Abstract
Background
Therapy for patients with liver cancer in the advanced stage remains a great challenge, and there are very few approved treatments. Although accumulated evidence demonstrates the importance of lncRNAs in liver cancer, data on the functional roles and molecular mechanisms of endogenous bornavirus-like nucleoprotein (EBLN3P) have been rarely reported.
Materials and Methods
The bioinformatics prediction software ENCORI was used to predict the putative binding sites of EBLN3P. The regulatory roles of EBLN3P and miR-144-3p in cell proliferation, migration and invasion ability were verified by the Cell Counting Kit-8, wound healing and Transwell assays, respectively. The interactions among EBLN3P, miR-144-3p and DOCK4 were explored by a luciferase assay and Western blotting. The expression of EBLN3P and microRNA (miR)-144-3p in liver cancer tissues was quantified by reverse transcription-quantitative PCR, and the expression of dedicator of cytokinesis 4 (DOCK4) was quantified by immunohistochemical analysis.
Results
The present results revealed that overexpression of EBLN3P or knockdown of miR-144-3p promoted liver cancer cell proliferation, migration and invasion. Bioinformatics analysis and a luciferase assay demonstrated that EBLN3P directly interacts with miR-144-3p to attenuate miR-144-3p binding to the 3ʹ-untranslated region of DOCK4. Furthermore, the mechanistic investigations showing that the miR-144-3p/DOCK4 regulatory loop was activated by knockdown of miR‐144-3p or overexpression of DOCK4 validate the roles of EBLN3P in promoting liver cancer cell proliferation, migration and invasion in vitro. Elevated levels of EBLN3P and DOCK4 and decreased miR-144-3p expression were observed in both liver cancer tissues and cell lines.
Conclusion
The present study is the first to demonstrate that EBLN3P may act as a ceRNA to modulate DOCK4 expression by competitively sponging miR-144-3p, leading to the regulation of liver cancer progression, which provides new insights for liver cancer diagnosis and treatment.
Keywords: competing endogenous RNAs, dedicator of cytokinesis 4, long noncoding RNAs, microRNAs, liver cancer, endogenous bornavirus-like nucleoprotein
Introduction
Liver cancer is one of the most common types of cancer in the world.1 Although its incidence and mortality rates have declined significantly in western countries, liver cancer is still a significant health threat in certain regions of the world, such as China and Japan.2,3 Surgery with radical resection is suitable for only a limited percentage of patients, whereas many patients are often diagnosed at later stages or experience postsurgical diseases such as relapse or metastasis, which worsens their prognoses.4 Comprehensive treatment of advanced liver cancer remains unsatisfactory, and thus, it is of vital importance to elucidate the detailed mechanisms of liver cancer progression.5
Long noncoding RNAs (lncRNAs), defined as genome transcripts consisting of >200 nucleotides that are not translated into proteins, are associated with various biological developmental processes, such as tumor proliferation and metastasis.6 Tumor gene expression can be positively or negatively modulated by lncRNAs, either through epigenetic transcriptional regulation or posttranscriptional regulation.7 Epigenetically, lncRNAs may interact with the transcription preinitiation complex at the promoter region or directly form base pairs with RNA and DNA.8 Posttranscriptionally, lncRNAs act as precursors of microRNAs (miRNAs/miRs) and competing endogenous RNAs (ceRNAs) to control cell fate.9 Among those regulatory RNAs, ceRNAs, which regulate gene expression via miRNA mediation, have been widely studied and recognized.10 It has been reported that the interaction of human epidermal growth factor receptor 2 (HER2) with miR-331-3p may underlie the oncogenic roles of the lncRNA Hox transcript antisense intergenic RNA (HOTAIR) in gastric cancer.11,12 Similarly, another study illustrated that lncRNA H19 has oncogenic functions, which can be attributed to its ceRNA activity through which it sequesters miR-138 and miR-200a in colorectal cancer (CRC).13 A previous study revealed that miR-144-3p is associated with abnormal expression in various tumors. It has been reported that miR-144-3p facilitates the progression of nasopharyngeal carcinoma via directly targeting phosphatase and tensin homolog (PTEN).14 However, the roles of lncRNA EBLN3P and miR-144-3p in liver cancer remain unclear.
In mammals, the Rho GTPase family plays critical roles in cell proliferation, cell motility, tumor cell malignant transformation, and cancer metastasis and invasion.15 Guanine nucleotide exchange factors (GEFs) “turn on” Rho GTPases by inducing the exchange of GDP for GTP.16 Rho GEFs include Dbl-related classical GEFs and atypical Dock family Rho GEFs.17 DOCK4 is a member of the Dock180 family of proteins, which mediates the outgrowth of patient-derived glioblastoma cells through activation of Rac.18 However, the expression profile and regulatory mechanism of DOCK4 in liver cancer remains unknown. In a previous study, the ceRNA network that may regulate DOCK4 expression was discovered by bioinformatics analysis and verified by molecular biology techniques. The data revealed that the pseudogene-derived lncRNA EBLN3P can act as a ceRNA that bind with miR-144-3p and further upregulates the protein expression of its target DOCK4. EBLN3P is a pseudogene of endogenous bornavirus-like nucleoprotein 3 (EBLN3) on chromosome 9. However, its expression and its effects in human disease have not been reported. The aim of the present study was to explore the biological effects of DOCK4 in liver cancer progression and its potential regulatory mechanisms. It was demonstrated that DOCK4 is highly expressed in liver cancer tissues and that the EBLN3P/miR-144-3p/DOCK4 regulatory loop axis may be a potential therapeutic target for liver cancer treatment.
Materials and Methods
RNA Extraction and Reverse Transcription-Quantitative PCR
Nuclear RNA and cytoplasmic RNA were separately isolated from cell lines using the Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek Corp) according to the manufacturer’s protocol. Total RNA was extracted from tissues and cell lines using an RNA extraction kit (Takara Biotechnology Co., Ltd.). Reverse transcription into cDNA was conducted with Prime Script RT Master Mix (Takara Biotechnology Co., Ltd.) according to the manufacturer’s protocol. qPCR was performed via a SYBR green-based fluorescence method (SYBR Premix Ex Taq Kit; Takara Biotechnology Co., Ltd.) and the MX3000P® qRT-PCR system (Agilent Technology, Inc.) according to the following parameters: 95°C for 10 min followed by 40 cycles of 95°C for 10 sec and 60°C for 50 sec. The relative expression levels were calculated using the 2−ΔΔCq method. GAPDH and U6 were used as the internal references for mRNA and miRNA expression, respectively. The primer sequences are listed in Table 1.
Table 1.
Primer Sequences for Quantitative Real-Time PCR
| Gene | Forward Sequence | Reverse Sequence |
|---|---|---|
| EBLN3P | 5ʹ-CAGACTAAAGGATCAAGCGAGA-3’ | 5ʹ-ATCAATTGCCACAGGTTGAAGA-3’ |
| hsa-miR-144-3p | 5ʹ- GCCCCTACAGTATAGATGATGTA −3’ | 5ʹ- GGATGCAGGTGCTGGAGGT −3’ |
| DOCK4 | 5ʹ- GGATACCTACGGAGCACGAG-3’ | 5ʹ- AGCCATCACACTTCTCCAGG-3’ |
| U6 | 5ʹ-CTCGCTTCGGCAGCACA-3’ | 5ʹ-AACGCTTCACGAATTTGCGT-3’ |
| GAPDH | 5ʹ-CCAGGTGGTCTCCTCTGA-3’ | 5ʹ-GCTGTAGCCAAATCGTTGT-3’ |
Western Blot Assay
The Western blot procedure was carried out as previously described.20 Cells were plated in each well of a 6-well plate and cultured in DMEM supplemented with 10% FBS. Then, the cells were washed with ice-cold phosphate-buffered saline (PBS) three times, and cell lysates were prepared with RIPA lysis buffer (Beyotime Institute of Biotechnology). Proteins were extracted from liver tissues using T-PER Tissue Protein Extraction Reagent (cat. no. 78,510; Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocol. Subsequently, the protein concentration was determined using a BCA assay kit (Bio-Rad Laboratories, Inc.). The proteins were loaded and electrophoresed on a 10% SDS-PAGE gel. Subsequently, the proteins were transferred onto nitrocellulose membranes (EMD Millipore). The membranes were blocked with 5% skimmed milk for 1 h and coincubated with an anti-DOCK4 primary antibody (1:10,000; cat. no. ab85723; Abcam) at 4°C overnight. The membranes were washed three times with TBST containing 0.1% Tween 20. Following coincubation with HRP-labeled goat anti-mouse/rabbit IgG (1:5000; Sigma-Aldrich; Merck KGaA) for 2 h at room temperature, the bands were analyzed by an ECL detection kit (Pierce; Thermo Fisher Scientific, Inc.).
Immunohistochemical (IHC) Analysis
Routine hematoxylin and eosin staining was performed prior to immunohistochemical analysis. Briefly, paraffin-embedded samples were cut into 3-μm sections and then dewaxed with xylene and rehydrated in graded ethanol. Antigen retrieval was conducted with citrate buffer, pH6.0 (1:300; cat. no. ZLI-9065; OriGene Technologies, Inc.) For antigen retrieval, the sections were heated at 97°C for 20 min. Following a brief proteolytic digestion and peroxidase blocking, the sections were incubated with a DOCK4 polyclonal antibody (1:500; cat. no. ab85723; Abcam) overnight at 4°C, and HRP/Fab polymer conjugate (1:1000; cat. no. PV-6000-D; OriGene Technologies, Inc.) was applied as the secondary antibody. Finally, the sections were stained with diaminobenzidine substrate and counterstained with hematoxylin. Two independent investigators semiquantitatively evaluated DOCK4-positive staining without prior knowledge of the clinicopathologic data. The final immunoreactivity scores (IRSs) were assessed based on the percentage of positively stained cells (0 points, 0–5% positive cells; 1 point, 6–25% positive cells; 2 points, 26–50% positive cells; 3 points, 51–75% positive cells; and 4 points, 76–100% positive cells) as well as staining intensity scores (0 points, no staining; 1 point, weak staining; 2 points, moderate staining; and 3 points, strong staining). A final IRS of >4 indicated strong positivity, while all other scores indicated weak positivity.
Cell Culture
The hepatocellular carcinoma cell lines (PLHC1, Hep3B, and SNU398), a hepatoblastoma cell line (HepG2) and a human hepatocyte cell line (THLE3) were obtained from the American Type Culture Collection and had been authenticated by short tandem repeat profiling. Dulbecco’s modified Eagle medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) was used to culture all cells at 37°C in 5% CO2.
Cell Transfection and Plasmid Construction
NC (negative control) mimics, miR-144-3p mimics, NC inhibitor and miR-144-3p inhibitor were acquired from Guangzhou RiboBio Co., Ltd. The EBLN3P overexpression plasmid pcDNA-EBLN3P, the EBLN3P knockdown siRNA plasmid si-EBLN3P and a corresponding negative control siRNA (si-NC) were synthesized by Shanghai GenePharma Co., Ltd. Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for transfection according to the manufacturer’s instructions. Cells were collected for further use after 48 h of transfection.
Cell Counting Kit-8 (CCK-8) Assay
The CCK-8 assay (Dojindo Molecular Technologies, Inc.) was used to evaluate cell proliferation capacity as previously described.19 Briefly, 2x104 cells were seeded in a 96-well plate in triplicate for each condition. At different time points (12, 24, 36, 48, 60 and 72 h), 10 μL of CCK-8 solution was added to each well for an additional 4 h of incubation at 37°C. The absorbance was recorded at a wavelength of 490 nm by a microplate reader (Thermo Fisher Scientific, Inc.).
Cell Colony Formation Assay
For the colony formation assay, cells were seeded in 6-well plates at a density of 1x102 cells/well and then cultured for 15 days under standard conditions. Thereafter, the cell colonies were washed with PBS twice, subsequently fixed in 70% methanol for 10 min and stained with 0.5% crystal violet for 5 min. Finally, images of the cell colonies were captured, and the colonies were counted.
Wound Healing Assay
The cells were seeded in 6-well plates and incubated until they reached 90% confluence in serum-free medium. A sterile pipette tip was used to create ~1-mm-wide wounds. The detached cells were gently washed off twice, and the medium was replaced with complete medium containing 1% FBS. Images of the wound were captured at 0, 24 and 48 h using a light microscope.
Transwell Assay
Cells were seeded on Matrigel-coated upper chambers for the invasion assay (BD Biosciences). Culture medium without FBS and culture medium containing 10% FBS was added to the upper and lower wells, respectively, and the cells were incubated for another 24 h. After wiping off the noninvaded cells, the filters were fixed in 90% ethanol and stained with crystal violet. Cells were counted in random fields perform each chamber by using an inverted microscope (Olympus Corporation). The procedure was carried out as described previously.21
Dual-Luciferase Reporter Assays
The assay was carried out as described previously.22 Wild-type and mutant EBLN3P and DOCK4 reporter plasmids (EBLN3P-WT-luc, EBLN3P-MUT-luc, DOCK4-WT-luc, DOCK4-MUT-luc) containing wild-type or mutant miR-144-3p mimics or NC binding site mimics, were synthesized by Shanghai GenePharma Co., Ltd. The synthesized reporter plasmids were cotransfected with miR-144-3p mimics or NC mimics with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) when the cells reached 70% confluence. Luciferase activity was analyzed via the dual-luciferase reporter assay system (Promega Corporation) after 48 h.
Clinical Tissue Sample Collection
Tumor specimens were collected from 74 patients with hepatocellular carcinoma (HCC), and 72 nonneoplastic hepatic tissues were collected from patients who underwent surgery at The First Hospital of Jilin University. The patients were selected based on the following criteria: pathological diagnosis of HCC; no history of anticancer therapy before tumor resection; and no diagnosis of any additional malignancies. Pathological staging was based on the UICC/AJCC TNM Classification (8th edition of 2016). Collection of clinical specimens was approved by the Ethics Committee of the First Hospital of Jilin University according to the Declaration of Helsinki (approval no. JLU01698). Informed consent was collected from all participants. The status of each HCC patient was validated by an outpatient or telephone interview in order to construct a 5-year Kaplan-Meier survival curve. Patients who did not survive due to diseases other than HCC were excluded from the present study.
Statistical Analysis
All experimental data are presented as the mean ± SD, and each experiment was conducted in triplicate. Statistical analyses were performed and graphs were generated with SPSS V17.0 (SPSS, Inc.) and GraphPad Prism V5.02 (GraphPad Software, Inc.) software. Comparisons between two groups were performed with the Student’s t-test, and ANOVA with Dunnett’s multiple comparisons test was used for multiple group comparisons. P<0.05 was considered to indicate a statistically significant difference. The association between the survival rate of patients with liver cancer and the DOCK4, miR-144-3p or EBLN3P expression level was investigated using Kaplan-Meier survival curves and the Log rank test. Additionally, the χ2 test was applied for association analysis of clinical case indicators.
Results
EBLN3P Knockdown Suppressed the Proliferation, Migration and Invasion of Liver Cancer Cells in vitro
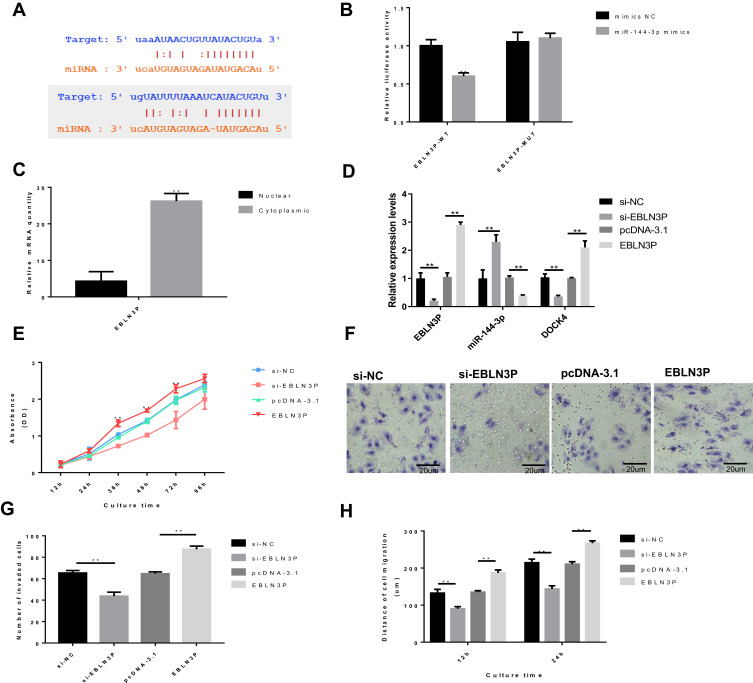
Bioinformatics analysis demonstrated that EBLN3P can directly interact with miR-144-3p and that miR-144-3p can directly interact with DOCK4 (Figure 1A). Furthermore, dual-luciferase reporter assays were performed to verify this hypothesis. miR-144-3p was overexpressed by transfecting hepatoblastoma HepG2 cells with miR-144-3p mimics. The luciferase assay results demonstrated that transfection of miR-144-3p mimics significantly weakened the luciferase signal of reporter plasmids containing EBLN3P-WT but had no influence on the activity of reporter plasmids containing EBLN3P-MUT (Figure 1B). Next, hepatoblastoma HepG2 cells were transfected with EBLN3P siRNA (si-EBLN3P) or the pcDNA3.1-EBLN3P overexpression vector (EBLN3P) to further investigate the possible impact of EBLN3P on the cellular behaviors of liver cancer cells. Negative control siRNA (si-NC) and the blank vector plasmid (pcDNA-3.1) were used as knockdown and overexpression controls, respectively. First, the nuclear and cytoplasmic expression levels of EBLN3P in HepG2 cells were explored via RT-qPCR, and the data revealed that the EBLN3P was mainly expressed in the cell cytoplasm in HepG2 cells (Figure 1C). Moreover, the expression levels of EBLN3P, miR-144-3p and DOCK4 were explored via RT-qPCR (Figure 1D). The RT-qPCR results indicated a negative association between EBLN3P and miR-144-3p expression and that the expression levels of DOCK4 were positively correlated with the expression levels of EBLN3P (Figure 1D). Together, these results demonstrated that EBLN3P serves as a ceRNA for miR-144-3p to inhibit miR-144-3p expression. Moreover, knockdown of EBLN3P notably decreased the proliferation, invasion and migration of HepG2 cells, which was enhanced by EBLN3P overexpression, as shown by the CCK-8 assay, transwell assay and wound healing assay, respectively (Figure 1EH). In contrast, overexpression of EBLN3P accelerated the proliferation, invasion and migration of HepG2 cells (Figure 1EH). Thus, these results implied that EBLN3P has the oncogenic potential to induce liver cancer cell proliferation, migration and invasion.
Figure 1.
EBLN3P knockdown inhibits the proliferation, migration and invasion of liver cancer cells in vitro. HepG2 cells were transfected with EBLN3P-targeted siRNA (si-EBLN3P), negative control siRNA (si-NC), pcDNA3.1-EBLN3P overexpression vector (pcDNA-EBLN3P) or blank vector plasmid (pcDNA-3.1). (A) Bioinformatics analysis demonstrated that EBLN3P can directly interact with miR-144-3p (up) and that miR-144-3p can directly bind to the 3ʹ-UTR regions of DOCK4 (down). (B) Dual-luciferase reporter assays were performed to verify the impact of miR-144-3p on the luciferase signal of reporter plasmids containing EBLN3P. (C) The nuclear and cytoplasmic expression levels of EBLN3P in HepG2 cells. (D) The expression levels of EBLN3P and miR-144-3p were explored via reverse transcription-quantitative PCR. (E) Cell proliferation was tested by the Cell Counting Kit-8 assay. (F) The number of invaded cells. (G) Cell invasion was detected by the Transwell assay. (H) Cell migration was detected by the wound healing assay. That data is shown as the mean ± SD. **P<0.01.
Abbreviations: siRNA, small interfering RNA; miR, microRNA
miR-144-3p Inhibits the Proliferation, Migration and Invasion of Liver Cancer Cells by Downregulating DOCK4 in vitro
DOCK4 has been demonstrated to promote tumor progression in lung adenocarcinoma (ADC) metastasis. However, its impact on liver cancer and interaction with miR-144-3p have not been studied yet. The potential target genes mediated by miR-144-3p in liver cancer cells were evaluated, and dedicator of cytokinesis protein 4 (DOCK4) was identified as one of the target genes of miR-144-3p through the bioinformatics software ENCORI. Next, a dual-luciferase reporter gene assay illustrated that the luciferase signal of cells cotransfected with miR-144-3p mimics and the wild-type DOCK4 vector was notably decreased compared with that of cells cotransfected with miR-144-3p mimics and the mutant DOCK4 vector (Figure 2A). This implied that miR-144-3p probably binds to the 3ʹ-UTR of DOCK4. Furthermore, RT-qPCR and Western blotting confirmed that overexpression of miR-144-3p significantly decreased DOCK4 expression, whereas knockdown of miR-144-3p significantly enhanced the expression of DOCK4 at both the mRNA and protein levels in HepG2 cells (Figure 2BD). Taken together, these results implied that miR-144-3p acts as an upstream regulator of DOCK4 expression. The CCK‐8 assay revealed a notable decrease in the proliferation rate in the miR-144-3p mimics group compared to the NC mimics group, while the proliferation rate was significantly increased in the miR-144-3p inhibitor group compared with the control group (Figure 2E). Analysis of migration and invasion further verified this pattern. The migration and invasion capacities of liver cancer cells transfected with miR-144-3p mimics were decreased, but those of cells transfected with the inhibitor were increased (Figure 2FH). Collectively, these data indicate that miR-144-3p acts as a tumor suppressor in liver cancer cells that can restrain multiple malignant behaviors of HepG2 cells, including cell proliferation, migration and invasion.
Figure 2.
miR-144-3p inhibits the proliferation, migration and invasion of liver cancer cells by downregulating DOCK4. (A) The miR-144-3p mimics and luciferase reporter plasmids containing wild-type or mutant DOCK4 3ʹ-UTR were cotransfected into HepG2 cells. The dual luciferase reporter gene assay was performed to verify the direct binding association between miR-144-3p and DOCK4. (B) HepG2 cells were transfected with NC mimics, miR-144-3p mimics, NC inhibitor, or miR-144-3p inhibitor, and then, reverse transcription-quantitative PCR was conducted to evaluate the relative expression levels of miR-144-3p and DOCK4. (C) Western blotting validated that the overexpression of miR-144-3p significantly decreased DOCK4 expression. (D) Corresponding analysis of DOCK4 expression. (E) Cell proliferation was tested by the Cell Counting Kit-8 assay. (F) Cell invasion was detected by the Transwell assay. (G) The number of invaded cells. (H) Cell migration was detected by the wound healing assay. The data are shown as the mean ± SD. **P<0.01.
Abbreviations: miR, microRNA; UTR, untranslated region.
EBLN3P Regulates Liver Cancer Cells via the miR-144-3p/DOCK4 Pathway
Rescue experiments were carried out to assess the effects of the EBLN3P-miR-144-3p-DOCK4 pathway on HepG2 cell activities. Overexpression of DOCK4 was achieved by transfecting cells with pcDNA3.1‐DOCK4 or miR-144-3p inhibitor (Figure 3A and B). The results demonstrated that knockdown of EBLN3P markedly decreased cell proliferation, migration and invasion. However, compared to transfected with si-EBLN3P alone, cotransfection with si-EBLN3P and miR-144-3p inhibitor or DOCK4 significantly increased cell proliferation, migration and invasion (Figure 3CF). Accordingly, the data suggests that EBLN3P exerts its regulatory role through the miR-144-3p/DOCK4 axis to promote the development of liver cancer cells.
Figure 3.
EBLN3P regulates liver cancer cells via the miR-144-3p/DOCK4 pathway. High mRNA (A) and protein (B) expression level of DOCK4 was achieved by transfecting cells with pcDNA3.1‐DOCK4 or miR-144-3p inhibitor. Knockdown of EBLN3P markedly reduced cell proliferation (C), invasion (D and E) and migration (F). However, compared to transfection with sh-EBLN3P alone, co-transfection with sh-EBLN3P and miR-144-3p inhibitor or DOCK4 significantly increased cell proliferation (C), invasion (D and E) and migration (F). The data are shown as the mean ± SD.**P < 0.01.
EBLN3P and DOCK4 are Upregulated While miR-144-3p is Downregulated in Liver Cancer Tissues and Cell Lines
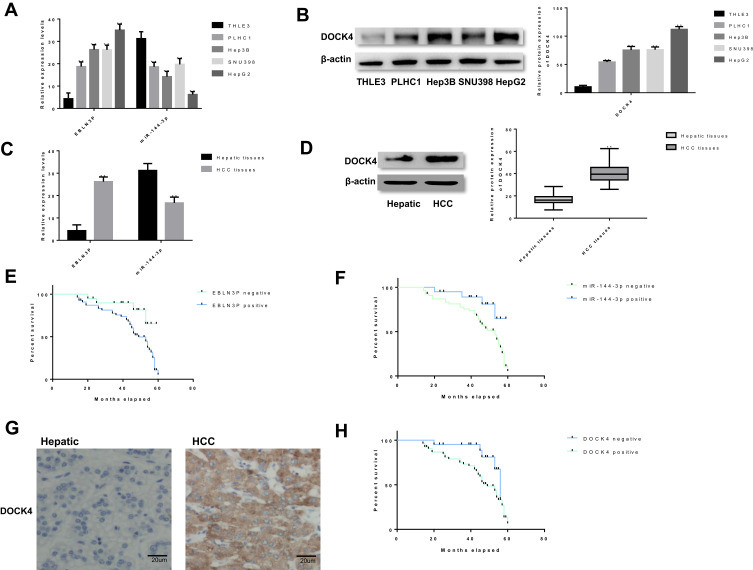
To investigate the regulatory role of EBLN3P in liver cancer, whether EBLN3P and miR-144-3p are dysregulated in liver cancer was investigated. The RTq-PCR and Western blotting results showed that EBLN3P and DOCK4 were expressed at higher levels in HCC cell lines (PLHC1, Hep3B, and SNU398) and the hepatoblastoma cell line (HepG2) than in the normal human hepatic astrocyte cell line (THLE3), while miR-144-3p was expressed in the reverse manner (Figure 4A and B). These data demonstrated that EBLN3P, miR-144-3p and DOCK4 may play an important role in regulating the development of liver cancer, including HCC and hepatoblastoma. To further explore the expressions of EBLN3P, miR-144-3p and DOCK4 in HCC, EBLN3P, miR-144-3p and DOCK4 expression levels were similarly assessed in HCC tissues and nonneoplastic hepatic tissues. As expected, EBLN3P and DOCK4 were expressed at higher levels and miR-144-3p was expressed at a lower level in HCC tissues than in hepatic tissues (Figure 4C and D). Moreover, the associations between EBLN3P and miR-144-3p expressions and the prognosis of patients with HCC were explored via Kaplan-Meier survival curves and the Log rank test. As shown in Figure 4E, F and H, patients with HCC and positive miR-144-3p expression had longer overall survival times than patients with negative miR-144-3p expression (P=0.004). Furthermore, patients with HCC and positive EBLN3P or DOCK4 expression had shorter overall survival times than patients with negative EBLN3P or DOCK4 expression (P=0.012; P=0.019). Furthermore, DOCK4 expression in HCC tissues was investigated via IHC analysis, as shown in Figure 4G and Table 2, and it was found that DOCK4 was expressed at a higher level in HCC tissues (54/74) than in hepatic tissues (14/72). Moreover, DOCK4 was revealed to be associated with distant metastasis (P<0.01) and tumor-node-metastasis stages (AJCC) (P<0.01).
Figure 4.
EBLN3P and DOCK4 were upregulated, while miR-144-3p was downregulated in liver cancer tissues and cell lines. (A) The expression levels of EBLN3P and miR-144-3p in liver cancer cell lines (PLHC1, Hep3B, SNU398 and HepG2) and the hepatocyte cell line (THLE3) were tested by RTq-PCR. (B) The Western blotting results showed that DOCK4 was expressed at a higher level in four liver cancer cell lines (PLHC1, Hep3B, SNU398 and HepG2) than in the normal human hepatic astrocyte cell line (THLE3). (C) The expression patterns of EBLN3P and miR-144-3p were measured in liver cancer samples and normal tissue via RTq-PCR. (D) The expression patterns of DOCK4 were measured in liver cancer samples and normal tissue via Western blotting. (E) The association between EBLN3P expression and the prognosis of patients with liver cancer was explored. (F) The association between miR-144-3p expression and the prognosis of patients with liver cancer was explored. (G) The expression patterns of DOCK4 were measured in liver cancer samples and normal tissue via immunohistochemistry. (H) The association between DOCK4 expression and the prognosis of patients with liver cancer was explored. The data are shown as the mean ± SD. **P<0.01.
Abbreviations: RT-qPCR, reverse transcription-quantitative PCR; miR, microRNA.
Table 2.
Expression Levels of DOCK4 and Summary of the Clinicopathological Characteristics of Liver Cancer Patients
| Variable | Patients, N |
DOCK4 (+) |
DOCK4(-) | P-value |
|---|---|---|---|---|
| Liver cancer tissues | 74 | 54 | 20 | <0.01 |
| Hepatic tissues | 72 | 14 | 58 | |
| Age (years) | ||||
| ≤60 | 42 | 30 | 12 | 0.726a |
| >60 | 32 | 24 | 8 | |
| Sex | ||||
| Male | 41 | 29 | 12 | 0.743a |
| Female | 33 | 25 | 8 | |
| HbsAg | ||||
| + | 67 | 49 | 18 | 0.872a |
| - | 7 | 5 | 2 | |
| Distant metastasis | ||||
| + | 44 | 37 | 7 | <0.01 |
| - | 30 | 17 | 13 | |
| Serum AFP (ng/mL) | ||||
| <400 | 46 | 33 | 13 | 0.162a |
| >400 | 28 | 21 | 7 | |
| TNM stage (AJCC) | ||||
| I–II | 49 | 32 | 17 | <0.01 |
| III–IV | 25 | 22 | 3 |
Notes: aNo statistical significance was found with the χ2 test/χ2 Goodness-of-Fit Test.
Abbreviations: DOCK, dedicator of cytokinesis protein; TNM, tumor-node-metastasis; AFP, alpha-fetoprotein.
Discussion
The role of lncRNAs in carcinogenesis was initially identified due to the differential expression of these lncRNAs in caner tissues compared with normal tissues.23–27 Thus far, accumulating evidence has indicated that the capacities of lncRNAs to regulate complex cellular behaviors, such as cell growth and metastasis, are commonly deregulated in cancer, including liver cancer.28–30 Although many of potential biomarkers have been reported, specific diagnostic biomarkers for liver cancer have not yet been confirmed. EBLN3P is a novel lncRNA located on chromosome 9: 37,079,935–37,086,874 forward strand. The present study demonstrated that EBLN3P is markedly upregulated in liver cancer tissues and cell lines. In vitro, functional assays indicated that the deprivation of EBLN3P suppresses liver cancer cell proliferation, migration and invasion, demonstrating the potential of EBLN3P as a therapeutic target for liver cancer intervention. Hence, the underlying mechanism in liver cancer cell lines was next explored.
The ceRNA theory was first proposed in 2011 and was subsequently extensively accepted in the field of noncoding RNA.10 lncRNAs may serve as ceRNAs by sponging miRNAs to hinder the downregulated pathway. Bioinformatics analyses revealed that there is a conserved miR-144-3p binding site in EBLN3P. Therefore, it was postulated that EBLN3P can act as a ceRNA to affect miR-144-3p. Subsequently, it was validated that miR-144-3p has a reciprocal suppressive effect on EBLN3P expression, and knockdown of miR-144-3p induced the proliferation, migration and invasion of liver cancer cells in vitro. Importantly, the dual-luciferase assay further showed that EBLN3P directly interacts with miR-144-3p to decrease its expression, suggesting that EBLN3P serves as an miRNA sponge that binds to and regulates miR-144-3p expression.
miRNAs control gene expression by binding to the 3′-UTR of the target gene, which causes mRNA cleavage or translational repression.31 Rho GTPases play critical roles in the initiation and progression of various tumors;32 however, the regulation of Rho GTPases in liver cancer remains largely unknown. Additionally, the role and molecular mechanism of DOCK4, an atypical Rho GEF,33 in liver cancer are unknown. This study provides the first evidence that DOCK4 is overexpressed in liver cancer and that its positive expression is associated with lymph node metastasis and a higher tumor-node-metastasis (TNM) stage. Patients with liver cancer and positive DOCK4 expression exhibited a shorter overall survival time than patients with negative DOCK4 expression. The present study demonstrated that DOCK4 promoted the proliferation of liver cancer cells in vitro. It was also found that DOCK4 expression in lymph node metastases was higher than that in primary liver cancer tissues and that DOCK4 increased the migration and invasion abilities of liver cancer cells in vitro and in vivo. As a member of the Dock family of GEFs, DOCK4, a typical Rac1 activator, has been reported to be a key component of the TGF-β/Smads pathway, which promotes lung ADC cell extravasation and metastasis.34 In accordance with previous reports, the present data showed that DOCK4 promoted the motility of liver cancer cells. In the present study, bioinformatics analysis revealed that DOCK4 was regulated via the EBLN3P/miR-144-3p axis, and the expression levels of DOCK4 were positively correlated with the expression levels of EBLN3P in liver cancer cells. Moreover, the dual-luciferase assays confirmed the direct interaction between miR-144-3p and DOCK4. Furthermore, rescue experiments showed that EBLN3P promoted proliferation and metastasis in liver cancer via the regulation of the miR-144-3p/DOCK4 signaling axis. Although this study reveals a novel mechanism whereby liver cancer cells are regulated by EBLN3P in vitro, further studies are required to fully understand the complexity of the mechanism. Future studies in model mice are needed to further verify the novel molecular mechanism of EBLN3 and value its potential as a therapeutic target.
In conclusion, the present study is the first to demonstrate that EBLN3P acts as a novel oncogene in liver cancer. Furthermore, EBLN3P acts as a ceRNA to regulate DOCK4 expression by competitively sponging miR-144-3p, thereby regulating the progression of liver cancer. Therefore, the present findings provide useful information for the identification of new biomarkers for early diagnosis and therapies for liver cancer progression.
Funding Statement
This study was supported by Jilin Province Science and Technology Development Projects (20160414032GH).
Abbreviations
ceRNA, competing endogenous RNA; DOCK4, dedicator of cytokinesis 4; HOTAIR, hox transcript antisense intergenic RNA; lncRNA, long noncoding RNA; miRNAs, microRNAs; RT‐qPCR, reverse transcription-quantitative PCR, siRNA, small interfering RNA.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Kulik L, El-Serag HB. Epidemiology and management of liver cancer. Gastroenterology. 2019;156:477–491 e471. doi: 10.1053/j.gastro.2018.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mak LY, Cruz-Ramon V, Chinchilla-Lopez P, et al. Global epidemiology, prevention, and management of liver cancer. american society of clinical oncology educational book. Am Society Clin Oncol. 2018:262–279. Annual Meeting. [DOI] [PubMed] [Google Scholar]
- 3.Zhang G, Li R, Deng Y, Zhao L. Conditional survival of patients with liver cancer: results from the surveillance, epidemiology, and end results registry. Expert Rev Gastroenterol Hepatol. 2018;12:515–523. doi: 10.1080/17474124.2018.1453806 [DOI] [PubMed] [Google Scholar]
- 4.Venerito M, Vasapolli R, Rokkas T, Malfertheiner P. Gastric cancer: epidemiology, prevention, and therapy. Helicobacter. 2018;23(Suppl 1):e12518. doi: 10.1111/hel.12518 [DOI] [PubMed] [Google Scholar]
- 5.Kamarajah SK, Frankel TL, Sonnenday C, Cho CS, Nathan H. Critical evaluation of the American Joint Commission on Cancer (AJCC) 8th edition staging system for patients with liver cancer (liver cancer): a surveillance, epidemiology, end results (SEER) analysis. J Surg Oncol. 2018;117:644–650. doi: 10.1002/jso.24908 [DOI] [PubMed] [Google Scholar]
- 6.Sarfi M, Abbastabar M, Khalili E. Long noncoding RNAs biomarker-based cancer assessment. J Cell Physiol. 2019;234:16971–16986. doi: 10.1002/jcp.28417 [DOI] [PubMed] [Google Scholar]
- 7.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang L, Wang Y, Chen J, et al. Long noncoding RNA PCAT1, a novel serum-based biomarker, enhances cell growth by sponging miR-326 in oesophageal squamous cell carcinoma. Cell Death Dis. 2019;10(7):513. doi: 10.1038/s41419-019-1745-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Dou P, Liu T, He S. Application of long noncoding RNAs in osteosarcoma: biomarkers and therapeutic targets. Cellular Physiol Biochem. 2017;42(4):1407–1419. doi: 10.1159/000479205 [DOI] [PubMed] [Google Scholar]
- 10.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X-H, Sun M, Nie F-Q, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Molecular Cancer 2014;13:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee HE, Park KU, Yoo SB, et al. Clinical significance of intratumoral HER2 heterogeneity in gastric cancer. Eur J Cancer. 2013;49:1448–1457. doi: 10.1016/j.ejca.2012.10.018 [DOI] [PubMed] [Google Scholar]
- 13.Liang W-C, Fu W-M, Wong C-W, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song L, Chen L, Luan Q, Kong Q. miR-144-3p facilitates nasopharyngeal carcinoma via crosstalk with PTEN. J Cell Physiol. 2019;234:17912–17924. doi: 10.1002/jcp.28424 [DOI] [PubMed] [Google Scholar]
- 15.Ungefroren H, Witte D, Lehnert H. The role of small GTPases of the Rho/Rac family in TGF‐β‐induced EMT and cell motility in cancer. Developmental Dynamics. 2018;247:451–461. doi: 10.1002/dvdy.24505 [DOI] [PubMed] [Google Scholar]
- 16.Jansen S, Gosens R, Wieland T, Schmidt M. Paving the Rho in cancer metastasis: rho GTPases and beyond. Pharmacol Ther. 2018;183:1–21. doi: 10.1016/j.pharmthera.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 17.Blangy A. RhoGEFs as therapeutic targets. Molecular Biology in Health and Disease. 2018. [Google Scholar]
- 18.Egnuni MT, Speirs V, Chakrabarty A, Wurdak H, Short S, Mavria G. Rho GTPase signaling and role of the Rac1 exchange factor DOCK4 in GBM invasion and vascular growth. Neuro-Oncology. 2018;20:i17. doi: 10.1093/neuonc/nox238.077 [DOI] [Google Scholar]
- 19.Zhang X, Wang X, Wang A, Li Q, Zhou M, Li T. CLDN10 promotes a malignant phenotype of osteosarcoma cells via JAK1/Stat1 signaling. J Cell Commun Signal. 2019;13:395–405. doi: 10.1007/s12079-019-00509-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Wang H, Li Q, Li T. CLDN2 inhibits the metastasis of osteosarcoma cells via down-regulating the afadin/ERK signaling pathway. Cancer Cell Int. 2018;18:160. doi: 10.1186/s12935-018-0662-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinoshita T, Nohata N, Hanazawa T, et al. Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. Br J Cancer. 2013;109:2636–2645. doi: 10.1038/bjc.2013.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Lu Z, Wang N, et al. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med. 2018;50:57. doi: 10.1038/s12276-018-0082-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu D, Chen K, Bai Y, et al. Screening of diagnostic markers for osteosarcoma. Mol Med Rep. 1020;14. [DOI] [PubMed] [Google Scholar]
- 24.Kong C, Hansen MF. Biomarkers in Osteosarcoma. Expert Opin Med Diagn. 2009;3:13–23. doi: 10.1517/17530050802608496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–532. doi: 10.1016/j.ctrv.2013.11.006 [DOI] [PubMed] [Google Scholar]
- 26.Bartonicek N, Maag JLV, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochimica Et Biophysica Acta (BBA) Gene Regulatory Mechanisms. 1839;10971109:2014. [DOI] [PubMed] [Google Scholar]
- 28.Pan Y, Li C, Chen J, et al. The emerging roles of long noncoding RNA ROR (lincRNA-ROR) and its possible mechanisms in human cancers. Cellular Physiol Biochem. 2016;40:219–229. doi: 10.1159/000452539 [DOI] [PubMed] [Google Scholar]
- 29.Souckova K, Ivkovic TC, Slaby O. Non-coding RNA therapy in cancer In: Precision Medicine for Investigators, Practitioners and Providers. Elsevier; 2020:211–220. [Google Scholar]
- 30.Vidovic D, Huynh TT, Konda P, et al. ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ. 2020;27:363–378. doi: 10.1038/s41418-019-0362-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pu M, Chen J, Tao Z, et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cellular Molecular Life Sci. 2019;76:441–451. doi: 10.1007/s00018-018-2940-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olayioye MA, Noll B, Hausser A. Spatiotemporal control of intracellular membrane trafficking by rho GTPases. Cells. 2019;8:1478. doi: 10.3390/cells8121478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundaravel S, Kuo W-L, Jeong JJ, et al. Loss of function of DOCK4 in myelodysplastic syndromes stem cells is restored by inhibitors of DOCK4 signaling networks. Clin Cancer Res. 2019;25:5638–5649. doi: 10.1158/1078-0432.CCR-19-0924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J-R, Tai Y, Jin Y, et al. TGF-β/Smad signaling through DOCK4 facilitates lung adenocarcinoma metastasis. Genes Dev. 2015;29:250–261. doi: 10.1101/gad.248963.114 [DOI] [PMC free article] [PubMed] [Google Scholar]