Abstract
Pollinators are important drivers of angiosperm diversification at both micro- and macroevolutionary scales. Both hummingbirds and bats pollinate the species-rich and morphologically diverse genus Vriesea across its distribution in the Brazilian Atlantic Forest. Here, we (i) determine if floral traits predict functional groups of pollinators as documented, confirming the pollination syndromes in Vriesea and (ii) test if genetic structure in Vriesea is driven by geography (latitudinal and altitudinal heterogeneity) or ecology (pollination syndromes). We analysed 11 floral traits of 58 Vriesea species and performed a literature survey of Vriesea pollination biology. The genealogy of haplotypes was inferred and phylogenetic analyses were performed using chloroplast (rps16-trnk and matK) and nuclear (PHYC) molecular markers. Floral traits accurately predict functional groups of pollinators in Vriesea. Genetic groupings match the different pollination syndromes. Species with intermediate position were found between the groups, which share haplotypes and differ morphologically from the typical hummingbird- and bat-pollinated flowers of Vriesea. The phylogeny revealed moderately to well-supported clades which may be interpreted as species complexes. Our results suggest a role of pollinators driving ecological isolation in Vriesea clades. Incipient speciation and incomplete lineage sorting may explain the overall low genetic divergence within and among morphologically defined species, precluding the identification of clear species boundaries. The intermediate species with mixed floral types likely represent a window into shifts between pollinator syndromes. This study reports the morphological-genetic continuum that may be typical of ongoing pollinator-driven speciation in biodiversity hotspots.
Keywords: Atlantic Forest, chiropterophily, floral evolution, ornithophily, pollination syndromes, speciation, Vriesea
In this study, we show floral traits to accurately predict hummingbird and bat pollinators in the Atlantic Forest genus Vriesea. Genetic groupings match the different pollination syndromes. We identify species with intermediate genetic position which differ morphologically from the typical hummingbird- and bat-pollinated Vriesea flowers by their mixed floral traits. We suggest a role of pollinators driving ecological isolation in Vriesea clades and that intermediate species likely represent a window into shifts between pollinators.
Introduction
Ever since the Origin of Species (Darwin 1859), biologists have debated the processes responsible for the generation of morphological and genetic diversity within and among species. Both geographic and ecological factors can shape genetic structure by affecting gene flow through isolation by distance or isolation by environment, respectively. Isolation by distance results from physical, geographic distance and barriers that restrict gene flow among populations (Wright 1943), whereas isolation by environment results from different ecological environments limiting gene flow (Wang and Bradburd 2014). Isolation by environment can be generated by diverse ecological processes, such as pollinator-driven genetic differentiation in plant populations (Johnson 2010). Pollinators are important drivers of angiosperm diversification at both micro- and macroevolutionary scales, either promoting selection of floral traits or speciation (Van der Niet et al. 2014).
Bromeliads are a highly diverse angiosperm clade in the Neotropics (Givnish 2017). Specifically, the genus Vriesea is one of the most conspicuous representatives of epiphytes in the Atlantic Forest, which constitutes its main centre of diversity (ca. 227 species; Stehmann et al. 2009; Ramos et al. 2019; Gouda et al. 2020, continuously updated), 85 % of which are found in no other biome (BFG - The Brazil Flora Group 2018). Key innovations such as leaves that form a rosette, absorptive trichomes, epiphytism and vertebrate pollination are likely linked to its rapid diversification in the last 6 million years (Givnish et al. 2014; Kessous et al. 2020). In addition, the genus is distributed across a broad range of habitats in the Atlantic Forest domain, from the restingas (sandy, coastal plains) to campos de altitude (high altitude fields, including cloud forests) and rocky outcrops (inselbergs) (Smith and Downs 1977; Costa et al. 2014, 2015; Givnish et al. 2014; Ramos et al. 2019; Kessous et al. 2020). The large latitudinal (ca. 5–30°S) and altitudinal (0–2892 m a.s.l.) amplitude of the Atlantic Forest also make it a prime study area for plant diversification (Antonelli and Sanmartín 2011; Alvares et al. 2013). Such extraordinary environmental heterogeneity has been shown to influence Vriesea species diversification and variation in morphological traits (Neves et al. 2019).
Based on flower morphology, two sections were recognized in Vriesea: V. sect. Vriesea, with red to yellow floral bracts, tubular flowers with exserted stamens; and V. sect. Xiphion (see Barfuss et al. 2016 for the correct application of the name Xiphion), with brown to green floral bracts, campanulate corollas with included stamens (Costa et al. 2014). These sections were originally described based on two main floral types: hummingbird- and bat-pollinated flowers, respectively (Mez 1896, 1934–1935). Since then, species have been classified into one or another section based on the labile character of the stamen position in relation to the corolla fauce (included vs. exserted) (Mez 1896, 1934–1935; Harms 1930; Smith and Downs 1977). This classification led to the controversial taxonomic placement of some Vriesea species with mixed floral traits. To date, no phylogeny supports the two sections as monophyletic (Costa et al. 2015; Gomes-da-Silva and Souza-Chies 2017; Machado et al. 2020).
The unique set of floral traits (morphology, colour, scent, rewards and phenology) that are associated to a particular group of pollinators define plant pollination syndromes (Fenster et al. 2004). Studies on pollination biology of Vriesea species from both sections have added information on flower shape, time of anthesis (diurnal or nocturnal), floral visitors and effective pollinators confirming the evident adaptation to hummingbird or bat pollinators (e.g. Sazima et al. 1999; Buzato et al. 2000). Based on the colours of the corolla and floral bracts and time of flower anthesis, more Vriesea species have the hummingbird (ca. 137 spp.) than bat pollination syndrome (ca. 90 spp.) (Costa et al. 2014; Gouda et al. 2020, continuously updated). This difference in species richness is also seen in the high diversity of Atlantic Forest plant-pollinating hummingbird species compared with that of bats (Grantsau 1989; Marinho-Filho and Sazima 1998). More importantly, bromeliads are the most representative family of hummingbird- and bat-pollinated flora in the Atlantic Forest, with genus Vriesea being the richest bat-pollinated lineage (Sazima et al. 1999; Buzato et al. 2000; Aguilar-Rodriguez et al. 2019).
Here, we test if floral traits accurately predict the different functional groups of pollinators in Vriesea by using data on known pollinators from the literature. We also test if genetic structure in Vriesea is mainly shaped by geography (latitudinal and altitudinal heterogeneity) or ecology (pollination syndromes). Our study provides a first general assessment of the role of pollinator interactions in shaping genetic structure and promoting floral traits diversity in Vriesea.
Materials and Methods
Morphological analyses of floral traits—predicting functional groups of pollinators and validating pollination syndromes
We sampled 76 accessions representing 58 Vriesea species, for which we extracted also the molecular data. Such sampling covers the morphological and geographic diversity within genus (Table 1; seeSupporting Information—Table S1). To visualize the morphological variation of floral traits in Vriesea and test if the species group according to the two different pollination syndromes (hummingbird or bat), we performed a non-metric multidimensional scaling (NMDS; Rabinovitz 1975) using Gower distance (Gower 1971). Non-metric multidimensional scaling is an ordination method which groups similar objects close to one another based on rank distances and has been used for similar kinds of data to build floral morphospaces (Ollerton et al. 2009; Chartier et al. 2014). In addition to the NMDS, we ran a cluster analysis using the same distance measure to depict morphological relatedness among species. Both analyses were performed in PAST 3.22 (Hammer et al. 2001). To test for significant differences among the resulting different clusters, which we expected to correspond to the two pollination syndromes, we ran a PERMANOVA using the R 3.5.0 (R Development Core Team 2018) package ‘vegan’ (Oksanen et al. 2018), based on 10 000 permutations. We used data on floral bract colour [mostly yellow tones, Fig. 1E; red tones, Fig. 1B and C; green, Fig. 1I; purple/brown, Fig. 1J; stramineous (with texture and colour of straw, dry and crumbly, Fig. 1G and L)], floral bract size related to flower length (shorter than midpoint of the flower, Fig. 1D; equal to midpoint to longer than flower, Fig. 1C), floral bract imbrication (not imbricate, Fig. 1E; imbricate, Fig. 1B), flower disposition along inflorescence rachis or branches (polystichous, Fig. 1D; distichous, Fig. 1E), time of day of flower anthesis (diurnal; nocturnal), position of flowers at anthesis in relation to floral bract (included with <1/3 of the flower exposed, Fig. 1D; exserted with half of more exposed, Fig. 1J), torsion of the flowers (not-secund, Fig. 1B; partially or totally secund, Fig. 1G, I and L), flower odour (absent; present), corolla colour (mostly yellow, Fig. 1A; white, Fig. 1J; pale-yellow, Fig. 1G; green; wine/purple, Fig. 1H), corolla shape (tubular, Fig. 1A; campanulate, Fig. 1G) and stamens position at anthesis (included, Fig. 1J and L; exserted, Fig. 1A, C and F), seeSupporting Information—Table S2. We compiled trait information from the taxonomic and/or phylogenetic works developed by Moura and Costa (2014), Costa et al. (2015), Gomes-da-Silva and Souza-Chies (2017), Neves et al. (2018) and Uribbe et al. (2020). Such works were based on vast sampling of each species both in the field and herbaria collections and therefore we are taking into account the existing intraspecific variation. All traits investigated are potentially involved in attraction and performance (effectiveness, efficiency, efficacy) of pollinators.
Table 1.
Taxa sampled for chloroplast (matK and rps16-trnK) and nuclear (PHYC) regions, assigned to their respective putative pollination syndrome. Species codes, haplotypes and locality information (country, federal state and municipality). RJ, Rio de Janeiro; ES, Espírito Santo; MG, Minas Gerais; SP, São Paulo; BA, Bahia; PR, Paraná; PE, Pernambuco; SC, Santa Catarina; RS, Rio Grande do Sul.
| Pollination syndrome | Species | Species code | Haplotypes cpDNA/PHYC | Locality |
|---|---|---|---|---|
| Outgroup—bats and hawkmoths (Martinelli et al. 1994) | Alcantarea imperialis | Ai | H1/Hn1 | Brazil, RJ, Teresópolis |
| A. regina | Ar | H2/Hn2 | Brazil, RJ, Rio de Janeiro | |
| Stigmatodon croceanus | Scr | H3/Hn3 | Brazil, RJ, Santa Maria Madalena | |
| S. harrylutheri | Sha | H4/Hn4 | Brazil, ES | |
| S. plurifolius | Spl | H5/Hn5 | Brazil, ES | |
| S. costae | Vco | H18/- | Brazil, RJ, Niterói | |
| Vriesea (‘Stigmatodon’) oligantha | Vol | H31/- | Brazil, MG | |
| Vriesea—hummingbirds | V. agostiniana | Vag | H8/Hn10 | Brazil, SP, Caraguatatuba |
| V. amethystina | Vam | H9/Hn11 | Brazil, RJ, Rio de Janeiro | |
| V. billbergioides | Vbi | H11/Hn13 | Brazil, RJ, Teresópolis | |
| V. botafogensis | Vbo | H13/Hn15 | Brazil, RJ, Rio de Janeiro | |
| V. cacuminis | Vcu | H14/Hn13 | Brazil, MG, Ibitipoca | |
| V. calimaniana | Vcl | H15/Hn16 | Brazil, ES, Venda Nova do Imigrante | |
| V. capixabae | Vcp | H6/Hn17 | Brazil, ES, Ibitirama | |
| V. carinata var. carinata | Vcr | H6/Hn18 | Brazil, ES, Santa Teresa | |
| V. carinata var. flavominiata | Vcrf | H16/Hn19 | Brazil, ES, Santa Maria de Jetibá | |
| V. carinata var. mangaratibensis | Vcrm | H6/Hn20 | Brazil, RJ, Mangaratiba | |
| V. duvaliana | Vdu | H6/Hn22 | Brazil, BA, Itacaré | |
| V. eltoniana | Vel | H19/Hn23 | Brazil, RJ, Arraial do Cabo | |
| V. ensiformis var. ensiformis | Vem | H6/Hn6 | Brazil, SP, Cananéia | |
| V. ensiformis var. ensiformis | Vem | H20/Hn25 | Brazil, PR, Guaraqueçaba | |
| V. ensiformis var. ensiformis | Vem | H6/Hn26 | Brazil, PE, Caruaru | |
| V. ensiformis var. ensiformis | Vem | H20/- | Brazil, SC, Corupá | |
| V. ensiformis var. bicolor | Venb | H6/Hn24 | Brazil, SP, Caraguatatuba | |
| V. erythrodactylon | Ver | H21/Hn27 | Brazil, ES, Santa Teresa | |
| V. flammea | Vfm | H17/Hn29 | Brazil, PR, Paranaguá | |
| V. flammea | Vfm | H17/Hn21 | Brazil, SP | |
| V. flava | Vfl | H23/Hn30 | Brazil, PR, Morretes | |
| V. fluviatilis | Vfu | H25/Hn32 | Brazil, RJ, Santa Maria Madalena | |
| V. fluviatilis | Vfu | H19/Hn6 | Brazil, RJ, Casimiro de Abreu | |
| V. gracilior | Vgc | H24/- | Brazil, ES, Santa Teresa | |
| V. gradata | Vgr | H26/Hn33 | Brazil, RJ, Petrópolis | |
| V. gutatta | Vgu | H27/- | Brazil, SC, Antônio Carlos | |
| V. heterostachys | Vhe | H6/Hn32 | Brazil, RJ, Teresópolis | |
| V. aff. heterostachys | Vhe | H6/Hn6 | Brazil, SP, Ilha do Cardoso | |
| V. inflata | Vif | H6/- | Brazil, SP, São Luis do Paraitinga | |
| V. aff. inflata | Vif | H6/Hn7 | Brazil, SP, Bananal | |
| V. interrogatoria | Vit | H28/Hn20 | Brazil, SP, Cunha | |
| V. lubbersii | Vlu | H47/- | Brazil, RJ, Valença | |
| V. maxoniana | Vma | H48/- | Bolívia, Santa Cruz | |
| V. modesta | Vmo | H6/Hn32 | Brazil, RJ, Santa Ma. Madalena | |
| V. neoglutinosa | Vne | H30/Hn36 | Brazil, RJ | |
| V. paraibica | Vpa | H32/- | Brazil, RJ, Nova Friburgo | |
| V. aff. procera | Vpr | H7/Hn8 | Brazil, RS, Torres | |
| V. psittacina | Vpi | H34/Hn6 | Brazil, RJ | |
| V. recurvata | Vrc | H35/- | Brazil, BA, Ilhéus | |
| V. repandostachys | Vrp | H6/- | Brazil, ES, Domingos Martins | |
| V. rhodostachys | Vrh | H36/Hn37 | Brazil, ES, Santa Teresa | |
| V. rhodostachys | Vrh | H37/Hn38 | Brazil, BA, Arataca | |
| V. rodigasiana | Vro | H38/- | Brazil, SP, Cananéia | |
| V. rubyae | Vru | H6/Hn39 | Brazil, RJ, Petrópolis | |
| V. rubyae | Vru | H39/Hn40 | Brazil, RJ, Angra dos Reis | |
| V. sandrae | Vsa | H40/- | Brazil, BA, Santa Terezinha | |
| V. saundersii | Vau | H43/Hn41 | Brazil, RJ, Rio de Janeiro | |
| V. scalaris var. scalaris | Vsc | H6/Hn42 | Brazil, ES, Santa Teresa | |
| V. scalaris var. scalaris | Vsc | H6/Hn42 | Brazil, PR, Paranaguá | |
| V. scalaris var. viridis | Vscv | H42/Hn42 | Brazil, PE, Taquaritinga do Norte | |
| V. seideliana | Vse | H6/Hn6 | Brazil, ES, Domingos Martins | |
| V. simplex | Vsi | H6/Hn43 | Brazil, ES, Santa Teresa | |
| V. simplex | Vsi | H6/- | Brazil, SP, São Luis do Paraitinga | |
| V. sucrei | Vsu | H44/- | Brazil, RJ, Arraial do Cabo | |
| V. sucrei | Vsu | H44/Hn45 | Brazil, ES, Domingos Martins | |
| V. taritubensis var. brevisepala | Vtab | H8/- | Brazil, RJ, Guapimirim | |
| V. taritubensis var. patens | Vtap | H8/Hn9 | Brazil, SP, São Luis do Paraitinga | |
| V. taritubensis var. taritubensis | Vta | H8/Hn6 | Brazil, RJ, Paraty | |
| V. teresopolitana | Vte | H6/- | Brazil, RJ, Teresópolis | |
| V. vagans | Vva | H6/Hn47 | Brazil, SP, Bananal | |
| Vriesea—bats | V. atra | Vat | H10/Hn12 | Brazil, RJ, Teresópolis |
| V. bituminosa | Vbt | H12/Hn14 | Brazil, RJ, Teresópolis | |
| V. fenestralis | Vfe | H22/Hn28 | Brazil, RJ, Nova Iguaçu | |
| V. fosteriana | Vfo | H13/Hn31 | Brazil, ES, Cachoeiro de Itapemirim | |
| V. gigantea | Vgi | H13/Hn12 | Brazil, ES, Domingos Martins | |
| V. grandiflora | Vgd | H13/- | Brazil, RJ | |
| V. hydrophora | Vhy | H13/Hn34 | Brazil, RJ, Teresópolis | |
| V. longicaulis | Vlo | H29/Hn35 | Brazil, RJ, Teresópolis | |
| V. longistaminea | Vlg | H46/- | Brazil, MG, Mariana | |
| V. minuta | Vmi | H13/- | Brazil, BA | |
| V. pabstii | Vpb | H13/Hn13 | Brazil, SP, Bananal | |
| V. platynema var. variegata | Vplv | H33/- | Brazil, PR | |
| V. pseudatra | Vps | H13/- | Brazil, RJ, Rio de Janeiro | |
| V. sazimae | Vsz | H41/- | Brazil, RJ, Itatiaia | |
| V. sincorana | Vsc | H43/Hn44 | Brazil, BA, Rio de Contas | |
| V. unilateralis | Vun | H45/Hn46 | Brazil, SP, Cunha |
Figure 1.
Floral morphological diversity of Vriesea species evidencing hummingbird (A–F) and bat (G–L) pollination syndromes. (A) Vriesea simplex, (B) V. duvaliana, (C) V. ensiformis var. bicolor, (D) V. cacuminis, (E) V. gracilior, (F) V. procera, (G) V. pseudatra, (H) V. sazimae, (I) V. unilateralis, (J) V. crassa, (K) V. sincorana, (L) V. longiscapa. Photos: B. Neves, F. P. Uribbe, R. L. Moura, A. F. Costa, T. Wendt, R. Sadala and Bromeliário imperialis via Bromeliad Photo Index.
Lastly, to determine if floral traits predict pollinators and this way validate the pollination syndromes in the genus, we compiled pollinators and floral visitors for 39 Vriesea species based on a literature survey and personal observations [seeSupporting Information—Table S3]. We expected to find separate clusters for each syndrome in the ordination analyses described above. Then we tested whether the species with known pollinators fall into their respective syndrome cluster, allowing for confirmation of syndromes.
Molecular analyses
DNA sampling.
We sampled 83 individuals including the 76 accessions of Vriesea from the morphological analyses of floral traits and seven outgroup accessions from the sister genera Alcantarea and Stigmatodon (Table 1; Barfuss et al. 2016). We collected young leaves from individuals in natural populations that were preserved in silica gel before DNA extraction using 2× CTAB (Doyle and Doyle 1987) with the modifications of Fay et al. (1998). We amplified and sequenced DNA for two chloroplast (rps16-trnk and matK; Crayn et al. 2000; Shaw et al. 2007) and one nuclear (PHYC; Barfuss et al. 2016) regions, as described in Kessous et al. (2020). We generated a total of 34 sequences for 21 accessions in this study [seeSupporting Information—Table S1]. Additionally, we included sequences generated by Barfuss et al. (2016) and Kessous et al. (2020) available in GenBank. For the 16 samples collected by others that were not georeferenced, we estimated geographical coordinates based jointly on locality information from collectors and records from speciesLink database (http://splink.cria.org.br/).
Haplotype and phylogenetic relationships.
We verified the sequences electropherograms in Chromas 2.33 (Chromas Technelysium, South Brisbane, Australia) and performed multiple sequence alignment in MAFFT 7.0 (Katoh and Standley 2013) following default settings. The final data sets had 83 accessions for the concatenated chloroplast markers with 1543 bp and 60 accessions for the nuclear marker with 664 bp. We analysed chloroplast and nuclear data sets separately to evaluate the evolutionary relationships among haplotypes and taxa.
To first explore genetic variation in Vriesea, we inferred the haplotype genealogy for both chloroplast and nuclear data sets. Such analysis can reveal genetic patterns other than that evidenced by the phylogenies, showing how haplotypes group. The use of this analysis is justified to our sampling by the recency of the genus Vriesea and the existence of several incipient species still in process of differentiation (Wendt et al. 2008; Zanella et al. 2016; Neri et al. 2017; Kessous et al. 2020). Also, such approach has been used in a multispecies context for another bromeliad genera (Krapp et al. 2014; Goetze et al. 2017). We excluded the mononucleotide repeat length variations due to ambiguous alignment and coded the indels longer than 1 bp as a single mutational event. We identified the haplotypes using DnaSP 5.10.01 (Librado and Rozas 2009) and built the haplotype network using the median-joining method (Bandelt et al. 1999) in Network 5 (available at http://www.fluxus-engineering.com).
To infer phylogenetic relationships among chloroplast haplotypes and among species for both chloroplast and nuclear data sets, we performed Bayesian analyses in MrBayes 3.2 (Ronquist et al. 2012) using the CIPRES server (Miller et al. 2010). We used the HKY nucleotide substitution model calculated in MEGA X (Kumar et al. 2018) for each molecular marker separately. Two runs of four Monte Carlo Markov Chain (MCMC) computations were run for 10 000 000 generations. We sampled trees every 1000 generations. The first 25 % of generations were discarded as burn-in. The consensus tree was drawn in FigTree 1.4.2 (Rambaut 2014). We considered well-supported clades those with posterior probabilities (PPs) above 0.95.
Genetic divergence.
To assess the consistency of the genetic relationships among taxa, we ran independent principal coordinate analysis (PCoA) for both chloroplast and nuclear data in the R package ‘ape’ (Paradis et al. 2004). We used the matrix of pairwise genetic distances amongst the Vriesea individuals computed in MEGA under a Maximum Composite Likelihood model, estimating variance using 1000 replicates of bootstrap.
Results
Morphological analyses of floral traits—predicting pollinators and validating pollination syndromes
The NMDS and cluster analysis of floral traits inferred morphological separation of the two pollination syndromes in Vriesea (Fig. 2). The first two NMDS axes retained 98 % of the variance of the original data variation (stress 0.14, axis 1 R2 = 0.90, axis 2 R2 = 0.08). We found significant differences among clusters corresponding to the different pollination syndromes (F(1, 74) = 381.56, P < 0.0001). The hummingbird-pollinated species have floral bracts that are mostly red or yellow, frequently equal or longer in length than the flower and are either imbricate or not; flowers are distichous or polystichous, with diurnal anthesis, frequently exserted from floral bracts with half or more exposed, not-secund and odourless; the corolla is yellow or rarely white especially in polystic flowers, is tubular and with mostly exserted stamens (Fig. 1A–F). The bat-pollinated species have floral bracts that are mostly green, purple or brown, frequently equal or longer than the flower, not imbricate; flowers are distichous, with nocturnal anthesis, frequently exserted from the floral bracts with half or more exposed, can be secund (with flowers shifting to an angle of 90° towards one side of the inflorescence) or partially secund (flowers shift <90°), present a garlic odour; corolla is frequently pale-yellow, campanulate, stamens frequently included (Fig. 1G–L).
Figure 2.
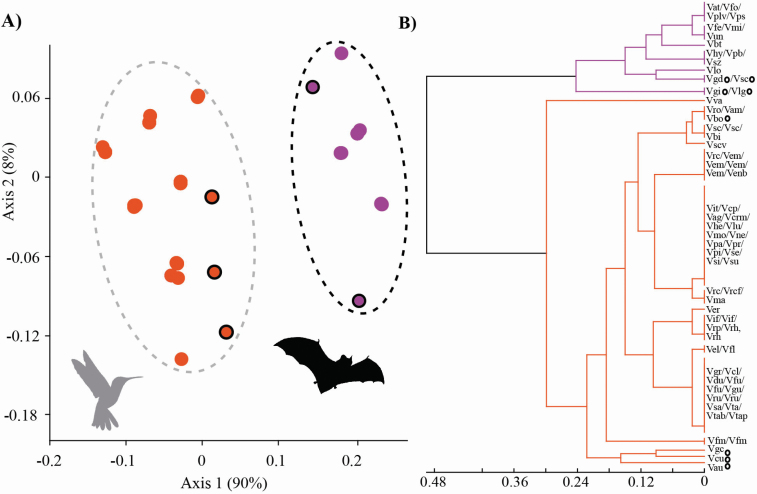
Morphological analyses supporting divergence among Vriesea pollination syndromes, hummingbird (orange) and bat (purple). (A) Non-metric multidimensional scaling and (B) cluster dendogram of 11 categorical reproductive characters related to floral bract and flower. Species with morphology distinct from the typical hummingbird- and bat-pollinated flowers (mixed floral traits) are circled in black. Silhouette images of pollinators downloaded from http://phylopic.org.
Species with mixed floral morphology showed intermediate position along NMDS axis 1 (Fig. 2). Such species clearly belong to their respective pollination syndrome cluster but are differentiated from the typical hummingbird (Fig. 1A) and bat (Fig. 1G) flowers. Among hummingbird-pollinated species: V. cacuminis has the petal apex erect, often presenting a small flower aperture and included stamens (Fig. 1D); V. saundersii presents the same characteristics as the former; additionally, it is highly similar to V. botafogensis which has a more typical hummingbird-pollinated flower due to its clearly exserted stamens, but shares overall peculiar morphology of inflorescence and rosette; V. gracilior has urceolate flowers with included stamens (Fig. 1E). Among bat-pollinated species: V. sincorana (Fig. 1K), V. gigantea, V. grandiflora and V. longistaminea have long exserted stamens that are arranged separately.
We compiled information of pollination biology from the literature for 23 species of the 58 Vriesea species sampled here [seeSupporting Information—Table S3]. All of the 23 species with known pollinators fell into their respective pollination syndrome cluster, including those with mixed floral traits (Fig. 1D, E and K; seeSupporting Information—Table S3). With this result, we showed floral traits to accurately predict pollinators in Vriesea confirming pollination syndrome assignments for all species we sampled.
Haplotype and phylogenetic relationships
The chloroplast data set comprised 1516 bp with a GC (guanine–cytosine) content of 29.9 % and 84 polymorphic sites (51 transitions, 26 transversions and 8 indels) and 48 haplotypes. The nuclear data set was 664 bp with a GC content of 49.1 % and 74 polymorphic sites (39 transitions, 37 transversions and 2 indels) and 47 haplotypes [seeSupporting Information—Table S1].
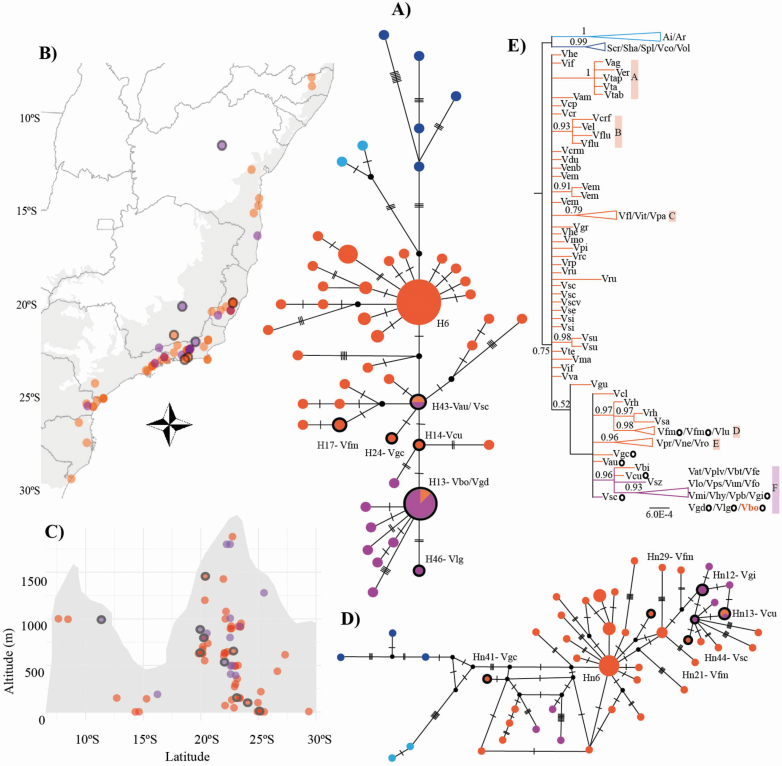
The chloroplast haplotype network reflected the different pollination syndromes in Vriesea (Fig. 3A) and did not resolve any groupings across latitudinal or elevational space (Fig. 3B and C). Vriesea presented two haplogroups pollinated either by hummingbirds and bats, which shared only two haplotypes (H13 and H43, Table 1; Fig. 3A). The haplotypes of each syndrome were resolved in a star-like pattern. The most frequent and central haplotype (H6) was exclusively shared among 21 hummingbird-pollinated species. The second most frequent (H13) was shared among seven bat-pollinated species and the hummingbird-pollinated V. botafogensis (Table 1; Fig. 3A; seeSupporting Information—Fig. S1). H43 was shared between species of both syndromes, V. saundersii and V. sincorana. Haplotypes of the species with mixed floral types had an intermediate position in the chloroplast network (Fig. 3A). Haplotypes H6 and H13 were found in individuals occurring from 8°S to 25°S and 16°S to 22°S, at altitudes from the sea level to 1019 m and 150 to 1801 m in the highlands, respectively. Such variation spans almost the entire geographic range of Atlantic Forest Vriesea species [seeSupporting Information—Fig. S4]. Most of the remaining plastid haplotypes were restricted to a single species.
Figure 3.
Results based on the cpDNA (matK and rps16-trnK) and PHYC data set for 83 taxa, including 76 Vriesea accessions. Coloured symbols represent hummingbird (orange) and bat (purple) syndromes in Vriesea. Blue symbols represent the outgroup genera Alcantarea and Stigmatodon. Vriesea species with floral morphology distinct from the typical hummingbird- and bat-pollinated flowers (mixed floral traits) are circled in black. (A) Chloroplast median-joining network showing genetic divergence among pollination groups. Each circle represents a haplotype with the size proportional to its total frequency. We indicate codes for haplotypes of intermediate species and the most frequent H6 and Hn6 (Table 1). Mutational steps are indicated with dashes and hypothetic haplotypes with black dots. (B) Map of Vriesea species sampling distribution along the Brazilian Atlantic Forest in grey and (C) across altitude. The mountain profile is illustrative. (D) PHYC median-joining network. (E) Chloroplast Bayesian phylogeny with posterior probabilities above 0.50 shown.
The nuclear haplotype network showed weak relationship among species with different pollination syndromes (Fig. 3D). We detected a star-like pattern for the hummingbird syndrome. The most frequent haplotype was shared among six hummingbird-pollinated species (Hn6). Hn13 was shared among species with both pollination syndromes (Table 1; Fig. 3D; seeSupporting Information—Fig. S1). The remaining haplotypes were restricted to a single species.
The chloroplast phylogeny inferred six moderately to well-supported clades, despite the backbone of the tree being unresolved (Fig. 3E). The genus was recovered as monophyletic (PP = 0.75) and the hummingbird-pollinated species (clades A, B, C, D and E) were supported (1.00, 0.93, 0.79, 0.98 and 0.96 PP, respectively), while clade F mainly included bat-pollinated species, but also the hummingbird-pollinated V. billbergioides, V. cacuminis and V. botafogensis (PP = 0.93; Table 1). The phylogenies of the chloroplast haplotypes and of the nuclear marker PHYC showed low resolution [seeSupporting Information—Figs S2andS3]. Finally, the outgroups, Stigmatodon and Alcantarea, were well-supported as distinct genetic groups in all analyses.
Genetic divergence
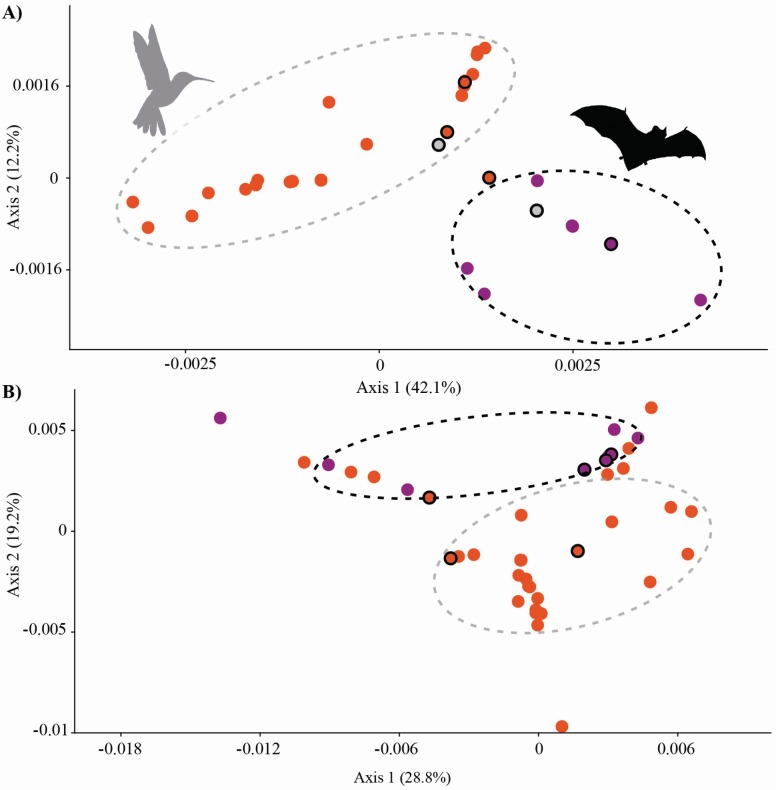
The PCoA revealed close genetic relationships of species within the distinct pollination syndromes (Fig. 4). In the chloroplast data, species from both hummingbird and bat pollination syndromes formed distinct genetic clusters (Fig. 4A). For PHYC, we detected an overlap among the pollination groups (Fig. 4B). We showed species with mixed floral traits to occupy an intermediate position along axis 1 of chloroplast PCoA plot (Fig. 4A). On the PHYC PCoA plot the pattern was diffuse and the species with mixed floral traits were scattered in morphospace, tending to concentrate in the middle of axis 2 (Fig. 4B).
Figure 4.
Principal coordinate analysis for (A) cpDNA and (B) PHYC data sets showing genetic divergence among hummingbird (orange dots) and bat (purple dots) pollination syndromes in Vriesea. Species with morphology distinct from the typical hummingbird- and bat-pollinated flowers (mixed floral traits) are circled in black. Grey dots represent overlap of species from both syndromes which have no genetic divergence. Silhouette images of pollinators downloaded from http://phylopic.org.
Discussion
Using morphological and molecular data we show hummingbird and bat pollinators to be closely associated with ecological isolation between Vriesea clades (Figs 2 and 3A). The selection imposed by the distinct pollinator groups likely promoted floral traits and species diversification, particularly within clades associated with each pollinator group or derived from shifts among pollinator groups. Also, we confirm floral traits to predict pollinator functional groups in Vriesea. We identify species with intermediate position between the different pollination syndrome groupings, which share haplotypes (H13, H43 in Fig. 3A and Hn13 in Fig. 3D), are phylogenetically related (clade F in Fig. 3E) and differ morphologically from the typical hummingbird- and bat-pollinated Vriesea flowers by their mixed floral traits (Fig. 1D, E and K). The phylogeny reveals moderately to well-supported clades that are congruent with haplotype relationships and reflect groupings of recognized species complexes (Fig. 3E). We suggest that incipient speciation and incomplete lineage sorting may cause the low genetic divergence detected in the markers surveyed, which also lead to species delimitation challenges in the genus. Future genomic sequencing may further refine the patterns reported here.
Prediction of pollination syndromes
Considering jointly the literature available for Vriesea species pollination biology [seeSupporting Information—Table S3] and our NMDS and clustering results based on 11 categorical traits (Fig. 2), we support floral morphology as a good predictor of functional groups of pollinators. Other studies have shown a similar predictive utility of pollination syndromes in other angiosperm groups (Rosas-Guerrero et al. 2014; Serrano-Serrano et al. 2017). Here, we focused on the two likely most effective functional groups of pollinators in Vriesea: hummingbirds and bats. Although another pollinators such as insects have also been shown to provide pollination services for Vriesea species, they are probably less effective as they carry less pollen, potentially drive more self-pollination which can affect seed viability and fly shorter distances than vertebrates (Fleming et al. 2009; Schmid et al. 2011; Paggi et al. 2013). Although a recent study showed hummingbirds as efficient as bees and ants in facilitating pollination per visit in V. neoglutinosa, fruit set was reduced when only insects are allowed to visit the flowers, insects potentially bring more self-pollen and seed quality was not examined in order to assess viability (Magalhães et al. 2018). Additionally, Vriesea flowers produce large volume of nectar which results in higher amount of sugar, reinforcing the strong association with vertebrate pollinators whose energetic needs are higher than insects (Sazima et al. 1995; Göttlinger et al. 2019). Some cases of bat-pollinated flowers have been reported to be visited by hummingbirds at dawn, but when hummingbirds feed on the small amount of nectar left by bats in withered flowers, it does not compete with the performance of primary pollinators (Sazima et al. 1995; Aguilar-Rodríguez et al. 2019).
The morphological characters analysed here are fundamental not only to define syndromes but also to define pollinator performance. Bergamo et al. (2019) showed that in long tubular flowers, floral bracts create colour contrast and enhance the petal signal more for hummingbirds than for bees, hence creating an avoidance mechanism against nectar robbing bees. We measured floral bract size related to flower length and its relative position, traits that are directly linked to bract/petals contrast. We also measured flower position and torsion following the hypothesis that in species with distichous and secund flowers, pollinators can easily reach a greater number of flowers since they all face the same side (Costa et al. 2014; Aguilar-Rodríguez et al. 2019). Flower torsion is especially important for bats when pollinating compound inflorescences, since the flowers from each branch are properly exposed. Further, stamen position influences pollinator effectiveness due to the placement of pollen deposition along the pollinator body. Rocca and Sazima (2013) showed that short-billed hummingbirds receive pollen on the top of the head and deposit it on the centre of the stigma, whereas long-billed species receive pollen on the proximal part of the bill and deposit it on the lower lobe of the stigma in V. rodigasiana. In this case, short-billed hummingbird-pollinated flowers had six times more pollen tubes formed than long-billed ones. In bat-pollinated species, the pollen of flowers with anthers located in the lower side of corolla, such as in V. bituminosa, is concentrated on bat chins, whereas the spread anthers of V. gigantea deposit pollen all over the face of bats (Sazima et al. 1995).
Ecological isolation drives genetic structure in Vriesea
Our results suggest an important role of ecological instead of geographical isolation in driving genetic structure in Vriesea (Fig. 3). The specialized association with different vertebrate pollinator groups has resulted in ecological isolation amongst clades within Vriesea. We found few shared haplotypes between syndromes in our chloroplast and nuclear data (Fig. 3A and D) and demonstrated clear floral specialization on both hummingbirds and bats for pollination (Fig. 1). It has been argued that strong plant–pollinator interactions lead to an increase in species diversity (Givnish et al. 2014; Lagomarsino et al. 2016; Serrano-Serrano et al. 2017). We hypothesize that the association with hummingbirds and bats is a main biotic driver of Vriesea diversification. In addition, pollinator shifts can open new adaptive space for species diversification and distinct mechanisms may increase speciation within each main pollinator group (which seems to be where most diversification in the genus occurs), such as (i) floral specialization on specific pollinator species; (ii) efficiency of pollen transfer and deposition by specific pollinator species which affect connectivity among populations resulting in allopatric speciation; (iii) different flowering time and pollinator behaviour when foraging; and (iv) different pollinator species distribution along altitude and habitat types according to their physiological preferences (Aguilar-Rodriguez et al. 2019; Kessler et al. 2020).
We found stronger structure for syndromes in the chloroplast DNA compared to the more diffuse pattern for the nuclear markers, and a proportionally low number of haplotypes for the chloroplast than nuclear data (Fig. 3A and D; Table 1). Maternally inherited markers such as those from the chloroplast are generally highly structured and present low variation (Petit et al. 2005; Goetze et al. 2017), at least when compared to nuclear DNA (Barfuss et al. 2016). Despite the high structure, haplotype sharing does occur within and among syndromes, as well as among species collected in distant localities, for both chloroplast and nuclear DNA, albeit infrequently. For example, haplotypes H6 and Hn12 are found in species occurring along a range of ca. 3000 km (Table 1; seeSupporting Information—Table S1). Such findings may reflect incipient speciation due to recent species diversification within the genus (crown age 4–2 Mya, Kessous et al. 2020), preventing sufficient time to accumulate genetic differences and causing various species complexes. With this, our findings suggest incomplete lineage sorting through the retention of ancestral polymorphisms. Incomplete lineage sorting often explains such disperse spatial patterns of shared genetic variation across species (Goetze et al. 2017). Also, interspecific gene flow has been shown by previous studies to occur in Vriesea (Zanella et al. 2016; Neri et al. 2017), as well as in its sister group Alcantarea (Lexer et al. 2016).
We did not detect a geographic pattern from our data (like shown by Krapp et al. (2014) for other bromeliad genera) and show that Vriesea species from each syndrome are widely distributed across latitude and altitude in the Atlantic Forest (Fig. 3B and C; seeSupporting Information—Fig. S4). Studies comparing hummingbird- and bat-pollinated plant assemblages in the Atlantic Forest at different altitudinal ranges showed higher species diversity of both Vriesea and pollinators in the lowlands (Sazima et al. 1999; Buzato et al. 2000). For bromeliads in general, it has been argued that bat-pollinated species are more diverse at humid mid-elevations and lowlands, whereas hummingbird-pollinated species are more diverse in mid-elevation to highlands, which coincides with the physiological demands of the different groups of pollinators (see review of Kessler et al. 2020). In addition, these authors discuss the shifts among pollination syndromes to occur predominantly in transition zones at mid-elevation areas. Such distribution pattern does not agree with the one of the intermediate species we recognize in this study, as they are distributed along whole range of Vriesea species (Fig. 3C). Considering the high species richness of Vriesea, studies on pollination biology to unveil such patterns remain scarce.
Regardless of whether there are distributional differences across altitude between the two syndromes, there are certainly differences in habitat types. Bat-pollinated Vriesea usually occur in open habitats as epiphytes in the forest canopies, or are rupiculous or saxicolous in the highlands, making it easier for bats to echolocalize the flowers in addition to the olfactory and visual attractants (Gonzalez-Terrazas et al. 2016), whereas hummingbird-pollinated species are more frequent epiphytes in the forest understory, in both highlands and lowlands, or terrestrial in the restingas (for occurrence and habitat information on Vriesea species, see Flora do Brasil 2020, continuously updated). The interaction of multiple biotic and abiotic factors drives diversification in species-rich Neotropical clades (Antonelli et al. 2018). As a first step to understand Vriesea diversification we here show a strong signal of pollinators in shaping genetic structure and likely contributing to its high species diversity.
Infrageneric relationships
We recovered some well-supported clades consisting exclusively of hummingbird-pollinated species and a large clade including mostly bat-pollinated species, but also the species with mixed morphological traits, despite the overall low resolution of the phylogeny (Fig. 3E). Additionally, these well-supported clades comprise species complexes. Clade A includes species of V. incurvata complex (Neves et al. 2018); clade B together with clade C, which is formed by species of V. paraibica group (Costa et al. 2009), comprise the ‘inflated group’. The ‘inflated group’ was recovered in the phylogeny produced by Gomes-da-Silva and Souza-Chies (2017), but without including V. fluviatilis (former V. gradata var. bicolor, Kessous and Costa 2017). The monophyly of V. corcovadensis group was also inferred (Gomes-da-Silva and Souza-Chies 2017; Machado et al. 2020) and we corroborate it here (clade D). Clade E includes species of the V. procera complex Uribbe et al. (2020). Clade F mainly includes Vriesea with bat pollination syndrome but also the mixed forms: the hummingbird-pollinated V. cacuminis and V. botafogensis, and the bat-pollinated V. gigantea, V. grandiflora, V. longistaminea and V. sincorana (Table 1; Fig. 3E). Differences mainly consist in the corolla aperture, shape and position of stamens at anthesis (see Fig. 1D, E and K; Costa et al. 2015).
Sanmartin-Gajardo and Sazima (2005) identified a putative transition from hummingbird to bat pollination in a species of Sinningieae (Gesneriaceae) with intermediate floral traits. Floral specialization on hummingbird syndromes may not be an evolutionary dead end, as transitions may occur to hawkmoth or bat, for example (Tripp and Manos 2008) and even the reverse, from bat to hummingbird (Lagomarsino et al. 2017). For bromeliads, Aguilar-Rodriguez et al. (2019) showed pollination by hummingbirds to be the ancestral syndrome, with bat pollination originating multiple times. The authors sampled only three Vriesea species (sensuBarfuss et al. 2016), but identified the genus as one of the most representative among the chiropterophilous in Bromeliaceae. Kessler et al. (2020) reported three shifts from hummingbird to bat and one shift from bat to hummingbird in Vriesea (sensuGomes-da-Silva and Souza-Chies 2017). To properly track these shifts in pollination syndromes in Vriesea, a robust phylogeny resolved at the shallow phylogenetic relationships is needed.
We suggest that species with mixed floral traits are a window into shifts between pollinator syndrome, constituting possible transitional floral types in Vriesea. Interestingly, despite the morphological differences, the species with mixed floral types clearly belong to their respective syndrome and may be a product of transitions as well as reversions. Species that are product of reversions to the ancestral pollination syndromes likely do not show the exact original floral traits, only their general appearance (Tripp and Manos 2008). Such transitions and reversions have a historical baggage and may likely be driven by selection pressures imposed by pollinators.
Concluding Remarks
Our results support the hypothesis that pollinators drive ecological isolation in Vriesea bromeliads, which show clear floral specialization towards hummingbirds and bats. In our assessment, we defined floral trait diversity and identified possible transitional floral types in Vriesea, generating insights on shifts between pollination syndromes to be further explored. Also, we demonstrated the utility of pollination syndromes in predicting functional pollinator groups in Vriesea. The morphological-genetic continuum we identified here may be typical of ongoing pollinator-driven speciation. Unveiling the agents and mechanisms behind the evolution of species complexes in biodiversity hotspots such as the Atlantic Forest is of high relevance to further understand the evolution of plants.
Supporting Information
The following additional information is available in the online version of this article—
Table S1. Taxa sampled for chloroplast (rps16-trnK and matK) and nuclear (PHYC) regions, with geographical coordinates, altitude, voucher information and GenBank accession numbers.
Table S2. Floral traits measured for the morphological analyses.
Table S3. Known pollinators and floral visitors of Vriesea species.
Figure S1. cpDNA (matK and rps16-trnK) and PHYC median-joining networks showing genetic divergence among Vriesea pollination groups.
Figure S2. Bayesian phylogeny of cpDNA haplotypes (matK and rps16-trnK) for 83 taxa, including 76 Vriesea accessions.
Figure S3. Bayesian phylogeny based on PHYC data set for 60 taxa, including 55 Vriesea accessions.
Figure S4. Map of Brazil with geographic distribution of (A) hummingbird- (orange) and (B) bat-pollinated (purple) Vriesea species along the Atlantic Forest (in grey).
Data Availability
All data are provided in Supporting Information and available at GenBank https://www.ncbi.nlm.nih.gov/genbank/.
Aknowledgements
We thank to Luis Fernando Gonçalves, Suara S. A. Jacques and Fernando P. Uribbe for fieldwork assistance, Camila Aguiar-Melo for help with analyses and Heloisa Alves de Lima Carvalho and Marina Moreira Muniz for discussion on pollination syndromes.
Sources of Funding
This research was funded by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001 (PROTAX II 88887.199918/2018-00, PDSE 88881.132750/2016-01 to B.N.; PROTAX II 88887.199917/2018-00 to I.M.K.; and DS 31001017014P9 to D.R.C.); the International Association of Plant Taxonomy grant to B.N.; the Swedish Research Council, the Swedish Foundation for Strategic Research, the Knut and Alice Wallenberg Foundation and the Royal Botanic Gardens, Kew to A.A.; the Swedish Research Council (2017-04980 to C.D.B.); the Museu Nacional of the Universidade Federal do Rio de Janeiro (UFRJ), the Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (478345/2013-5, 305704/2018-4) and the Programa Institucional de Internacionalização CAPES/PrInt UFRJ to A.F.C.; and the Swedish Foundation for International Cooperation in Research and Higher Education-STINT/CAPES to C.D.B. and A.F.C.
Contributions by the Authors
B.N., F.S. and A.F.C. designed the study. B.N., I.M.K., R.L.M. and D.R.C. collected, identified and processed samples at the molecular lab. B.N., I.M.K. and F.S. analysed data. All authors contributed to data interpretation and discussion. B.N. wrote the manuscript with contributions of all authors.
Conflict of Interest
None declared.
Literature Cited
- Aguilar-Rodríguez PA, Krömer T, Tschapka M, García-Franco JG, Escobedo-Sarti J, MacSwiney GMC. 2019. Bat pollination in Bromeliaceae. Plant Ecology and Diversity 12:1–19. [Google Scholar]
- Alvares CA, Stape JL, Sentelhas PC, De Moraes JL, Sparovek G. 2013. Köppen’s climate classification map for Brazil. Meteorologische Zeitschrift 22:711–728. [Google Scholar]
- Antonelli A, Ariza M, Albert J, Andermann T, Azevedo J, Bacon C, Faurby S, Guedes T, Hoorn C, Lohmann LG, Matos-Maraví P, Ritter CD, Sanmartín I, Silvestro D, Tejedor M, Ter Steege H, Tuomisto H, Werneck FP, Zizka A, Edwards SV. 2018. Conceptual and empirical advances in Neotropical biodiversity research. PeerJ 6:e5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonelli A, Sanmartín I. 2011. Why are there so many plant species in the Neotropics? Taxon 60:403–414. [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16:37–48. [DOI] [PubMed] [Google Scholar]
- Barfuss MH, Till W, Leme EM, Pinzón JP, Manzanares JM, Halbritter H, Samuel R, Brown GK. 2016. Taxonomic revision of Bromeliaceae subfam. Tillandsioideae based on a multi-locus DNA sequence phylogeny and morphology. Phytotaxa 279:1–97. [Google Scholar]
- Bergamo PJ, Wolowski M, Telles FJ, De Brito VLG, Varassin IG, Sazima M. 2019. Bracts and long-tube flowers of hummingbird-pollinated plants are conspicuous to hummingbirds but not to bees. Biological Journal of the Linnean Society 126:533–544. [Google Scholar]
- BFG - The Brazil Flora Group. 2018. Brazilian Flora 2020: innovation and collaboration to meet target 1 of the Global Strategy for Plant Conservation (GSPC). Rodriguésia 69:1513–1527. [Google Scholar]
- Buzato S, Sazima M, Sazima I. 2000. Hummingbird-pollinated floras at three Atlantic Forest sites. Biotropica 32:824–841. [Google Scholar]
- Chartier M, Jabbour F, Gerber S, Mitteroecker P, Sauquet H, von Balthazar M, Staedler Y, Crane PR, Schönenberger J. 2014. The floral morphospace–a modern comparative approach to study angiosperm evolution. The New Phytologist 204:841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa AF Gomes-da-Silva J, Wanderley MGL. 2014. Vriesea (Bromeliaceae, Tillandsioideae): taxonomic history, and morphology of the Brazilian lineages. The Journal of the Torrey Botanical Society 141:338–352. [Google Scholar]
- Costa AF, Gomes-da-Silva J, Wanderley MGL. 2015. Vriesea (Bromeliaceae, Tillandsioideae): a cladistic analysis of eastern Brazilian species based on morphological characters. Rodriguésia 66:429–440. [Google Scholar]
- Costa AF, Rodrigues PJFP, Wanderley MGL. 2009. Morphometric analysis of Vriesea paraibica Wawra complex (Bromeliaceae). Botanical Journal of the Linnean Society 159:163–181. [Google Scholar]
- Crayn DM, Terry RG, Smith JAC, Winter K. 2000. Molecular systematic investigations in Pitcairnioideae (Bromeliaceae) as a basis for understanding the evolution of crassulacean acid metabolism (CAM). In: Wilson KL, Morrison DA, eds. Monocots: systematics and evolution. Melbourne, Australia: CSIRO, 569–579. [Google Scholar]
- Darwin C. 1859. The origin of species by means of natural selection, or the preservation of favoured races in the struggle for life, 1st edn. London: John Murray. [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19:11–15. [Google Scholar]
- Fay MF, Olmstead RG, Richardson JE, Santiago E, Prance GT, Chase MW. 1998. Molecular data support the inclusion of Duckeodendron cestroides in Solanaceae. Kew Bulletin 53:203–212. [Google Scholar]
- Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD. 2004. Pollination syndromes and floral specialization. Annual Review of Ecology, Evolution and Systematics 35:375–403. [Google Scholar]
- Fleming TH, Geiselman C, Kress WJ. 2009. The evolution of bat pollination: a phylogenetic perspective. Annals of Botany 104:1017–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora do Brasil 2020. [continuously updated]. Jardim Botânico do Rio de Janeiro http://floradobrasil.jbrj.gov.br/ (1 November 2019).
- Givnish TJ. 2017. A New World of plants. Science 358:1535–1536. [DOI] [PubMed] [Google Scholar]
- Givnish TJ, Barfuss MH, Van Ee B, Riina R, Schulte K, Horres R, Gonsiska PA, Jabaily RS, Crayn DM, Smith JA, Winter K, Brown GK, Evans TM, Holst BK, Luther H, Till W, Zizka G, Berry PE, Sytsma KJ. 2014. Adaptive radiation, correlated and contingent evolution, and net species diversification in Bromeliaceae. Molecular Phylogenetics and Evolution 71:55–78. [DOI] [PubMed] [Google Scholar]
- Goetze M, Zanella CM, Palma-Silva C, Büttow MV, Bered F. 2017. Incomplete lineage sorting and hybridization in the evolutionary history of closely related, endemic yellow-flowered Aechmea species of subgenus Ortgiesia (Bromeliaceae). American Journal of Botany 104:1073–1087. [DOI] [PubMed] [Google Scholar]
- Gomes-da-Silva J, Souza-Chies TT. 2017. What actually is Vriesea? A total evidence approach in a polyphyletic genus of Tillandsioideae (Bromeliaceae, Poales). Cladistics 34:181–199. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Terrazas TP, Martel C, Milet-Pinheiro P, Ayasse M, Kalko EK, Tschapka M. 2016. Finding flowers in the dark: nectar-feeding bats integrate olfaction and echolocation while foraging for nectar. Royal Society Open Science 3:160199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttlinger T, Schwerdtfeger M, Tiedge K, Lohaus G. 2019. What do Nectarivorous bats like? Nectar composition in Bromeliaceae with special emphasis on bat-pollinated species. Frontiers in Plant Science 10:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouda EJ, Butcher D, Gouda K. 2020. [continuously updated]. Encyclopaedia of Bromeliads https://botu07.bio.uu.nl/bcg/encyclopedia/brome/ (1 May 2019).
- Gower JC. 1971. A general coefficient of similarity and some of its properties. Biometrics 27:857–871. [Google Scholar]
- Grantsau R. 1989. Os beija-flores do Brasil, 1st edn. Rio de Janeiro, Brazil: Editora Expressão e Cultura. [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4:9. [Google Scholar]
- Harms H. 1930. Bromeliaceae. In: Engler HGA, Prantl KAE, eds. Die naturlichen Pflanzenfamilien, Vol. 15a, 2nd edn. Leipzig, Germany: Wilhem Engelman, 65–159. [Google Scholar]
- Johnson SD. 2010. The pollination niche and its role in the diversification and maintenance of the southern African flora. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 365:499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M, Abrahamczyk S, Krömer T. 2020. The role of hummingbirds in the evolution and diversification of Bromeliaceae: unsupported claims and untested hypotheses. Botanical Journal of the Linnean Society 192:592–608. [Google Scholar]
- Kessous IM, Costa AF. 2017. A new name and new status in Vriesea (Bromeliaceae) from Brazil. Novon: A Journal for Botanical Nomenclature 25:434–435. [Google Scholar]
- Kessous IM, Neves B, Couto DR, Paixao-Souza B, Pederneiras LC, Barfuss MHJ, Salgueiro F, Moura RL, Costa AF. 2020. Historical biogeography of a Brazilian lineage of Tillandsioideae (Subtribe Vrieseinae, Bromeliaceae): the Paranaean Sea hypothesized as the main vicariant event. Botanical Journal of the Linnean Society 192:625–641. [Google Scholar]
- Krapp F, Pinangé DSB, Benko-Iseppon AM, Leme EM, Weising K. 2014. Phylogeny and evolution of Dyckia (Bromeliaceae) inferred from chloroplast and nuclear sequences. Plant Systematics and Evolution 300:1591–1614. [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagomarsino LP, Condamine FL, Antonelli A, Mulch A, Davis CC. 2016. The abiotic and biotic drivers of rapid diversification in Andean bellflowers (Campanulaceae). New Phytologist 210:1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagomarsino LP, Forrestel EJ, Muchhala N, Davis CC. 2017. Repeated evolution of vertebrate pollination syndromes in a recently diverged Andean plant clade. Evolution 71:1970–1985. [DOI] [PubMed] [Google Scholar]
- Lexer C, Marthaler F, Humbert S, Barbará T, de la Harpe M, Bossolini E, Paris M, Martinelli G, Versieux LM. 2016. Gene flow and diversification in a species complex of Alcantarea inselberg bromeliads. Botanical Journal of the Linnean Society 181:505–520. [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Machado TM, Loiseau O, Paris M, Weigand A, Versieux LM, Stehmann JR, Lexer C, Salamin N. 2020. Systematics of Vriesea (Bromeliaceae): phylogenetic relationships based on nuclear gene and partial plastome sequences. Botanical Journal of the Linnean Society 192:656–674. [Google Scholar]
- Magalhães AFP, Maruyama PK, Tavares LAF, Martins RL. 2018. The relative importance of hummingbirds as pollinators in two bromeliads with contrasting floral specializations and breeding systems. Botanical Journal of the Linnean Society 188:316–326. [Google Scholar]
- Marinho-Filho JS, Sazima I. 1998. Brazilian bats and conservation biology: a first survey. In: Kunz TH, Racey PA, eds. Bat biology and conservation. Washington, DC: Smithsonian Institute, 282–294. [Google Scholar]
- Martinelli G. 1994. Reproductive biology of Bromeliaceae in the Atlantic rainforest of southeastern Brazil. PhD Thesis, University of St Andrews, Scotland, UK. [Google Scholar]
- Mez C. 1896. Bromeliaceae. In: De Candolle ALPP, De Candolle ACP, eds. Monographiae phanerogamarum, Vol. 9 Paris: G. Masson, 990 p. [Google Scholar]
- Mez C. 1934. –1935. Bromeliaceae. In: Engler HGA, ed. Das pflanzenreich IV.32 (Heft 100, 1–4). Berlim, Germany: Wilhem Engelmen, 667 p. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, 1–8. [Google Scholar]
- Moura RL, Costa AF. 2014. Taxonomic notes on Vriesea sect. Xiphion (Bromeliaceae) with descriptions of three new species. Systematic Botany 39:791–803. [Google Scholar]
- Neri J, Wendt T, Palma-Silva C. 2017. Natural hybridization and genetic and morphological variation between two epiphytic bromeliads. AoB PLANTS 10:plx061; doi: 10.1093/aobpla/plx061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves B, Uribbe FP, Jacques SSA, Zanella CM, Costa AF. 2018. Species boundaries in the Vriesea incurvata (Bromeliaceae) complex after a broad morphometric and taxonomic study. Systematic Botany 43:870–888. [Google Scholar]
- Neves B, Zanella CM, Kessous IM, Uribbe FP, Salgueiro F, Bered F, Antonelli A, Bacon CD, Costa AF. 2019. Drivers of bromeliad leaf and floral bract variation across a latitudinal gradient in the Atlantic Forest. Journal of Biogeography 47:261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2018. Vegan: community ecology package. R Package v. 2.5-2. https://CRAN.R-project.org/package=vegan. [Google Scholar]
- Ollerton J, Alarcón R, Waser NM, Price MV, Watts S, Cranmer L, Hingston A, Peter CI, Rotenberry J. 2009. A global test of the pollination syndrome hypothesis. Annals of Botany 103:1471–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paggi GM, Silveira LCT, Zanella CM, Bruxel M, Bered F, Kaltchuk-Santos E, Palma-Silva C. 2013. Reproductive system and fitness of Vriesea friburgensis, a self-sterile bromeliad species. Plant Species Biology 28:169–176. [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG. 2005. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Molecular Ecology 14:689–701. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2018. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; https://www.Rproject.org (1 December 2018). [Google Scholar]
- Rabinovitz GB. 1975. An introduction to nonmetric multidimensional scaling. American Journal of Political Science 19:343–390. [Google Scholar]
- Rambaut A. 2014. FigTree, version 1.4.2 software. Edinburgh, UK: Institute of Evolutionary Biology, University of Edinburgh. [Google Scholar]
- Ramos FN, Mortara SR, Monalisa-Francisco N, Elias JPC, Neto LM, Freitas L, Kersten R, Amorim AM, Matos FB, Nunes-Freitas AF, Alcantara S, Alexandre MHN, de Almeida-Scabbia RJ, de Almeida OJG, Alves FE, de Oliveira Alves RM, Alvim FS, de Andrade ACS, de Andrade S, Aona LYS, Araujo AC, de Araújo KCT, Ariati V, Assis JC, de Azevedo CO, Barbosa BF, Barbosa DEF, Barbosa FDR, de Barros F, Basilio GA, Bataghin FA, Bered F, Bianchi JS, Blum CT, Boelter CR, Bonnet A, Brancalion PHS, Breier TB, Brion CT, Buzatto CR, Cabral A, Cadorin TJ, Caglioni E, Canêz L, Cardoso PH, de Carvalho FS, Carvalho RG, Catharino ELM, Ceballos SJ, Cerezini MT, César RG, Cestari C, Chaves CJN, Citadini-Zanette V, Coelho LFM, Coffani-Nunes JV, Colares R, Colletta GD, Corrêa NM, da Costa AF, da Costa GM, Costa LMS, Costa NGS, Couto DR, Cristofolini C, da Cruz ACR, Del Neri LA, di Pasquo M, Dos Santos Dias A, Dias LDCD, Dislich R, Duarte MC, Fabricante JR, Farache FHA, de Faria APG, Faxina C, Ferreira MTM, Fischer E, Fonseca CR, Fontoura T, Francisco TM, Furtado SG, Galetti M, Garbin ML, de Gasper AL, Goetze M, Gomes-da-Silva J, Gonçalves MFA, Gonzaga DR, Silva ACGE, Guaraldo AC, Guarino ESG, Guislon AV, Hudson LB, Jardim JG, Jungbluth P, Kaeser SDS, Kessous IM, Koch NM, Kuniyoshi YS, Labiak PH, Lapate ME, Santos ACL, Leal RLB, Leite FS, Leitman P, Liboni AP, Liebsch D, Lingner DV, Lombardi JA, Lucas E, Luzzi JDR, Mai P, Mania LF, Mantovani W, Maragni AG, Marques MCM, Marquez G, Martins C, Martins LDN, Martins PLSS, Mazziero FFF, Melo CA, de Melo MMF, Mendes AF, Mesacasa L, Morellato LPC, Moreno VS, Muller A, Murakami MMDS, Cecconello E, Nardy C, Nervo MH, Neves B, Nogueira MGC, Nonato FR, de Oliveira-Filho AT, de Oliveira CPL, Overbeck GE, Marcusso GM, Paciencia MLB, Padilha P, Padilha PT, Pereira ACA, Pereira LC, Pereira RAS, Pincheira-Ulbrich J, Pires JSR, Pizo MA, Pôrto KC, Rattis L, Reis JRM, Reis SGD, da Rocha-Pessôa TC, Rocha CFD, Rocha FS, Rodrigues ARP, Rodrigues RR, Rogalski JM, Rosanelli RL, Rossado A, Rossatto DR, Rother DC, Ruiz-Miranda CR, Saiter FZ, Sampaio MB, Santana LD, Santos JSD, Sartorello R, Sazima M, Schmitt JL, Schneider G, Schroeder BG, Sevegnani L, Júnior VOS, da Silva FR, da Silva MJ, Silva MPP, Silva RG, Silva SM, Singer RB, Siqueira G, Soares LE, de Sousa HC, Spielmann A, Tonetti VR, Toniato MTZ, Ulguim PSB, van den Berg C, van den Berg E, Varassin IG, da Silva IBV, Vibrans AC, Waechter JL, Weissenberg EW, Windisch PG, Wolowski M, Yañez A, Yoshikawa VN, Zandoná LR, Zanella CM, Zanin EM, Zappi DC, Zipparro VB, Zorzanelli JPF, Ribeiro MC. 2019. ATLANTIC EPIPHYTES: a data set of vascular and non-vascular epiphyte plants and lichens from the Atlantic Forest. Ecology 100:e02541. [DOI] [PubMed] [Google Scholar]
- Rocca MA, Sazima M. 2013. Quantity versus quality: identifying the most effective pollinators of the hummingbird-pollinated Vriesea rodigasiana (Bromeliaceae). Plant Systematics and Evolution 299:97–105. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Guerrero V, Aguilar R, Martén-Rodríguez S, Ashworth L, Lopezaraiza-Mikel M, Bastida JM, Quesada M. 2014. A quantitative review of pollination syndromes: do floral traits predict effective pollinators? Ecology Letters 17:388–400. [DOI] [PubMed] [Google Scholar]
- SanMartin-Gajardo I, Sazima M. 2005. Chiropterophily in Sinningieae (Gesneriaceae): Sinningia brasiliensis and Paliavana prasinata are bat-pollinated, but P. sericiflora is not. Not yet? Annals of Botany 95:1097–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazima M, Buzato S, Sazima I. 1995. Bat pollination of Vriesea in southeastern Brazil. Bromelia 2:29–37. [Google Scholar]
- Sazima M, Buzato S, Sazima I. 1999. Bat-pollinated flower assemblages and bat visitors at two Atlantic Forest sites in Brazil. Annals of Botany 83:705–712. [Google Scholar]
- Schmid S, Schmid VS, Zillikens A, Steiner J. 2011. Diversity of flower visitors and their role for pollination in the ornithophilous bromeliad Vriesea friburgensis in two different habitats in southern Brazil. Ecotropica 17:91–102. [Google Scholar]
- Serrano-Serrano ML, Rolland J, Clark JL, Salamin N, Perret M. 2017. Hummingbird pollination and the diversification of angiosperms: an old and successful association in Gesneriaceae. Proceedings Royal Society B 284:e20162816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL. 2007. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany 94:275–288. [DOI] [PubMed] [Google Scholar]
- Smith LB, Downs RJ. 1977. Tillandsioideae (Bromeliaceae). Flora Neotropica, monograph 14, part 2. New York, NY: Hafner Press, 663–1942, fig. 213–467. [Google Scholar]
- Stehmann JR, Forzza RC, Salino A, Sobral M, Costa DP, Kamino LHY. 2009. Plantas da Floresta Atlântica, 1st edn. Rio de Janeiro, Brazil: Jardim Botânico do Rio de Janeiro. [Google Scholar]
- Tripp EA, Manos PS. 2008. Is floral specialization an evolutionary dead-end? Pollination system transitions in Ruellia (Acanthaceae). Evolution 62:1712–1737. [DOI] [PubMed] [Google Scholar]
- Uribbe FP, Neves B, Jacques SSA, Costa AF. 2020. Morphological variation in the Vriesea procera complex (Bromeliaceae, Tillandsioideae) in the Brazilian Atlantic Rainforest, with recognition of new taxa. Systematic Botany 45:53–68. [Google Scholar]
- Van der Niet T, Peakall R, Johnson SD. 2014. Pollinator-driven ecological speciation in plants: new evidence and future perspectives. Annals of Botany 113:199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IJ, Bradburd GS. 2014. Isolation by environment. Molecular Ecology 23:5649–5662. [DOI] [PubMed] [Google Scholar]
- Wendt T, Coser TS, Matallana G, Guilherme FAG. 2008. An apparent lack of prezygotic reproductive isolation among 42 sympatric species of Bromeliaceae in southeastern Brazil. Plant Systematics and Evolution 275:31–41. [Google Scholar]
- Wright S. 1943. Isolation by distance. Genetics 28:114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanella CM, Palma-Silva C, Goetze M, Bered F. 2016. Hybridization between two sister species of Bromeliaceae: Vriesea carinata and V. incurvata. Botanical Journal of Linnean Society 181:491–504. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are provided in Supporting Information and available at GenBank https://www.ncbi.nlm.nih.gov/genbank/.