE PGD Consortium Steering Committee. Hum Reprod 1999;14:3138–3148. [DOI] [PubMed] [Google Scholar]
- Geraedts J, Handyside A, Harper J, Liebaers I, Sermon K, Staessen C, Thornhill A, Viville S, Wilton L; ESHRE PGD Consortium Steering Committee. ESHRE Preimplantation Genetic Diagnosis (PGD) Consortium: data collection II (May 2000). Hum Reprod 2000;15:2673–2683. [DOI] [PubMed] [Google Scholar]
- Goossens V, Harton G, Moutou C, Scriven PN, Traeger-Synodinos J, Sermon K, Harper JC. ESHRE PGD Consortium data collection VIII: cycles from January to December 2005 with pregnancy follow-up to October 2005. Hum Reprod 2008;23:2629–2645. [DOI] [PubMed] [Google Scholar]
- Goossens V, Harton G, Moutou C, Traeger-Synodinos J, Van Rij M, Harper JC. ESHRE PGD Consortium data collection IX: cycles from January to December 2006 with pregnancy follow-up to October 2007.Hum Reprod 2009;24:1786–1810. [DOI] [PubMed] [Google Scholar]
- Goossens V, Traeger-Synodinos J, Coonen E, De Rycke M, Moutou C, Pehlivan T, Derks-Smeets IA, Harton G. ESHRE PGD Consortium data collection XI: cycles from January to December 2008 with pregnancy follow-up to October 2009. Hum Reprod 2012;27:1887–1911. [DOI] [PubMed] [Google Scholar]
- Harper JC, Boelaert K, Geraedts J, Harton G, Kearns WG, Moutou C, Muntjewerff N, Repping S, SenGupta S, Scriven PN. et al. ESHRE PGD Consortium data collection V: cycles from January to December 2002 with pregnancy follow-up to October 2003. Hum Reprod 2006;21:3–21. [DOI] [PubMed] [Google Scholar]
- Harper JC, Coonen E, De Rycke M, Fiorentino F, Geraedts J, Goossens V, Harton G, PehlivanBudak T, Renwick P, Sengupta S. et al. What next for preimplantation genetic screening (PGS)? A position statement from the ESHRE PGD Consortium steering committee. Hum Reprod 2010. a;25:821–823. [DOI] [PubMed] [Google Scholar]
- Harper JC, Coonen E, De Rycke M, Harton G, Moutou C, Pehlivan T, Traeger-Synodinos J, Van Rij M, Goossens V. ESHRE PGD Consortium: data collection X: cycles from January to December 2007 with pregnancy follow-up to October 2008. Hum Reprod 2010. b;25:2685–2707. [DOI] [PubMed] [Google Scholar]
- Harper JC, de Die-Smulders C, Goossens V, Harton G, Moutou C, Repping S, Scriven PN, SenGupta S, Traeger-Synodinos J, Van Rij MC. et al. ESHRE PGD Consortium data collection VII: cycles from January to December 2004 with pregnancy follow-up to October 2005. Hum Reprod 2008;23:741–755. [DOI] [PubMed] [Google Scholar]
- Harper JC, Wilton L, Traeger-Synodinos J, Goossens V, Moutou C, SenGupta S, PehlivanBudak T, Renwick P, De Rycke M, Geraedts J, Harton G. The ESHRE PGD Consortium: 10 years of data collection. Hum Reprod Update 2012;18:234–247. [DOI] [PubMed] [Google Scholar]
- Moutou C, Goossens V, Coonen E, De Rycke M, Kokkali G, Renwick P, SenGupta SB, Vesela K, Traeger-Synodinos J. ESHRE PGD Consortium data collection XII: cycles from January to December 2009 with pregnancy follow-up to October 2010. Hum Reprod 2014;29:880–903. [DOI] [PubMed] [Google Scholar]
- Sermon K, Moutou C, Harper J, Geraedts J, Scriven P, Wilton L, Magli MC, Michiels A, Viville S, De Die C. ESHRE PGD Consortium data collection IV: May–December 2001. Hum Reprod 2005;20:19–34. [DOI] [PubMed] [Google Scholar]
- Sermon KD, Michiels A, Harton G, Moutou C, Repping S, Scriven PN, SenGupta S, Traeger-Synodinos J, Vesela K, Viville S. et al. ESHRE PGD Consortium data collection VI: cycles from January to December 2003 with pregnancy follow-up to October 2004. Hum Reprod 2007;22:323–336. [DOI] [PubMed] [Google Scholar]
- Zegers-Hochschild F, Adamson D, Dyer S, Racowsky C, de Mouzon J, Sokol R, Rienzi L, Sunde A, Schmidt L, Cooke ID, Simpson JL, van der Poel S. The international glossary on infertility and fertility care. Hum Reprod 201732, 1786–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.