Abstract
Background
Colorectal cancer (CRC) is one of the most common cancers worldwide, and more than half of CRC patients have CRC liver metastasis (CRCLM). Mounting evidence indicates that high mobility group protein A1(HMGA1) is overexpressed in many cancer types, but its role in CRCLM has been obscure.
Material/Methods
Using immunohistochemistry, we assessed the expression of HMGA1 in 73 patients with CRCLM, and compared HMGA1 mRNA in 17 pairs of CRCs, CRCLM tissues, and normal liver tissues. The clinical significance of HMGA1 was evaluated by analyzing its correlation with the clinicopathological factors and overall survival (OS) rates. The function of HMGA1 in CRC invasion was investigated and the underlying mechanism of HMGA1-induced invasion was explored with in vitro experiments.
Results
In CRCLMs, the high-HMGA1 and low-HMGA1 patients accounted for 53.42% and 46.58% of all patients, respectively. High HMGA1 expression in CRCLM was significantly associated with low OS rates. In vitro experiments demonstrated that HMGA1 promoted glucose transporter 3 (GLUT3) transcription and expression in CRC cells. GLUT3 was required in HMGA1-involved invasion, and GLUT3 expression was associated with poor prognosis of CRCLM.
Conclusions
High HMGA1 and GLUT3 expression in CRCLM was significantly correlated with poor prognosis of CRCLM. HMGA1 promoted CRC invasion by elevating GLUT3 transcription and expression.
MeSH Keywords: Colorectal Neoplasms, Glucose Transporter Type 3, HMGA1a Protein, Neoplasm Metastasis, Prognosis
Background
Colorectal cancer (CRC) is one of the most common cancers worldwide, with 1.8 million new cases in 2018 and approximately one-third of patients presenting with metastatic disease [1]. The liver is the most common metastatic destination of CRC metastasis because of the hepatic artery and portal drainage [2]. The 5-year overall survival (OS) rate of patients diagnosed with synchronous liver metastases of colorectal cancer (CRCLM) is less than 15% [3]. CRC metastatic lesions in the liver account for two-thirds of deaths from colorectal cancer [4]. Although the 5-year OS rate is 58% after liver resection for CRCLM, more than half of patients develop tumor recurrence after surgical resection [5]. Distant metastasis is still the major cause of death in patients with colorectal cancer, and markers for better distinguishing the heterogeneous groups are needed to develop more precise therapeutic strategies.
High mobility group proteins (HMGs) regulate the transcription of numerous genes by interacting with transcription factors and modulating the structure of chromatin [6]. The HMG family can be classified into 3 subgroups based on the DNA-binding motif. HMGA members have DNA-binding AT hooks, HMGB members have HMG-boxes, and HMGN members have nucleosomal-binding domains [7]. As architectural transcription factors, HMG proteins do not have transcriptional activity [8], but they can regulate the transcription of target genes by binding with the structures. In HMG family, HMGA1 is the most well-studied member and mounting evidence shows that HMGA1 is overexpressed in many cancer types and promotes the occurrence and metastasis of cancer, including the ovarian cancer, breast cancer, and pancreatic cancer [9–12]. In CRC, HMGA1 was reported to be upregulated in CRC and microarray analysis showed it is involved in CRC metastasis [13,14], but the correlation between HMGA1 and CRC hepatic metastasis has not been previously reported.
In the present study, we assessed the expression of HMGA1 in 73 patients with CRCLM using immunohistochemistry (IHC), and compared HMGA1 mRNA in 17 pairs of CRCs, CRCLM tissues, and normal liver tissues. The clinical significance of HMGA1 was evaluated by analyzing its correlation with the clinicopathological factors and the OS rates. The function of HMGA1 in CRC invasion was investigated and the underlying mechanism of HMGA1-induced invasion was explored.
Material and Methods
Ethics
All the specimens were obtained with written consent of patients. The entire study was approved and supervised by the Ethics Committee of YIDU Central Hospital and Linyi Central Hospital (approval no. 2019053512).
Patients and follow-ups
A total of 103 patients underwent radical surgery of CRCLM from 2010 to 2018 in YIDU Central Hospital and Linyi Central Hospital. From these 103 patients, we enrolled 73 patients (29 males and 44 females) who had received standard adjuvant therapy and follow-ups. The duration of follow-up ranged from 3 to 78 months, with an average follow-up of 17.6 months. The fresh tissues were obtained during surgery without interfering with the diagnosis, and the paraffin-embedded tissues were obtained from the Pathology Department.
Cells and agents
Human CRC cell line SW480 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA), and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin. FGF2 were from PeproTech Company. Antibodies were as follows: HMGA1 antibodies (Abcam, Cat#ab129153), Glucose transporter 3 (GLUT3) (Santa Cruz Biotechnology, Cat#sc-74399), and β-actin antibody (Sigma-Aldrich, Cat#A5316).
Real-time quantitative PCR
Total mRNA was extracted from 17 pairs of CRCs, CRCLM tissues, and normal liver tissues with Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The total of 1 μg of mRNA was reversely transcribed into cDNA using the SuperScript cDNA Kit (Thermo Fisher Scientific) following the instructions. SYBR green method with PCR master mix (Thermo Fisher Scientific) was applied for quantitative PCR, with GPADH as an internal control. Gene expression levels were normalized by HMGA1 level in normal liver as 1.0. The primers were as follows:
HMGA1: GGAAAAGGACGGCACTGAGA;
TGGTGGTTTTCCGGGTCTTG.
GLUT3: CAGCGAGACCCAGAGATGC;
GACCCCAGTGTTGTAGCCAA.
GAPDH: GAGTCAACGGATTTGGTCGT;
GACAAGCTTCCCGTTCTCAG.
Immunohistochemistry and score
IHC analysis of the 73 cases of CRCLMs was performed as described in a previous study [15]. The results of IHC scores were semi-quantified according to the IHC score, which was the combination of staining intensity and positive cell percentage. The scores for percentage of positive tumor cells were defined as: score 1 (0–25% positive tumor cells), 2 (25–50% positive tumor cells), and 3 (more than 50% positive tumor cells). The scores for staining intensity were defined as 1 (no staining), 2 (weak staining), and 3 (strong yellow). The final score was the product of these 2 scores multiplied, ranging from 0 to 9. All the patients were divided into low and high HMGA1 or GLUT3 groups according to the cut-off, which was defined by the ROC curve based on a previous study [16]. In our study, the cut-offs of HMGA1 and GLUT3 were 2.5 and 3.5, respectively.
Overexpression and knockdown
The pcDNA3.1 vector carrying GLUT3 was transfected into U118 cells for GLUT3 overexpression. Two independent siRNAs of HMGA1 and GLUT3 were used for the knockdown of HGMA1 and GLUT3. All transfections were achieved by lipofectamine 2000 according to the manual. The siRNAs were purchased from Santa Cruz Biotech (Santa Cruz, CA, USA).
Luciferase assay
Luciferase assay of GLUT3 was performed with the Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) according to a previous study [17]. The human GLUT3 promoter region ranged from nucleotides −1007 to +1 (relative to the transcription initiation site) and was cloned into the KpnI/HindIII sites of the PGL3-basic dual luciferase reporter as luciferase reporters. PGL3-basic plasmid was set as the empty control. siRNA of HMGA1 was transfected into U118 cells for knockdown using lipofectamine 2000. At 48 h after siHMGA1 transfection, the cells were treated with the Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Immunoblotting assays
Western blot analysis was performed to detect the protein expressions according to our previous methods [18]. In brief, RIPA lysis buffer was used to lyse the cells. The total cell lysates were centrifuged at 10 000 g for 30 min and the supernatants were mixed with the loading buffer and boiled for 10 min. A total of 10 μg protein was subjected to SDS-PAGE electrophoresis and then transferred to polyvinylidene difluoride membranes (Thermo Fisher). After incubation in 5% bovine serum albumin, the membranes were immersed with primary antibodies overnight and the corresponding secondary antibodies for 1 h. ECL agents were finally applied to visualize the proteins.
Invasion assays
The transwell assay was applied for the invasion assay, as previously described [18]. In brief, 3×104 cells were passaged into the matrigel-precoated chamber and cultured for 12 h for adhesion. We added 10 ng/ml FGF2 into the lower chamber as a chemoattractant. After 24-h culture, the invaded cells in the lower chamber were fixed and stained. Cell numbers were counted from at least 5 random visual fields under microscope. The data were from 3 independent experiments.
Statistical analysis
All statistical analyses were performed using SPSS 24.0. Correlations between HMGA1 expression levels and clinicopathological features were analyzed with the chi-square test. The correlation between HMGA1 and GLUT3 mRNA were analyzed with Spearman correlation analysis. OS curves were displayed with Kaplan-Meier method and the statistical significances were calculated with the log-rank test. For cellular experiments, data were compared using the t test. P less than 0.05 was considered as statistically significant.
Results
Expression of HMGA1 in CRCLMs
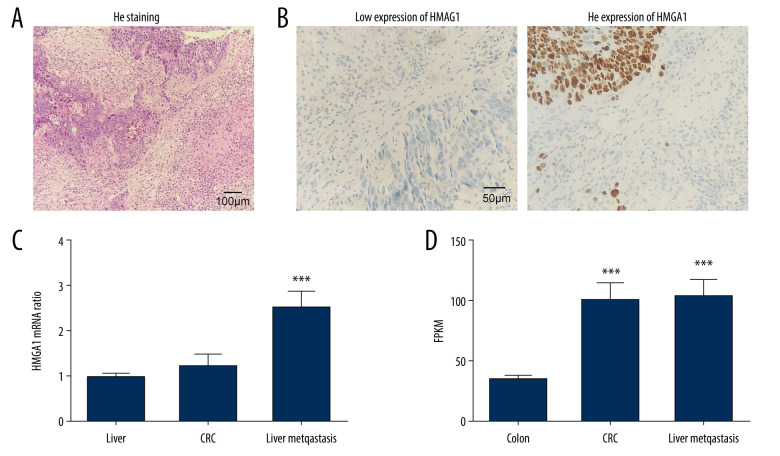
We detected HMGA1 expression in 73 CRCLMs by IHC. All cases were pathologically diagnosed as synchronous liver metastasis of CRC (Figure 1A). According to the IHC score, the CRCLMs were divided into high-HMGA1 and low-HMGA1 groups (Figure 1B), which accounted for 53.42% and 46.58% of patients, respectively (Table 1). The CRCLM tissues, normal liver tissues, and primary tumors of CRC from 17 patients with synchronous CRCLM were used for mRNA extraction and HMGA1 mRNA detection. HMGA1 expression in CRCLMs were significantly higher than in CRCs and normal liver tissues, suggesting that HMGA1 promotes liver metastasis (Figure 1C). Interestingly, HMGA1 levels in CRCLMs and CRCs were significantly higher than in normal colon tissues in a high-throughput sequencing (GSE144259), but the HMGA1 level in CRCLMs and CRCs were not significantly different (Figure 1D).
Figure 1.
Expression of HMGA1 in CRCLM. (A) The metastasis of CRC was pathologically confirmed by HE staining. (B) HMGA1 expression in 73 CRCLMs was detected using IHC. Patients were categorized into HMGA1 low and high expression groups. (C) HMGA1 expression was detected with qRT-PCR in the 17 synchronous CRCLM tissues. (D) HMGA1 mRNA in normal colon tissues, CRC, and CRCLM was compared with mRNA sequencing.
Table 1.
Basic characteristics of patients with CRCLM.
| Factors | Number | Percentage |
|---|---|---|
| Sex | ||
| Male | 29 | 39.73% |
| Female | 44 | 60.27% |
| Age | 0 | |
| <60 | 15 | 20.55% |
| ≥60 | 58 | 79.45% |
| Differentiation | 0 | |
| Good | 44 | 60.27% |
| Moderate+poor | 29 | 39.73% |
| Lymph node invasion | 0 | |
| No (N0) | 46 | 63.01% |
| Yes (N1/2) | 27 | 36.99% |
| Number of CRCLM | 0 | |
| <3 | 38 | 52.05% |
| ≥3 | 35 | 47.95% |
| CRCLM distribution | 0 | |
| Unilobar | 46 | 63.01% |
| Bilobar | 27 | 36.99% |
| HMGA1 | ||
| Low | 39 | 53.42% |
| High | 34 | 46.58% |
| GLUT3 | ||
| Low | 37 | 50.68% |
| High | 36 | 49.32% |
Clinical significance of HMGA1 in CRC metastasis
We evaluated the correlations between HMGA1 in CRCLMs and other clinicopathological factors by the chi-square test to assess its clinical significance. The enrolled clinicopathological factors included sex, age, tumor differentiation, lymph node invasion, liver metastases number, and distribution. However, we found no factors that were significantly associated with HMGA1 expression (Table 2).
Table 2.
Correlations between HMGA1 and CRCLM characteristics.
| Factors | HMGA1 | P* | |
|---|---|---|---|
| Low | High | ||
| Sex | |||
| Male | 13 | 16 | 0.236 |
| Female | 26 | 18 | |
| Age | |||
| <60 | 8 | 7 | 0.994 |
| ≥60 | 31 | 27 | |
| Differentiation | |||
| Good | 25 | 19 | 0.632 |
| Moderate+poor | 14 | 15 | |
| Lymph node invasion | |||
| No (N0) | 23 | 23 | 0.444 |
| Yes (N1/2) | 16 | 11 | |
| Number of CRCLM | |||
| <3 | 22 | 16 | 0.425 |
| ≥3 | 17 | 18 | |
| CRCLM distribution | |||
| Unilobar | 26 | 20 | 0.489 |
| Bilobar | 13 | 14 | |
| GLUT3 | |||
| Low | 24 | 13 | 0.047 |
| High | 15 | 21 | |
Chi-square test.
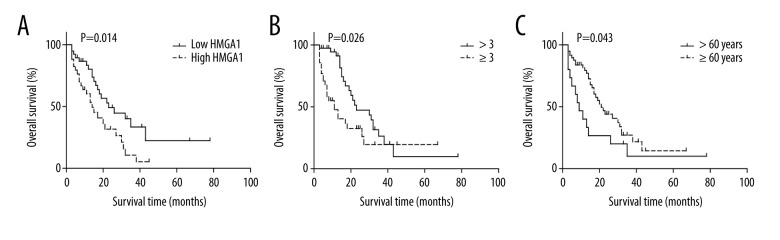
The prognostic value of the relationships between all the above factors and the OS rates was analyzed with univariate analysis using the log-rank test (Table 3). In univariate analysis, HMGA1 had a significant correlation with the OS rate of CRCLM and indicated a poor prognosis (P=0.014). The median survival times of patients with low and high HMGA1 levels in CRCLM were 34.0 and 17.7 months, respectively (Figure 2A). Besides HMGA1, CRCLM number was also associated with a poor prognosis of CRCLM (Figure 2B). Intriguingly, we also demonstrated that younger patients usually had poorer prognosis compared with older patients (≥60 years old) (Figure 2C).
Table 3.
The prognostic value of HMGA1 and other factors.
| Factors | Univariate | Multivariate | |||
|---|---|---|---|---|---|
| Medium OS time | P* | HR | 95% CI | P** | |
| Sex | |||||
| Male | 21.4 | 0.659 | |||
| Female | 28.2 | ||||
| Age | |||||
| <60 | 18.4 | 0.043 | 1 | ||
| ≥60 | 27.5 | 0.42 | 0.22–0.83 | 0.012 | |
| Differentiation | |||||
| Good | 29.9 | 0.232 | |||
| Moderate+poor | 19.4 | ||||
| Lymph node invasion | |||||
| No (N0) | 30.2 | 0.449 | |||
| Yes (N1/2) | 20.2 | ||||
| Number of CRCLM | |||||
| <3 | 29.7 | 0.026 | 1 | ||
| ≥3 | 21.8 | 3.10 | 0.53–18.23 | 0.212 | |
| CRCLM distribution | |||||
| Unilobar | 26.1 | 0.695 | |||
| Bilobar | 27.8 | ||||
| HMGA1 | |||||
| Low | 34 | 0.014 | 1 | ||
| High | 17.7 | 2.37 | 1.26–4.44 | 0.007 | |
| GLUT3 | |||||
| Low | 29.5 | 0.039 | 1 | ||
| High | 22.6 | 0.58 | 0.10–3.46 | 0.554 | |
Log-rank test;
Cox-regression model.
Figure 2.
The association between HMGA1 expression, metastatic lesion number, patients age, and OS rate. High expression of HMGA1 (A), presence of more than 3 tumors (B) and age under 60 years were significantly associated with lower OS rates.
Using a Cox-regression model, we performed multivariate analysis to determine the independent prognostic factors (Table 3). HMGA1 (P=0.007) and patient age (P=0.012) were confirmed as independent prognostic factors of CRCLM, indicating poor prognosis. Due to the significant interaction with HMGA1, GLUT3 was not identified as an independent biomarker (P=0.054).
HMGA1 promoted GLUT3 expression in CRCLMs
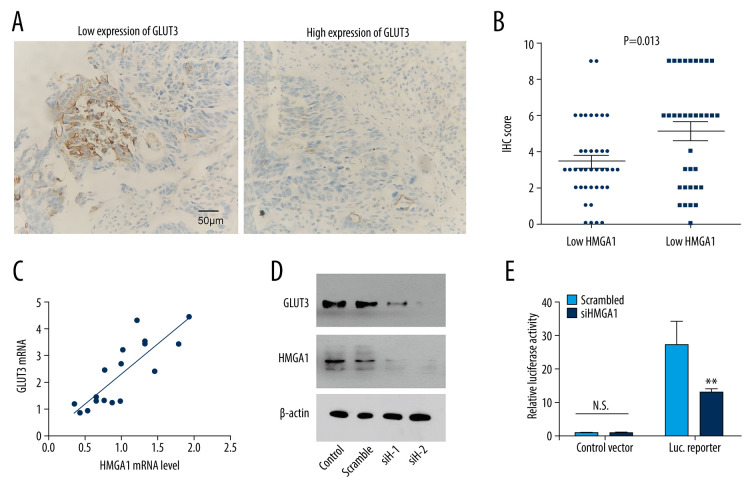
The expression of many proteins involved in tumor progression were reported to be regulated by HMGA1. Here, we assessed the influence of HMGA1 on GLUT3 expression because GLUT3 is an essential protein involved in tumor invasion and metastasis [19]. GLUT3 expression was first detected with IHC and semi-qualified with IHC scores in CRCLMs (Figure 3A). Consequently, patients with high CRCLM HMGA1 also had elevated GLUT3 expression, indicating that HMGA1 and GLUT3 had a close association in CRCLM (Figure 3B). Results of chi-square testing showed a significant positive association between HMGA1 and GLUT3 (Table 2). The mRNA of GLUT3 was detected in the 17 fresh tissues of CRCLM, which showed that GLUT3 mRNA was upregulated with HMGA1 mRNA (Figure 3C). All the above results suggest there is a relationship between HMGA1 and GLUT3. Our in vitro experiments showed the change of GLUT3 expression after silencing HMGA1 with 2 siRNAs. As a result, GLUT3 expression was decreased after HMGA1 knockdown in SW480 cells (Figure 3D), indicating that HMGA1 regulated GLUT3 expression. Moreover, luciferase assay results showed the effect of HMGA1 on GLUT3 transcription (Figure 3E). After HMGA1 knockdown, the luciferase activity of GLUT3 reporter plasmid was dramatically decreased, suggesting that HMGA1 induced GLUT3 expression by promoting its transcription.
Figure 3.
HMGA1 induced the expression and transcription of GLUT3 in CRCLM. (A) Representative IHC images of GLUT3 high and low expression. (B) Patients with high HMGA1 expression had higher IHC scores of GLUT3. The t test showed a statistically significant difference. (C) In the qPCR of 17 synchronous CRCLMs, GLUT3 expression changed along with the HMGA1 mRNA. Spearman correlation analysis showed a statistically significant difference. (D) HMGA1 knockdown substantially decreased the expression of GLUT3. In SW480 cells, HMGA1 was silenced by 2 independent siRNAs. (E) HMGA1 knockdown decreased the transcription of GLUT3. The transcription of GLUT3 was detected with luciferase in SW480 cells after silencing HMGA1.
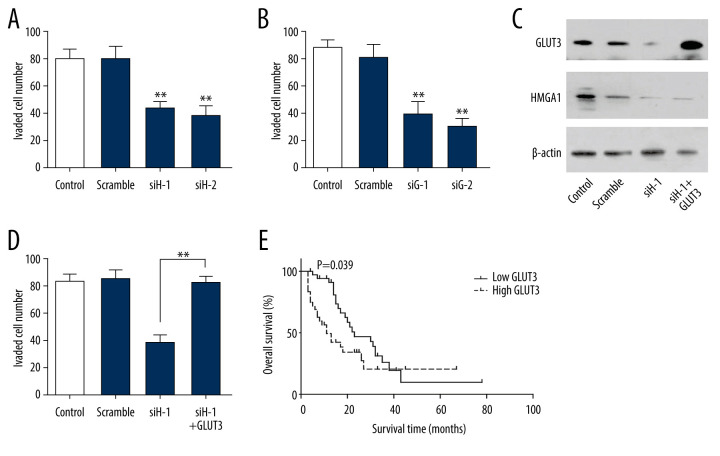
GLUT3 was required in HMGA1-induced invasion
The influence of HGMA1 on tumor invasion was detected by silencing HMGA1. In SW480 cells, HMGA1 knockdown substantially impaired the invasion ability (Figure 4A, Supplementary Figure 1A), indicating the essential role of GLUT3 in CRC invasion. Similar results were observed when GLUT3 was silenced (Figure 4B, Supplementary Figure 1B), which suggested that GLUT3 was also involved in CRC invasion. To investigate the role of GLUT3 in HMGA1-induced CRC, we knocked down HMGA1 and overexpressed GLUT3 for rescue (Figure 4C). The results showed that GLUT3 overexpression extensively reversed the invasion attenuation caused by HMGA1 knockdown (Figure 4D, Supplementary Figure 1C), indicating that GLUT3 was required in HMGA1-induced CRC invasion. We also evaluated the prognostic value of GLUT3 expression by univariate analysis. Similar to HMGA1, high expression of GLUT3 was significantly associated with the OS rate of CRCLM (Figure 4E). The median survival times of low and high GLUT3 were 29.5 and 22.6 months, respectively (Table 3)
Figure 4.
GLUT3 was required in HMGA1-induced invasion. (A, B) HMGA1 (A) or GLUT3 (B) knockdown significantly impaired the invasion of SW480. The invasion of SW480 was detected with transwell assay.(C) SW480 cells were transfected with GLUT3 plasmid for rescue assay in the presence of HMGA1 knockdown. (D) GLUT3 overexpression rescued the invasion decrease caused by HMGA1 knockdown. (E) High GLUT3 in CRCLM was significantly associated with poorer prognosis.
Discussion
The recurrence of metastasis after radical surgery is the main risk for patients with CRC and more than 50% patients had disease recurrence within 2 years after CRCLM resection. Patients with CRCLM need more accurate risk stratification and prediction of prognosis. For example, somatic mutations of RAS/TP53 and APC/PIK3CA are important biomarkers associated with outcome after CRCLM resection [20]. Here, we demonstrated that HMGA1 was upregulated in CRCLMs, and it was a prognostic biomarker of CRCLM. These results could be used to stratify the patients with CRCLM more precisely and provide more evidence for the individual treatment of CRC.
HMGA1 is widely accepted to play an important role in neoplastic transformation in many cancer types, such as thyroid, lung, breast, bladder, prostate, colon, pancreas, and hematopoietic system [21]. Moreover, it is interesting to note that HMGA1 expression is low or undetectable in normal tissues [21]. In CRC, HMGA1 was significantly elevated in CRCs compared to adjacent, nonmalignant tissues, and drive metabolic alterations that contributed to CRC carcinogenesis [22]. A study using comprehensive gene expression profiles verified that HMGA1 is an important oncogene [23]. However, no direct evidence has elucidated the role of HMGA1 in CRC progression and prognosis, and no studies have investigated the relationship between HMGA1 and CRCLM. The present study is the first to show that HMGA1 promotes invasion and hepatic metastasis of CRC by elevating GLUT3 expression. Our results strengthen evidence showing the oncogenic role of HMGA1 in CRC and also provide new insights into its role in CRC progression. However, our conclusions lack the support of in vivo experiments. Further experiments could be performed to elucidate the molecular mechanism by which GLUT3 affects metastasis.
Numerous data support the tumorigenic potency of HMGA1, and the molecular mechanism is complicated. As a transcription factor, HMGA1 usually influences cellular processes by regulating downstream effectors. The expression and transcription of many target genes involved in tumor progression were reported to be regulated by HMGA1, including Kinesin-like protein KIFC1 (KIFC1), c-Myc, and Forkhead box protein M1 (FOXM1) [17,24,25]. The present study demonstrates that HMGA1 can promote the expression of GLUT3, but we did not screen for the binding sequence of HMGA1. In fact, all HMG proteins interact with chromatin structures by their functional motif rather than recognizing specific nucleotide sequences of DNA or chromatin [26]. In the present study, we found that GLUT3 was the effector responsible for HMGA1-induced invasion and metastasis, suggesting that therapies targeting GLUT3 signaling may suppress the CRC metastasis and help prolong survival time.
GLUT3 is a facilitative glucose transporter and it also mediates the uptake of various other monosaccharides, including the galactose, mannose, xylose, and fucose [27,28]. Increased sugar uptake has been widely proved to promote oncogenesis in many cancers. GLUT3 is a known driver of a cancer stem cell phenotype, enabling cancer cells to survive glucose deprivation [29]. GLUT3 was previously reported to be involved in tumorigenesis, progression, or prognosis of several cancer types, including glioblastoma, hepatocellular carcinoma, bladder cancer, and lung cancer [30–32]. In CRC, GLUT3-YAP-dependent signaling was important in the metastasis of metastatic colorectal cancer [19]. GLUT3 addiction has been regarded as a druggable vulnerability in some special tumor types such as glioblastoma [33]. Our findings show that GLUT3 is essential in CRC invasion and is correlated with the prognosis of CRCLM, which also indicates the potency of GLUT3 as a druggable target of CRCLM.
Conclusions
In CRCLMs, the high-HMGA1 and low-HMGA1 patients accounted for 53.42% and 46.58% of all patients, respectively. High HMGA1 expression in CRCLM was significantly associated with low OS rates. In vitro experiments demonstrated that HMGA1 promoted the GLUT3 transcription and expression in CRC cells. GLUT3 was required in HMGA1-involved invasion, and GLUT3 expression was also related with the poor prognosis of CRCLM. All these results demonstrate that high HMGA1 and GLUT3 expression in CRCLM is significantly correlated with poor prognosis of CRCLM. HMGA1 promotes CRC invasion via elevating GLUT3 transcription and expression.
Supplementary Data
(A) Representative transwell images of Figure 4A. (B) Representative transwell images of Figure 4B. (C) Representative transwell images of Figure 4D.
Footnotes
Conflicts of interest
None.
Source of support: Departmental sources
References
- 1.Kopetz S. New therapies and insights into the changing landscape of colorectal cancer. Nat Rev Gastroenterol Hepatol. 2019;16(2):79–80. doi: 10.1038/s41575-018-0100-z. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Tumor metastasis: Mechanistic insights and clinical challenges. Nat Med. 2006;12(8):895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 3.Liu H, Xu Y, Zhang Q, et al. Prognostic significance of TBL1XR1 in predicting liver metastasis for early stage colorectal cancer. Surg Oncol. 2017;26(1):13–20. doi: 10.1016/j.suronc.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Kopetz S, Chang GJ, Overman MJ, et al. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol. 2009;27(22):3677–83. doi: 10.1200/JCO.2008.20.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Xu Y, Zhang Q, et al. Correlations between TBL1XR1 and recurrence of colorectal cancer. Sci Rep. 2017;7:44275. doi: 10.1038/srep44275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Martino M, Fusco A, Esposito F. HMGA and cancer: A review on patent literatures. Recent Pat Anticancer Drug Discov. 2019;14(3):258–67. doi: 10.2174/1574892814666190919152001. [DOI] [PubMed] [Google Scholar]
- 7.Catez F, Hock R. Binding and interplay of HMG proteins on chromatin: Lessons from live cell imaging. Biochim Biophys Acta. 2010;1799(1–2):15–27. doi: 10.1016/j.bbagrm.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Reeves R. Nuclear functions of the HMG proteins. Biochim Biophys Acta. 2010;1799(1–2):3–14. doi: 10.1016/j.bbagrm.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fedele M, Fusco A. HMGA and cancer. Biochim Biophys Acta. 2010;1799(1–2):48–54. doi: 10.1016/j.bbagrm.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Tian X, Song J, Zhang X, et al. MYC-regulated pseudogene HMGA1P6 promotes ovarian cancer malignancy via augmenting the oncogenic HMGA1/2. Cell Death Dis. 2020;11(3):167. doi: 10.1038/s41419-020-2356-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Z, Wang W, Wang Y, et al. Long noncoding RNA LINC00963 promotes breast cancer progression by functioning as a molecular sponge for microRNA-625 and thereby upregulating HMGA1. Cell Cycle. 2020;19(5):610–24. doi: 10.1080/15384101.2020.1728024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolb S, Fritsch R, Saur D, et al. HMGA1 controls transcription of insulin receptor to regulate cyclin D1 translation in pancreatic cancer cells. Cancer Res. 2007;67(10):4679–86. doi: 10.1158/0008-5472.CAN-06-3308. [DOI] [PubMed] [Google Scholar]
- 13.Xian L, Georgess D, Huso T, et al. HMGA1 amplifies Wnt signalling and expands the intestinal stem cell compartment and Paneth cell niche. Nat Commun. 2017;8:15008. doi: 10.1038/ncomms15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi Y, Sawada G, Sato T, et al. Microarray analysis reveals that high mobility group A1 is involved in colorectal cancer metastasis. Oncol Rep. 2013;30(3):1488–96. doi: 10.3892/or.2013.2602. [DOI] [PubMed] [Google Scholar]
- 15.Liu N, Zhang J, Sun S, et al. Expression and clinical significance of fibroblast growth factor 1 in gastric adenocarcinoma. Onco Targets Ther. 2015;8:615–21. doi: 10.2147/OTT.S79204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu YF, Liu ZL, Pan C, et al. HMGB1 correlates with angiogenesis and poor prognosis of perihilar cholangiocarcinoma via elevating VEGFR2 of vessel endothelium. Oncogene. 2019;38(6):868–80. doi: 10.1038/s41388-018-0485-8. [DOI] [PubMed] [Google Scholar]
- 17.Ha TK, Her NG, Lee MG, et al. Caveolin-1 increases aerobic glycolysis in colorectal cancers by stimulating HMGA1-mediated GLUT3 transcription. Cancer Res. 2012;72(16):4097–109. doi: 10.1158/0008-5472.CAN-12-0448. [DOI] [PubMed] [Google Scholar]
- 18.Tian X, Yang C, Yang L, et al. PTPRF as a novel tumor suppressor through deactivation of ERK1/2 signaling in gastric adenocarcinoma. Onco Targets Ther. 2018;11:7795–803. doi: 10.2147/OTT.S178152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo CC, Ling HH, Chiang MC, et al. Metastatic colorectal cancer rewrites metabolic program through a Glut3-YAP-dependent signaling circuit. Theranostics. 2019;9(9):2526–40. doi: 10.7150/thno.32915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Ougolkov A, Yamashita K, et al. beta-Catenin and ras oncogenes detect most human colorectal cancer. Clin Cancer Res. 2003;9(8):3073–79. [PubMed] [Google Scholar]
- 21.Sumter TF, Xian L, Huso T, et al. The High Mobility Group A1 (HMGA1) transcriptome in cancer and development. Curr Mol Med. 2016;16(4):353–93. doi: 10.2174/1566524016666160316152147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams MD, Zhang X, Belton AS, et al. HMGA1 drives metabolic reprogramming of intestinal epithelium during hyperproliferation, polyposis, and colorectal carcinogenesis. J Proteome Res. 2015;14(3):1420–31. doi: 10.1021/pr501084s. [DOI] [PubMed] [Google Scholar]
- 23.Grade M, Hummon AB, Camps J, et al. A genomic strategy for the functional validation of colorectal cancer genes identifies potential therapeutic targets. Int J Cancer. 2011;128(5):1069–79. doi: 10.1002/ijc.25453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao XP, Cao Y, Zhao H, et al. HMGA1 promoting gastric cancer oncogenic and glycolytic phenotypes by regulating c-myc expression. Biochem Biophys Res Commun. 2019;516(2):457–65. doi: 10.1016/j.bbrc.2019.06.071. [DOI] [PubMed] [Google Scholar]
- 25.Zanin R, Pegoraro S, Ros G, et al. HMGA1 promotes breast cancer angiogenesis supporting the stability, nuclear localization and transcriptional activity of FOXM1. J Exp Clin Cancer Res. 2019;38(1):313. doi: 10.1186/s13046-019-1307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reeves R. Molecular biology of HMGA proteins: Hubs of nuclear function. Gene. 2001;277(1–2):63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 27.Seatter MJ, De la Rue SA, Porter LM, Gould GW. QLS motif in transmembrane helix VII of the glucose transporter family interacts with the C-1 position of D-glucose and is involved in substrate selection at the exofacial binding site. Biochemistry. 1998;37(5):1322–26. doi: 10.1021/bi972322u. [DOI] [PubMed] [Google Scholar]
- 28.Deng D, Sun P, Yan C, et al. Molecular basis of ligand recognition and transport by glucose transporters. Nature. 2015;526(7573):391–96. doi: 10.1038/nature14655. [DOI] [PubMed] [Google Scholar]
- 29.Flavahan WA, Wu Q, Hitomi M, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16(10):1373–82. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo MH, Chang WW, Yeh BW, et al. Glucose transporter 3 is essential for the survival of breast cancer cells in the brain. Cells. 2019;8(12):1568. doi: 10.3390/cells8121568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao H, Hao Y, Zhou X, et al. Prognostic value of glucose transporter 3 expression in hepatocellular carcinoma. Oncol Lett. 2020;19(1):691–99. doi: 10.3892/ol.2019.11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ali A, Levantini E, Fhu CW, et al. CAV1 – GLUT3 signaling is important for cellular energy and can be targeted by Atorvastatin in non-small cell lung cancer. Theranostics. 2019;9(21):6157–74. doi: 10.7150/thno.35805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cosset E, Ilmjarv S, Dutoit V, et al. Glut3 addiction is a druggable vulnerability for a molecularly defined subpopulation of glioblastoma. Cancer Cell. 2017;32(6):856–68.e5. doi: 10.1016/j.ccell.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Representative transwell images of Figure 4A. (B) Representative transwell images of Figure 4B. (C) Representative transwell images of Figure 4D.