Abstract
Purpose
Endometrial cancer (EC) is an aggressive tumor in females and the development of EC is considered to regulate by some long non-coding RNAs (lncRNAs). Therefore, this study aimed to investigate the regulatory mechanism of lncRNA LINC01106 on EC.
Methods
The expression of lncRNA LINC01106, miR-449a and MET in EC tissues and cells was detected by qRT-PCR. Through MTT, wound healing and transwell invasion assays, the proliferation, migration and invasion of EC cells were detected, respectively. The xenograft tumor model was constructed in nude mice to confirm the inhibiting effect of LINC01106 knockdown on EC in vivo. The interactions between miR-449a and LINC01106/MET were predicted by Starbase/Targetscan software and verified by the dual-luciferase reporter assay or RNA immunoprecipitation assay. Western blot assay was performed to determine the protein level of MET.
Results
LncRNA LINC01106 expression was highly up-regulated in EC tissues and cells. The proliferation, migration and invasion of EC cells in vitro were inhibited by the transfection of sh-LINC01106. The growth of tumor xenograft was suppressed by injection of sh-LINC01106. MiR-449a was a target of LINC01106and was negatively modulated by LINC01106. MiR-449a overexpression suppressed the proliferation, migration and invasion of EC cells. In addition, MET was identified as a target gene of miR-449a. Both the high expression of miR-449a and low expression of MET reversed the inhibiting effects of LINC01106 knockdown on Ishikawa cells.
Conclusion
Silencing of LINC01106 inhibits the occurrence and development of EC via regulating the miR-449a/MET axis. This study provides a possible therapeutic strategy for EC.
Keywords: endometrial cancer, lncRNA LINC01106, miR-449a, MET
Introduction
Endometrial cancer (EC) is a malignant tumor that occurs in females and severely affects the health of patients.1 The mortality of EC is exceeding high at the advanced stage.2,3 Although various therapies for treating EC have been improved, the therapeutic effects remain disillusionary.4,5 In consequence, further molecular researches on the potential mechanism of EC to explore promising therapeutic targets in EC are the urgent problems to be solved.
Long non-coding RNAs (LncRNAs) are participated in modulating the tumorigenesis and progression of many human cancers,6–8 including EC.9–11 For instance, overexpression of LINC-ROR is reported to promote the growth of EC.9 Up-regulation of LINC01016 accelerates the malignant biological behaviors of EC cells, such as increasing the proliferation and invasion abilities of EC cells.10 LINC01410 overexpression can also facilitate the proliferation and invasion of EC cells.11 Notably, the emerging evidences have displayed that LINC01106 is related to the occurrence and progression of several cancers, such as nasopharyngeal carcinoma,12 lung adenocarcinoma13 and colorectal cancer.14 However, whether LINC01106 involves in regulating EC progression has not been fully elucidated.
MicroRNAs (miRNAs) have been reported to exhibit their anti-tumor roles on EC.15–17 Ni et al have demonstrated that miR-638 knockdown accelerates the development of EC.15 Zhao et al have revealed that overexpression of miR-195 suppresses the abilities of migration and invasion of EC cells.16 Li et al have found the migration and invasion of EC in vitro were inhibited by miR-34c.17 Interestingly, miR-449a has been also disclosed to function as a suppressor in the development of EC.18,19 Up-regulation of miR-449a inhibits the proliferation, migration and invasion of EC cells, thus eventually retarding the growth of EC.20 Nevertheless, we are still unclear whether the suppressive effects of miR-449a on EC progression is regulated by LINC01106.
As an oncogene, metprotoncogene (MET) plays a crucial role in tumorigenesis and the expression of MET is observed to up-regulate in various human malignancies, such as ovarian cancer,21,22 colorectal cancer23 and non-small cell lung cancer.24,25 Growing data have revealed that MET is also taken part in modulating the progression of EC.26–28 More importantly, miR-449a can restrain the growth of hepatocellular carcinoma through modulating MET.29 However, the possible roles of miR-449a/MET axis on the occurrence and development of EC remain to be further explored.
In the current study, we explored the effect of lncRNA LINC01106 knockdown on the progression of EC and the regulatory mechanism between LINC01106 and the miR-449a/MET axis in EC. Our findings are aimed to provide a promising therapeutic target for EC.
Materials and Methods
Tissues Collection
From 2016 to 2018, 64 EC patients (<60, n = 28; ≥60, n = 36) were selected in our hospital. The age range of the 64 patients was 35 to 66 years (mean age, 46.1±4.9 years). All EC patients were confirmed via histopathological examination. The inclusion criteria included first-time diagnosis and no prior history of radiotherapy, chemotherapy or other adjuvant therapies. The exclusion criteria included the presence of other types of malignant tumor and patients who had received EC treatment before admission. The tumor and adjacent normal tissues (within 2 cm of the tumors) of the patients were collected by surgery. Each patient in this study obtained the written informed consent. The protocols of this study were reviewed and approved by ethical committee of Qingdao Traditional Chinese Medicine Hospital (approval number: 2018-09-15).
Cell Grouping and Transfection
We procured the human endometrial-stromal cell line (hESC) from Otwo Biotech, Ltd (Shenzhen, China) and the human EC cell lines (KLE, Ishikawa and HEC-1A) from Procell Life Science & Technology, Ltd (Wuhan, China). The cells were cultured in DMEM medium containing 10% fetal bovine serum (FBS) at 37°C with 5% CO2. The shRNA-negative control (sh-NC) and shRNA-LINC01106-1/-2 (sh-LINC01106-1/-2) were procured from Sangon Biotech, Inc (Shanghai, China). Overexpression-MET (pcDNA-MET) and its negative control (pcDNA-NC), miR-449a mimics, miR-449a inhibitor and their negative control (miR-NC) were all obtained from Ribo Biotech, Ltd (Guangzhou, China). The above agents were transfected into the cells using a Lipofectamine RNAiMAX kit (Invitrogen, Carlsbad, CA, USA). After treatment for 48 h, the cells were harvested to perform the subsequent trails.
Quantitative Reverse-Transcription PCR (qRT-PCR)
Total RNA was extracted from EC tissues and cells using a TRIzol kit (Invitrogen, Inc.). The GoScript reverse-transcription system (Promega, Madison, WI, USA) was used to reversely transcribe the extracted RNA into cDNA. The thermocycling conditions were: 94°C for 10 min, followed by 40 cycles at 94°C for 10 sec, 60°C for 20 sec and 72°C for 1 min. GADPH or U6 was used as the internal reference. Gene expression was quantified using the 2−ΔΔCt method.
MTT Assay
The viability of EC cells was detected by MTT assay. The transfected Ishikawa and HEC-1A cells were seeded into a 96-well plate (2 × 105 cells per well). Subsequently, the cells were incubated for 24, 48, 72 and 96 h, followed by adding 20 µL MTT (GENECHEM, Inc, Shanghai, China) to each well to incubate another 2 h at 37°C. The viability (OD450) was analyzed using a Multiskan Spectrum microplate reader (Thermo Fisher Scientific).
Wound Healing Assay
Ishikawa and HEC-1A cells were seeded into 6-well plates and grown until 100% confluence in DMEM. Then, the wounds on cell monolayer layers were created using a pipette tip. After that, the cells were incubated for 24 h in a serum-free medium. At 0 h and 24 h, the measurement and imaging of wound closure were measured by AxioVision v4.7 software (Carl Zeiss Meditec, Dublin, CA, USA) under a microscope (magnification, x 200). We calculated the wound healing rate (%) in accordance with the formula: (0 h gap distance – gap distance at 24 h)/0 h gap distance × 100%.
Transwell Invasion Assay
Transwell chamber (Becton, Dickinson and Company, FL, NJ, USA) with Matrigel coating was used to detect cell invasion. Briefly, the upper chamber was seeded with 2 × 105 cells (Ishikawa and HEC-1A) per well. Then, DMEM containing 10% FBS was added into the lower chamber. After 3 h of incubation at 37°C, the cells in the lower chamber were stained with 0.5% crystal violet (Merck KGaA, Darmstadt, Germany) for 15 min. A light microscope (magnification, x 400) was used to count the stained cells in five randomly selected fields.
Tumor Xenografts in Nude Mice
The study was firstly obtained the approval of the ethical committee of Qingdao Traditional Chinese Medicine Hospital (approval number: 2018-10-15) and performed in the Animal Experimental Center of our hospital. All animal experiments were undertaken in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The healthy female BALB/c nude mice (weighing 22 ± 2 g) were procured from Cavens Lab, Ltd (Changzhou, China) and fed in a sterile environment under a 12 h cycle (12 h for light and 12 h for dark) at 20°C. Ishikawa cells transfected with sh-LINC01106-1 or sh-NC (2 × 106 cells/100 ul, s.c.) were injected into the right flanks of the nude mice (n = 5). Tumor volumes were measured every other week. Four weeks later, mice were anesthetized with pentobarbital sodium (50 mg/kg) and then sacrificed by cervical dislocation. The tumor xenograft was separated from mice and weighted.
RNA Immunoprecipitation (RIP) Assay
RIP assay was performed using a MagnaRIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA). In brief, the pcDNA-MS2/pcDNA-MS2-LINC01106 and pBobi-MS2-GFP obtained from OBiO Technology (Shanghai, China) were co-transfected into the Ishikawa and HEC-1A cells for incubating 48 h. Subsequently, the cells were lysed with RIP lysis buffer. The extracts of cells were incubated with RIP buffer magnetic beads with anti-GFP or Mice IgG (Cell Signaling Technology, Danvers, MA, USA) at 4 °C for overnight, followed by washing with RIP wash buffer. After that, the eluate was collected. The expression of miRNAs (miR-525-3p, miR-524-3p, miR-449a, miR-744-5p, miR-151a-3p, miR-34c-5p, miR-34a-5p and miR-3167) was detected by qRT-PCR.
Dual Luciferase Reporter (DLR) Assay
The cloning 3ʹUTR sequences containing the binding site were cloned into pGL3 vector to establish the wild-type vector. The Phusion Site-Directed Mutagenesis Kit (Thermo Fisher Scientific) was used to construct the mutant-type vector. Wild-type/mutant-type vector and miR-449a mimics/miR-NC were co-transfected into the cells at 37°C for 48 h. Then, Ishikawa and HEC-1A cells were lysed. The collecting supernatant was used to measure relative luciferase activity by a Dual-Luciferase Reporter Assay System (Promega).
Western Blot Analysis
Following extracting proteins from Ishikawa and HEC-1A cells by RIPA buffer containing protease inhibitor, the BCA Protein Assay Kit (Abcam, Cambridge, MA, USA) was used to detect the protein concentrations. A total of 40 µg proteins were separated by 10% SDS-PAGE. Then, the resolving proteins were transferred into PVDF membranes. At room temperature, the blocking was performed using 5% bovine serum albumin (BSA). After blocking, membranes were incubated overnight at 4°C with primary antibodies against MET (1:1000; Abcam) and β-actin (1:1000; Abcam). Then, tris-buffered saline Tween-20 (TBST) was used to wash the membranes for at least three times. Subsequently, at room temperature, the secondary antibody HRP-conjugated anti-mice IgG (1:5000; Santa Cruz, Waltham, MA, USA) was added to incubate for 1 h. β-actin was used as the internal reference. The membranes were visualized by ECL chemiluminescent substrate reagent kit (Invitrogen), and quantified by ImageLab software (Bio-Rad, Hercules, CA, USA).
Statistical Analysis
SPSS software (version 20.0, USA) was used to perform statistical analyses. Data were expressed as the mean ± standard deviation. Student’s t-test was used to assess the differences between the two groups. One-way ANOVA was used to evaluate the differences among multiple groups. After ANOVA analysis, the pairwise comparison was performed using Tukey’s multiple comparisons test. Pearson’s correlation analysis was used to determine the correlation between the expression levels of miR-449a and LINC01106/MET in EC tissues. P-value less than 0.05 indicated a statistically significant difference. All experiments were conducted in triplicate in at least three independent experiments.
Results
LncRNA LINC01106 is Highly Expressed in EC Tissues and Cells
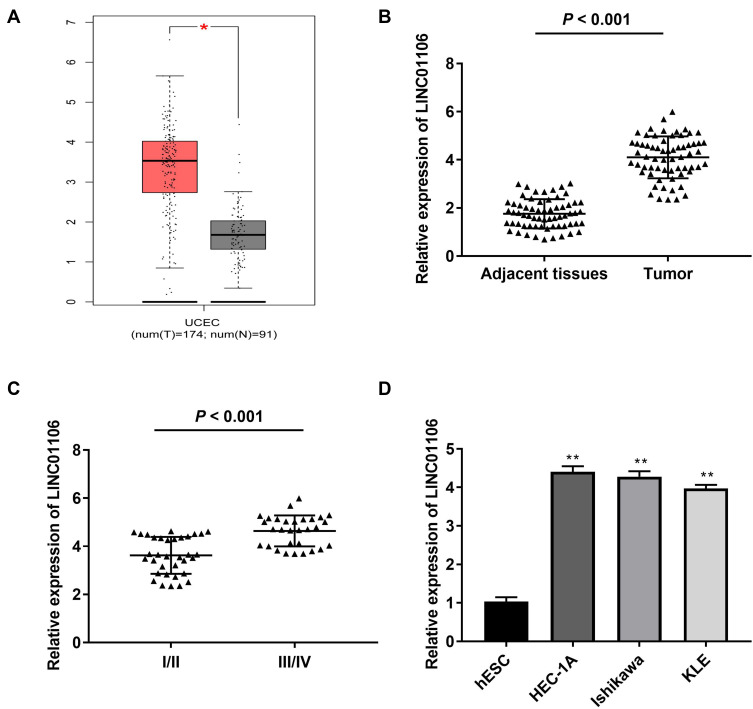
Based on TCGA database, LINC01106 expression in uterine corpus endometrial carcinoma (UCEC) and normal tissues was analyzed. We discovered that LINC01106 expression was highly up-regulated in UCEC tissues in contrast to that in normal tissues (Figure 1A, P < 0.05). Subsequently, we detected the expression of LINC01106 in EC and adjacent tissues from 64 patients to further verify the above results. The results of qRT-PCR uncovered that LINC01106 expression was dramatically increased in tumor tissues compared to that in adjacent tissues (Figure 1B, P < 0.001). Similarly, LINC01106 expression was significantly up-regulated in stage III/IV of EC compared to that in stage I/II (Figure 1C, P < 0.001). As shown in Table 1, the high expression and low expression of LINC01106 exhibited significant differences in tumor stage and metastasis. Meanwhile, the expression of LINC01106 in the EC cell lines (HEC-1A, Ishikawa and KLE) and the human endometrial-stromal cell line (hESC) was detected. qRT-PCR results revealed that LINC01106 expression in EC cell lines (HEC-1A Ishikawa and KLE) was dramatically increased in contrast to that in the hESC cells (Figure 1D, P < 0.01). Additionally, the expression of LINC01106 in Ishikawa and HEC-1A cell lines was relatively higher among these three cell lines. Therefore, Ishikawa and HEC-1A cell lines were chosen to perform the subsequent experiments (Figure 1D, P < 0.01). We speculated that LINC01106 might play a pathogenic role in EC.
Figure 1.
LncRNA LINC01106 is highly expressed in EC/UCEC tissues and EC cells. (A) The expression of lncRNA LINC01106 in UCEC tissues and normal tissues was analyzed in TCGA. *P < 0.05. (B) The expression of lncRNA LINC01106 in EC tissues (n = 64) and adjacent tissues (n = 64) was detected by qRT-PCR. P < 0.001. (C) The expression of lncRNA LINC01106 at different WHO grades was detected by qRT-PCR. P < 0.001. (D) The expression of lncRNA LINC01106 in HEC-1A, Ishikawa, KLE and hESC cells was detected by qRT-PCR. **P < 0.01 vs the hESC cells group.
Table 1.
Correlations Between lncRNA LINC01106 Expression and Clinicopathological Characteristics in EC
| Characteristics | Total | LINC01106 Expression | P-value | |
|---|---|---|---|---|
| Low (32) | High (32) | |||
| Age | 0.614 | |||
| <60 years | 36 | 17 | 19 | |
| ≥60 years | 28 | 15 | 13 | |
| Resection degree | 0.802 | |||
| Total resection | 31 | 16 | 15 | |
| Subtotal resection | 33 | 16 | 17 | |
| Diameter | 0.798 | |||
| <5cm | 25 | 13 | 12 | |
| ≥5cm | 39 | 19 | 20 | |
| Metastasis | 0.012* | |||
| NO | 34 | 22 | 12 | |
| YES | 30 | 10 | 20 | |
| WHO Grade | 0.045* | |||
| I/II | 34 | 21 | 13 | |
| III/IV | 30 | 11 | 19 | |
Notes: WHO, World Health Organization. In metastasis: *P = 0.012; In WHO Grade: *P = 0.045.
LncRNA LINC01106 Knockdown Inhibits the Proliferation, Migration and Invasion of EC Cells in vitro and Suppresses the Growth of Tumor Xenograft in vivo
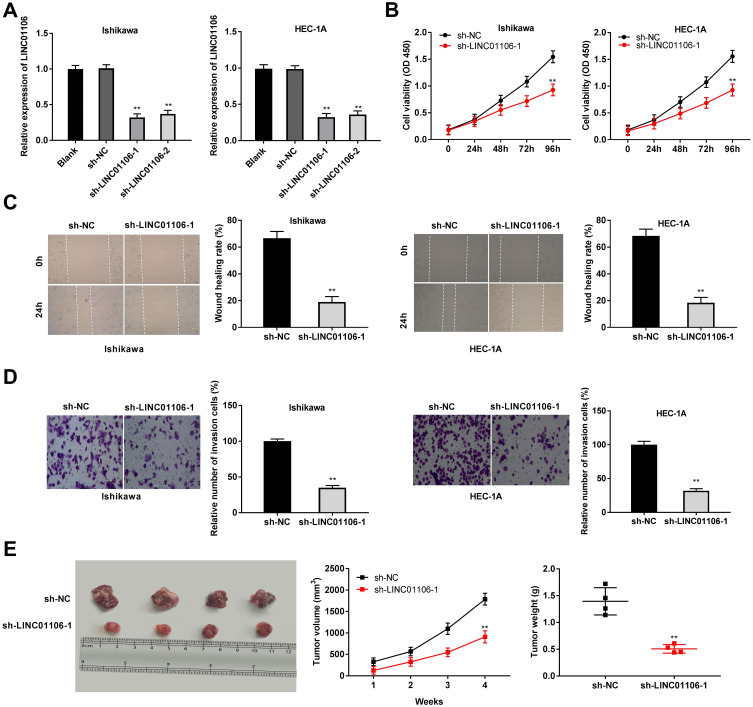
After transfection of sh-LINC01106-1/-2/NC into Ishikawa and HEC-1A cells, the efficiency of transfection was detected by qRT-PCR. We found that LINC01106 expression was significantly down-regulated by the transfection of sh-LINC01106-1/-2 (Figure 2A, P < 0.01), suggesting that sh-LINC01106-1/-2 was successfully transfected into EC cells. We chose sh-LINC01106 to perform the following trails due to its relatively high silencing efficiency. MTT assay revealed that the viability of Ishikawa and HEC-1A cells was obviously declined in the sh-LINC01106-1 group compared with the sh-NC group (Figure 2B, P < 0.01). Wound healing assay uncovered that the wound healing rate of Ishikawa cells was reduced by LINC01106 knockdown. Simultaneously, the wound healing rate of HEC-1A cells presented the similar results (Figure 2C, P < 0.01). In line with the above results, transwell invasion assay indicated that the number of invasion EC cells was lessened by LINC01106 knockdown (Figure 2D, P < 0.01). In addition, we also investigated the effect of LINC01106 inhibition on the growth of tumor xenograft in vivo. We found that tumor volume was dramatically smaller and tumor weight was obviously lighter in the sh-LINC01106-1 group compared to those in the sh-NC group (Figure 2E, P < 0.01). These results suggested that silence of LINC01106 inhibited proliferation, migration and invasion of EC cells in vitro and suppressed the growth of tumor xenograft in vivo.
Figure 2.
LncRNA LINC01106 knockdown inhibits proliferation, migration and invasion of EC cells in vitro and suppresses the growth of EC in vivo. (A) The expression of LINC01106 in EC cells was detected by qRT-PCR. (B) The viability (OD450) of EC cells was measured by MTT assay. (C) The wound healing rate of EC cells was measured by wound healing assay. (D) The number of invasion cells was measured by transwell invasion assay. (E) The tumor volumes were monitored at different time points and tumor weights were measured 4 weeks later. **P < 0.01 vs the sh-NC group.
LncRNA LINC01106 Targets miR-449a
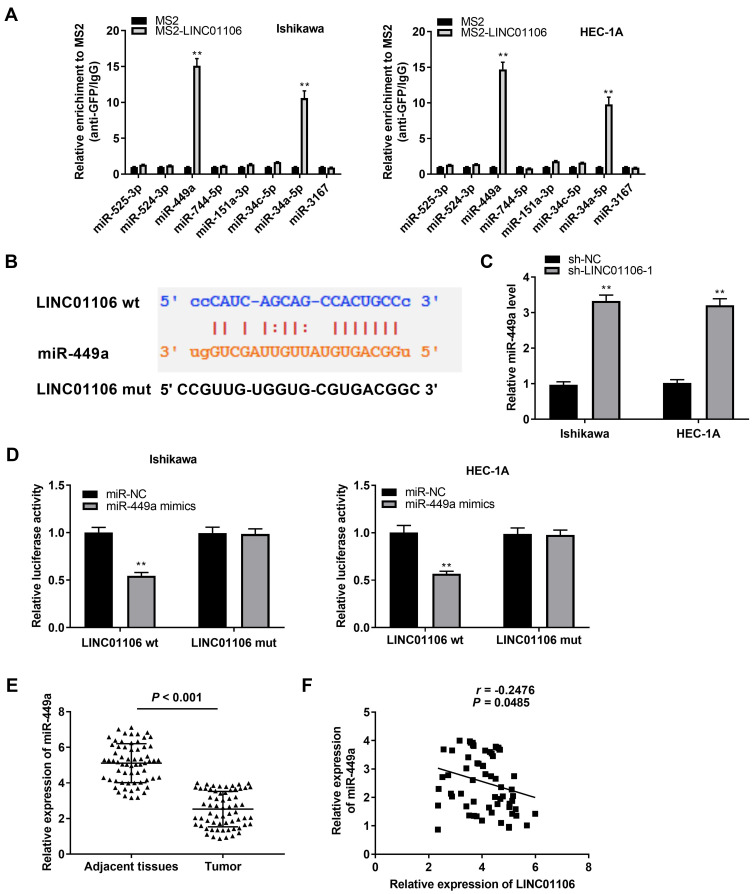
Based on the Lncbase database and Starbase database, eight miRNAs were chosen to screen out the target of LINC01106 by RIP assay. The results demonstrated that the LINC01106 immunoprecipitates obtained from Ishikawa and HEC-1A cells were notably enriched with miR-449a and miR-34a-5p compared to those in the empty vector (MS2) group (Figure 3A, P < 0.01). MiR-449a was chosen to further verify whether it is a target of LINC01106. Subsequently, we used the Starbase software to predict the potential binding site between LINC01106 and miR-449a (Figure 3B) and detected the expression of miR-449a after the transfection of sh-LINC01106-1/NC into Ishikawa and HEC-1A cells. qRT-PCR results disclosed that transfection of sh-LINC01106-1 significantly up-regulated miR-449a expression (Figure 3C, P < 0.01). To further determine the relationship between LINC01106 and miR-449a, DLR assay was performed both in Ishikawa and HEC-1A cells. We discovered the luciferase activity in the LINC01106-WT/miR-449a mimics group was obviously decreased in contrast to that in the LINC01106-WT/miR-NC group (Figure 3D, P < 0.01). Additionally, miR-449a expression was reduced in tumor tissues in comparison to the adjacent tissues (Figure 3E, P < 0.001). Besides, t a negative correlation between LINC01106 and miR-449a was exhibited in EC tissues (Figure 3F, P = 0.0485, r = −0.2476). On account of the above data, we speculated that miR-449a was a target of LINC01106 and was negatively modulated by LINC01106.
Figure 3.
MiR-449a is the direct target of lncRNA LINC01106. (A) Eight miRNAs selected from starbase database was detected by dual luciferase reporter assay and confirmed by qRT-PCR. **P < 0.01 vs the MS2 group. (B) The predicted complementary binding site of lncRNA LINC01106 and miR-449a. (C) The expression of miR-449a after transfection of sh-LINC01106-1/NC in EC cells was detected by qRT-PCR. **P < 0.01 vs the sh-NC group. (D) The luciferase activity in EC cells co-transfected with pGL3-LINC01106 WT/pGL3-LINC01106 MUT and miR-449a mimics/NC was determined by dual luciferase reporter assay. **P < 0.01 vs the miR-NC group. (E) The expression of miR-449a in tumor tissues (n = 64) and adjacent tissues (n = 64) was detected by qRT-PCR. P < 0.001. (F) The correlation between miR-449a and LINC01106. P = 0.0485, r = −0.2476.
Overexpression of miR-449a Inhibits the Proliferation, Migration and Invasion of EC Cells in vitro
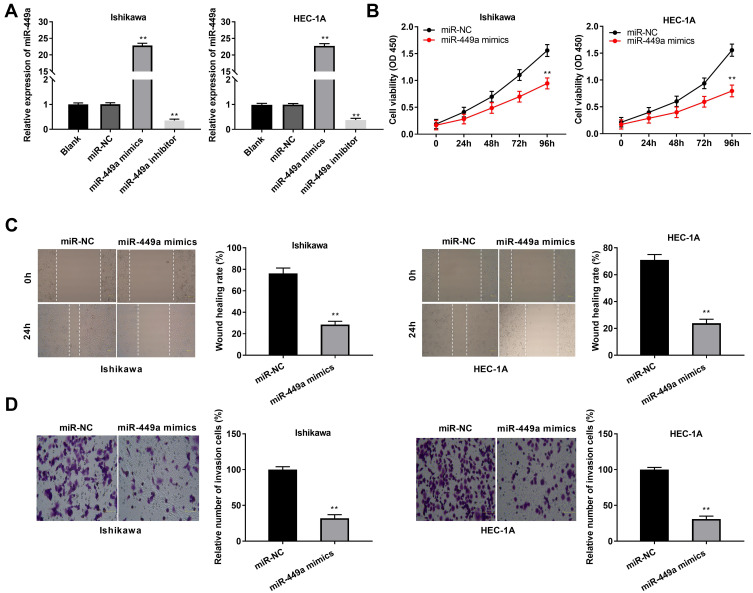
To investigate the role of miR-449a on the biological functions of EC cells (Ishikawa and HEC-1A cells), the transfection efficiency of miR-449a was detected. The results of qRT-PCR demonstrated that miR-449a expression was visibly up-regulated by miR-449a mimics, whereas was down-regulated by miR-449a inhibitor (Figure 4A, P < 0.01), which demonstrated that miR-449a mimics/inhibitor was transfected into EC cells successfully. MTT assay showed that overexpression of miR-449a could suppress the cell viability in both two cell lines (Figure 4B, P < 0.01). Wound healing assay and transwell invasion assay demonstrated that the wound healing rate of EC cells and the number of invasion cells were both inhibited by miR-449a overexpression (Figure 4C and D, P < 0.01). These data implied that overexpression of miR-449a could inhibit the proliferation, migration and invasion of EC cells.
Figure 4.
MiR-449a suppresses proliferation, migration and invasion of EC cells. (A) The expression of miR-449a after transfection of miR-449a mimics/inhibitor/NC in EC cells was detected by qRT-PCR. (B) The viability (OD450) of EC cells was measured by MTT assay. (C) The wound healing rate of EC cells was measured by wound healing assay. (D) The number of invasion cells was measured by transwell invasion assay. **P < 0.01 vs the miR-NC group.
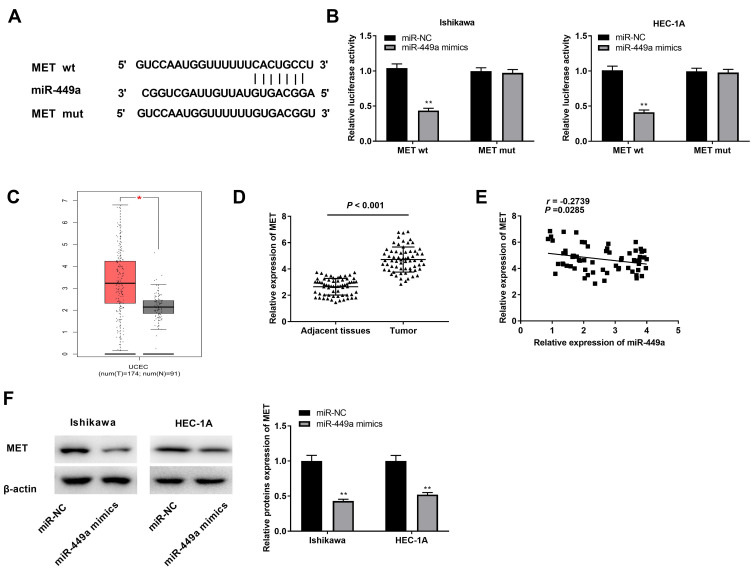
MiR-449a Targets MET
Through Targetscan software, the potential binding site between miR-449a and MET was depicted (Figure 5A). Through dual luciferase reporter assay, in contrast to the MET-WT/miR-NC group, a decreased luciferase activity was exhibited in the MET-WT/miR-449a mimics group, which proved MET was a target gene of miR-449a (Figure 5B, P < 0.01). On the basis of TCGA database, we analyzed the status of MET expression in UCEC and normal tissues. The results presented that the expression of MET was highly increased in UCEC tissues compared to that in normal tissues (Figure 5C, P < 0.05). The expression of MET in EC and adjacent tissues was used to further verify the above results. The results of qRT-PCR uncovered that MET expression was higher in tumor tissues than that in adjacent tissues (Figure 5D, P < 0.001). Moreover, miR-449a expression was negatively correlated with MET in EC tissues (Figure 5E, P = 0.0285, r = −0.2739). Western blot assay demonstrated that the protein expression of MET was down-regulated by miR-449a overexpression (Figure 5F, P < 0.01). In brief, we identified MET was a target gene of miR-449a.
Figure 5.
Identification of MET as the target gene of miR-449a. (A) The predicted complementary binding site of MET and miR-449a. (B) The luciferase activity in EC cells co-transfected with pGL3-MET WT/pGL3-MET MUT and miR-449a mimics/NC was determined by dual luciferase reporter assay. **P < 0.01 vs the miR-NC group. (C) The expression of MET in UCEC tissues and normal tissues was analyzed in TCGA. *P < 0.05. (D) The expression of miR-449a in tumor tissues (n = 64) and adjacent tissues (n = 64) was detected by qRT-PCR. P < 0.001. (E) The correlation between miR-449a and MET. P = 0.0285, r = −0.2739. (F) The protein expression of MET in EC cells was detected by Western blot assay. **P < 0.01 vs the miR-NC group.
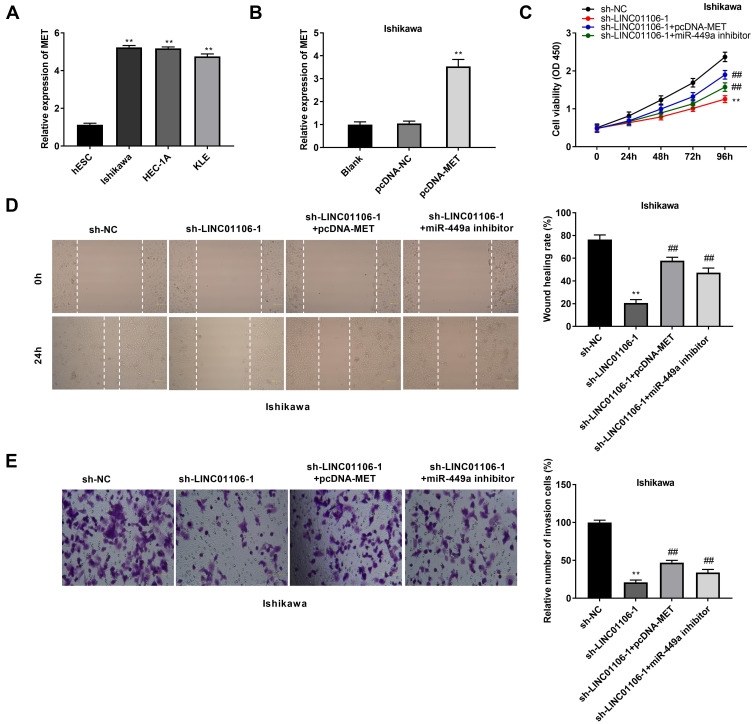
LncRNA LINC01106 Knockdown Suppresses the Proliferation, Migration and Invasion of EC Cells by Regulating the miR-449a/MET Axis in vitro
The expression of MET in the EC cell lines (HEC-1A, Ishikawa and KLE) and the human endometrial-stromal cell line (hESC) was detected by qRT-PCR. We found that MET expression was dramatically up-regulated in the EC cell lines compared to the hESC cells (Figure 6A, P < 0.01). Then, qRT-PCR was performed to detect the transfection efficiency of pcDNA-MET/NC in Ishikawa cells. The results demonstrated that MET expression in the pcDNA-MET group was highly increased in comparison to the pcDNA-NC group (Figure 6B, P < 0.01) and we confirmed pcDNA-MET was transfected into Ishikawa cell. MTT assay demonstrated that the viability of Ishikawa cells was significantly declined in the sh-LINC01106-1 group in contrast to the sh-NC group. Interestingly, compared with the sh-LINC01106-1 group, cell viability was partly promoted in both the sh-LINC01106-1 + pcDNA-MET group and the sh-LINC01106-1 + miR-449a inhibitor group (Figure 6C, P < 0.01). We also found that the wound healing rate of EC cells and the number of invasion cells were both declined in the sh-LINC01106-1 group in comparison to those in the sh-NC group. Nevertheless, the inhibiting effects of sh-LINC01106-1 on the migration and invasion of Ishikawa cells were both reversed by up-regulation of MET and down-regulation of miR-449a (Figure 6D and E, P < 0.01). These results implied that silencing of LINC01106 could suppress the proliferation, migration and invasion of EC cells in vitro through regulating the miR-449a/MET axis.
Figure 6.
LINC01106 knockdown inhibits proliferation, migration and invasion of EC cells in vitro by regulating miR-449a/MET. (A) The expression of MET in HEC-1A, Ishikawa, KLE and hESC cells was detected by qRT-PCR. **P < 0.01 vs the hESC cells group. (B) The expression of MET after transfection of pcDNA-MET/NC into Ishikawa cells was detected by qRT-PCR. **P < 0.01 vs the pcDNA-NC group. (C) The viability (OD450) of Ishikawa cells was measured by MTT assay. (D) The wound healing rate of Ishikawa cells was measured by wound healing assay. (E) The number of invasion cells was measured by transwell invasion assay. **P < 0.01 vs the sh-NC group, ##P < 0.01 vs the sh-LINC01106-1 group.
Discussion
EC, as one of the most serious cancers of women, usually contributes to the highest mortality among female-related cancers.2,3 Many lncRNAs are taken part in the occurrence of EC.11,30 LINC01410 is highly expressed both in EC tissues and cells and the up-regulation of LINC01410 is closely related to tumor stage.11 LINC01220 expression is dramatically up-regulated in EC tissues and its high expression is strongly associated with tumor stage.30 Consistent with the above two findings, we revealed that the expression of LINC01106 was obviously elevated in EC tissues and cells, and high expression of LINC01106 presented a notable connection with the tumor stage. In addition, we also demonstrated that LINC01106 was correlated with tumor metastasis. Therefore, we speculated that LINC01106 might be an onco-lncRNA in EC.
Also, some lncRNAs have been found to act as important regulators in cell proliferation, migration and invasion and apoptosis in EC.10,14,30 For instance, LINC01220 silencing suppresses the proliferation and facilitates the apoptosis of EC cells.30 The proliferation and invasion of EC cells are restrained by LINC01410 knockdown.11 Suppression of LINC01016 inhibits the proliferation, migration and invasion of EC cells.10 In this study, we discovered that LINC01106 inhibition suppressed the proliferation, migration and invasion of EC cells. In line with our results, Guo et al have reported that silence of LINC01106 suppresses the proliferation and migration of colorectal cancer cells.31 However, the previous study only investigated the regulatory mechanism of LINC01106 knockdown on the proliferation and migration in colorectal cancer cells. Our results further confirmed that knockdown of LINC01106 also attenuates the invasion ability of EC cell. The above results implied that LINC01106 knockdown retarded the occurrence and development of EC.
MiR-449a is participated in the progression of tumors and serves as anti-tumor role in several tumor types.32–34 Ding et al have uncovered that the expression of miR-449a is down-regulated in non-small cell lung cancer (NSCLC), whereas overexpression of miR-449a reduces the proliferation and facilitates the apoptosis of NSCLC cells.32 Li et al have disclosed that miR-449a expression is obviously decreased in gastric cancer (GC) cells, while overexpression of miR-449a can suppress the proliferation of GC cells.33 Similarly, Feng et al have found that the expression of miR-449a is dramatically reduced in colitis-associated colorectal cancer (CAC) tissues and miR-449a up-regulation restrains the viability of colorectal cancer cells, thus inhibiting the progression of CAC.34 In this study, we demonstrated that miR-449a expression was obviously decreased in EC tissues and cells. The viability, the migration and invasion of EC cells could be inhibited by miR-449a overexpression. In line with these results, some researches have revealed that the expression of miR-449a is markedly decreased in EC tissues19 and miR-449a overexpression can diminish cell proliferation, metastasis and invasion in EC.20 At the same time, LINC01106 was identified to target miR-449a and miR-449a expression was negatively modulated by LINC01106. We inferred LINC01106 silencing attenuated the progression of EC by regulating miR-449a.
The emerging evidence suggests that MET is an important factor in the development of cancers,23,24,29 including EC.26–28 Yang et al have presented that MET is highly expressed in ovarian cancer tissues and cells.22 Zhang et al have reported that the proliferation, migration and invasion of NSCLC cells are mediated by MET.25 In the current study, we discovered MET expression in EC tissues was significantly increased. In line with this result, Xiao has showed that MET expression is dramatically up-regulated in EC tissues.28 Meanwhile, MET was proved to be a target gene of miR-449a and there was an inverse relation between miR-449a and MET. The results displayed that MET was negatively regulated by miR-449a. Similar to our results, Cheng et al have displayed that miR-449a inhibits the growth of hepatocellular carcinoma through regulating MET.29 Simultaneously, we revealed that both up-regulation of MET and down-regulation of miR-449a reversed the suppressive effect of LINC01106 knockdown on the proliferation, migration, and invasion of Ishikawa cells. The results suggested that LINC01106 knockdown inhibited EC progression via regulating the miR-449a/MET axis.
To be concluded, the current study uncovered that knockdown of LINC01106 could restrain the proliferation, migration and invasion of EC cells through regulating the miR-449a/MET axis in vitro. However, this study did not confirm the interactions among LINC01106, miR-449a and MET in EC tissues in vivo, which may be a limitation of the present study. Further experiments will be performed to elucidate these issues in the future. In a word, we hope these findings will still provide a new strategy for treating EC.
Funding Statement
No funding was received.
Data Sharing Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Informed Consent
The protocols of this study were reviewed and approved by the ethical committee in Qingdao Traditional Chinese Medicine Hospital (approval number: 2018-09-15). Each patient in this study obtained the written informed consent. And animal experiment was obtained the approval of the ethical committee in our hospital (approval number: 2018-10-15) and performed in The Animal Experimental Center of our hospital.
Consent for Publication
The participant has consented to the submission of the study to the journal.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interest.
References
- 1.Dahlgren E, Friberg LG, Johansson S, et al. Endometrial carcinoma; ovarian dysfunction — a risk factor in young women. Eur J Obstet Gynecol Reprod Biol. 1991;41(2):143–150. doi: 10.1016/0028-2243(91)90092-Y [DOI] [PubMed] [Google Scholar]
- 2.Treeck O, Skrzypczak M, Schuler-Toprak S, Weber F, Ortmann O. Long non-coding RNA CCAT1 is overexpressed in endometrial cancer and regulates growth and transcriptome of endometrial adenocarcinoma cells. Int J Biochem Cell Biol. 2020;122:105740. doi: 10.1016/j.biocel.2020.105740 [DOI] [PubMed] [Google Scholar]
- 3.Lv Y, Chen S, Wu J, et al. Upregulation of long non-coding RNA OGFRP1 facilitates endometrial cancer by regulating miR-124-3p/SIRT1 axis and by activating PI3K/AKT/GSK-3beta pathway. Artif Cells Nanomed Biotechnol. 2019;47(1):2083–2090. doi: 10.1080/21691401.2019.1617727 [DOI] [PubMed] [Google Scholar]
- 4.Humber CE, Tierney J, Symonds PR, Collingwood M, Green J. Chemotherapy for advanced, recurrent or metastatic endometrial carcinoma. Cochrane Database Syst Rev. 2005;5(4):CD003915. [DOI] [PubMed] [Google Scholar]
- 5.Boll D, Verhoeven RHA, Van DA, et al. Incidence and survival trends of uncommon corpus uteri malignancies in the Netherlands, 1989–2008. Int J Gynecol Cancer. 2012;22(4):599–606. doi: 10.1097/IGC.0b013e318244cedc [DOI] [PubMed] [Google Scholar]
- 6.Yang QS, Li B, Xu G, et al. Long noncoding RNA LINC00483/microRNA‐144 regulates radiosensitivity and epithelial–mesenchymal transition in lung adenocarcinoma by interacting with HOXA10. J Cell Physiol. 2019;234(7):11805–11821. doi: 10.1002/jcp.27886 [DOI] [PubMed] [Google Scholar]
- 7.Sun CB, Wang HY, Han XQ, et al. LINC00511 promotes gastric cancer cell growth by acting as a ceRNA. World J Gastrointest Oncol. 2020;12(4):394–404. doi: 10.4251/wjgo.v12.i4.394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Q, Cheng J, Zhang JD, Zhang YL, Luo SX. The expression and functional mechanism of long non-coding RNA LINC00339 in colorectal cancer. Zhonghua Yi Xue Za Zhi. 2019;99(24):1881–1886. doi: 10.3760/cma.j.issn.0376-2491.2019.24.009 [DOI] [PubMed] [Google Scholar]
- 9.Xu X-Y, Zhang J, Qi Y-H, Kong M, Hu -J-J. Linc-ROR promotes endometrial cell proliferation by activating the PI3K-Akt pathway. Eur Rev Med Pharmacol Sci. 2018;22(8):2218–2225. [DOI] [PubMed] [Google Scholar]
- 10.Pan X, Li D, Huo J, Kong F, Yang H, Ma X. LINC01016 promotes the malignant phenotype of endometrial cancer cells by regulating the miR-302a-3p/miR-3130-3p/NFYA/SATB1 axis. Cell Death Dis. 2018;9(3):303. doi: 10.1038/s41419-018-0291-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu M, Ding N, Zhuang S, Li Y. LINC01410/miR-23c/CHD7 functions as a ceRNA network to affect the prognosis of patients with endometrial cancer and strengthen the malignant properties of endometrial cancer cells. Mol Cell Biochem. 2020;1–11. [DOI] [PubMed] [Google Scholar]
- 12.Li XX, Liang XJ, Zhou LY, et al. Analysis of differential expressions of long non-coding RNAs in nasopharyngeal carcinoma using next-generation deep sequencing. J Cancer. 2018;9(11):1943–1950. doi: 10.7150/jca.23481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Z, Wen S, Zhang Y, Shi X, Wang G. Identification of dysregulated long non-coding RNAs/microRNAs/mRNAs in TNM I stage lung adenocarcinoma. Oncotarget. 2017;8(31):51703–51718. doi: 10.18632/oncotarget.18512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun F, Liang W, Tang K, Hong M, Qian J. Profiling the lncRNA-miRNA-mRNA ceRNA network to reveal potential crosstalk between inflammatory bowel disease and colorectal cancer. PeerJ. 2019;7:e7451. doi: 10.7717/peerj.7451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni J, Liang S, Shan B, Tian W, Wang H, Ren Y. Methylationassociated silencing of miR638 promotes endometrial carcinoma progression by targeting MEF2C. Int J Mol Med. 2020;45(6):1753–1770. doi: 10.3892/ijmm.2020.4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao X, Dai L, Yue Q, et al. MiR-195 inhibits migration, invasion and epithelial-mesenchymal transition (EMT) of endometrial carcinoma cells by targeting SOX4. J Biosci. 2019;44(6):146. [PubMed] [Google Scholar]
- 17.Li F, Chen H, Huang Y, Zhang Q, Zheng F. MiR-34c plays a role of tumor suppressor in HEC-1-B cells by targeting E2F3 protein. Oncol Rep. 2015;33(6):3069–3074. doi: 10.3892/or.2015.3894 [DOI] [PubMed] [Google Scholar]
- 18.Hu Y, Wu A-Y, Xu C, et al. MicroRNA-449a inhibits tumor metastasis through AKT/ERK1/2 inactivation by targeting steroid receptor coactivator (SRC) in endometrial cancer. J Cancer. 2019;10(2):547–555. doi: 10.7150/jca.27748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye W, Xue J, Zhang Q, et al. MiR-449a functions as a tumor suppressor in endometrial cancer by targeting CDC25A. Oncol Rep. 2014;32(3):1193–1199. doi: 10.3892/or.2014.3303 [DOI] [PubMed] [Google Scholar]
- 20.Wu AY, Hu Y, Cang W, et al. Suppressive effect of microRNA-449a on the NDRG1/PTEN/AKT axis regulates endometrial cancer growth and metastasis. Exp Cell Res. 2019;382(2):111468. doi: 10.1016/j.yexcr.2019.06.013 [DOI] [PubMed] [Google Scholar]
- 21.Zhang M, Wang Y, Matyunina LV, Akbar A, McDonald JF. The ability of miRNAs to induce mesenchymal-to-epithelial transition (MET) in cancer cells is highly dependent upon genetic background. Cancer Lett. 2020;480:15–23. doi: 10.1016/j.canlet.2020.03.023 [DOI] [PubMed] [Google Scholar]
- 22.Yang S, Li Z, Luo R. miR-34c targets MET to improve the anti-tumor effect of cisplatin on ovarian cancer. Onco Targets Ther. 2020;13:2887–2897. doi: 10.2147/OTT.S239425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du F, Li X, Feng W, et al. SOX13 promotes colorectal cancer metastasis by transactivating SNAI2 and c-MET. Oncogene. 2020;39(17):3522–3540. doi: 10.1038/s41388-020-1233-4 [DOI] [PubMed] [Google Scholar]
- 24.Salgia R, Sattler M, Scheele J, Stroh C, Felip E. The promise of selective MET inhibitors in non-small cell lung cancer with MET exon 14 skipping. Cancer Treat Rev. 2020;87:102022. doi: 10.1016/j.ctrv.2020.102022 [DOI] [PubMed] [Google Scholar]
- 25.Zhang T, Liu C, Yu Y, et al. TBL1XR1 is involved in c-Met-mediated tumorigenesis of human nonsmall cell lung cancer. Cancer Gene Ther. 2019. [DOI] [PubMed] [Google Scholar]
- 26.Wagatsuma S, Konno R, Sato S, Yajima A. Tumor angiogenesis, hepatocyte growth factor, and c-Met expression in endometrial carcinoma. Cancer. 1998;82(3):520–530. doi: [DOI] [PubMed] [Google Scholar]
- 27.He Y, Peng Z, Pan X, Wang H, Ouyang Y. Expression and correlation of C-met and estrogen receptor in endometrial carcinomas. J Sichuan Univ Med Sci Ed. 2003;34(1):78–79, 88. [PubMed] [Google Scholar]
- 28.Xiao L. The expression and significance of hepatocyte growth factor and c-Met receptor in endometrial carcinoma. J Chongqing Med Univ. 2004;29(6):789–791. [Google Scholar]
- 29.Cheng J, Wu LM, Deng XS, et al. MicroRNA-449a suppresses hepatocellular carcinoma cell growth via G1 phase arrest and the HGF/MET c-Met pathway. Hepatobiliary Pancreat Dis Int. 2018;17(4):336–344. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Kong C, Wu C, Wang Y, Ying X. Knocking down of LINC01220 inhibits proliferation and induces apoptosis of endometrial carcinoma through silencing MAPK11. Biosci Rep. 2019;39(7):BSR20181794. doi: 10.1042/BSR20181794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo K, Gong W, Wang Q, et al. LINC01106 drives colorectal cancer growth and stemness through a positive feedback loop to stimulate Hedgehog activation. SSRN. 2019;45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding M, Qiu TF, Zhou PG. microRNA-449a suppresses non-small cell lung cancer. Cell Biochem Biophys. 2015;71(2):1255–1259. doi: 10.1007/s12013-014-0339-0 [DOI] [PubMed] [Google Scholar]
- 33.Li X, Li H, Zhang R, et al. MicroRNA-449a inhibits proliferation and induces apoptosis by directly repressing E2F3 in gastric cancer. Cell Physiol Biochem. 2015;35(5):2033–2042. doi: 10.1159/000374010 [DOI] [PubMed] [Google Scholar]
- 34.Feng Y, Dong YW, Song YN, et al. MicroRNA449a is a potential predictor of colitisassociated colorectal cancer progression. Oncol Rep. 2018;40(3):1684–1694. doi: 10.3892/or.2018.6566 [DOI] [PubMed] [Google Scholar]