Abstract
Rationale
Prediction of patients at risk for mortality can help triage patients and assist in resource allocation.
Objectives
Develop and evaluate a machine learning-based algorithm which accurately predicts mortality in COVID-19, pneumonia, and mechanically ventilated patients.
Methods
Retrospective study of 53,001 total ICU patients, including 9166 patients with pneumonia and 25,895 mechanically ventilated patients, performed on the MIMIC dataset. An additional retrospective analysis was performed on a community hospital dataset containing 114 patients positive for SARS-COV-2 by PCR test. The outcome of interest was in-hospital patient mortality.
Results
When trained and tested on the MIMIC dataset, the XGBoost predictor obtained area under the receiver operating characteristic (AUROC) values of 0.82, 0.81, 0.77, and 0.75 for mortality prediction on mechanically ventilated patients at 12-, 24-, 48-, and 72- hour windows, respectively, and AUROCs of 0.87, 0.78, 0.77, and 0.734 for mortality prediction on pneumonia patients at 12-, 24-, 48-, and 72- hour windows, respectively. The predictor outperformed the qSOFA, MEWS and CURB-65 risk scores at all prediction windows. When tested on the community hospital dataset, the predictor obtained AUROCs of 0.91, 0.90, 0.86, and 0.87 for mortality prediction on COVID-19 patients at 12-, 24-, 48-, and 72- hour windows, respectively, outperforming the qSOFA, MEWS and CURB-65 risk scores at all prediction windows.
Conclusions
This machine learning-based algorithm is a useful predictive tool for anticipating patient mortality at clinically useful timepoints, and is capable of accurate mortality prediction for mechanically ventilated patients as well as those diagnosed with pneumonia and COVID-19.
Keywords: Mortality prediction, COVID-19, SARS-CoV-2, Machine learning, Artificial intelligence
Highlights
-
•
Mortality predictions have not previously been evaluated for COVID-19 patients.
-
•
Machine learning may be a useful predictive tool for anticipating patient mortality.
-
•
Prediction can be estimated at clinically useful windows up to 72 h in advance.
1. Introduction
Infection prevention and control recommendations from the World Health Organization (WHO) stress that early detection, effective triage, and isolation of potentially infectious patients are essential to prevent unnecessary exposures to COVID-19 [1]. However, the rapid spread of COVID-19 has outpaced US healthcare facilities’ ability to administer diagnostic tests to guide the quarantine and triage COVID-19 patients [[2], [3], [4], [5]]. The outbreak significantly affects the availability of necessary hospital resources (i.e. respirators [6] and mechanical ventilators [[7], [8], [9], [10], [11], [12]]). COVID-19 can be lethal, with a variable case fatality rate considered to be between that of severe acute respiratory syndrome (SARS; 9.5% [13]) and influenza (0.1%) [[14], [15], [16]] and the potential to develop into severe respiratory diseases [[17], [18], [19]]. During this period of unprecedented health crisis, clinicians must prioritize care for at-risk individuals to maximize limited resources. Mortality prediction tools aid in triage and resource allocation by providing advance warning of patient deterioration. Our prior work has validated machine-learning (ML) algorithms for their ability to predict mortality and patient stability in a variety of settings and on diverse patient populations [[20], [21], [22], [23], [24]].
2. Theory
Of particular interest during the COVID-19 pandemic is mortality prediction of COVID-19 patients, as well as those who have developed respiratory complications such as pneumonia and conditions requiring mechanical ventilation. Some prior studies predicting mortality in the mechanically vented subpopulation have used a logistic regression model. When applied on day 21 [25] or 14 [26,27] of mechanical ventilation, this provides a probability of 1-year mortality. These studies were designed to determine the long-term prognosis of patients receiving prolonged mechanical ventilation. Here we present a mortality prediction tool applied to intensive care unit (ICU) patients requiring mechanical ventilation as well as those diagnosed with pneumonia, with mortality prediction windows of 12, 24, 48 and 72 h prior to death. We apply this algorithm for the same mortality prediction windows in COVID-19 patients.
3. Materials and methods
3.1. Data sources
Patient records were collected from the Medical Information Mart for Intensive Care (MIMIC) dataset, an openly available dataset developed by the MIT Lab for Computational Physiology, comprising de-identified health data associated with ~60,000 intensive care unit admissions [28]. It includes demographics, vital signs, laboratory tests, medications, and more. Data collection was passive with no impact on patient safety. MIMIC data has been de-identified in compliance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule.
Patient records of COVID-19 polymerase chain reaction (PCR) positive patients were collected from a community hospital and formatted in the same manner as the MIMIC dataset. A total of 114 patient encounters were collected between 12 March and 12 April 2020. Data collection was passive with no impact on patient safety. Dascena establishes de-identification by removing all protected health information (PHI) identifiers and by jittering all timestamps (including date of birth (DOB)) randomly either forwards or backwards in time. Studies performed on de-identified patient data constitute non-human subjects research, and thus this study has been determined by the Pearl Institutional Review Board to be Exempt according to FDA 21 CFR 56.104 and 45CFR46.104(b) (4): (4) Secondary Research Uses of Data or Specimens under study number 20-DASC-119.
3.1.1. Data processing
For the MIMIC and community hospital datasets, we included only records for patients aged 18 years or older. We excluded patient records for which there were no raw data or no discharge or death dates. We then filtered for length of stay (LOS) for the different look aheads of 12, 24, 48, and 72 h. Table 1 lists the number of patients for each inclusion criterion from the MIMIC dataset. Inclusion criteria for the community hospital dataset are listed in Table 2. We minimally processed raw electronic health record (EHR) data to generate features. Following imputation of missing values, we averaged one value for each measurement each hour for up to 3 h preceding prediction time. We also calculated differences between the current hour and the prior hour and between the prior hour and the hour before that. We concatenated these values from each measurement into a feature vector. For the MIMIC dataset, pneumonia patients were identified by International Classification of Diseases (ICD) codes, while those requiring mechanical ventilation and their corresponding start times were determined by chart measurements indicative of a mechanical ventilation setting. In the community hospital dataset, COVID-19 patients were identified with positive SARS-Cov2 PCR tests.
Table 1.
Inclusion criteria for patients in the MIMIC dataset. *Required measurements include Age, Heart Rate, Respiratory Rate, Peripheral Oxygen Saturation (SpO2), Temperature, Systolic Blood Pressure, Diastolic Blood Pressure, White Blood Cell Counts, Platelets, Lactate, Creatinine, and Bilirubin.
| Criterion | Encounters |
|---|---|
| ICU stays in MIMIC | 61,532 |
| ICU stays with patients aged ≥ 18 years, any measurements present* | 53,001 |
| Length of stay filtering for all patients 12 h | 50,695 |
| Length of stay filtering for all patients 24 h | 40,959 |
| Length of stay filtering for all patients 48 h | 26,576 |
| Length of stay filtering for all patients 72 h | 18,275 |
| Length of stay filtering for mechanically ventilated patients 12 h | 24,934 |
| Length of stay filtering for mechanically ventilated patients 24 h | 21,414 |
| Length of stay filtering for mechanically ventilated patients 48 h | 16,085 |
| Length of stay filtering for mechanically ventilated patients 72 h | 12,368 |
| Length of stay filtering for pneumonia patients 12 h | 8879 |
| Length of stay filtering for pneumonia patients 24 h | 7678 |
| Length of stay filtering for pneumonia patients 48 h | 5600 |
| Length of stay filtering for pneumonia patients 72 h | 4169 |
Table 2.
Inclusion criteria for patients in the community hospital dataset. *Required measurements include Age, Heart Rate, Respiratory Rate, Peripheral Oxygen Saturation (SpO2), Temperature, Systolic Blood Pressure, Diastolic Blood Pressure, White Blood Cell Counts, Platelets, Lactate, Creatinine, and Bilirubin.
| Criterion | Encounters |
|---|---|
| COVID positive stays in community hospital | 114 |
| COVID positive stays with patients aged ≥ 18 years, any measurements present* | 114 |
| Length of stay filtering for all COVID positive patients 12 h | 114 |
| Length of stay filtering for all COVID positive patients 24 h | 112 |
| Length of stay filtering for all COVID positive patients 48 h | 110 |
| Length of stay filtering for all COVID positive patients 72 h | 103 |
Data were discretized into 1 h intervals, beginning at the time of the first recorded patient measurement and hourly measurements were required for each input variable. Measurements were averaged to produce a single value in cases when multiple observations of the same patient measurement were taken within a given hour. This ensures that the measurement rate was the same across patients and across time. Missing values were imputed by carrying forward the most recent past measurement in cases where no measurement of a clinical variable was available for a given hour. For some patients with infrequent measurements of one or more vital signs, this simple imputation resulted in many consecutive hours with identical values.
Our publication on the use of gradient boosted trees for sepsis detection and prediction describes the data processing in detail [29]. Predictions were generated for all experiments using the following variables: Age, Heart Rate, Respiratory Rate, Peripheral Oxygen Saturation (SpO2), Temperature, Systolic Blood Pressure, Diastolic Blood Pressure, White Blood Cell Counts, Platelets, Lactate, Creatinine, and Bilirubin, over an interval of 3 h and their corresponding differentials in that interval.
3.2. Gold standard
The outcome of interest was in-hospital patient mortality, determined retrospectively for each patient. In the MIMIC dataset, we used the expire_flag field to identify the last stays of those patients. Similarly, the community hospital dataset contains a deceased flag that is either true or false to determine mortality.
3.2.1. The machine learning algorithm
The classifier was created using the XGBoost method for fitting “boosted” decision trees. We applied the XGBoost package for Python32 to the patient age and vital sign measurements and their temporal changes, where temporal changes included hourly differences between each measurement beginning 3 h before prediction time. Gradient boosting, which XGBoost implements, is an ensemble learning technique that combines results from multiple decision trees to create prediction scores. Each tree splits the patient population into smaller and smaller groups, successively. Each branch splits the patients who enter it into two groups, based on whether their value of some covariate is above or below some threshold—for instance, a branch might divide patients according to whether their temperature is above or below 100 °F. After some number of branches, the tree ends in a set of “leaves.” Each patient is in exactly one leaf, according to the values of his or her measurements. Each “leaf” of the tree is predicted to have the same risk of mortality. The covariate involved in each split and the threshold value are selected by an algorithm designed to trade off fit to the training data and accuracy on out-of-sample data by using cross-validation to avoid “over-fitting.” We restricted tree depth to a maximum of six branching levels, set the learning rate parameter of XGBoost to 0.1, and restricted the tree ensembles to 1000 trees to limit the computational burden.
Hyperparameter optimization was performed using cross-validated grid search. We included a hyperparameter for the early stopping of the iterative tree-addition procedure to prevent overfit of the model on the training data and optimized across this hyperparameter using fivefold cross-validation. Due to computational and time constraints, hyperparameter optimization was performed across a sparse parameter grid, where the candidate hyperparameter values were chosen to span large ranges of viable parameter space. Cross-validated grid search was conducted to determine the optimal combination of candidate hyperparameters. While XGBoost has a large number of trainable parameters, computational and time constraints limited the set of parameters to be tuned to just those parameters with the largest impact on performance on the training data and most relevant to the prediction task.
To validate the boosted tree predictor when training and testing was performed on data from the same institution, we used fivefold cross-validation. For each model, four-fifths of the patients were randomly selected to train the model and the remaining one-fifth were used as a hold-out set to test the predictions. To account for the random selection of the training set, reported performance metrics are the average performance of the five separately trained models arising from fivefold cross-validation, each of which was trained on four-fifths of the data and tested on the remaining fifth. For AUROC, we also reported the standard deviation of the five AUROC values obtained from cross-validation.
For patients who died, we modeled mortality 12, 24, 48, and 72 h before death to evaluate the performance with a variety of lead times. For mechanically ventilated encounters, the time point was the start of ventilation for positive and negative class. Predictors were trained independently for each distinct lookahead time. In 12, 24, 48 and 72 h long lookahead predictions following a 3-h window of measurements, patients must have data for, respectively, 15, 27, 51 or 75 respective hours preceding the time of in-hospital mortality or the time of discharge. Accordingly, we selected patients with the appropriate stays for the training and testing of each lookahead.
3.3. Comparison to rule-based methods
To calculate the AUROC for rule-based predictors of mortality, we calculated quick Sepsis Related Organ Failure Assessment (qSOFA), Modified Early Warning Score (MEWS) and CURB-65 scores for patients in the MIMIC database. qSOFA has also been used to predict poor outcomes in pneumonia patients, including the need for mechanical ventilation, and has been shown to either match or outperform other outcome predictors such as SOFA, CRB, CRB-65 and the pneumonia severity index (PSI) [30,31]. Among more generally used mortality prediction scores, qSOFA has been shown to have similar predictive performance to that of Acute Physiology, Age, Chronic Health Evaluation (APACHE) II or SOFA, as evidenced by a lack of statistical difference between AUROC [32]. The MEWS and CURB-65 scores have also been validated for mortality prediction in general patient populations [33,34] and those with community-acquired pneumonia [35] or COVID-19 [36], respectively. Scores were calculated using the entire dataset. We calculated the qSOFA score using systolic blood pressure, respiratory rate, and Glasgow Coma Scale (GCS) from EHR data. MEWS was calculated using systolic blood pressure, heart rate, respiratory rate, and temperature. GCS was used as a proxy for evaluating AVPU. CURB-65 scores were computed using age, BUN, respiratory rate, as well as systolic and diastolic blood pressure. A GCS of less than or equal to 14 was used as a proxy for confusion. Comparator score calculations for patients in the community hospital dataset were modified based on available data.
4. Results
XGBoost model training and testing was performed on the MIMIC dataset. Patient demographic information for all ICU encounters as well as each subpopulation are presented in Table 3, Table 4, Table 5. Patient demographic information for all encounters from the community hospital data set are listed in Table 6.
Table 3.
Patient demographic information for MIMIC dataset for all encounters (53,001).
| Characteristic | MIMIC (%) | |
|---|---|---|
| Age | 18–29 | 4.70 |
| 30–39 | 5.25 | |
| 40–49 | 10.65 | |
| 50–59 | 17.52 | |
| 60–69 | 20.99 | |
| >70 | 40.90 | |
| Gender | Male | 43.68 |
| Female | 56.32 | |
| In-hospital Death | Yes | 9.59 |
| No | 90.41 |
Table 4.
Patient demographic information for MIMIC dataset for all pneumonia encounters (9,166).
| Characteristic | MIMIC (%) | |
|---|---|---|
| Age | 18–29 | 2.97 |
| 30–39 | 4.34 | |
| 40–49 | 9.33 | |
| 50–59 | 17.17 | |
| 60–69 | 21.01 | |
| >70 | 45.18 | |
| Gender | Male | 53.99 |
| Female | 46.01 | |
| In-hospital Death | Yes | 11.41 |
| No | 88.59 |
Table 5.
Patient demographic information for MIMIC dataset for all mechanically ventilated encounters (25,895).
| Characteristic | MIMIC (%) | |
|---|---|---|
| Age | 18–29 | 4.50 |
| 30–39 | 4.38 | |
| 40–49 | 10.16 | |
| 50–59 | 17.91 | |
| 60–69 | 22.39 | |
| >70 | 40.66 | |
| Gender | Male | 59.1 |
| Female | 40.9 | |
| In-hospital Death | Yes | 15.73 |
| No | 84.27 |
Table 6.
Patient demographic information for community hospital dataset encounters.
| Characteristic | Community Hospital (%) | |
|---|---|---|
| Age | 18–29 | 7.00 |
| 30–39 | 10.5 | |
| 40–49 | 8.77 | |
| 50–59 | 15.79 | |
| 60–69 | 23.68 | |
| >70 | 34.21 | |
| Gender | Male | 58.77 |
| Female | 41.33 | |
| In-hospital Death | Yes | 21.0 |
| No | 79.0 |
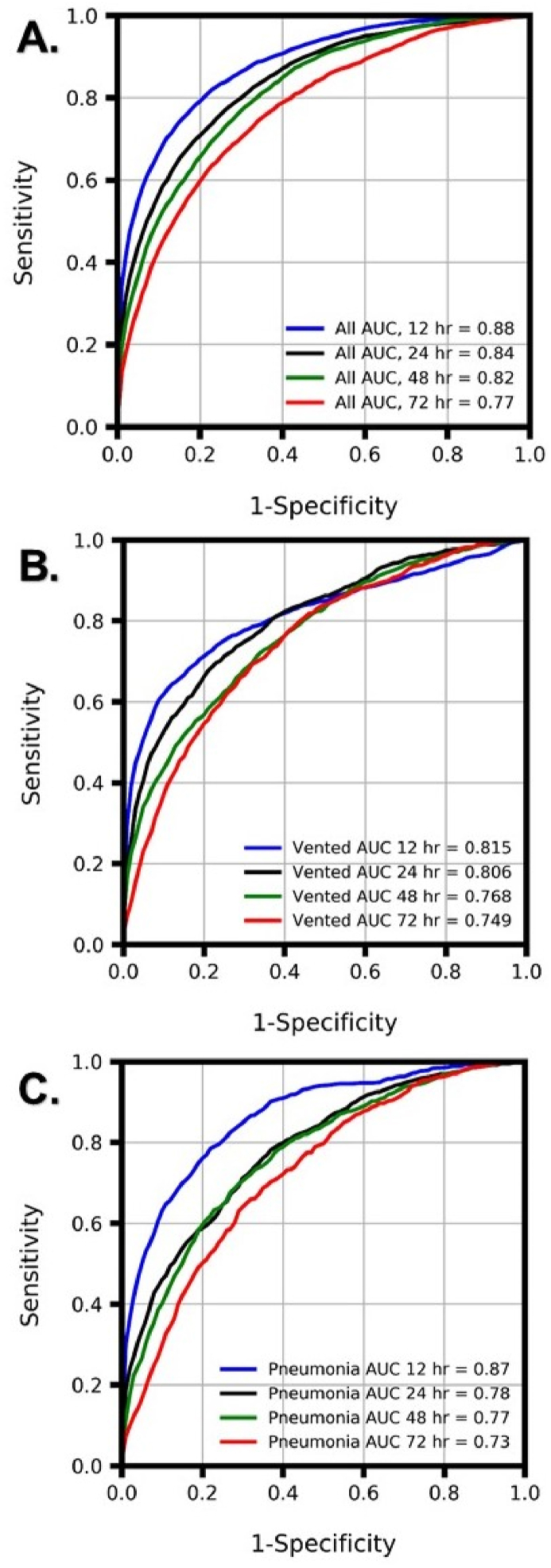
The XGBoost ML algorithm predicted mortality in all ICU patients as well as mechanically ventilated and pneumonia patients more accurately than qSOFA, MEWS and CURB-65 at all prediction windows ( Table 7, Table 8 and Supplementary Table S5). When trained and tested on the MIMIC dataset, the XGBoost predictor obtained AUROCs of 0.82, 0.81, 0.77, and 0.75 for mortality prediction on mechanically ventilated patients at 12-, 24-, 48-, and 72- hour windows, respectively, and AUROCs of 0.87, 0.78, 0.77, and 0.73 for mortality prediction on pneumonia patients at 12-, 24-, 48-, and 72- hour windows, respectively (Fig. 1). Feature importance statistics are listed in Table S1, Table S2, Table S3, Table S4.
Table 7.
Comparison of AUROC, average precision (APR), sensitivity, specificity, F1, diagnostic odds ratio (DOR), positive and negative likelihood ratios (LR+ and LR‒), accuracy and recall obtained by the machine learning algorithm (MLA) and the qSOFA score for mortality prediction at 12-, 24-, 48-, and 72- hour windows on pneumonia patients using the MIMIC dataset. Standard deviations are listed in parenthesis. For AUROC and APR the operating point was set near a sensitivity of 0.800.
| MLA: Pneumonia | qSOFA: Pneumonia | MEWS: Pneumonia | CURB-65: Pneumonia | ||
|---|---|---|---|---|---|
| 12 h | AUROC | 0.865 (0.0027) | 0.719 | 0.792 | 0.595 |
| APR | 0.594 (0.0075) | 0.242 | 0.405 | 0.154 | |
| Sensitivity | 0.800 (0.0000) | 0.933 | 0.884 | 0.973 | |
| Specificity | 0.761 (0.0101) | 0.304 | 0.472 | 0.169 | |
| F1 | 0.467 (0.0094) | 0.280 | 0.323 | 0.255 | |
| DOR | 12.74 (0.715) | 6.126 | 6.831 | 7.397 | |
| LR+ | 3.35 (0.143) | 1.342 | 1.674 | 1.171 | |
| LR- | 3.35 (0.143) | 1.342 | 1.674 | 1.171 | |
| Accuracy | 0.766 (0.0088) | 0.385 | 0.524 | 0.272 | |
| Recall | 0.804 (0.0000) | 0.933 | 0.884 | 0.973 | |
| 24 h | AUROC | 0.783 (0.0017) | 0.721 | 0.779 | 0.612 |
| APR | 0.442 (0.0070) | 0.267 | 0.346 | 0.159 | |
| Sensitivity | 0.802 (0.0000) | 0.932 | 0.906 | 0.974 | |
| Specificity | 0.594 (0.0324) | 0.285 | 0.439 | 0.142 | |
| F1 | 0.349 (0.0160) | 0.271 | 0.313 | 0.247 | |
| DOR | 5.99 (0.727) | 5.484 | 7.552 | 6.211 | |
| LR+ | 1.99 (0.144) | 1.304 | 1.614 | 1.136 | |
| LR- | 1.99 (0.144) | 1.304 | 1.614 | 1.136 | |
| Accuracy | 0.621 (0.0283) | 0.367 | 0.498 | 0.248 | |
| Recall | 0.802 (0.0000) | 0.932 | 0.906 | 0.974 | |
| 48 h | AUROC | 0.769 (0.0074) | 0.681 | 0.747 | 0.606 |
| APR | 0.407 (0.0099) | 0.264 | 0.334 | 0.178 | |
| Sensitivity | 0.803 (0.0000) | 0.917 | 0.866 | 0.975 | |
| Specificity | 0.580 (0.0308) | 0.238 | 0.394 | 0.122 | |
| F1 | 0.374 (0.0158) | 0.284 | 0.317 | 0.271 | |
| DOR | 5.67 (0.701) | 3.454 | 4.211 | 5.318 | |
| LR+ | 1.92 (0.138) | 1.203 | 1.429 | 1.110 | |
| LR- | 1.92 (0.138) | 1.203 | 1.429 | 1.110 | |
| Accuracy | 0.612 (0.0264) | 0.335 | 0.462 | 0.245 | |
| Recall | 0.803 (0.0000) | 0.917 | 0.866 | 0.975 | |
| 72 h | AUROC | 0.726 (0.0047) | 0.645 | 0.668 | 0.592 |
| APR | 0.333 (0.0168) | 0.227 | 0.275 | 0.185 | |
| Sensitivity | 0.801 (0.0030) | 0.933 | 0.867 | 0.970 | |
| Specificity | 0.507 (0.0137) | 0.202 | 0.333 | 0.098 | |
| F1 | 0.357 (0.0070) | 0.296 | 0.315 | 0.281 | |
| DOR | 4.16 (0.307) | 3.542 | 3.250 | 3.541 | |
| LR+ | 1.63 (0.052) | 1.169 | 1.300 | 1.075 | |
| LR- | 1.63 (0.052) | 1.169 | 1.300 | 1.075 | |
| Accuracy | 0.553 (0.0120) | 0.315 | 0.416 | 0.233 | |
| Recall | 0.807 (0.0000) | 0.933 | 0.867 | 0.970 |
Table 8.
Comparison of AUROC, average precision (APR), sensitivity, specificity, F1, diagnostic odds ratio (DOR), positive and negative likelihood ratios (LR+ and LR‒), accuracy and recall obtained by the machine learning algorithm (MLA) and the qSOFA score for mortality prediction at 12-, 24-, 48-, and 72- hour windows on mechanically ventilated patients using the MIMIC dataset. Standard deviations are listed in parenthesis. For AUROC and APR the operating point was set near a sensitivity of 0.800.
| MLA: Mechanically Ventilated | qSOFA: Mechanically Ventilated | MEWS: Mechanically Ventilated | CURB-65: Mechanically Ventilated | ||
|---|---|---|---|---|---|
| 12 h | AUROC | 0.815 (0.0030) | 0.731 | 0.808 | 0.620 |
| APR | 0.598 (0.0055) | 0.276 | 0.417 | 0.173 | |
| Sensitivity | 0.803 (0.0016) | 0.969 | 0.845 | 0.988 | |
| Specificity | 0.647 (0.0241) | 0.232 | 0.630 | 0.098 | |
| F1 | 0.394 (0.0159) | 0.280 | 0.400 | 0.253 | |
| DOR | 7.54 (0.902) | 9.414 | 9.287 | 9.237 | |
| LR+ | 2.29 (0.167) | 1.261 | 2.285 | 1.096 | |
| LR- | 2.29 (0.167) | 1.261 | 2.285 | 1.096 | |
| Accuracy | 0.668 (0.0211) | 0.331 | 0.659 | 0.218 | |
| Recall | 0.802 (0.0000) | 0.969 | 0.845 | 0.988 | |
| 24 h | AUROC | 0.806 (0.0030) | 0.729 | 0.789 | 0.626 |
| APR | 0.506 (0.0098) | 0.274 | 0.357 | 0.179 | |
| Sensitivity | 0.803 (0.0017) | 0.970 | 0.810 | 0.987 | |
| Specificity | 0.634 (0.0101) | 0.244 | 0.611 | 0.109 | |
| F1 | 0.392 (0.0060) | 0.289 | 0.381 | 0.260 | |
| DOR | 7.06 (0.299) | 10.384 | 6.714 | 9.343 | |
| LR+ | 2.20 (0.060) | 1.283 | 2.084 | 1.108 | |
| LR- | 2.20 (0.060) | 1.283 | 2.084 | 1.108 | |
| Accuracy | 0.658 (0.0087) | 0.344 | 0.638 | 0.230 | |
| Recall | 0.802 (0.0000) | 0.970 | 0.810 | 0.987 | |
| 48 h | AUROC | 0.768 (0.0034) | 0.715 | 0.753 | 0.611 |
| APR | 0.488 (0.0048) | 0.312 | 0.357 | 0.209 | |
| Sensitivity | 0.804 (0.0019) | 0.977 | 0.826 | 0.977 | |
| Specificity | 0.553 (0.0091) | 0.182 | 0.546 | 0.085 | |
| F1 | 0.398 (0.0046) | 0.322 | 0.403 | 0.298 | |
| DOR | 5.08 (0.198) | 9.266 | 5.710 | 3.840 | |
| LR+ | 1.80 (0.038) | 1.194 | 1.818 | 1.067 | |
| LR- | 1.80 (0.038) | 1.194 | 1.818 | 1.067 | |
| Accuracy | 0.595 (0.0076) | 0.315 | 0.592 | 0.233 | |
| Recall | 0.803 (0.0000) | 0.977 | 0.826 | 0.977 | |
| 72 h | AUROC | 0.749 (0.0053) | 0.657 | 0.665 | 0.601 |
| APR | 0.406 (0.0115) | 0.261 | 0.278 | 0.213 | |
| Sensitivity | 0.805 (0.0000) | 0.977 | 0.943 | 0.983 | |
| Specificity | 0.558 (0.0135) | 0.153 | 0.261 | 0.070 | |
| F1 | 0.413 (0.0068) | 0.327 | 0.347 | 0.308 | |
| DOR | 5.22 (0.288) | 7.683 | 5.799 | 4.322 | |
| LR+ | 1.82 (0.056) | 1.154 | 1.276 | 1.057 | |
| LR- | 1.82 (0.056) | 1.154 | 1.276 | 1.057 | |
| Accuracy | 0.601 (0.0111) | 0.297 | 0.380 | 0.230 | |
| Recall | 0.805 (0.0000) | 0.977 | 0.943 | 0.983 |
Fig. 1.
Comparison of area under the receiver operating characteristic (AUROC) curves for XGBoost models. AUROCs for the boosted tree predictor are presented for 12-, 24-, 48-, and 72-h mortality prediction with training and testing performed on MIMIC data from (A) all ICU patients as well as subpopulations of (B) mechanically ventilated (vented) ICU patients and (C) pneumonia ICU patients.
Detailed performance metrics for the XGBoost predictor on pneumonia and mechanically ventilated patients are presented in Table 7, Table 8 and on COVID-19 patients in Table 9. All predictor training and testing was performed on the MIMIC data set. The diagnostic odds ratio (DOR) is a measure for comparing diagnostic accuracy between tools and is calculated as (True Positive/False Negative)/(False Positive/True Negative). DOR represents the ratio of the odds of a true positive prediction of mortality in patients who died within a certain prediction window to the odds of a false positive prediction of mortality in patients who did not die within a certain prediction window. For all prediction windows, the XGBoost predictor had a higher DOR than qSOFA.
Table 9.
Comparison of AUROC, average precision (APR), sensitivity, specificity, F1, diagnostic odds ratio (DOR), positive and negative likelihood ratios (LR+ and LR‒), accuracy and recall obtained by the ML algorithm (MLA) and the qSOFA, MEWS and CURB-65 scores for mortality prediction at 12-, 24-, 48-, and 72- hour windows on 114 COVID-19 PCR Positive Patients from the community hospital data set. Standard deviations are listed in parenthesis. For AUROC and APR the operating point was set near a sensitivity of 0.800. n/a (not applicable).
| MLA | qSOFA | MEWS | CURB-65 | ||
|---|---|---|---|---|---|
| 12 h | AUROC | 0.910 (0.0024) | 0.791 | 0.769 | 0.780 |
| APR | 0.795 (0.0054) | 0.510 | 0.514 | 0.369 | |
| Sensitivity | 0.826 (0.0000) | 1.000 | 1.000 | 1.000 | |
| Specificity | 0.804 (0.0239) | 0.000 | 0.022 | 0.500 | |
| F1 | 0.638 (0.0228) | 0.338 | 0.343 | 0.505 | |
| DOR | 19.89 (2.912) | n/a | n/a | n/a | |
| LR+ | 4.29 (0.506) | 1.000 | 1.023 | 2.000 | |
| LR- | 4.29 (0.506) | 1.000 | 1.023 | 2.000 | |
| Accuracy | 0.809 (0.0191) | 0.204 | 0.221 | 0.602 | |
| Recall | 0.826 (0.0000) | 1.000 | 1.000 | 1.000 | |
| 24 h | AUROC | 0.903 (0.0059) | 0.840 | 0.780 | 0.764 |
| APR | 0.754 (0.0127) | 0.563 | 0.515 | 0.354 | |
| Sensitivity | 0.826 (0.0000) | 0.826 | 1.000 | 1.000 | |
| Specificity | 0.816 (0.0054) | 0.822 | 0.033 | 0.444 | |
| F1 | 0.649 (0.0054) | 0.655 | 0.346 | 0.479 | |
| DOR | 21.03 (0.770) | 21.969 | n/a | n/a | |
| LR+ | 4.48 (0.134) | 4.647 | 1.034 | 1.800 | |
| LR- | 4.48 (0.134) | 4.647 | 1.034 | 1.800 | |
| Accuracy | 0.818 (0.0043) | 0.823 | 0.230 | 0.558 | |
| Recall | 0.826 (0.0000) | 0.826 | 1.000 | 1.000 | |
| 48 h | AUROC | 0.862 (0.0088) | 0.792 | 0.724 | 0.802 |
| APR | 0.684 (0.0156) | 0.478 | 0.444 | 0.384 | |
| Sensitivity | 0.818 (0.0000) | 1.000 | 0.955 | 1.000 | |
| Specificity | 0.773 (0.0334) | 0.000 | 0.022 | 0.522 | |
| F1 | 0.598 (0.0297) | 0.328 | 0.321 | 0.506 | |
| DOR | 15.80 (3.051) | n/a | 0.477 | n/a | |
| LR+ | 3.69 (0.555) | 1.000 | 0.976 | 2.093 | |
| LR- | 3.69 (0.555) | 1.000 | 0.976 | 2.093 | |
| Accuracy | 0.782 (0.0268) | 0.196 | 0.205 | 0.616 | |
| Recall | 0.818 (0.0000) | 1.000 | 0.955 | 1.000 | |
| 72 h | AUROC | 0.873 (0.0034) | 0.722 | 0.797 | 0.751 |
| APR | 0.649 (0.0209) | 0.364 | 0.452 | 0.320 | |
| Sensitivity | 0.819 (0.0190) | 1.000 | 0.857 | 1.000 | |
| Specificity | 0.760 (0.0181) | 0.000 | 0.611 | 0.467 | |
| F1 | 0.576 (0.0171) | 0.318 | 0.486 | 0.467 | |
| DOR | 14.64 (2.376) | n/a | 9.429 | n/a | |
| LR+ | 3.43 (0.261) | 1.000 | 2.204 | 1.875 | |
| LR- | 3.43 (0.261) | 1.000 | 2.204 | 1.875 | |
| Accuracy | 0.771 (0.0146) | 0.189 | 0.658 | 0.568 | |
| Recall | 0.810 (0.0000) | 1.000 | 0.857 | 1.000 |
These results suggest that the XGBoost predictor is capable of predicting mortality in pneumonia, mechanically ventilated, and COVID-19 patients and outperforms the qSOFA, MEWS and CURB-65 mortality risk scores.
5. Discussion
Accurate mortality prediction can assist with the allocation of limited hospital resources and optimize patient management. Additionally, advanced mortality prediction can facilitate decision making with family and caregivers. The commonly used MEWS [37], the APACHE [38], Simplified Acute Physiology Score (SAPS II) [39], Sepsis-Related Organ Failure Assessment (SOFA) [40], and the quick SOFA (qSOFA) score [41] provide a rough estimate of mortality prediction, however the specificity and sensitivity of these tools are limited for COVID and mechanically ventilated populations [42]. Machine learning (ML) has been previously broadly applied to predictive tasks within the biosciences [[43], [44], [45], [46]]. ML-based tools for mortality prediction have been applied to sepsis [47,48] cardiac arrest [49], coronary artery disease [50], and extubation [51] patient populations, and have been implemented in a broad range of clinical settings, including the emergency department (ED) [48] and the intensive care unit (ICU) [[52], [53], [54], [55]].
Studies of mortality prediction on pneumonia and mechanically ventilated patients are particularly relevant for COVID-19 related lung complications. We have demonstrated that machine learning algorithms are useful predictive tools for anticipating patient mortality at clinically useful windows of 12, 24, 48, and 72 h in advance and have validated mortality prediction accuracy for COVID-19, pneumonia, mechanically ventilated, and all ICU patients (Fig. 1), demonstrating that for all prediction types and windows, our ML algorithm outperforms the qSOFA, MEWS and CURB-65 severity scores (Table 7, Table 8, Table 9).
A meta-analysis of studies focusing on predicting mortality in pneumonia patients showed that of the three commonly used prognostic scores which predicted mortality, the Pneumonia Severity Index (PSI) had the highest AUROC of 0.81. However, this index was used for predicting 30-day mortality specifically among patients with community acquired pneumonia [56]. When trained and tested on the MIMIC dataset, the XGBoost predictor obtained AUROCs of 0.87, 0.78, 0.77, and 0.73 for mortality prediction on pneumonia patients at 12-, 24-, 48-, and 72- hour windows, respectively (Fig. 1, Table 7).
When trained and tested on the community hospital dataset, the XGBoost predictor obtained AUROCs of 0.91, 0.90, 0.86, and 0.87 for mortality prediction on COVID-19 PCR positive patients at 12-, 24-, 48-, and 72- hour windows, respectively (Table 9). The algorithm outperformed the qSOFA, MEWS and CURB-65 risk scores at all prediction windows (Table 9). This ML algorithm can be used to automatically monitor patient populations without incurring additional data entry or impeding clinical workflow, and patient alerts can be set to desired thresholds for sensitivity and specificity of alerting as needed in different care settings. As a clinical decision support tool, the machine learning algorithm presented in this study may assist clinicians in navigating the complexities surrounding COVID-19 related resource allocation. During a pandemic, accurate triage of patients is essential for improving patient outcomes, effectively utilizing clinical care teams, and efficiently allocating resources. The benefit of our approach is that when our machine learning algorithm is implemented in clinical ICU settings, healthcare providers can potentially identify patients at risk of significant COVID-19 related decompensation before they deteriorate, thus facilitating effective resource allocation and identifying those patients most likely to benefit from increased care.
There are several limitations to our study. The ML algorithm developed on the MIMIC dataset used only data from the ICU. Therefore, further research is required to evaluate performance of the algorithm in other patient care settings. Further, because the algorithm only utilized laboratory data and vital signs as inputs, it did not account for actions undertaken by the care team. These actions could signify aggressive treatment or withdrawal of treatment and could cause changes to algorithm inputs, potentially leading to variations in the algorithm's prediction score. On one hand, incorporating care team actions into algorithm inputs could be useful feedback to the care team in the sense that it may aid them in determining whether a given intervention was harmful or beneficial. On the other hand, accounting for actions undertaken by the care team may complicate the interpretation of what it means to “anticipate” mortality, given that the current state of knowledge of the care team is unknown. Finally, because this is a retrospective study, we cannot determine the performance of the mortality prediction algorithm in a prospective clinical setting. Prospective validation is required to determine how clinicians may respond to risk predictions as well as whether predictions can affect patient outcomes or resource allocation.
6. Conclusion
The ML algorithm presented in this study is a useful predictive tool for anticipating patient mortality at clinically useful windows up to 72 h in advance, and capable of accurate mortality prediction for COVID-19, pneumonia, and mechanically ventilated patients.
Patient and public involvement statement
Patients and the public were not involved in the design and conduct of the study, choice of outcome measures, or recruitment to the study due to the nature of data collection.
Dissemination declaration
Transparency declaration: RD affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as originally planned (and, if relevant, registered) have been explained.
Funding sources
N/A.
Ethical approval
Data has been deidentified and, as such, does not constitute human subjects research.
Conflicts of interest
All authors who have affiliations listed with Dascena (San Francisco, California, USA) are employees or contractors of Dascena.
Trial registration
This study has been registered on ClinicalTrials.gov under study number NCT04358510.
Provenance and peer review
Not commissioned, externally peer reviewed.
Guarantor
The Guarantor is the one or more people who accept full responsibility for the work and/or the conduct of the study, had access to the data, and controlled the decision to publish
CRediT authorship contribution statement
Logan Ryan: Methodology, Investigation. Carson Lam: Conceptualization, Methodology, Formal analysis, Writing - original draft, Supervision. Samson Mataraso: Conceptualization, Methodology, Formal analysis, Writing - original draft, Supervision. Angier Allen: Investigation, Writing - original draft. Abigail Green-Saxena: Writing - original draft. Emily Pellegrini: Writing - original draft. Jana Hoffman: Conceptualization, Methodology, Formal analysis, Writing--Original Draft, Supervision. Christopher Barton: Supervision. Ritankar Das: had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis, Conceptualization, Methodology, Formal analysis, Writing - original draft, Supervision.
Acknowledgments
N/A.
Contributor Information
Logan Ryan, Email: lryan@dascena.com.
Carson Lam, Email: clam@dascena.com.
Samson Mataraso, Email: samson@dascena.com.
Angier Allen, Email: aallen@dascena.com.
Abigail Green-Saxena, Email: abigail@dascena.com.
Emily Pellegrini, Email: emilypellegrini@dascena.com.
Jana Hoffman, Email: jana@dascena.com.
Christopher Barton, Email: chris@dascena.com.
Andrea McCoy, Email: amccoy@caperegional.com.
Ritankar Das, Email: ritankar@dascena.com.
Supplemental Material
Table S1.
Feature Importance 12 h outlook using MIMIC dataset
| Rank | Feature | Importance (f_score) |
|---|---|---|
| 1 | SpO2_-2 | 81 |
| 2 | HR | 77 |
| 3 | SysABP_diff_-2 | 76 |
| 4 | DiasABP_-0 | 68 |
| 5 | SysABP_-0 | 68 |
| 6 | WBC_-1 | 66 |
| 7 | HR_-2 | 64 |
| 8 | Platelets_-1 | 64 |
| 9 | RespRate_diff_-1 | 63 |
| 10 | SpO2_diff_-1 | 62 |
Table S2.
Feature Importance 24 h outlook using MIMIC dataset
| Rank | Feature | Importance (f_score) |
|---|---|---|
| 1 | SpO2_diff_-1 | 92 |
| 2 | DiasABP_-1 | 69 |
| 3 | Platelets_-1 | 68 |
| 4 | DiasABP_diff_-1 | 64 |
| 5 | Temp_-1 | 61 |
| 6 | RespRate_-2 | 60 |
| 7 | DiasABP_diff_-2 | 60 |
| 8 | WBC_-1 | 60 |
| 9 | HR_diff_-1 | 57 |
| 10 | RespRate_diff_-1 | 56 |
Table S3.
Feature Importance 48 h outlook using MIMIC dataset
| Rank | Feature | Importance (f_score) |
|---|---|---|
| 1 | WBC_-2 | 48 |
| 2 | DiasABP_diff_-2 | 44 |
| 3 | HR_-2 | 43 |
| 4 | DiasABP_-1 | 41 |
| 5 | RespRate_-1 | 41 |
| 6 | Temp_diff_-2 | 38 |
| 7 | Temp_diff_-1 | 36 |
| 8 | Temp_-0 | 35 |
| 9 | SpO2_diff_-2 | 35 |
| 10 | WBC_-1 | 34 |
Table S4.
Feature Importance 72 h outlook using MIMIC dataset
| Rank | Feature | Importance (f_score) |
|---|---|---|
| 1 | SysABP_diff_-1 | 61 |
| 2 | Platelets_-0 | 46 |
| 3 | SpO2_-0 | 46 |
| 4 | DiasABP_-2 | 45 |
| 5 | Lactate_-0 | 45 |
| 6 | RespRate_-0 | 45 |
| 7 | SysABP_-1 | 44 |
| 8 | Creatinine_-1 | 42 |
| 9 | Temp_-2 | 42 |
| 10 | RespRate_diff_-1 | 37 |
Table S5.
Comparison of AUROC, average precision (APR), sensitivity, specificity, F1, diagnostic odds ratio (DOR), positive and negative likelihood ratios (LR+ and LR‒), accuracy and recall obtained by the machine learning algorithm (MLA) and the qSOFA score for mortality prediction at 12-, 24-, 48-, and 72- hour windows on all ICU patients using the MIMIC dataset. Standard deviations are listed in parenthesis. For AUROC and APR the operating point was set near a sensitivity of 0.800.
| MLA: All ICU |
qSOFA: All ICU |
MEWS: All ICU |
CURB-65: All ICU |
||
|---|---|---|---|---|---|
| 12 h | AUROC | 0.862 (0.0012) | 0.760 | 0.833 | 0.652 |
| APR | 0.553 (0.0018) | 0.225 | 0.392 | 0.131 | |
| Sensitivity | 0.801 (0.0000) | 0.949 | 0.897 | 0.984 | |
| Specificity | 0.750 (0.0046) | 0.373 | 0.559 | 0.185 | |
| F1 | 0.378 (0.0040) | 0.236 | 0.290 | 0.198 | |
| DOR | 12.09 (0.297) | 11.085 | 11.054 | 14.088 | |
| LR+ | 3.21 (0.059) | 1.513 | 2.033 | 1.208 | |
| LR- | 3.21 (0.059) | 1.513 | 2.033 | 1.208 | |
| Accuracy | 0.755 (0.0041) | 0.426 | 0.590 | 0.260 | |
| Recall | 0.801 (0.0000) | 0.949 | 0.897 | 0.984 | |
| 24 h | AUROC | 0.819 (0.0018) | 0.742 | 0.804 | 0.636 |
| APR | 0.432 (0.0052) | 0.223 | 0.339 | 0.136 | |
| Sensitivity | 0.800 (0.0000) | 0.939 | 0.896 | 0.978 | |
| Specificity | 0.671 (0.0036) | 0.357 | 0.524 | 0.178 | |
| F1 | 0.338 (0.0023) | 0.245 | 0.292 | 0.210 | |
| DOR | 8.18 (0.131) | 8.629 | 9.473 | 9.692 | |
| LR+ | 2.43 (0.026) | 1.462 | 1.882 | 1.189 | |
| LR- | 2.43 (0.026) | 1.462 | 1.882 | 1.189 | |
| Accuracy | 0.684 (0.0032) | 0.416 | 0.562 | 0.258 | |
| Recall | 0.800 (0.0000) | 0.939 | 0.896 | 0.978 | |
| 48 h | AUROC | 0.789 (0.0016) | 0.706 | 0.760 | 0.616 |
| APR | 0.408 (0.0024) | 0.232 | 0.297 | 0.158 | |
| Sensitivity | 0.801 (0.0000) | 0.945 | 0.882 | 0.977 | |
| Specificity | 0.619 (0.0110) | 0.282 | 0.437 | 0.137 | |
| F1 | 0.356 (0.0063) | 0.269 | 0.301 | 0.242 | |
| DOR | 6.55 (0.315) | 6.781 | 5.771 | 6.791 | |
| LR+ | 2.10 (0.063) | 1.316 | 1.565 | 1.132 | |
| LR- | 2.10 (0.063) | 1.316 | 1.565 | 1.132 | |
| Accuracy | 0.641 (0.0097) | 0.364 | 0.492 | 0.241 | |
| Recall | 0.801 (0.0000) | 0.945 | 0.882 | 0.977 | |
| 72 h | AUROC | 0.746 (0.0026) | 0.655 | 0.685 | 0.603 |
| APR | 0.356 (0.0017) | 0.218 | 0.256 | 0.182 | |
| Sensitivity | 0.802 (0.0015) | 0.929 | 0.872 | 0.963 | |
| Specificity | 0.546 (0.0049) | 0.247 | 0.388 | 0.111 | |
| F1 | 0.361 (0.0025) | 0.295 | 0.321 | 0.270 | |
| DOR | 4.86 (0.114) | 4.299 | 4.296 | 3.216 | |
| LR+ | 1.77 (0.020) | 1.233 | 1.423 | 1.083 | |
| LR- | 1.77 (0.020) | 1.233 | 1.423 | 1.083 | |
| Accuracy | 0.583 (0.0043) | 0.347 | 0.459 | 0.236 | |
| Recall | 0.801 (0.0000) | 0.929 | 0.872 | 0.963 |
References
- 1.World Health Organization . Jan 11, 2020. Clinical management of severe acute respiratory infection when Novel coronavirus (2019-nCoV) infection is suspected: interim Guidance. [Google Scholar]
- 2.Zhai Y., Wang Y., Zhang M., Gittell J.H., Jiang S., Chen B., Cui F., He X., Zhao J., Wang X. From isolation to coordination: how can telemedicine help combat the COVID-19 outbreak? medRxiv preprint. 2020. [DOI]
- 3.Johnson S.R. 2020. Flu, Coronavirus Challenge Hospitals' Ability to Withstand Surge of Patients.https://www.modernhealthcare.com/safety-quality/flu-coronavirus-challenge-hospitals-ability-withstand-surge-patients Available from. [Google Scholar]
- 4.Specht L. 2020 March 10. What Does the Coronavirus Mean for the U.S. Health Care System? Some Simple Math Offers Alarming Answers. Available from https://www.statnews.com/2020/03/10/simple-math-alarming-answers-covid-19/ [Google Scholar]
- 5.Li R., Rivers C., Tan Q., Murray M.B., Toner E., Lipsitch M. The demand for inpatient and ICU beds for COVID-19 in the US: lessons from Chinese cities. medRxiv. 2020:20033241. doi: 10.1101/2020.03.09.20033241. [DOI] [Google Scholar]
- 6.Soucheray S. Hospitals scramble to keep up with CDC N95, mask guidance. 2020. http://www.cidrap.umn.edu/news-perspective/2020/03/hospitals-scramble-keep-cdc-n95-mask-guidance Available from.
- 7.Rubinson L., Vaughn F., Nelson S. Mechanical ventilators in US acute care hospitals. Disaster Med. Public Health Prep. 2010;4(3):199–206. doi: 10.1001/dmp.2010.1. [DOI] [PubMed] [Google Scholar]
- 8.Johns Hopkins Bloomberg School of Public Health. Center for Health Security . Johns Hopkins Bloomberg School of Public Health; Baltimore, MD: February 14, 2020. Ventilator Stockpiling and Availability in the US.http://www.centerforhealthsecurity.org/resources/COVID-19/200214-VentilatorAvailability-factsheet.pdf [Google Scholar]
- 9.Malatino E.M. Strategic National Stockpile: overview and ventilator assets. Respir. Care. 2008;53(1):91–95. [PubMed] [Google Scholar]
- 10.Mehrabi A., Dillon P., Kelly K. Experimental studies on performance of ventilators stored in the Strategic National Stockpile. J Emerg Manag. 2018 Sep/Oct;16(5):321–336. doi: 10.5055/jem.2018.0381. [DOI] [PubMed] [Google Scholar]
- 11.Huang H.C., Araz O.M., Morton D.P. Stockpiling ventilators for influenza pandemics. Emerg. Infect. Dis. 2017;23(6):914–921. doi: 10.3201/eid2306.161417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ajao A., Nystrom S.V., Koonin L.M. Assessing the capacity of the healthcare system to use additional mechanical ventilators during a large-scale public health emergency. Disaster Med. Public Health Prep. 2015;9(6):634–641. doi: 10.1017/dmp.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A novel coronavirus emerging in China—key questions for impact assessment. N. Engl. J. Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 14.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016 Aug;14(8):523. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fauci AS Lane, Hc Redfield R.R. Covid-19—navigating the uncharted. N. Engl. J. Med. 2020 doi: 10.1056/NEJMe2002387. published online Feb 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajgor D.D., Lee M.H., Archuleta S., Bagdasarian N., Quek S.C. The many estimates of the COVID-19 case fatality rate. Lancet Infect. Dis. 2020 Jul;20(7):776–777. doi: 10.1016/S1473-3099(20)30244-9. Epub 2020 Mar 27. PMID: 32224313; PMCID: PMC7270047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. J. Am. Med. Assoc. 2020 Mar 17;323(11):1061–1069. doi: 10.1001/jama.2020.1585. PMID: 32031570; PMCID: PMC7042881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb 15;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. Epub 2020 Jan 24. Erratum in: Lancet. 2020 Jan 30;: PMID: 31986264; PMCID: PMC7159299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020 Feb 15;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. Epub 2020 Jan 30. PMID: 32007143; PMCID: PMC7135076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohamadlou H., Panchavati S., Calvert J., Lynn-Palevsky A., Le S., Allen A., Pellegrini E., Green-Saxena A., Barton C., Fletcher G., Shieh L. Multicenter validation of a machine-learning algorithm for 48-h all-cause mortality prediction. Health Inf. J. 2020 Sep;26(3):1912–1925. doi: 10.1177/1460458219894494. Epub 2019 Dec 30. PMID: 31884847. [DOI] [PubMed] [Google Scholar]
- 21.Calvert J., Mao Q., Hoffman J.L., Jay M., Desautels T., Mohamadlou H., Chettipally U., Das R. Using electronic health record collected clinical variables to predict medical intensive care unit mortality. Annals of medicine and surgery. 2016 Nov 1;11:52–57. doi: 10.1016/j.amsu.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desautels T., Calvert J., Hoffman J., Mao Q., Jay M., Fletcher G., Barton C., Chettipally U., Kerem Y., Das R. Using transfer learning for improved mortality prediction in a data-scarce hospital setting. Biomed. Inf. Insights. 2017 Jun 8;9 doi: 10.1177/1178222617712994. 1178222617712994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calvert J., Mao Q., Rogers A.J., Barton C., Jay M., Desautels T., Mohamadlou H., Jan J., Das R. A computational approach to mortality prediction of alcohol use disorder inpatients. Comput. Biol. Med. 2016 Aug 1;75:74–79. doi: 10.1016/j.compbiomed.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 24.Das R., Wales D.J. Vol. 4. Royal Society open science; 2017 Jul 26. p. 170175. (Machine learning landscapes and predictions for patient outcomes). 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carson S.S., Kahn J.M., Hough C.L., Seeley E.J., White D.B., Douglas I.S., Cox C.E., Caldwell E., Bangdiwala S.I., Garrett J.M., Rubenfeld G.D. A multicenter mortality prediction model for patients receiving prolonged mechanical ventilation. Crit. Care Med. 2012 Apr;40(4):1171. doi: 10.1097/CCM.0b013e3182387d43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hough C.L., Caldwell E.S., Cox C.E., Douglas I.S., Kahn J.M., White D.B., Seeley E.J., Bangdiwala S.I., Rubenfeld G.D., Angus D.C., Carson S.S. Development and validation of a mortality prediction model for patients receiving 14 days of mechanical ventilation. Crit. Care Med. 2015 Nov;43(11):2339. doi: 10.1097/CCM.0000000000001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim W.Y., Jo E.J., Eom J.S., Mok J., Kim M.H., Kim K.U., Park H.K., Lee M.K., Lee K. Validation of the Prognosis for Prolonged Ventilation (ProVent) score in patients receiving 14 days of mechanical ventilation. J. Crit. Care. 2018 Apr 1;44:249–254. doi: 10.1016/j.jcrc.2017.11.029. [DOI] [PubMed] [Google Scholar]
- 28.Johnson A.E., Pollard T.J., Shen L., Li-wei H.L., Feng M., Ghassemi M., Moody B., Szolovits P., Celi L.A., Mark R.G. MIMIC-III, a freely accessible critical care database. Scientific data. 2016 May 24;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao Q., Jay M., Hoffman J.L. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open. 2018;8(1) doi: 10.1136/bmjopen-2017-017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolditz M., Scherag A., Rohde G., Ewig S., Welte T., Pletz M., CAPNETZ Study Group Comparison of the qSOFA and CRB-65 for risk prediction in patients with community-acquired pneumonia. Intensive Care Med. 2016 Dec 1;42(12):2108–2110. doi: 10.1007/s00134-016-4517-y. [DOI] [PubMed] [Google Scholar]
- 31.Zhou H., Lan T., Guo S. Prognostic prediction value of qSOFA, SOFA, and admission lactate in septic patients with community-acquired pneumonia in emergency department. Emergency Medicine International. 2020;2020 doi: 10.1155/2020/7979353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J.Y., Chen Y.X., Guo S.B., Mei X., Yang P. Predictive performance of quick Sepsis-related Organ Failure Assessment for mortality and ICU admission in patients with infection at the. Am. J. Emerg. Med. 2016 Sep 1;34(9):1788–1793. doi: 10.1016/j.ajem.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 33.Subbe C.P., Kruger M., Rutherford P., Gemmel L. Validation of a modified early warning score in medical admissions. QJM: Int. J. Med. October 2001;94(Issue 10):521–526. doi: 10.1093/qjmed/94.10.521. [DOI] [PubMed] [Google Scholar]
- 34.Burch V.C., Tarr G., Morroni C. Modified early warning score predicts the need for hospital admission and inhospital mortality. Emerg. Med. J. 2008 Oct 1;25(10):674–678. doi: 10.1136/emj.2007.057661. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z.X., Yong Y., Tan W.C., Shen L., Ng H.S., Fong K.Y. Prognostic factors for mortality due to pneumonia among adults from different age groups in Singapore and mortality predictions based on PSI and CURB-65. Singap. Med. J. 2018 Apr;59(4):190–198. doi: 10.11622/smedj.2017079. Epub 2017 Aug 14. PMID: 28805234, PMCID: PMC5915635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satici C., Demirkol M.A., Altunok E.S., Gursoy B., Alkan M., Kamat S., Demirok B., Surmeli C.D., Calik M., Cavus Z., Esatoglu S.N. Performance of pneumonia severity index and CURB-65 in predicting 30-day mortality in patients with COVID-19. Int. J. Infect. Dis. 2020 Sep 1;98:84–89. doi: 10.1016/j.ijid.2020.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subbe C., Slater A., Menon D., Gemmell L. Validation of physiological scoring systems in the accident and emergency department. Emerg. Med. J. 2006;23(11) doi: 10.1136/emj.2006.035816. 841e845. PMID:17057134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knaus W.A., Wagner D.P., Draper E.A. The Apache III prognostic system. Chest. 1991;100(6):1619–1636. doi: 10.1378/chest.100.6.1619. [DOI] [PubMed] [Google Scholar]
- 39.Le Gall J.R., Lemeshow S., Saulnier F. A new simplified acute physiology score (SAPS II) based on a European/North American multicenter study. J. Am. Med. Assoc. 1993;270(24) doi: 10.1001/jama.270.24.2957. 2957e2963. PMID:8254858. [DOI] [PubMed] [Google Scholar]
- 40.Ferreira F.L. Serial evaluation of the SOFA score to predict outcome in critically ill patients. J. Am. Med. Assoc. 2001;286(14):1754–1758. doi: 10.1001/jama.286.14.1754. [DOI] [PubMed] [Google Scholar]
- 41.Singer M., Deutschman C.S., Seymour C.W. The third international consensus definitions for sepsis and septic shock (sepsis-3) J. Am. Med. Assoc. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siontis G.C., Tzoulaki I., Ioannidis J.P. Predicting death: an empirical evaluation of predictive tools for mortality. Arch. Intern. Med. 2011;171(19) doi: 10.1001/archinternmed.2011.334. 1721e1726. PMID:21788535. [DOI] [PubMed] [Google Scholar]
- 43.Ballard A.J., Das R., Martiniani S., Mehta D., Sagun L., Stevenson J.D., Wales D.J. Energy landscapes for machine learning. Phys. Chem. Chem. Phys. 2017;19(20):12585–12603. doi: 10.1039/c7cp01108c. [DOI] [PubMed] [Google Scholar]
- 44.Das R., Wales D.J. Energy landscapes for a machine-learning prediction of patient discharge. Phys. Rev. 2016 Jun 17;93(6) doi: 10.1103/PhysRevE.93.063310. [DOI] [PubMed] [Google Scholar]
- 45.Barton C., Chettipally U., Zhou Y., Jiang Z., Lynn-Palevsky A., Le S., Calvert J., Das R. Evaluation of a machine learning algorithm for up to 48-hour advance prediction of sepsis using six vital signs. Comput. Biol. Med. 2019 Jun 1;109:79–84. doi: 10.1016/j.compbiomed.2019.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calvert J., Hoffman J., Barton C., Shimabukuro D., Ries M., Chettipally U., Kerem Y., Jay M., Mataraso S., Das R. Cost and mortality impact of an algorithm-driven sepsis prediction system. J. Med. Econ. 2017 Jun 3;20(6):646–651. doi: 10.1080/13696998.2017.1307203. [DOI] [PubMed] [Google Scholar]
- 47.Vieira S.M., Mendonça L.F., Farinha G.J. Modified binary PSO for feature selection using SVM applied to mortality prediction of septic patients. Appl. Soft Comput. 2013;13(8):3494–3504. [Google Scholar]
- 48.Taylor R.A., Pare J.R., Venkatesh A.K. Prediction of in-hospital mortality in emergency department patients with sepsis: a local big data-driven, machine learning approach. Acad. Emerg. Med. 2016;23(3):269–278. doi: 10.1111/acem.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acharya U.R., Fujita H., Sudarshan V.K. An integrated index for detection of sudden cardiac death using discrete wavelet transform and nonlinear features. Knowl-Based Syst. 2015;83:149–158. [Google Scholar]
- 50.Motwani M., Dey D., Berman D.S. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur. Heart J. 2017;38(7):500–507. doi: 10.1093/eurheartj/ehw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsieh M.H., Hsieh M.J., Chen C.M., Hsieh C.C., Chao C.M., Lai C.C. Comparison of machine learning models for the prediction of mortality of patients with unplanned extubation in intensive care units. Sci. Rep. 2018;8(1):17116. doi: 10.1038/s41598-018-35582-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pirracchio R., Petersen M.L., Carone M. Mortality prediction in intensive care units with the Supper ICU Learner Algorithm (SICULA): a population-based study. Lancet Resp Med. 2015;3(1):42–52. doi: 10.1016/S2213-2600(14)70239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson A.E., Pollard T.J., Mark R.G. vol. 68. 2017. Reproducibility in critical care: a mortality prediction case study. (Proceedings of the 2nd Machine Learning for Healthcare Conference). Boston, MA. [Google Scholar]
- 54.Delahanty R.J., Kaufman D., Jones S.S. Development and evaluation of an automated machine learning algorithm for in-hospital mortality risk adjustment among critical care patients. Crit. Care Med. 2018;46(6):e481–e488. doi: 10.1097/CCM.0000000000003011. [DOI] [PubMed] [Google Scholar]
- 55.Marafino B.J., Park M., Davies J.M. Validation of prediction models for critical care outcomes using natural language processing of electronic health record data. JAMA Netw Open. 2018;1(8) doi: 10.1001/jamanetworkopen.2018.5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chalmers J.D., Singanayagam A., Akram A.R., Mandal P., Short P.M., Choudhury G., Wood V., Hill A.T. Severity assessment tools for predicting mortality in hospitalised patients with community-acquired pneumonia. Systematic review and meta-analysis. Thorax. 2010 Oct 1;65(10):878–883. doi: 10.1136/thx.2009.133280. [DOI] [PubMed] [Google Scholar]