Highlights:
-
•
Patients with coronavirus disease 2019 (COVID-19) had altered inflammatory markers.
-
•
High sensitivity C-reactive protein-(pre)albumin ratios positively correlated with severe COVID-19.
-
•
Prognostic nutritional index negatively correlated with risk of severe COVID-19.
-
•
Nomogram combining the three factors reliably predicted the progression of COVID-19.
-
•
High sensitivity C-reactive protein-prealbumin and -albumin ratios helped estimate duration of hospitalization.
Keywords: COVID-19, High sensitivity C-reactive protein-albumin ratio, Prognostic nutritional index, High sensitivity C-reactive protein-prealbumin ratio, Nomogram, Severity
Abstract
Background
Patients with severe coronavirus disease 2019 (COVID-19) develop acute respiratory distress and multi-system organ failure and are associated with poor prognosis and high mortality. Thus, there is an urgent need to identify early diagnostic and prognostic biomarkers to determine the risk of developing serious illness.
Methods
We retrospectively analyzed 114 patients with COVID-19 at the Jinyintan Hospital, Wuhan based on their clinical and laboratory data. Patients were categorized into severe and mild to moderate disease groups. We analyzed the potential of serological inflammation indicators in predicting the severity of COVID-19 in patients using univariate and multivariate logistic regression, receiver operating characteristic curves, and nomogram analysis. The Spearman method was used to understand the correlation between the serological biomarkers and duration of hospital stay.
Results
Patients with severe disease had reduced neutrophils and lymphocytes; severe coagulation dysfunction; altered content of biochemical factors (such as urea, lactate dehydrogenase); elevated high sensitivity C-reactive protein levels, neutrophil–lymphocyte, platelet-lymphocyte, and derived neutrophil–lymphocyte ratios, high sensitivity C-reactive protein-prealbumin ratio (HsCPAR), systemic immune-inflammation index, and high sensitivity C-reactive protein-albumin ratio (HsCAR); and low lymphocyte-monocyte ratio, prognostic nutritional index (PNI), and albumin-to-fibrinogen ratio. PNI, HsCAR, and HsCPAR correlated with the risk of severe disease. The nomogram combining the three parameters showed good discrimination with a C-index of 0.873 and reliable calibration. Moreover, HsCAR and HsCPAR correlated with duration of hospital stay.
Conclusion
Taken together, PNI, HsCAR, and HsCPAR may serve as accurate biomarkers for the prediction of disease severity in patients with COVID-19 upon admission/hospitalization.
1. Introduction
Coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a pandemic that poses a major threat to global public health. As of July 3, 2020, there have been 10,871,361 confirmed cases of COVID-19 across 188 countries with 521,298 deaths [1]. Although the majority of patients with COVID-19 exhibit mild or moderate symptoms, 19% of patients develop severe symptoms or critical illness [2]. Yang et al. have reported the 28-day rate of mortality of critically ill patients with COVID-19 to be 61.5% [3]. Some COVID-19 patients experience mild symptoms in the early stages of disease and deteriorate over time. Without timely intervention and treatment, such cases of COVID-19 are associated with poor prognosis and high mortality [4]. The treatment of severe COVID-19 patients has become a major challenge for the prevention and control of COVID-19. Thus, it is imperative to identify reliable predictors of COVID-19 severity and enable treatment in the early stages of disease along with effectively allocating critical-care resources. Multiple studies have investigated the predictors of severe COVID-19, such as C-reactive protein (CRP), interleukin 6 (IL-6), and lymphocyte (LYM) counts [5], [6], [7]. However, the role of combinations of inflammatory markers in determining disease severity remains to be understood. Severe or critically ill COVID-19 patients exhibit extreme systemic inflammation and poor nutritional status [8], [9], [10]. Serological biomarkers, including neutrophil–lymphocyte ratio (NLR), platelet-lymphocyte ratio (PLR), lymphocyte-monocyte ratio (LMR), derived neutrophil–lymphocyte ratio (dNLR), high sensitivity C-reactive protein-albumin ratio (HsCAR), albumin-to-fibrinogen ratio (AFR), prognostic nutritional index (PNI), systemic immune-inflammation index (SII), and high sensitivity C-reactive protein-prealbumin ratio (HsCPAR), are indicators of systematic inflammation and nutritional status and function as prognostic factors for malignancies and inflammatory disease [11], [12], [13]. However, how these serological biomarkers change in the peripheral blood and their clinical values in COVID-19 remain unclear. In this study, we have investigated the potential of these serological biomarkers in predicting the severity of COVID-19.
2. Materials and methods
2.1. Study design and participants
We enrolled 114 COVID-19 patients admitted to the Jinyintan Hospital in Wuhan between February 10 and March 7, 2020 in this study. All the patients were diagnosed based on the recommendations by the World Health Organization. The primary outcome was the development of severe COVID-19 during hospitalization. Mild to moderate or severe-critically ill cases of COVID-19 were diagnosed according to the Guidance for Corona Virus Disease 2019 (7th edition) by the National Health Commission of China [14]. Of these 114 cases, 58 and 56 patients were severe (including 5 critically ill patients) and mild to moderate cases, respectively. This study was approved by the Jinyintan Hospital Ethics Committee and complied with the Declaration of Helsinki (approval no. KY-2020-79.01). The data used in this study were anonymous, thereby waiving the requirement for informed consent.
2.2. Data collection
We collected the information and analyzed parameters from patients, such as gender, age, comorbidities, and duration between hospital admission and discharge. Results of laboratory tests, such as hematological and coagulation indicators, biochemical indices, and inflammatory markers, were also collected and evaluated upon admission. The haematological tests including white blood cells (WBC, ×109/L; normal range 4–10), platelet (PLT, ×109/L; normal range 125–350), neutrophils (NEU, ×109/L; normal range 1.8–6.3), lymphocyte (LYM, ×109/L; normal range 1.1–3.2) and monocytes (MON, ×109/L; normal range 0.1–0.6) were detected on the Mindray BC-6900 hematology analyzer (Mindray, Shenzhen, China). The coagulation profile including prothrombin time (PT, S; normal range 10.5–13.5), international normalized ratio (INR; normal range 0.8–1.2), activated partial thromboplastin time (APTT, S; normal range 21–37), thrombin time (TT, S; normal range 13–21), fibrinogen (Fbg, g/L; normal range 2.0–4.0), fibrinogen degradation product (FDP, μg/L; normal range 0–5) , and D-dimer (mg/L; normal range 0–1.5), were determined by CS-5100 automated blood coagulation analyzer (Sysmex, Kobe, Japan). Serum biochemical parameters, including alanine aminotransferase (ALT, U/L; normal range 9–50), aspartate aminotransferase (AST, U/L; normal range 15–40), total protein (TP, g/L; normal range 65–85), total bilirubin (TBIL, μmol/L; normal range 0–26), albumin (ALB, g/L; normal range 40–55), alkaline phosphatase (ALP, U/L; normal range 45–125), γ-glutamyltransferase (GGT, U/L; normal range 7–45), urea (mmol/L; normal range 3.6–9.5), creatinine (CREA, μmol/L; normal range 57–97), uric acid (UA, μmol/L; normal range 208–428), glucose (GLU, mmol/L; normal range 3.9–6.1), creatine kinase (CK, U/L; normal range 50–310), lactate dehydrogenase (LDH, U/L; normal range 120–250), prealbumin (PA, mg/L; normal range 200–430), total cholesterol (CHOL, mmol/L; normal range 3.3–5.2), triglyceride (TG, mmol/L; normal range 0.51–1.70), low-density lipoprotein cholesterol (LDL-C, mmol/L; normal range 2.10–3.37), high density liptein cholesterol (HDL- C, mmol/L; normal range 1.29–1.55), and high sensitivity C-reactive protein (HsCRP, mg/dL; normal range 0–5), were assayed using Abbott Architect c16000 automatic biochemistry analyzer (Abbott Diagnostics, North Chicago, USA). All laboratory tests were done in the clinical laboratory of Wuhan Jinyintan Hospital with standard procedures, and a detailed list of variables can be found in Table 2. Adults with severe COVID-19 were classified based on the following conditions: (1) respiratory distress, respiration rate (RR) ≥ 30 times/minute; (2) resting state with oxygen saturation ≤ 93%; (3) arterial blood oxygen partial pressure (PaO2)/oxygen concentration (FiO2) ≤ 300 mm Hg; and (4) lung imaging showed significant lesion progression > 50% within 24–48 h. Patients were considered critically ill if any of the following criteria were met: (1) respiratory failure requiring mechanical ventilation; (2) shock; and (3) a combination of organ failure and ICU monitoring.
Table 2.
Baseline characteristics and clinical data from patients with COVID-19 at admission. Abbreviations: WBC, white blood cell; PLT, platelet; NEU, neutrophil; LYM, lymphocyte; MON, monocyte; PT, prothrombin time; INR, international normalized ratio; APTT, acivated partial thromboplastin time; TT, thrombin time; Fbg, fibrinogen; FDP, fibrinogen degradation product; ALT, alanine aminotransferase; AST, aspartate aminotransferase; TP, total protein; TBIL, total bilirubin; ALB, albumin; ALP, alkaline phosphatase; GGT, γ-glutamyltransferase; CREA, creatinine; UA, uric acid; GLU, glucose; CK, creatine kinase; LDH, lactate dehydrogenase; PA, prealbumin; CHOL, total cholesterol; TG, triglyceride; LDL-C, low-density lipoprotein cholesterol; HDL- C , high density liptein cholesterol; HsCRP, high sensitivity C-reactive protein.
| Variables | All patients (n = 114) | Severe (including critically ill) (n = 58) | Mild to moderate (n = 56) | Z or X2value/p value |
|---|---|---|---|---|
| Age (y), median (IQR) | 62.00(51.00–70.00) | 64.00(49.75–73.00) | 60.50(52.25–68.75) | −0.632/0.527 |
| Sex (male/female) | 64/50 | 34/24 | 30/26 | 0.295/0.587 |
| Any comorbidity, n (%) | 73(64.04%) | 32(53.45%) | 40(71.42%) | 3.236/0.072 |
| Diabetes, n (%) | 16(14.04%) | 5(8.62%) | 11(19.64%) | 2.869/0.090 |
| Hypertension, n (%) | 37(32.46%) | 15(25.86%) | 22(39.29%) | 2.342/0.126 |
| Cardiovascular disease, n (%) | 17(14.91) | 6(10.34%) | 11(19.64%) | 1.941/0.164 |
| Gout, n (%) | 4(3.51%) | 1(1.72%) | 3(5.38%) | 1.111/0.292 |
| Chronic pulmonary disease, n (%) | 10(8.77%) | 8(13.79%) | 2(3.57%) | 3.720/0.054 |
| Chronic liver disease, n (%) | 4(3.51%) | 1(1.72%) | 3(5.38%) | 1.111/0.292 |
| Malignancy, n (%) | 4(3.51%) | 3(5.17) | 1(1.79%) | 0.965/0.326 |
| Others, n (%) | 12(10.53%) | 8(13.79) | 4(7.14%) | 1.264/0.261 |
| length of hospital stay (d) | 10.00(7.00–15.00) | 12.50(9.00–17.00) | 9.00(7.00–13.75) | −3.082/0.002 |
| Clinical Laboratory Data, median (IQR) | ||||
| WBC (×109/L) | 5.60(4.72–8.28) | 6.05(4.90–9.33) | 5.51(4.47–7.73) | −1.808/0.071 |
| PLT (×109/L) | 209.50(160.00–256.75) | 196.00(157.50–262.00) | 217.5(173.25–255.75) | −1.043/0.297 |
| NEU (×109/L) | 3.99(2.98–6.37) | 4.83(3.46–7.52) * | 3.76(2.64–4.89) | −3.157/0.002 |
| LYM (×109/L) | 0.99(0.68–1.48) | 0.85(0.56–1.19) * | 1.29(0.80–1.66) | −3.673/0.000 |
| MON (×109/L) | 0.39(0.27–0.49) | 0.36(0.26–0.48) | 0.41(0.29–0.52) | −1.126/0.260 |
| PT (S) | 11.60(10.90–12.45) | 11.80(11.00–12.75) * | 11.30(10.80–12.00) | −2.387/0.017 |
| INR | 0.99(0.93–1.05) | 1.01(0.93–1.09) * | 0.98(0.93–1.03) | −2.070/0.038 |
| APTT (S) | 26.45(23.05–29.63) | 27.05(23.98–30.45) | 25.80(22.68–28.18) | −1.814/0.070 |
| TT (S) | 16.70(16.08–17.60) | 16.85(15.98–17.90) | 16.65(16.10–17.30) | −0.953/0.341 |
| Fbg (g/L) | 4.20(2.70–5.73) | 5.00(3.65–6.10) * | 3.45(2.70–5.01) | −2.432/0.015 |
| D-dimer (mg/L) | 0.61(0.39–1.75) | 0.79(0.48–3.02) * | 0.52(0.36–0.95) | −2.775/0.006 |
| FDP (μg/L) | 2.05(1.28–7.76) | 2.75(1.60–14.25) * | 1.80(0.92–3.19) | −2.685/0.007 |
| ALT (U/L) | 32.50(22.00–54.25) | 32.00(22.00–50.25) | 35.00(22.00–54.75) | −0.150/0.881 |
| AST (U/L) | 34.00(26.00–56.00) | 38.00(26.75–64.25) * | 28.00(24.25–44.50) | −2.056/0.040 |
| TP (g/L) | 64.45(59.5–68.95) | 64.75(59.40–68.30) | 64.30(59.50–71.20) | −0.879/0.380 |
| TBIL (μmol/L) | 11.45(8.62–14.88) | 11.80(8.80–15.55) | 10.75(8.43–14.05) | −0.867/0.386 |
| ALB (g/L) | 31.40(26.33–36.45) | 29.05(22.73–32.35) | 34.65(30.03–39.83) | −4.838/0.000 |
| ALP (U/L) | 72.50(54.00–96.00) | 71.00(58.00–94.50) | 75.00(50.00–96.75) | −0.323/0.747 |
| GGT (U/L) | 32.50(22.00–72.50) | 33.50(23.75–79.25) | 32.00(21.00–62.00) | −0.981/0.327 |
| Urea (mmol/L) | 4.80(3.50–6.33) | 5.00(3.58–7.45) * | 4.60(3.50–5.35) | −2.013/0.044 |
| CREA (μmol/L) | 66.45(55.43–80.73) | 69.10(54.45–84.73) | 65.85(56.05–77.23) | −0.658/0.511 |
| UA (μmol/L) | 239.00(190.00–289.50) | 221.50(163.75–288.25) | 252.00(202.25–297.00) | −1.757/0.079 |
| GLU (mmol/L) | 6.15(5.60–8.33) | 6.30(5.70–7.65) | 5.80(5.50–8.60) | −0.723/0.470 |
| CK (U/L) | 84.50(53.75–137.25) | 94.00(54.00–203.50) | 76.50(50.00–114.25) | −1.457/0.145 |
| LDH (U/L) | 291.50(224.50–390.50) | 369.00(268.75–426.00) * | 253.50(202.25–323.00) | −4.608/0.000 |
| PA (mg/L) | 127.00(84.75–183.00) | 88.50(60.75–144.25) * | 166.00(118.00–242.25) | −5.127/0.000 |
| CHOL (mmol/L) | 3.70(3.16–4.39) | 3.63(3.15–4.04) | 3.89(3.15–4.88) | −1.607/0.108 |
| TG (mmol/L) | 1.32(1.04–1.80) | 1.23(1.00–1.70) | 1.45(1.12–1.88) | −1.919/0.055 |
| LDL-C (mmol/L) | 2.12(1.74–2.63) | 2.10(1.74–2.42) | 2.20(1.72–2.81) | −1.009/0.313 |
| HDL-C (mmol/L) | 0.98(0.78–1.19) | 0.88(0.74–1.14) * | 1.04(0.80–1.24) | −2.384/0.017 |
| HsCRP (mg/dL) | 23.20(5.03–80.58) | 54.60(20.50–156.25) * | 12.30(2.28–24.13) | −5.232/0.000 |
*p < 0.05 vs Mild to moderate patients. Data are median and interquartile range.
2.3. New serological markers
Previous studies showed severe COVID-19 patients were more likely to have higher levels of neutrophils, HsCRP, Fbg and MON, and lower levels of LYM, PLT, ALB and PA [15], [16], [17], [18], [19], [20]. The new serological biomarkers listed in Table 1 were derived from a combination of two or more of the above indicators, including NLR, PLR, LMR, dNLR, HsCAR, AFR, PNI, SII, and HsCPAR. Table 1 lists the formulas used to calculate these indices.
Table 1.
New serological markers and their formulas.
| Indexes | Formulas |
|---|---|
| NLR | NEU counts (109)/LYM counts (109) |
| PLR | PLT counts (109)/LYM counts (109) |
| LMR | LYM counts (109)/MON counts (109) |
| dNLR | NEU counts (109)/[WBC counts (109)-NEU counts (109)] |
| HsCAR | HsCRP (mg/L)/ALB (g/L) |
| AFR | ALB (g/L)/Fbg (g/L) |
| PNI | 10*ALB (g/L) + 5*LYM counts (109) |
| SII | PLT counts (109)*NEU counts (109)/LYM counts (109) |
| HsCPAR | HsCRP (mg/L)/PA (g/L) |
2.4. Statistical analysis
Statistical analysis was performed using SPSS 23.0 and GraphPad Prism 8.0. Categorical variables have been represented as percentages and continuous variables have been denoted as mean ± standard deviation or median (interquartile range). For normally and non-uniformly distributed data, the means of continuous variables were compared using the independent t-test and Mann-Whitney U test, respectively. Categorical variables were compared using the χ2 test. Correlation analysis was performed using the Spearman method. The independent factors of severe COVID-19 were selected by univariate and multivariate logistic regression analyses. The relative risk was assessed using ninety-five percent confidence intervals (95% CI) of odds ratio (OR). We used the R Project for Statistical Computing to generate receiver operating characteristic (ROC) curves, nomogram, and calibration and decision curves, and calculate Harrell’s concordance index (C-index). The predictive nomogram was a statistical model useful for risk assessment of severe COVID-19 and was developed using the selected independent factors. The discrimination and predictive abilities of the nomogram were assessed with C-index, where a larger index reflected a more accurate prediction of severe COVID-19. Calibration and decision curves were also generated to assess the predictive accuracy and agreement between predicted and observed severity and to assess the clinical usefulness of this nomogram respectively. p < 0.05 were considered statistically significant (*p < 0.05, **p < 0.01, and ***p < 0.001).
3. Results
3.1. Clinical and laboratory characteristics of patients with COVID-19
Based on the progression of COVID-19 during hospitalization, the 114 confirmed patients were categorized into the mild to moderate (56 cases) and severe (58 cases) groups. Patients enrolled in severe group were those deteriorated from not severe to severe COVID-19 during hospitalization, and those who did not progress to severe illness until discharge were sorted into the mild to moderate group. Patients in severe group should met the criteria in Section 2.1. Patient age [64.00(49.75–73.00) vs 60.50(52.25–68.75), P = 0.527], gender [34/24 vs 30/26, P = 0.587], or percentage of comorbidity were similar between the two groups. We observed substantial differences in the clinical and laboratory findings between the groups of patients (Table 2 ). As expected, the duration of hospital stay for severe disease group was longer than that for patients with mild to moderate disease. All laboratory parameters were obtained upon admission. LYM counts decreased in the severe disease group, while there were no differences in the WBC, PLT, and MON between the two groups. Patients in severe disease group exhibited high levels of indicators of coagulation, such as PT, INR, Fbg, FDP, and D-dimer (p < 0.05), whereas APTT and TT were comparable between the two groups. Biochemical testing of the enrolled patients were routinely performed in our laboratory. The levels of AST, Urea, and LDH were markedly higher in patients with severe disease than those in mild to moderate patients. Levels of ALB, PA, and HDL-C were lower in patients with severe disease than those in mild to moderate cases. Moreover, severe disease group had higher HsCRP levels than mild to moderate patients.
3.2. Novel serological indicators for the severity of disease
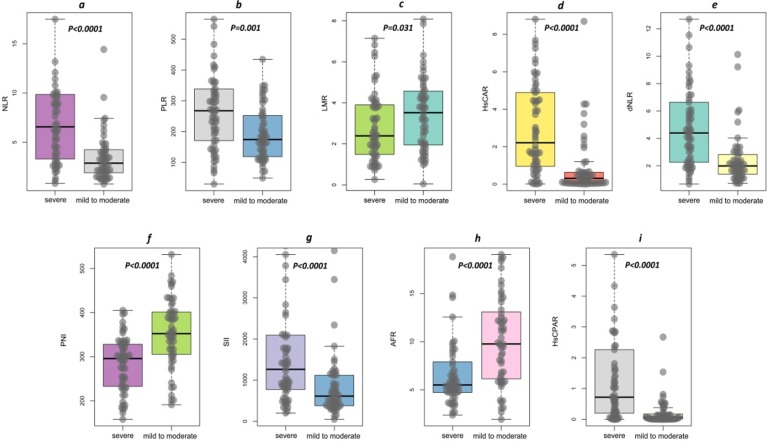
The levels of new serological biomarkers, including NLR (6.57[3.27–9.91] vs 3.03[1.86–4.38]), PLR (267.71[166.76–342.84] vs 173.48[116.17–252.97]), HsCAR (2.21[0.91–4.92] vs 0.33[0.05–0.64]), dNLR (4.39[2.25–6.69] vs 2.02[1.38–2.95]), SII (1263.52[752.80–2094.42] vs 618.35[373.58–1123.39]), and HsCPAR (0.719[0.197–2.291] vs 0.073[0.008–0.227]) were elevated in severe disease group (p < 0.0001; Fig. 1 a, b, d, e, g, h) compared to those in mild to moderate patients. Severe disease group exhibited lower levels of LMR (2.39[1.47–3.92] vs 3.62[1.95–4.59], p = 0.031, Fig. 1c), PNI (295.83[232.16–328.95] vs 352.78[304.89–403.45], P < 0.0001, Fig. 1f), and AFR (5.51[4.73–8.05] vs 9.82[6.09–13.92], P < 0.0001, Fig. 1i).
Fig. 1.
Novel serological indicators in COVID-19 patients. Patients with severe form of COVID-19 had higher levels of NLR (a), PLR (b), HsCAR (d), dNLR (e), SII (g), and HsCPAR (i), and lower levels of LMR (c), PNI (f), and AFR (h) when compared to those in mild to moderate patients. Statistical significance was determined using the Mann-Whitney U test. Abbreviations: NLR, neutrophil–lymphocyte ratio; PLR, platelet-lymphocyte ratio; LMR, lymphocyte-monocyte ratio; dNLR, derived neutrophil–lymphocyte ratio; HsCAR, high sensitivity C-reactive protein-albumin ratio; AFR, albumin-to-fibrinogen ratio; PNI, prognostic nutritional index; SII, systemic immune-inflammation index; HsCPAR, high sensitivity C-reactive protein-prealbumin ratio.
3.3. Correlation between new serological indicators and risk of developing severe COVID-19
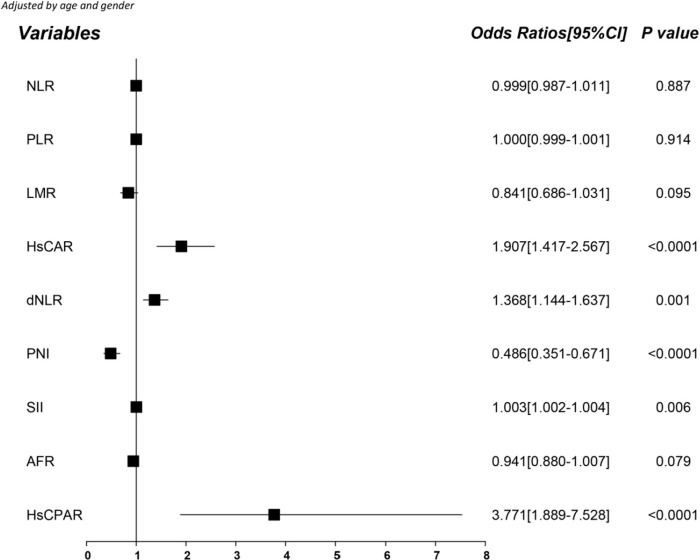
Since the serological indicators showed preference for patient groups, we wanted to verify if these indicators could serve as independent risk factors for severe COVID-19. Thus, we determined the crude Odds ratios (ORs) using univariate logistic regression analysis and adjusted the ORs for age and gender based on logistic regression analysis. HsCAR, dNLR, SII, and HsCPAR positively correlated with developing severe COVID-19, whereas PNI negatively correlated with the risk of developing severe disease (Fig. 2 ). The potential of predicting risk based on NLR, PLR, LMR, and AFR was unclear. Multivariate logistic regression analysis showed that HsCAR (OR: 1.452, 95% confidence interval [CI]: 1.093–1.930, P = 0.010), PNI (OR: 0.990, 95% CI: 0.983–0.996, P = 0.003) and HsCPAR (OR: 3.581, 95% CI: 1.425–8.997, P = 0.007) correlated with the risk of developing severe COVID-19.
Fig. 2.
Forest plot for the age and gender-adjusted serological indicators based on logistic regression analysis.
3.4. ROC curves for the potential of serological indicators in predicting severity of COVID-19
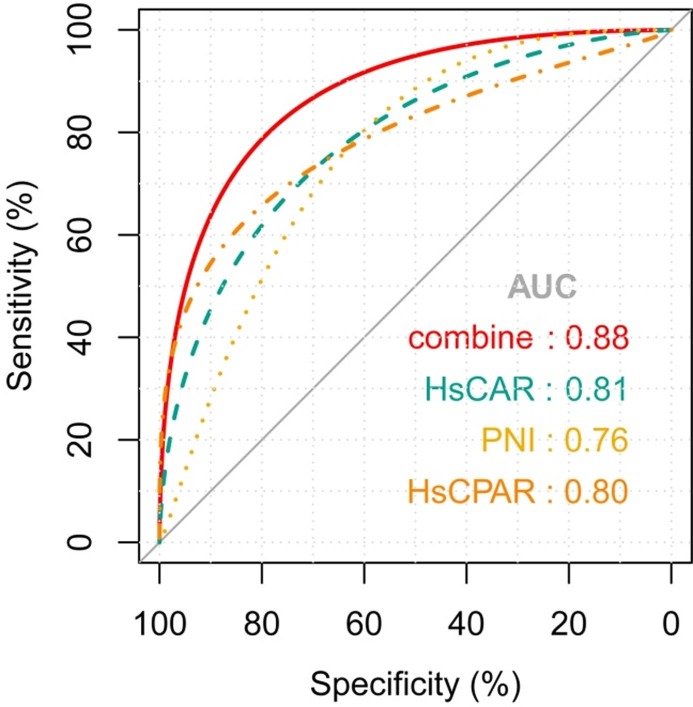
To determine the potential of the serological indicators in predicting the severity of COVID-19, we generated ROC curves for each of the nine serological indicators. We also determined the area under ROC curve (AUC), sensitivity, specificity, positive and negative predictive values, true and false positive rates, true and false negative rates, accuracy, precision, and Youden index. Table 3 lists the quantitative values for these parameters. HsCAR and HsCPAR had AUCs > 0.80, indicating good diagnostic performance. HsCAR had the highest negative predictive value, accuracy, and Youden index, while HsCPAR had the lowest false positive rate and highest specificity, positive predictive value, true negative rate, and precision. PNI had the lowest false negative rate and highest sensitivity and true positive rate. Thus, HsCAR, HsCPAR, and PNI were used to predict the severity of COVID-19. The best cut-off values of HsCAR, HsCPAR, and PNI were 0.71, 0.58, and 340.05, respectively. Combining HsCAR, HsCPAR, and PNI, ROC curve analysis showed that the AUC was 0.88 (95% CI, 0.81–0.93); this was higher than for the ROC curves for HsCAR (z statistic: 2.652, p = 0.008), HsCPAR (z statistic: 2.719, p = 0.007), and PNI (z statistic: 4.079, p < 0.0001; Fig. 3 ).
Table 3.
Diagnostic performance and parameters of the nine serological indicators.
| Index | NLR | PLR | LMR | HsCAR | dNLR | PNI | SII | AFR | HsCPAR |
|---|---|---|---|---|---|---|---|---|---|
| AUC | 0.75 | 0.68 | 0.62 | 0.81 | 0.76 | 0.76 | 0.72 | 0.72 | 0.80 |
| Best Cut-off Value | 5.08 | 229.09 | 2.56 | 0.71 | 3.39 | 340.05 | 809.02 | 6.51 | 0.58 |
| Sensitivity (%) | 63.79 | 63.79 | 56.90 | 82.76 | 63.79 | 86.21 | 72.41 | 68.97 | 58.62 |
| Specificity (%) | 82.14 | 71.43 | 66.07 | 80.36 | 85.71 | 60.71 | 67.86 | 73.21 | 92.86 |
| Negative Predictive Value (%) | 68.66 | 65.57 | 59.68 | 81.82 | 69.57 | 80.95 | 70.37 | 69.49 | 68.42 |
| Positive Predictive Value (%) | 78.72 | 69.81 | 63.46 | 81.36 | 82.22 | 69.44 | 70.00 | 72.73 | 89.47 |
| True Positive Rate (%) | 63.79 | 63.79 | 56.90 | 82.76 | 63.79 | 86.21 | 72.41 | 68.97 | 58.62 |
| False Positive Rate (%) | 17.86 | 28.57 | 33.93 | 19.64 | 14.29 | 39.29 | 32.14 | 26.79 | 7.14 |
| True Negative Rate (%) | 82.14 | 71.43 | 66.07 | 80.36 | 85.71 | 60.71 | 67.86 | 73.21 | 92.86 |
| False Negative Rate (%) | 36.21 | 36.21 | 43.10 | 17.24 | 36.21 | 13.79 | 27.59 | 31.03 | 41.38 |
| Accuracy (%) | 72.81 | 67.54 | 61.40 | 81.58 | 74.56 | 73.68 | 70.18 | 71.05 | 75.44 |
| Precision (%) | 78.72 | 69.81 | 63.46 | 81.36 | 82.22 | 69.44 | 70.00 | 72.73 | 89.47 |
| Youden Index | 145.94 | 135.22 | 122.97 | 163.12 | 149.51 | 146.92 | 140.27 | 142.18 | 151.48 |
Abbreviations: AUC, area under ROC curve; NLR, neutrophil–lymphocyte ratio; PLR, platelet-lymphocyte ratio; LMR, lymphocyte-monocyte ratio; dNLR, derived neutrophil–lymphocyte ratio; HsCAR, high sensitivity C-reactive protein-albumin ratio; AFR, albumin-to-fibrinogen ratio; PNI, prognostic nutritional index; SII, systemic immune-inflammation index; HsCPAR, high sensitivity C-reactive protein-prealbumin ratio.
Fig. 3.
ROC curves for HsCAR, HsCPAR, PNI, and a combination of the three indices for predicting the severity of COVID-19. HsCARAUC = 0.81 (95% CI, 0.73–0.88), p < 0.0001. HsCPARAUC = 0.80 (95% CI, 0.72–0.87), p < 0.0001. PNIAUC = 0.76 (95% CI, 0.67–0.84), p < 0.0001. Combination AUC = 0.827 (95% CI, 0.81–0.93), P < 0.0001. Abbreviations: ROC, receiver operating characteristic curves; AUC, area under ROC curve; HsCAR, high sensitivity C-reactive protein-albumin ratio; PNI, prognostic nutritional index; HsCPAR, high sensitivity C-reactive protein-prealbumin ratio.
3.5. Establishment of a prediction model
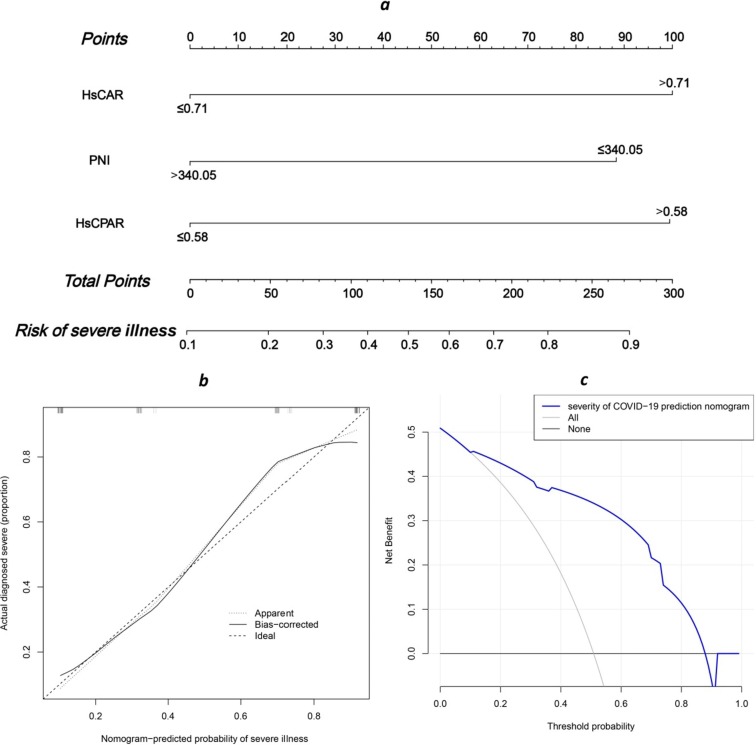
Based on the univariate and multivariate logistic regression analyses and performance of the ROC curves, HsCAR, HsCPAR, and PNI were used to generate the nomogram (Fig. 4 a). Each variable was assigned a score, and the total score was computed by summing individual scores, reflecting the probabilities of severe illness. Harrell’s C-index for the prediction nomogram was 0.873 (95% CI: 0.808–0.938), suggesting the potential of the model to predict disease severity. The calibration (Fig. 4b) and decision curves (Fig. 4c) also demonstrated that the nomogram was capable of predicting severe disease in patients with COVID-19. (See Fig. 5. )
Fig. 4.
Nomogram for the risk of severe illness. (a) The HsCAR, HsCPAR, and PNI combination nomogram. (b) Calibration curves of nomograms to show the correlation between the predicted probability and actual diagnostic results. (c) Decision curve analysis of the nomogram to predict severe illness. Abbreviations: HsCAR, high sensitivity C-reactive protein-albumin ratio; PNI, prognostic nutritional index; HsCPAR, high sensitivity C-reactive protein-prealbumin ratio.
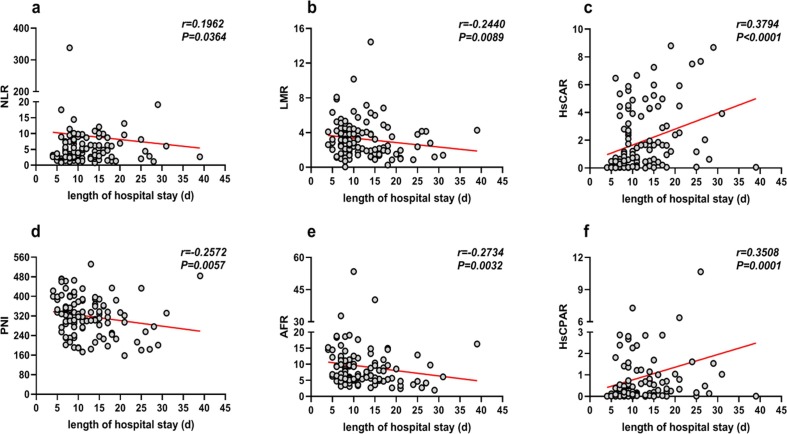
Fig. 5.
The correlation between duration of hospital stay for COVID-19 patients and novel serological markers. Abbreviations: NLR, neutrophil–lymphocyte ratio; LMR, lymphocyte-monocyte ratio; HsCAR, high sensitivity C-reactive protein-albumin ratio; PNI, prognostic nutritional index; AFR, albumin-to-fibrinogen ratio; HsCPAR, high sensitivity C-reactive protein-prealbumin ratio.
3.6. The correlation between duration of hospital stay and novel serological indicators
Finally, we determined the correlation between the serological markers and duration of hospital stay using Spearman's rank correlation coefficient. NLR, LMR, HsCAR, PNI, AFR, and HsCPAR positively correlated with the length of hospital stay for COVID-19 patients (Fig. 4). However, there was no correlation between PLR, dNLR, and SII, and duration of hospital stay. We adjusted the variables for age and gender using univariate linear regression analysis. HsCAR (β = 0.346, p < 0.0001), PNI (β = -0.195, p = 0.040), and HsCPAR (β = 0.246, p = 0.011) correlated with duration of hospital stay. Subsequently, the three independent predictors were included as a part of the multivariate linear regression analysis to conclude that HsCAR (β = 0.302, p = 0.003) and HsCPAR (β = 0.197, p = 0.038) were independently associated with duration of hospital stay, whereas PNI was not.
4. Discussion
Despite global efforts to contain the COVID-19 outbreak since December 2019, single-day confirmed cases have been on the rise [21]. Moreover, there is no specific drug or vaccine currently available for COVID-19. The progression to severe illness results in high rates of mortality (40%) in patients [22]. Timely intervention and control of disease progression are important in the prevention and control of COVID-19. Although the clinical and epidemiological features of this disease have been reported [23], [24], [25], few studies have focused on the risk factors for the development of severe disease in patients with COVID-19, especially based on laboratory data at admission. Studies have confirmed the presence of hematological abnormalities, such as the content of LYM, CRP, and ALB, in COVID-19 patients. These factors are important indicators of inflammation and immune response. Thus, we analyzed the potential of these novel inflammation markers in predicting the development of severe disease in COVID-19 patients. We then constructed a prediction model to determine severe illness during the early stages of COVID-19 in patients.
A total of 114 patients diagnosed with COVID-19 were included in this study. Among these, 58 cases developed severe illness during hospitalization, while the remaining 56 patients exhibited mild to moderate disease until discharge from the hospital. We found that patients who developed severe illness during hospitalization had lower levels of LYM, ALB, PA, and HDL-C, consistent with previous studies [26], [27], [28]. Decreased ALB, PA, and HDL-C suggested the inhibition of protein synthesis in the liver that strongly correlated with the severity and poor prognosis of COVID-19 [28]. Moreover, patients who developed severe disease also exhibited dysfunctional coagulation on admission, as evidenced by high levels of PT, INR, Fbg, FDP, and D-dimer. The levels of AST, UREA, LDH, and HsCRP in patients with severe illness were elevated as compared to those in patients who exhibited mild to moderate disease until discharge. This indicated serious liver and renal damage in patients during hospitalization. Combining these indices will facilitate the identification of factors associated with predicting the progression of mild to severe COVID-19 in patients. New serological indicators, including the variables of inflammation, nutrition, and coagulation, may better demonstrate the interactions between the virus and host. Virus-induced inflammation and immune response and nutritional status of the host play critical roles in disease progression [29], [30]. In this study, we observed significant differences between patients with severe and mild to moderate disease based on the serological indicators. Logistic regression, ROC curve, and test performance analyses showed that lower PNI and higher HsCAR and HsCPAR on admission correlated with higher ORs for severe COVID-19. LYM, ALB, PA, and HsCRP comprise the nutritional and inflammatory markers and the data for these markers can be easily obtained from serum tests and even readily available in routine laboratories upon admission to the hospital. Recent reports have shown that these indicators determine the prognosis of patients with COVID-19 [31], [32], [33]. As is well known, white blood cells play vital roles in the interaction between the immune system and SARS-CoV-2. Indeed, in our study, higher NEU and lower LYM were found at early stage in patients who would progress to severe disease. NLR and dNLR, derived from NEU and LYM, have high AUCs of 0.75 and 0.76. However, the overall performance of HsCAR, HsCPAR and PNI were better than NLR and dNLR in predicting severity of COVID-19. The univariate and multivariate logistic regression analyses also suggested the ORs of HsCAR, HsCPAR and PNI were the three most notable independent risk factors. In other diseases, such as systemic inflammatory response syndrome and tumors, these markers also have good predictive and prognostic value [34], [35]. We speculate that the degree of systemic inflammation, the nutritional status, and lymphocyte counts may determine whether a patient would progress to severe COVID-19 upon admission. Consistent with other studies, our results indicated that patients with higher inflammation and malnutrition at admission might be at higher risk for severe illness. Furthermore, we generated an prognostic nomogram with high C-index of 0.873 using these markers. This model was reliable and predicted the risk of severe illness during the early stages of disease, as confirmed by the decision and calibration curves. The model integrating HsCAR, HsCPAR and PNI, calculated according to values of LYM, ALB, PA, and HsCRP, was expected to provide an accurate, cost effective, fast and convenient clinical tool to stratify COVID-19 severities on admission. And our model may be used to prioritize the patients to be treated immediately and determine the course of personalized therapy.
Several studies have focused on the risk factors associated with severe illness in patients with COVID-19[36], [37], [38], [39]. However, very few studies have comprehensively investigated the potential of these novel factors related to inflammation or nutrition in patients with COVID-19, and even fewer studies have used nomograms to determine their potential in predicting severe illness on admission. Liu et al. [40] constructed a nomogram using NLR and CRP with a C-index of 0.784, which was lower than that for the model in this study. Although Zhou et al. [41] established a model with a relatively high C-index of 0.863, it was based on clinical data from patients that requires detailed history and testing, and the data collected may not be objective. However, HsCAR and HsCPAR were also demonstrated to be the best objective indicators for predicting the occurrence of severe COVID-19 and duration of hospital stay for patients in our study. Moreover, although 366 COVID-19 patients were enrolled in Zhou’s study, the percentage of severe COVID-19 patient was only 11.75% (43/323). And severe patients were older than non-severe patients. These factors may affect the accuracy of the model. Because our hospital was the designated hospital to treat COVID-19 patients in Wuhan at the beginning of the outbreak, the ratio of severe COVID-19 was relatively higher than other areas. Viral load and treatment options also determine the duration of hospital stay and severity of disease in a patient [39], [42], [43], [44]. Unfortunately, these data were not available for this study.
Notably, this study has some limitations. First, the relatively small patient cohort may have influenced the statistical significance of the data in this study. Second, we did not analyze the correlation between the inflammatory makers and viral loads. Third, this study was based on the duration of hospital stay, not the time elapsed between onset of symptoms to discharge from the hospital. The required data is difficult to obtain if patients do not visit the hospital once they notice symptoms.
In conclusion, HsCAR, PNI, and HsCPAR are novel systemic inflammatory markers for the severity of COVID-19 in patients. The nomogram and model generated by combining the three factors may accurately and easily predict severe illness in COVID-19 patients on admission. Moreover, HsCAR and HsCPAR might effectively estimate the duration of hospital stay for patients with COVID-19. Thus, multi-center studies based on larger cohorts will help further validate the model developed and help clinicians in predicting disease progression in patients with COVID-19.
CRediT authorship contribution statement
Guohui Xue: Conceptualization, Methodology, Formal analysis, Validation, Visualization, Writing - original draft. Xing Gan: Investigation, Resources, Software, Writing - review & editing. Zhiqiang Wu: Visualization, Resources. Dan Xie: Visualization. Yan Xiong: Visualization, Software. Lin Hua: Visualization. Bing Zhou: Validation. Nanjin Zhou: . Jie Xiang: Writing - review & editing, Resources, Conceptualization. Junming Li: Conceptualization, Project administration, Funding acquisition, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by grants from the Health and Family Planning Commission of Jiangxi Province (grant number No. 20164033 and No. 20174011). We would like to thank Editage (www.editage.cn) for English language editing.
References
- 1.Coronavirus 2019-nCoV, CSSE. Coronavirus 2019-nCoV Global Cases by Johns Hopkins CSSE. https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6 (accessed July 3 2020).
- 2.Wu Z., McGoogan J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 COVID-19 Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;32313:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3.Yang X., Yu Y., Xu J. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arabi Y.M., Murthy S., Webb S. COVID-19: a novel coronavirus and a novel challenge for critical care. Intensive Care Med. 2020;465:833–836. doi: 10.1007/s00134-020-05955-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poggiali E., Zaino D., Immovilli P. Lactate dehydrogenase and C-reactive protein as predictors of respiratory failure in CoVID-19 patients. Clin. Chim. Acta. 2020;509:135–138. doi: 10.1016/j.cca.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han H., Ma Q., Li C. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 2020;9(1):1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamasaki Y., Ooka S., Tsuchida T. The peripheral lymphocyte count as a predictor of severe COVID-19 and the effect of treatment with ciclesonide. Virus Res. 2020:198089. doi: 10.1016/j.virusres.2020.198089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammock B.D., Wang W., Gilligan M.M. Eicosanoids: the Overlooked Storm in COVID-19? Am. J. Pathol. 2020 doi: 10.1016/j.ajpath.2020.06.010. S0002-9440(20)30332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X., Li Y., Ge Y. Evaluation of Nutritional Risk and its Association With Mortality Risk in Severe and Critically Ill COVID-19 Patients. JPEN J. Parenter. Enteral Nutr. 2020 doi: 10.1002/jpen.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lidoriki I., Frountzas M., Schizas D. Could nutritional and functional status serve as prognostic factors for COVID-19 in the elderly? Med. Hypotheses. 2020;144 doi: 10.1016/j.mehy.2020.109946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L., Kong X., Yan C. The Research Progress on the Prognostic Value of the Common Hematological Parameters in Peripheral Venous Blood in Breast Cancer. Onco. Targets Ther. 2020;13:1397–1412. doi: 10.2147/OTT.S227171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J.H., Zhai E.T., Yuan Y.J. Systemic immune-inflammation index for predicting prognosis of colorectal cancer. World J. Gastroenterol. 2017;23(34):6261–6272. doi: 10.3748/wjg.v23.i34.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tai H., Zhu Z., Mei H. Albumin-to-Fibrinogen Ratio Independently Predicts 28-Day Mortality in Patients with Peritonitis-Induced Sepsis. Mediators Inflamm. 2020;2020:7280708. doi: 10.1155/2020/7280708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The General Office of the National Health Commission, The Office of the State Administration of Traditional Chinese Medicine. Notice on the issuance of COVID-19 diagnosis and treatment plan trial sixth edition. http://yzs.satcm.gov.cn/zhengcewenjian/2020-02-19/13221.html (accessed 19 Feb 2020).
- 15.Liu Y., Yang Y., Zhang C. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang A.P., Liu J.P., Tao W.Q. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int. Immunopharmacol. 2020;84 doi: 10.1016/j.intimp.2020.106504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S., Luo H., Wang Y. Clinical characteristics and risk factors of patients with severe COVID-19 in Jiangsu province, China: a retrospective multicentre cohort study. BMC Infect. Dis. 2020;20(1):584. doi: 10.1186/s12879-020-05314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y., Xue Y., Mao L. Prealbumin as a Predictor of Prognosis in Patients With Coronavirus Disease 2019. Front. Med. (Lausanne) 2020;7:374. doi: 10.3389/fmed.2020.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattiuzzi C., Lippi G. Serum prealbumin values predict the severity of coronavirus disease 2019 (COVID-19) J. Med. Virol. 2020 doi: 10.1002/jmv.26385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Violi F., Cangemi R., Romiti G.F. Is Albumin Predictor of Mortality in COVID-19? Antioxid. Redox Signal. 2020 doi: 10.1089/ars.2020.8142. [DOI] [PubMed] [Google Scholar]
- 21.Shereen M.A., Khan S., Kazmi A. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiersinga W.J., Rhodes A., Cheng A.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020 doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 23.Tu H., Tu S., Gao S., Shao A., Sheng J. Current epidemiological and clinical features of COVID-19; a global perspective from China. J. Infect. 2020;81(1):1–9. doi: 10.1016/j.jinf.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Y., Xu H., Yang M. Epidemiological characteristics and clinical features of 32 critical and 67 noncritical cases of COVID-19 in Chengdu. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y., Zheng F., Sun D. Epidemiology and clinical course of COVID-19 in Shanghai, China. Emerg. Microbes Infect. 2020;9(1):1537–1545. doi: 10.1080/22221751.2020.1787103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan J., Wang X., Chi J. Correlation between the variables collected at admission and progression to severe cases during hospitalization among patients with COVID-19 in Chongqing. J. Med. Virol. 2020 doi: 10.1002/jmv.26082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng Z., Ma Y., Zeng H. Simple nomogram based on initial laboratory data for predicting the probability of ICU transfer of COVID-19 patients: Multicenter retrospective study. J. Med. Virol. 2020 doi: 10.1002/jmv.26244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W., Li C., Wang Z. Decreased serum albumin level indicates poor prognosis of COVID-19 patients: hepatic injury analysis from 2,623 hospitalized cases. Sci China Life Sci. 2020:1–10. doi: 10.1007/s11427-020-1733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beck M.A., Levander O.A. Host nutritional status and its effect on a viral pathogen. J. Infect. Dis. 2000;182(Suppl 1):S93–S96. doi: 10.1086/315918. [DOI] [PubMed] [Google Scholar]
- 30.Mehta P., McAuley D.F., Brown M. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Violi F., Cangemi R., Romiti G.F. Is Albumin Predictor of Mortality in COVID-19? Antioxid. Redox Signal. 2020 doi: 10.1089/ars.2020.8142. [DOI] [PubMed] [Google Scholar]
- 32.Guo X., Liu W., Zhong J. Laboratory Findings in SARS-CoV-2 Infections in Zigong, China: Key Points for Serum Prealbumin. Clin. Lab. 2020;66(6) doi: 10.7754/Clin.Lab.2020.200419. [DOI] [PubMed] [Google Scholar]
- 33.Violi F., Ceccarelli G., Cangemi R. Hypoalbuminemia, Coagulopathy and Vascular Disease in Covid-19. Circ. Res. 2020 doi: 10.1161/CIRCRESAHA.120.317173. [DOI] [PubMed] [Google Scholar]
- 34.Xu H., Hu L., Wei X. The Predictive Value of Preoperative High-Sensitive C-Reactive Protein/Albumin Ratio in Systemic Inflammatory Response Syndrome After Percutaneous Nephrolithotomy. J. Endourol. 2019;33(1):1–8. doi: 10.1089/end.2018.0632. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z., Wang Y., Zhang X. Pretreatment prognostic nutritional index as a prognostic factor in lung cancer: Review and meta-analysis. Clin. Chim. Acta. 2018;486:303–310. doi: 10.1016/j.cca.2018.08.030. [DOI] [PubMed] [Google Scholar]
- 36.Li X., Xu S., Yu M. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J. Allergy Clin. Immunol. 2020;146(1):110–118. doi: 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He F., Luo Q., Lei M. Risk factors for severe cases of COVID-19: a retrospective cohort study. Aging (Albany NY) 2020;12 doi: 10.18632/aging.103803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J., Zhang S., Wu Z. Clinical outcomes of COVID-19 in Wuhan, China: a large cohort study. Ann. Intensive Care. 2020;10(1):99. doi: 10.1186/s13613-020-00706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X., Zhou H., Zhou Y. Risk factors associated with disease severity and length of hospital stay in COVID-19 patients. J. Infect. 2020;81(1):e95–e97. doi: 10.1016/j.jinf.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J., Liu Y., Xiang P. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J. Transl. Med. 2020;18(1):206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y., He Y., Yang H. Development and validation a nomogram for predicting the risk of severe COVID-19: A multi-center study in Sichuan, China. PLoS ONE. 2020;15(5) doi: 10.1371/journal.pone.0233328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Argyropoulos K.V., Serrano A., Hu J. Association of initial viral load in SARS-CoV-2 patients with outcome and symptoms. Am. J. Pathol. 2020 doi: 10.1016/j.ajpath.2020.07.001. S0002-9440(20)30328-X. [DOI] [PubMed] [Google Scholar]
- 43.Zheng S., Fan J., Yu F. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magleby R., Westblade L.F., Trzebucki A. Impact of SARS-CoV-2 Viral Load on Risk of Intubation and Mortality Among Hospitalized Patients with Coronavirus Disease 2019. Clin. Infect. Dis. 2020:ciaa851. doi: 10.1093/cid/ciaa851. [DOI] [PMC free article] [PubMed] [Google Scholar]