Abstract
Intercellular communication is fundamental to the survival and maintenance of all multicellular systems, whereas dysregulation of communication pathways can drive cancer progression. Extracellular vesicles (EVs) are mediators of cell-to-cell communication that regulate a variety of cellular processes involved in tumor progression. Overexpression of a specific plasma membrane enzyme, hyaluronan synthase 3 (HAS3), is one of the factors that can induce EV shedding. HAS3, and particularly its product hyaluronan (HA), are carried by EVs and are known to be associated with the tumorigenic properties of cancer cells. To elucidate the specific effects of cancerous, HAS3-induced EVs on target cells, normal human keratinocytes and melanoma cells were treated with EVs derived from GFP-HAS3 expressing metastatic melanoma cells. We found that the HA receptor CD44 participated in the regulation of EV binding to target cells. Furthermore, GFP-HAS3-positive EVs induced HA secretion, proliferation and invasion of target cells. Our results suggest that HAS3-EVs contains increased quantities of IHH, which activates the target cell hedgehog signaling cascade and leads to the activation of c-Myc and regulation of claspin expression. This signaling of IHH in HAS3-EVs resulted in increased cell proliferation. Claspin immunostaining correlated with HA content in human cutaneous melanocytic lesions, supporting our in vitro findings and suggesting a reciprocal regulation between claspin expression and HA synthesis. This study shows for the first time that EVs originating from HAS3 overexpressing cells carry mitogenic signals that induce proliferation and epithelial-to-mesenchymal transition in target cells. The study also identifies a novel feedback regulation between the hedgehog signaling pathway and HA metabolism in melanoma, mediated by EVs carrying HA and IHH.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03399-5) contains supplementary material, which is available to authorized users.
Keywords: Extracellular vesicles, IHH, Hedgehog signaling, c-Myc, Claspin, Proliferation, Melanoma, Cancer, Hyaluronan, Hyaluronan synthase
Introduction
Cancer cells depend on bidirectional communication between other cells and the extracellular environment for sustained growth, invasion and metastasis. Extracellular vesicles (EVs) represent one of the numerous pathways for cellular communication [1]. EVs are membrane-enclosed particles that are shed from the cell surface, carry bioactive molecules [2] and contribute to, for example, the communication between tumor cells and the extracellular matrix [3]. During the past decade, extensive research has demonstrated a role for EVs as effective intercellular messengers in cancer [4–6]. EVs can stimulate malignant transformation of target cells either by paracrine signaling or by fusion and internalization with the recipient cells [5]. The importance of EVs in cancer progression was first observed in 1980 when vesicles from highly metastatic melanoma cells (B16 mouse-F10) enabled poorly metastatic cells (mouse B16-F1) to metastasize to the lungs [7].
EVs are secreted from cells through different mechanisms including (1) exocytosis of multivesicular bodies via fusion with the plasma membrane, (2) direct budding from the plasma membrane, and (3) via the tips of filopodia or other plasma membrane protrusions [8]. We have previously shown that overexpression of HAS3 triggers growth of filopodia [9], which enhances EV shedding [10]. Hyaluronan synthases (HAS1-3) are plasma membrane enzymes responsible for the synthesis of hyaluronan (HA), which is involved in numerous cellular functions, such as proliferation and adhesion [11, 12]. Expression of HAS2 [13] and HAS3 [14, 15] are associated with malignant transformation and aggressive tumor growth. However, the specific features of HAS3-induced EVs and their role in cancer progression have yet to be studied.
Enhanced proliferation, accompanied by altered activity and expression of cell cycle proteins, is one of the hallmarks of cancer cells [16]. The commitment to a new round of cell division (G1/S phase) is the point where cell cycle molecules are altered in cancer cells enabling uncontrolled proliferation [17]. c-Myc is one of the prominent proto-oncogenes. When deregulated it enables oncogenic potential by promoting uncontrolled cell cycle and proliferation by directly regulating the cyclin-dependent kinases (CDKs) [18, 19]. Selective dependence on interphase CDKs explains the increased rounds of division in tumor cells [20]. c-Myc’s involvement in proliferation is reflected by its action in regulating growth-promoting genes [21] and CDK4 [19, 22].
Claspin is a nuclear protein involved in DNA replication and cellular homeostasis [23]. Expression of claspin in normal cells is hardly detectable at the G0/G1 phase, while it spikes at the S/G2 phase and drops during mitosis [24]. Claspin has a dual role in DNA replication; it can either promote or inhibit the process, depending on the circumstances. For example, while claspin initiates DNA replication during the G1/S phase, it can also inhibit DNA replication and thereby cell proliferation during DNA damage and repair. Claspin protein level directly corresponds to an increased or decreased proliferation rate [24, 25]. Apart from being an upstream regulator of claspin, c-Myc itself can also be regulated by numerous signal transduction pathways like hedgehog, Wnt, Notch, IGF and NFкB [21]. Constitutive activation of these pathways in cancer facilitates over-expression of c-Myc, as the cell traverses through cell cycle progression [16, 21].
Here we focus on hedgehog signaling (HH), a conserved pathway that plays an important role in embryonic development and in the progression of numerous cancers [26–28]. Binding of HH ligands such as Indian hedgehog (IHH), desert hedgehog (DHH) and sonic hedgehog (SHH) to the smoothened, frizzled class receptor (Smo) stimulates HH signaling and a cascade of downstream intracellular events that regulates cell survival, proliferation and differentiation [29]. It has been shown that HH ligands can be carried via EVs [30, 31]. In this study, we treated normal keratinocytes and melanoma cells with EVs originating from metastatic melanoma cells overexpressing GFP-HAS3 and investigated their possible effects on the target cell properties. We observed that GFP-HAS3 EVs carried IHH, HAS3, EGF, EGFR and CD44. Moreover, IHH in HAS3-EVs activated the HH signaling pathway, which subsequently triggered c-Myc activation, cell proliferation and epithelial-to-mesenchymal transition. c-Myc-induced proliferation of target cells was mediated by the regulation of claspin. This study shows for the first time that EVs originating from HAS3 overexpressing cells carry mitogenic signals that induce sustained proliferation and EMT in target cells.
Results
Characterization of EVs secreted by MV3 human metastatic melanoma cells
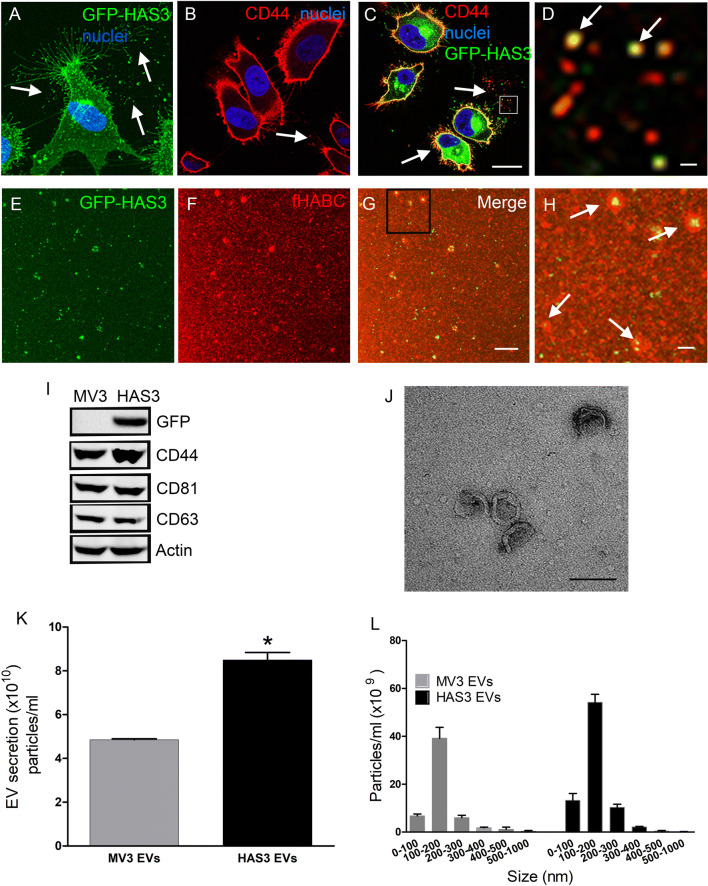
EVs were isolated from MV3 human melanoma cells stably expressing GFP-HAS3, which has been characterized previously [32, 33]. To trap the GFP-HAS3-positive EVs for visualization in situ we cultured the MV3 cells in type I collagen matrix. Live GFP-HAS3 expressing cells are grown inside a 3D gel of type I collagen matrix showed a high number of GFP-HAS3-positive filopodia and EVs shedding into the surrounding matrix (Fig. 1a). EVs shed from GFP-HAS3 expressing MV3 cells were termed as HAS3-EVs. Immunostaining of the 3D cultures demonstrated that both doxycycline-induced and uninduced cells secreted CD44-positive EVs (arrows in Fig. 1b, c), and most of the HAS3-EVs displayed an association with CD44 immunostaining (Fig. 1d). The HA-positivity of HAS3-EVs was analyzed in isolated EV preparations with confocal microscopy (Fig. 1e–h). Staining with the HA probe, fluorescently labelled HA binding complex (fHABC), indicated that EVs of different sizes were coated with HA (arrows in Fig. 1h). HAS3-EVs carried GFP-HAS3, while uninduced cells with native-EVs (hereafter termed as MV3-EVs) were negative, as indicated with GFP immunostaining (Fig. 1i). EVs from both the induced and uninduced cells carried the HA receptor, CD44 and EV markers, such as CD81 and CD63, as well as actin (Fig. 1i). To analyze the EV secretion from these cells, we utilized NTA analysis, which showed a 75% increase in the total particle counts in the HAS3-EVs group, compared to MV3-EVs (Fig. 1k). No difference was detected in the size distribution between MV3- and HAS3-EVs, and their average diameter was 150 nm (Fig. 1l). TEM analysis of isolated preparations confirmed a typical structure of EVs (Fig. 1j). The EV secretion and size distribution were tested in one more melanoma cell line, C8161 with doxycycline induced GFP-HAS3. EVs from uninduced cells will be addressed as C8161-EVs while the EVs from induced cells will be addressed as C8161 HAS3-EVs. The NTA analysis revealed a 58% increase in the total particle counts in C8161-HAS3 EVs group compared to C8161-EVs (Suppl. Figure 2a) while the size distribution between the two groups remained unchanged (Suppl. Figure 2b).
Fig. 1.
Characterization of EVs secreted by MV3 human metastatic melanoma cancer cells with stable, inducible expression of GFP-HAS3. a 3D projection of live GFP-HAS3 expressing cell grown in 3D collagen gel. Confocal sections of CD44-immunostained uninduced (b) and induced (c) MV3 cells and a higher magnification image of an area indicated by a white box in (c) is shown in (d). Panels e–h show HA staining of EV preparations isolated from GFP-HAS3 expressing cells, and h shows a higher magnification image of the area indicated by a black box in (g). i Western blotting of GFP-HAS3 detected with GFP antibody and other typical EV markers. Nanoparticle tracking analysis of EV levels (j) and size distribution (k) secreted from uninduced (MV3-EVs) and induced (HAS3-EVs) MV3 cells, and l transmission electron microscopy of EVs isolated from GFP-HAS3 expressing cells. Data represent mean ± S.E. from three independent experiments; *P < 0.05 (one-way ANOVA, Tukey’s test). Arrows in all panels indicate EVs. Scale bars represent 20 µm in (c), 2 µm in (d) and (h), 10 µm in (g) and 200 nm in (j)
Binding of HAS3-EVs to target cells and their effect on HA secretion, proliferation and invasion
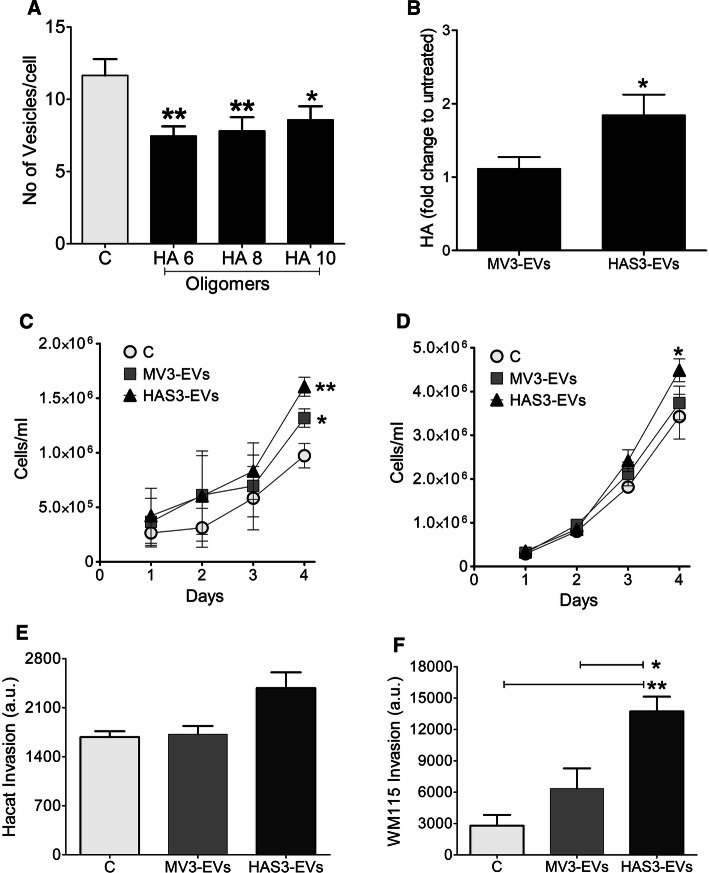
When a normal keratinocyte cell line, HaCaT, was treated with both EVs and HA oligomers of different lengths (HA6, HA8, HA10) (Suppl. Figure 1j, k), binding of EVs to the cell surface significantly decreased (Fig. 2a). HA oligomers are known to inhibit HA and CD44 interaction [34] and this showed that CD44 is one of the potential binding partners aiding in EV interaction with the cells. Treating HaCaT cells with HAS3-EVs increased HA secretion levels of the cells when compared to cells that were treated with MV3-EVs (Fig. 2b). Since increased cell proliferation is one of the earlier traits in cancer progression, we analyzed the effect of MV3- and HAS3-EVs on the proliferation rate of HaCaT and WM115, a primary melanoma cell line. Over a period of 4 days, both HaCaT (Fig. 2c) and WM115 (Fig. 2d) proliferation was significantly increased with MV3-EVs treatment, with a relatively higher rate in HAS3-EVs treatment. The proliferation rate of HaCaT cells when treated with EVs from a different melanoma cell line, C8161-GFP-HAS3 was observed to follow the same pattern. HaCaT cells treated with C8161 HAS3-EVs had significantly higher proliferation rate on day 4, compared to cells treated with C8161-EVs (Suppl. Figure 2c). We also assessed if the same trend was observed in cells invasion, after EV treatment using type I collagen as the matrix. While there was no significant change in the invasion of HaCaT cells (Fig. 2e), both MV3- and HAS3-EVs treatment-induced invasion in WM115 cells, with HAS3-EVs giving a statistically significant increase (Fig. 2f). The results showed that HAS3-EVs can promote oncogenic properties in the target cells, such as proliferation and invasion. Next, we investigated which factors could influence these changes.
Fig. 2.
Binding of HAS3-EVs and their effect on target cell HA secretion, proliferation and invasion. a Effect of short HA oligosaccharides (6–10 monosaccharide units) on binding of EVs originating from HAS3-induced MV3 cells to HaCaT cells. b Effect of melanoma cell-derived EV treatment on HA secretion levels of HaCaT cells. HA secretion was quantified and normalized to RNA content of the cells. Effect of melanoma-derived EVs on the proliferation rate (c, d) and invasion (e, f) in HaCaT (c, e) and WM115 (d, f) cells, respectively. The data represent mean ± S.E. of four independent experiments. *P value < 0.05, **P value < 0.01, one-way ANOVA (Tukey’s test)
Downstream effects and key players of HAS3-EVs in target cells
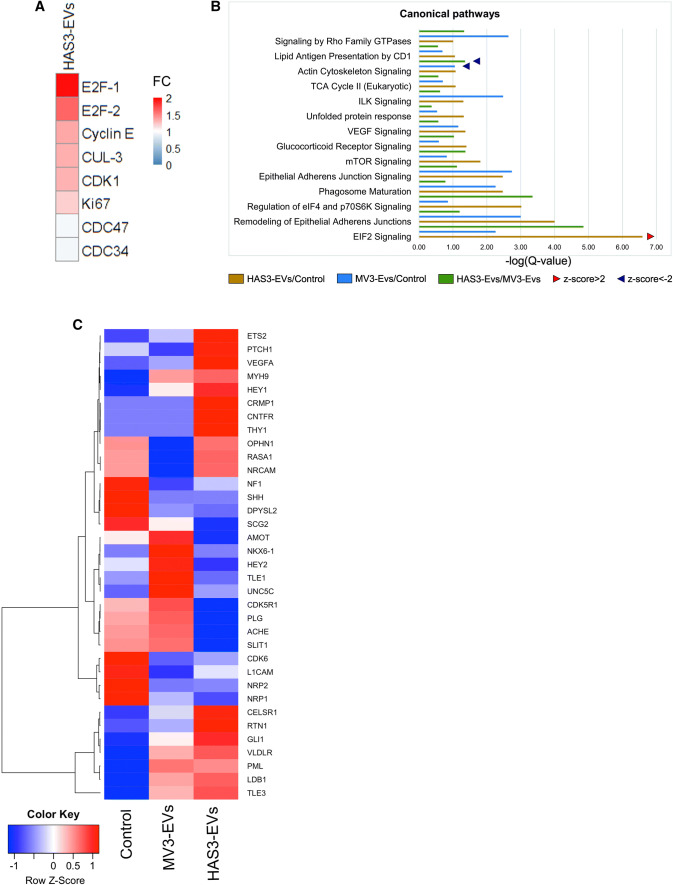
Due to the increase in cell proliferation after MV3- and HAS3-EVs treatment, we next analyzed changes in cell cycle proteins using an antibody array to target 60 key proteins relevant for cell proliferation. Interestingly, HAS3-EVs treatment induced an increased level of proteins such as cyclin E, E2F1, E2F2, CDK1, Ki97 and cullin-3, which are involved in DNA transcription and cell division (Fig. 3a). These results gave the intriguing indication that EVs treatment causes changes in the cell proliferation rate, which prompted us to employ complementary high-throughput RNA sequencing and label-free quantitative proteomic techniques to identify key role players. From whole cell lysates of HaCaT treated with or without MV3- and HAS3-EVs, differentially expressed proteins were analyzed using quantitative proteomics. Pathway analysis was performed using IPA software, comparing MV3- and HAS3-EVs treatments with the untreated control (Fig. 3b; Suppl. Table 1). The top-ranked pathways included rearranged cytoskeleton, increased cell proliferation and remodeled epithelial adherens junctions, all of which were positively associated with the HAS3-EVs group, and to a lesser extent with MV3-EVs (Fig. 3b). This expression pattern shows that EVs have an impact on various processes in the cells promoting them towards uncontrolled proliferation and possibly EMT. Overall, from both protein array and mass spectrometry analyses, we could discern that E2F signaling and levels of other regulatory proteins involved in cell cycle progression were increased upon HAS3-EVs treatment.
Fig. 3.
Downstream effects and keys players of HAS3-EVs in target cells. a Antibody array analysis showing the cell cycle protein expression of HaCaT cells, treated with HAS3-EVs compared to untreated control. The protein expression values of HaCaT cells treated with HAS3-EVs were charted based on fold change compared to untreated control. Six replicates of each protein on the array (FC fold change), b Differentially enriched canonical pathways of HaCaT cells proteome shared amid groups treated with MV3-EVs and HAS3-EVs, as compared to control. Arrow heads point to pathway activation (z-score > 2) or inhibition (z-score < -2), as predicted by IPA algorithms. c A heat map of row-wise scaled group mean mRNA expression of control HaCaT cells or cells treated with MV3- or HAS3-EVs (N = 3 per group) for genes involved in hedgehog signaling
Next, we wanted to identify the key driving factors through which HAS3-EVs affect these cellular processes. Using Western blotting, we compared the levels of c-Myc, NFκB-p65, CD44, EGFR and MAPK, which are involved in HA signaling and cell proliferation [35–39], in HaCaT cells treated with or without MV3- and HAS3-EVs. Furthermore, the levels of EMT markers such as E-cadherin and Slug were also analyzed (Suppl. Figure 1c). Although we could observe changes in many of the oncogenic and signaling proteins promoting cell proliferation, an increase in c-Myc expression in the HAS3-EVs group was profound and statistically significant. Additionally, the whole-cell proteome analysis of HaCaT cells showed claspin (Swiss-Prot ID: Q9HAW4, Suppl. Table 1) as one of the top hits in HAS3-EVs treatment. Claspin expression was significantly increased with HAS3-EVs (1.95-fold change) but not with MV3-EVs (0.86-fold change) (Suppl. Table 1). Thus, claspin could be one of the differentially expressed proteins between MV3- and HAS3-EVs treatment and we were interested to understand its role in HAS3-EVs induced cell proliferation. Involvement of claspin in cell cycle progression was also highlighted in the network analysis of the extracted proteome (CLSPN in Suppl. Figure 3). Interestingly, our RNA sequencing analysis showed that HaCaT cells treated with HAS3-EVs exhibited an enrichment of hedgehog signaling (HH) pathway genes such as GLI1 and PTCH1 when compared to untreated control and MV3-EVs treatment (Fig. 3c). Based on the above-described results, significantly elevated protein expression of claspin and mRNA levels of GLI1 and PTCH1 pinpoints that hedgehog signaling and claspin could be involved in HAS3-EVs mediated increased cell proliferation. Though not directly shown in our mass spectrometry or RNA-sequencing analyses, differential expression of c-Myc was observed with HAS3-EVs treatment with Western blotting. We could not find commonly affected key players in mRNA and protein expression in the treated HaCaT cells, thus differentially expressed targets that were related to cell proliferation were sorted out to characterize them further in our in vitro experiments. Therefore, we analyzed the possible connection between hedgehog signaling with c-Myc and claspin in HAS3-EVs mediated increase in cell proliferation.
HAS3-EVs act through c-Myc and claspin in target cells to regulate proliferation
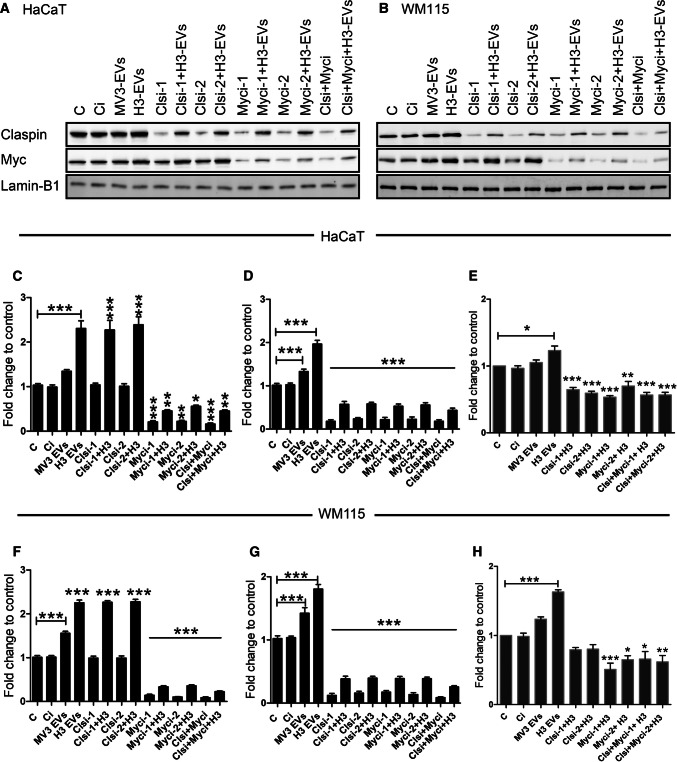
It has been previously reported that c-Myc and claspin regulate cell proliferation [24, 40] but their association with HH signaling and melanoma progression is largely unknown. Therefore, we studied the role of c-Myc and claspin on HaCaT and WM115 cell proliferation by siRNA-mediated knockdown. When treated with HAS3-EVs, c-Myc and claspin protein levels were significantly elevated, in both HaCaT (Fig. 4a, c, d) and WM115 cells (Fig. 4b, f, g). MV3-EVs treatment in both the cell types only led to minor changes in the expression of these proteins. Knocking down c-Myc and claspin with siRNAs significantly decreased their expression, while adding HAS3-EVs partially rescued it (Fig. 4a–d, f–g). Although knocking down c-Myc decreased the levels of claspin, the reciprocal effect was not observed. These results indicate that claspin expression was dependent on c-Myc activation for its expression while c-Myc protein expression was claspin-independent. Furthermore, HAS3-EVs had a direct role in c-Myc and claspin activation. Since c-Myc [21] and claspin [24] play a role in cell proliferation, we next studied the change in cell number 48 h post siRNA transfection. HAS3-EVs treated cells exhibited a higher proliferation rate compared to control cells (Fig. 4e, h). Also, HAS3-EVs treated cells with claspin and c-Myc knockdown showed a significantly lower proliferation rate (Fig. 4e, h). These results demonstrate that the increase in proliferation mediated by HAS3-EVs acted through c-Myc and claspin. Next, we aimed to resolve the specific activation mechanism of c-Myc by HAS3-EVs.
Fig. 4.
HAS3-EVs act through c-Myc and claspin in target cells to regulate proliferation. Western blots showing the expression of c-Myc and claspin in EVs- and siRNA-treated cells in HaCaT (a, c, d) and WM115 (b, f, g). Cell proliferation rate of HaCaT (e) and WM115 (h) cells with EVs- and siRNA-treatments. The statistical significance tests were done in comparison to control. The data represent mean ± S.E. of four independent experiments. *P value < 0.05, **P value < 0.01, ***P value < 0.001, one-way ANOVA (Tukey’s test). C control, Ci control siRNA (scrambled siRNA), H3 or H3-EVs HAS3-EVs, Clsi and Myci siRNAs for claspin and c-Myc
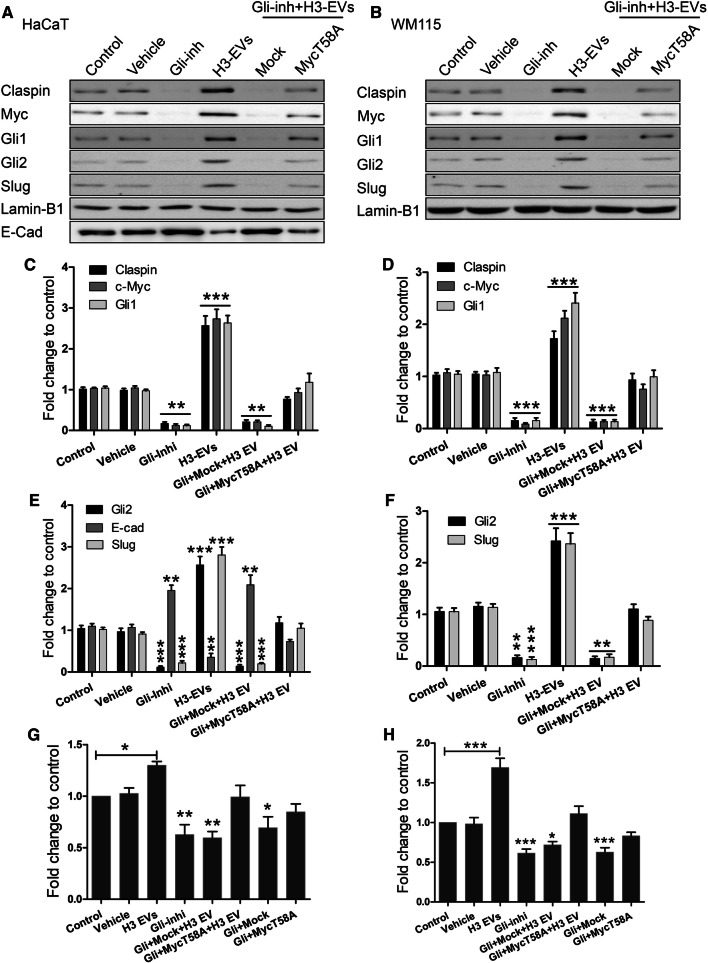
HAS3-EVs induce c-Myc and EMT via HH signaling
Since HH signaling was enhanced by HAS3-EVs treatment (Fig. 3c), we investigated the direct effect of HH pathway members on c-Myc activation. To this end, the central HH pathway transcription factors, Gli-1 and -2 were targeted using a specific inhibitor called GANT58 [41]. Furthermore, the protein levels of Gli-1 and -2, along with claspin, c-Myc and EMT regulators such as slug and E-cadherin, were analyzed with Western blotting. Constitutively overexpressing c-Myc with a point mutation T58A activates its oncogenic functions and diminishes its ability to accelerate apoptosis [42, 43]. T58A mutation has also increased c-Myc protein stability to twofold [44]. We wanted to see whether Gli inhibition has any effect on c-Myc regulation of claspin and increased cell proliferation. To test it, we used constitutively activated c-Myc T58A, thus checking whether the mutant could reverse the effects of Gli inhibition. HaCaT (Fig. 5a, c, e) and WM115 cells (Fig. 5b, d, f) treated with HAS3-EVs showed an increase in Gli-1 and -2, c-Myc and claspin protein levels. Gli inhibitor effectively reversed the effect of HAS3-EVs by decreasing the expression of Gli-1 and -2, c-Myc and claspin. Moreover, c-Myc T58A but not mock transfection partially rescued the effect of HAS3-EVs on these protein levels (Fig. 5a–f). We also measured the proliferation rate as an end result of these treatments; the Gli inhibitor with or without HAS3-EVs decreased the proliferation, while c-Myc T58A treatment along with HAS3-EVs increased proliferation similar to control cells (Fig. 5g, h). These results suggest that HH signaling induced by HAS3-EVs activates c-Myc and claspin to regulate cell proliferation. In addition to enhanced proliferation rate, EMT is also a hallmark for cancer. We, therefore, studied an epithelial marker (E-cadherin) and mesenchymal marker (slug), to map out the extent of HAS3-EV treatment. As it can be seen from the results, HAS3-EVs treatment decreased E-cadherin (Fig. 5a, d) and increased slug levels (Fig. 5 a, d, b, f) (Suppl. Figure 1b–i). WM115 did not express E-cadherin endogenously. The results thus illustrate that HH signaling from HAS3-EVs is responsible for the increased proliferation rate and EMT via activation of c-Myc.
Fig. 5.
HAS3-EVs induce c-Myc and EMT by HH signaling. Protein expression of HH and EMT markers were tested in HaCaT (a, c, e) and WM115 cells (b, d, f). Both HaCaT and WM115 cells were treated with a Gli inhibitor, GANT58 and a constitutively expressing c-Myc T58A plasmid, along with HAS3-EVs. The impact of the treatments on proliferation was also tested in both HaCaT (g) and WM115 (h) cells. All the statistical significance tests were done in comparison to control. The data represent mean ± S.E. of four independent experiments. *P value < 0.05, **P value < 0.01, ***P value < 0.001, one-way ANOVA (Tukey’s test). H3 or H3-EVs HAS3-EVs, Clsi and Myci siRNAs for claspin and c-Myc, E-cad E-cadherin, Gli-inhi or Gli Gli inhibitor
IHH is the specific factor for HAS3-EV mediated c-Myc activation and claspin correlates with HA in human cutaneous melanocytic lesions
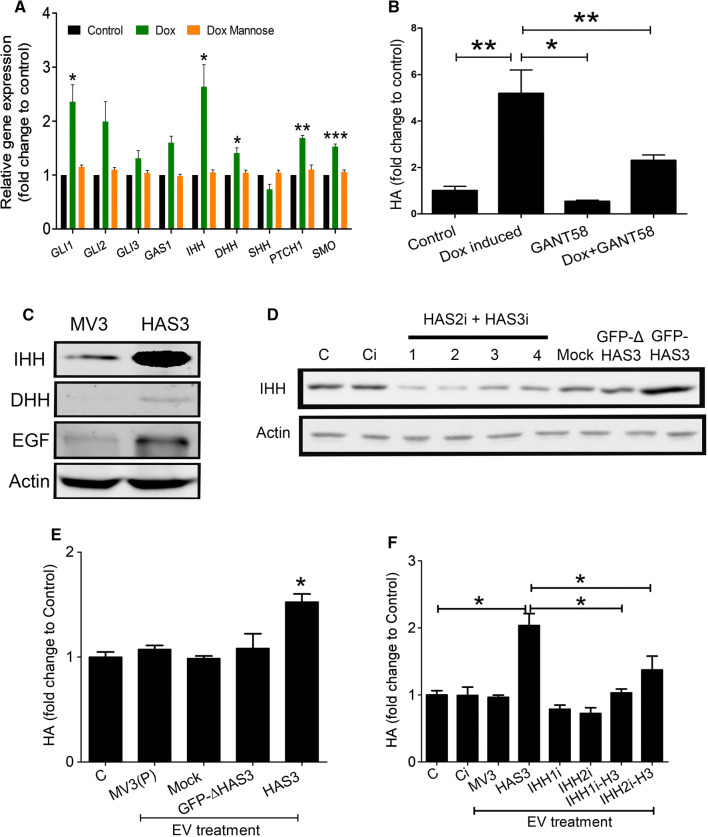
Next, we wanted to identify which mitogen was responsible for the activation of the HH pathway and how it was associated with HAS3-EVs. There are three principal HH ligands—IHH, DHH and SHH, all of which act through Gli proteins [3]. Using doxycycline inducible GFP-HAS3 overexpressing MV3 cells, we analyzed the expression of HH genes by qPCR. Among the different genes that were tested, IHH, DHH, GLI1, PTCH1 and SMO expression were significantly increased in MV3 cells with GFP-HAS3 overexpression (Fig. 6a). This observation gave additional support to our RNA sequencing results, where we identified GLI1 and PTCH1 gene expression to be elevated with HAS3-EVs treatment in HaCaT cells (Fig. 3d). Additionally, the GFP-HAS3 overexpressing MV3 cells were also treated with mannose which reduces UDP-GlcNAc (UDP-N-acetyl glucosamine), a substrate used by HAS1-3 to synthesize HA [10]. Interestingly, mannose addition with GFP-HAS3 overexpression effectively reversed the increased expression of IHH, DHH, GLI1, PTCH1 and SMO genes (Fig. 6a). Thus, it was clear that expression of HH signaling genes was associated with HA synthesis. To examine the putative relationship between HA synthesis and HH signaling, we again utilized MV3 cells with GFP-HAS3 overexpression by doxycycline induction. Of note, the addition of Gli inhibitor reduced HA secretion in the media when GFP-HAS3 expression was induced with doxycycline. Gli inhibitor did not influence HA secretion in the absence of doxycycline induction (Fig. 6b). We then analyzed the protein expression of IHH and DHH in MV3- and HAS3-EVs, alongside another well-known mitogen, epithelial growth factor (EGF). The results showed that HAS3-EVs contain a significantly higher IHH protein level, along with EGF, when compared with MV3-EVs. In fact, IHH level in HAS3-EVs was even more remarkable than EGF. However, DHH protein level was not different between the two groups (Fig. 6c). Therefore, we identified IHH to be one of the major mitogens present in HAS3-EVs.
Fig. 6.
Presence of IHH in HAS3-EVs and feedback between HA and HH pathways. a q-PCR showing the RNA expression of HH genes in MV3-GFP-HAS3 cells. HA production was induced with doxycyclin (Dox) and inhibited with mannose. b HA levels were detected in MV3-GFP-HAS3 cells with or without doxycycline and GANT58. c Western blots showing expression of IHH, DHH and EGF mitogens in MV3- and HAS3-EV lysates. Actin was used as the loading control. d Western blots showing expression of IHH in EVs collected from MV3 parental cells treated with HAS2 + HAS3i cocktail, mock, GFP-ΔHAS3 [45] and GFP-HAS3 plasmid. e Effect of EVs collected from MV3 parental cells treated with mock, GFP-ΔHAS3 and GFP-HAS3 plasmid on HA secretion in HaCaT cells. f HA level in HaCaT cells after treatment with EVs collected from MV3-GFP-HAS3 cells treated with IHHi and dox induction (IHHi and IHHi-HAS3 EVs). HA secretion was normalized on RNA levels. Data represent mean ± S.E. of four independent experiments. *P value < 0.05, **P value < 0.01, one-way ANOVA (Tukey’s test). C control, Ci control siRNA (scrambled siRNA), HAS3 or H3 or H3-EVs HAS3-EVs, MV3 (P) EVs EVs from MV3 parental cells, IHHi IHH siRNAs, HAS2i + HAS3i HAS2 and HAS3 siRNAs, Mock pcDNA3 vector
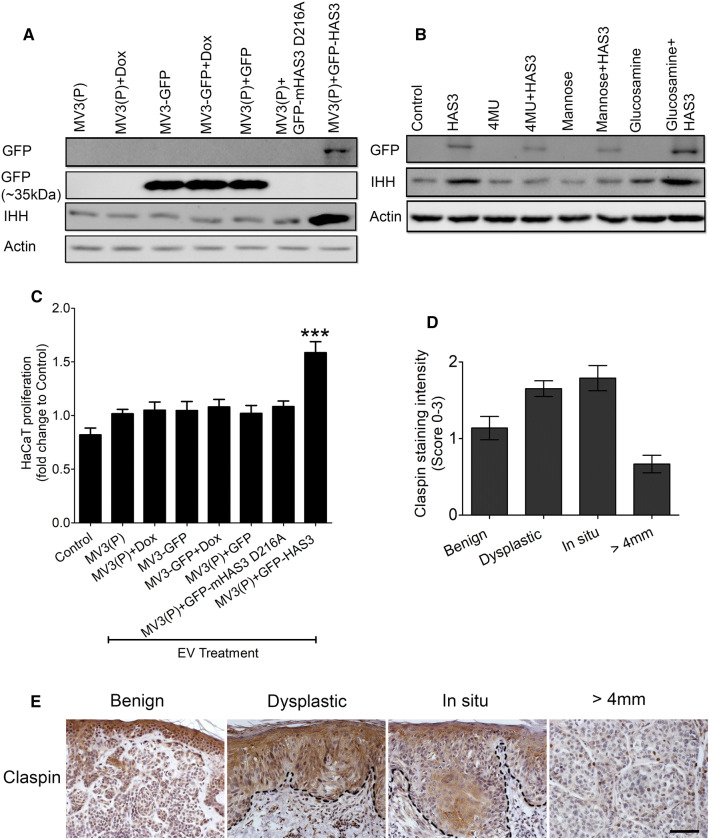
We next investigated whether HAS3 presence in plasma membrane and HA synthesis are associated with increased IHH secretion in EVs. MV3 parental cells were transiently transfected with an inactive GFP-ΔHAS3, which contains only the first 86 amino acids at N-terminus. Thus, the mutant protein cannot synthesize HA due to the absence of catalytic domain [45]. GFP-ΔHAS3 transfected cells showed a positive band near 35 kDa, whereas wildtype GFP-HAS3 was seen near 100 kDa in Western blotting (Suppl. Figure 3b). HA secretion was also not increased in GFP-ΔHAS3 transfected cells when compared to wildtype GFP-HAS3 group (Suppl. Figure 3c, d). Interestingly GFP-ΔHAS3 EVs did not show an increase in IHH protein level, unlike the wildtype GFP-HAS3 EVs (Fig. 6d). EVs collected from the GFP-ΔHAS3 transfected or parental MV3 cells did not increase HA secretion in HaCaT cells, when compared to cells treated with GFP-HAS3 EVs (Fig. 6e). Also, in MV3 parental cells, inhibition of endogenous HAS2 and HAS3 using a cocktail of siRNAs reduced IHH secretion in EVs (Fig. 6d). HAS2 and HAS3 inhibition also reduced HA secretion in MV3 cells (Suppl. Figure 4b). To further demonstrate the specificity of IHH as a stimulus in HAS3-EVs, siRNAs against IHH were used in MV3-GFP-HAS3 cells before isolating EVs. HaCaT cells were then treated with EVs from MV3-GFP-HAS3 cells with or without IHH siRNAs (or IHHi) and HA secretion was analyzed. The results showed a significant reduction in HA level in cells treated with IHHi-HAS3 EVs compared to cells treated with normal GFP-HAS3 EVs (Fig. 6f). This shows that IHH secretion in EVs is associated with HA synthesis and the released IHH in EVs influence HA synthesis in target cells. We have previously shown that HAS3 presence in plasma membrane and its release in EVs depends on its ability to synthesize HA. We have also demonstrated that HA synthesis by HAS3 is affected by D216A point mutation on its catalytic domain or by altering the availability of its substrates, UDP-GlcUA and UDP-GlcNAc using chemicals like 4MU, mannose and glucosamine [33, 46]. Though the point mutation was studied in mHAS3, it is important to pinpoint that there are 97% conserved amino acids shared between mHAS3 and human HAS3 protein sequences [47]. First, we transfected MV3 cells with GFP-mHAS3 D216A and treated MV3-HAS3 cells with 4MU, mannose and glucosamine. EVs were collected and examined for GFP and IHH immunostaining. As shown in (Fig. 7a), mHAS3 D216A was not present in EVs and did not increase IHH secretion, when compared to GFP-HAS3 transfection. While reducing UDP-GlcUA by 4MU and UDP-GlcNAc by mannose significantly blocked GFP-HAS3 presence in EVs, increasing UDP-GlcNAc by glucosamine enhanced GFP-HAS3 in EVs (Fig. 7b). IHH secretion was also increased in glucosamine treatment with GFP-HAS3 group (Fig. 7b). Glucosamine also had an effect without GFP-HAS3 overexpression, which could be partially explained by the fact that this chemical increases O-GlcNAc modification and activation of endogenous HAS3 and HAS2 [33, 48]. As the next step, we also analyzed changes in HaCaT cell proliferation with MV3-EVs containing GFP-mHAS3 D216A or GFP-HAS3. The results showed that only GFP-HAS3 EVs induced an increase in HaCaT proliferation (Fig. 7c), which is in line with our earlier observation (Fig. 2c). In all these experiments, doxycycline did not show any adverse effects (Suppl. Figure 2d–g). The above results show that HA synthesis is associated with the release of HAS3 and IHH in EVs. In addition, both HA synthesis and HH signaling have a positive feedback regulation on each other.
Fig. 7.
HA synthesis is associated with IHH secretion in HAS3-EVs. a Western blotting analysis of MV3 cells with GFP and GFP-mHAS3 D216A transfection. MV3 and MV3-GFP cells treated with or without doxycycline were also used as controls. The GFP band for HAS3 is near ~ 100 kDa while the band for only GFP is near ~ 35 kDa. b Western blotting analysis of MV3-HAS3 cells with or without 4MU, mannose and glucosamine treatments. Uninduced group was used as control. c 96 h HaCaT cell proliferation with or without treatment of EVs obtained from the described MV3 cell lines. Data represent mean ± S.E. of three independent experiments; **P value < 0.001, one-way ANOVA (Tukey’s test). In panels a and c, representative blots from 3 independent experiments are shown. d, e Melanoma tissue sections were stained to check for claspin immunoexpression, scale bar 100 µm. Data represent mean ± S.E. of 3–4 independent experiments in (a) and (b) and mean ± S.E. of 12–16 tissue sections score in (d). *P value < 0.05, **P value < 0.01, one-way ANOVA (Tukey’s test) in (a) and (b)
As proof of concept, we next wanted to know how claspin protein expression, as the downstream effector of HA-IHH-c-Myc signaling axis, correlates with HA content in different stages of cutaneous melanocytic lesions. Immunohistochemical staining indicated that keratinocytes and melanocytic cells were positive for intracellular claspin while it was almost negative in stroma (Fig. 7d). The staining intensity was moderate or weak in dysplastic melanocytic lesions and in situ melanomas, while the staining intensity in invasive melanomas was negative or weak (Fig. 7e). Interestingly, this staining pattern of claspin in different stages of melanoma corresponded to HA staining in melanoma progression as shown before [33, 49]. This suggests that during melanomagenesis claspin expression follows changes in HA content. Whether or not IHH and c-Myc follows a similar pattern of correlation to HA content during melanomagenesis remains to be studied in the future.
Discussion
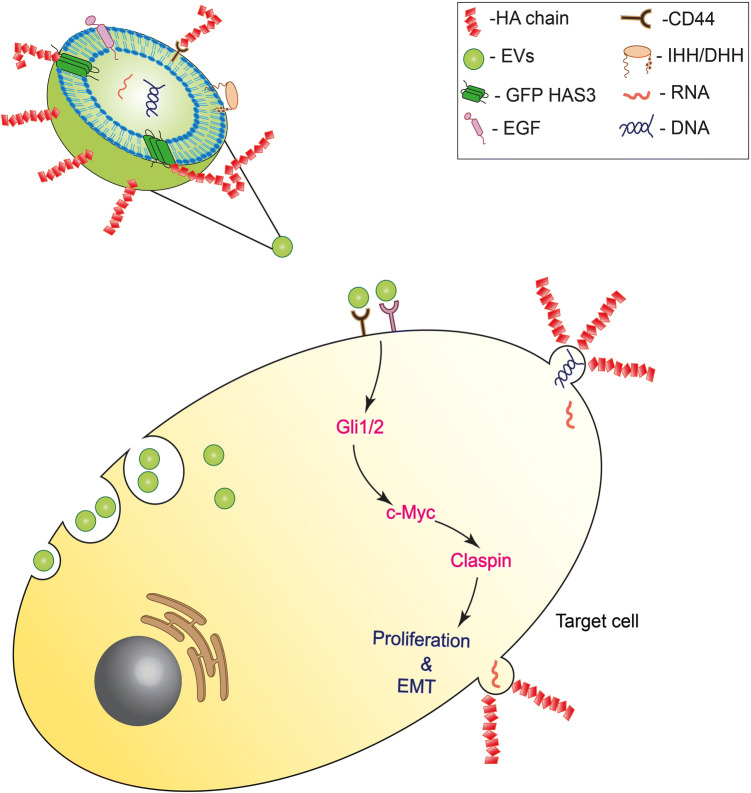
Cutaneous melanoma is one of the deadliest cancers with nearly 80% mortality rate. It is a cancer with an aggressive clinical course which has recently been reported to have acquired resistance to existing therapies [50]. New targets for therapies and the development of new biomarkers depends on a better understanding of the signaling sequence and the biological events occurring in the tumor cells. Tumor cells secrete more EVs than normal cells and carry genetic and cellular information allowing communication with neighboring cells and the tumor microenvironment, either via paracrine or autocrine signaling [51]. EMT and pre–metastatic niche formation depend on tumor-derived EVs [2, 52, 53]. In this study, we demonstrated that EVs released from HAS3 overexpressing metastatic melanoma cells (MV3) interacted with keratinocyte (HaCaT) and melanoma (WM115) cells and induced proliferation and expression of EMT markers, similar to that in tumorigenesis (Fig. 8). HaCaT cells were used in this study since their low endogenous HA secretion facilitates analyses of transformative changes by EVs secreted from a melanoma cell line such as MV3 with high levels of HA. The MV3 cells released more EVs due to HAS3 overexpression and carried HAS3 and HA coat on the vesicles. Incubating HaCaT and WM115 cells with HAS3-EVs resulted in a higher proliferation rate than cells with no treatment, or cells treated with MV3-EVs. HAS3-EVs increased the expression of c-Myc, an oncogenic factor activated in many different cancers types [21, 54]. IHH present in HAS3-EVs acted as a stimulus upstream to c-Myc and we showed that the signaling axis extended to the activation of claspin. The IHH-c-Myc-claspin axis may help a normal keratinocyte (HaCaT) and a melanoma cell line (WM115) to gain tumorigenic potential, such as increased proliferation; this effect may be due to an enhanced G1/S phase transition and EMT. Since c-Myc and hedgehog signaling are independently known to increase tumorigenic progression of the cells [21, 27], the implicated cross-talk between the two different pathways, here acting as one, is of significance (Fig. 8). However, our data cannot exclude the possibility that c-Myc and IHH could have additional, unknown downstream effectors and this possibility remains to be investigated in the future. We have also shown that HA synthesis in melanoma cells is associated with the release of HAS3 and IHH in EVs and there is a positive feedback regulation between HA and HH pathways. Thus, we suggest that HA synthesis could be one of the stimulating factors for IHH secretion in melanoma cells. Future studies should focus on understanding the molecular mechanisms behind the feedback relationship shared by HA and HH pathways.
Fig. 8.
A schematic presentation of the mechanisms triggered by HAS3-induced EVs in target cells. HAS3-EVs from melanoma cells are possibly interacting with receptors such as CD44 present on the surface of recipient cells. As shown in the figure, direct fusion with the plasma membrane and endocytosis are also possible mechanisms of interaction. IHH present in the EVs activates the HH signaling cascade in the recipient cells, leading to the activation of c-Myc and claspin expression. This signaling cascade leads to an increase in proliferation and EMT in the recipient cells
As a proto-oncogene, c-Myc plays an important role in the pathobiology of numerous cancers including melanoma. Being at the crossroads of numerous growth signaling pathways, c-Myc activates the transcription of several genes involved in cell growth, among which the E2F family of transcription factors have also been reported [55]. E2Fs are key factors in cell cycle progression with a direct control over cyclins and CDKs [56, 57]. Fernandez et al., reported that Myc interacts with E2F promoter and overexpression of Myc induces the expression of E2F genes [55, 58]. E2Fs are known to directly bind to and induce transcription of growth-promoting genes such as DHFR, DNA polymerase α, CDC6, ORC1, thymidine kinase, cyclin E, cyclin A and numerous other genes relevant for proliferation and differentiation [59–61]. Our antibody array results were complimentary to these findings; we found a twofold increase in E2F1 and > 1.5-fold increase in E2F2 in HaCaT cells treated with HAS3-EVs (Fig. 3a). Additionally, Cyclin E, CDK1 and the cell proliferation marker Ki67 were all increased upon HAS3-EVs treatment. c-Myc has been shown to regulate the expression of E2F transcription factors [55], which in turn regulate promoter activity of claspin and chk1 [62]. Our study showed that HAS3-EVs treated HaCaT and WM115 cells exhibited high levels of c-Myc and claspin and knocking them down decreased the proliferation rate. Furthermore, knocking down c-Myc decreased claspin expression while knocking down of claspin did not influence c-Myc levels. Bertolli et al., have shown that oncogenic stimulation causes replication stress, and oppositely, sustained E2F-dependent transcription allows the cells to decrease replicative stress and remove the block for cell proliferation [63]. During oncogenic transformation, cells could utilize claspin, because it helps to circumvent replicative stress [64]. However, in this study, we have shown that an increase in c-Myc and possibly E2F transcription factors are decisive factors in aiding enhanced cell proliferation due to IHH stimulus. Claspin plays a role downstream from c-Myc, perhaps regulating the replication stress due to increased proliferation. Nevertheless, the exact molecular mechanism behind claspin’s role remains to be explored.
Although we only investigated hedgehog signaling in this study, both hedgehog and MAPK pathways could be involved upstream of c-Myc, stimulating HAS3-EVs-mediated target cell proliferation. Crosstalk between MAPK and Gli was reported previously [65]. In this study, we observed that HAS3-EVs carried IHH and along with it, DHH and EGF as well. The high levels of IHH in HAS3-EVs were surprising, given that most of the reported studies on melanoma and other cancers have largely implicated the role of SHH in the process [66–68]. However, IHH is a well-known stimulus for self-renewal and enhanced proliferation in several cancers, including colorectal [69], lung [70], pancreatic [71], gastric [72], and T cell acute lymphoblastic leukemia [73]. Our results also point out that knocking down IHH in MV3-GFP-HAS3 cells negatively impacts the HAS3-EVs ability to enable HA secretion in HaCaT cells and downregulation of HAS2 and HAS3 in MV3 parental cells decreases IHH secretion in EVs. Thus, IHH could be an important mitogen in melanoma progression that has not been studied and it is interesting that HA signaling contributes to this phenomenon. The inhibition of Gli1/2 activity and the rescuing of c-Myc by transient transfection of MYC-T58A [74] demonstrated an involvement of the HH pathway in the HAS3-EVs treated cells. Activation of the HH pathway and increased c-Myc expression not only upregulated proliferation but also initiated EMT, which is important for cancer metastasis. The role of the HH pathway in EMT is well documented in cancer cells undergoing metastasis or tumor progression [75–79]. HH pathway signaling has been shown to have pathological effects in many cancer types [3]. Activation of the HH pathway increases cyclin D1 and B1 expression leading to an increase in proliferation in basal cell carcinoma [80].
Since MV3 is a metastatic melanoma cell line, it was of interest to analyze the relationship of HH gene expression and HA synthesis. Our results show that both the expression of HH genes and HAS3-mediated HA synthesis may form a positive feedback loop regulation. A similar positive feedback regulation between HA and other cancer promoting factors, such as ZEB1 [81] and Akt [82], has been identified, for example in breast cancer. Gli-1 and Gli-3 were shown to bind directly to the HAS2 promoter and regulate its transcription [83]. Although only HAS2 regulation is known so far, our results suggest that the HH pathway has a direct impact on HA synthesis, possibly by HAS3 regulation. We have previously shown that HA content increases in in situ melanoma but then declines in more advanced and invasive stages of melanoma, suggesting that HA may be more important in the early stages of cancer progression [33, 49]. In this work, as an end target of the HA-IHH-c-Myc-claspin signaling cascade, we have found that claspin follows a similar tissue staining pattern as HA during melanoma progression. However, it is unknown whether c-Myc and HH pathway factors Gli-1 and 2 also follow the same pattern of expression in different stages of melanoma.
About 40–50% of melanoma patients have mutations in valine at position 600 (Val600) of BRAF protein, which leads to aberrant activation of MAPK pathway [84]. Since MEK acts downstream to BRAF in the MAPK pathway, a combination of BRAF and MEK inhibitors such as vemurafenib, dabrafenib, trametinib and cobimetinib have improved the effect of treatment of patients with advanced and metastatic melanoma. Other classes of therapeutic drugs with improved efficacy that have gained attention in recent years are immunotherapeutic drugs such as checkpoint inhibitors and oncolytic viruses. Some of these drugs approved for advanced stages of melanoma include nivolumab and pembrolizumab (PD-1 inhibitors), ipilimumab (anti-CTLA4 antibody) and the oncolytic virus talimogene laherparepvec (TVEC). The increase in progression-free survival for these type of immunotherapy ranges from 2.9 to 11.5 months [85]. However, the challenge with these targeted therapeutics are the adverse effects and failure to improve patient survival to a significant extent [86, 87]. Also, almost 50% of patients do not respond to the immunotherapy option [85]. This demonstrates the urgent need to screen for novel biomarkers, to identify and target specific mutations and signaling pathways. EVs not only act as biomarkers in this regard but can also show how patients respond to a particular treatment. In peripheral blood, a gene signature of 15 differentially expressed genes, which includes MYC and CDK2, is predictive and prognostic to anti-CTLA4 immunotherapy treatment and 1 year overall survival in melanoma patients [88]. Tumor-derived EVs are abundant in plasma [89] and some of the differentially expressed genes described above could be applicable in melanoma EVs. Another study on melanoma patients pinpointed that lymphatic exudates contained enriched EVs with tumor-specific markers like tetraspanins, ICAM-1, Rab-GTPases and integrins. In fact, the tumor-specific markers are more enriched in lymph than in plasma, suggestive that lymph could be well suited for biomarker analysis in melanoma [90]. BRAF V600E oncogenic mutation is detected in lymph-derived melanoma EVs, and the increase in mutation frequency in patients correlates with risk of relapse [91]. These results indicate that EVs can be a powerful tool to assess melanoma progression and to predict therapy response. As shown in our study, one of the mitogens from hedgehog signaling pathway, IHH, is significantly enriched in melanoma derived HAS3-EVs. Considering the fact that HA content increases at the initial stages of melanoma [33, 49], and that there is a direct feedback regulation between HA synthesis and hedgehog signaling, one could speculate that melanoma EVs with enriched hedgehog mitogens like IHH may be a good predictor for the progression and therapy response at early stages of melanoma. In addition, it would be interesting to see if inhibitors of hedgehog signaling combined with the available immune therapeutics improve the survival of melanoma patients. Since HA coated EVs originate from HAS3 expressing cells [10], from primary cells such as mesothelial cells [92] and from human mesenchymal stem cells [93], their role as messengers in the extracellular space can be acknowledged. As shown in this study, HA-coated EVs are associated with the release of tumor promoting IHH mitogen from melanoma and these EVs could influence its surrounding microenvironment to melanoma progression. Future studies should focus on the mechanistic details on how HA synthesis mediates the release of IHH in EVs and how the secreted IHH promotes melanoma progression.
Materials and methods
Cell lines and culture conditions
MV3 parental cells, and the MV3 and C8161 cells stably overexpressing doxycycline-inducible GFP-HAS3 [32], the MV3 cells stably overexpressing GFP (MV3-GFP) and WM115 cells were cultured in DMEM medium with high glucose (4500 mg/l) (Euroclone, Milan, Italy). HaCaT cells were cultured in DMEM with low glucose (1000 mg/l) (Sigma-Aldrich, USA). All cells were supplemented with inactivated 10% fetal bovine serum (FBS) (HyClone, Thermo Scientific, Epsom, UK), 4 mM l-glutamine (Sigma-Aldrich, USA), 50 µg/ml streptomycin sulfate and 50 units/ml penicillin (Sigma-Aldrich, USA). The MV3 stable cell line was maintained in 50 µg/ml hygromycin and it was induced with 0.25 µg/ml doxycycline during experiments. To check the adverse effects of doxycycline on EV secretion and hyaluronan synthesis, MV3 and MV3-GFP cells were used (Suppl. Figure 2d–g).
EV-depleted FBS used for EV isolation experiments was prepared by ultracentrifugation at 110,000 × g for 16 h followed by sterile filtering with 0.22 µm filters (Minsart, Sartorius, Stedim, Biotech, Goettingen, Germany).
EV isolation
MV3 and C8161 cells (3.2 × 106) were plated in 15 cm cell culture dishes and incubated for 24 h to reach 50% confluency. The complete media was replaced with EV-free media (16 ml/dish) and collected after 48 h of incubation. To remove cell debris, the media was centrifuged at 1000 × g for 10 min at 4° C. Next, the supernatant was transferred into new tubes and centrifuged at 1200 × g for 20 min at 4° C. Finally, the supernatant was ultra-centrifuged at 110.000 × g for 90 min at 4° C and the pellet was dissolved in PBS (sterile filtered in 0.22 µM filter). The EV preparations were stored at − 80° C.
EVs isolated from MV3 cells stably overexpressing doxycycline-inducible GFP-HAS3 without dox induction are called as “MV3-EVs” and EVs collected after doxycycline induction are called as “HAS3-EVs”. EVs isolated from C8161 cells stably overexpressing doxycycline-inducible GFP-HAS3 without dox induction are called as “C8161-EVs” and EVs collected after doxycycline induction are called as “C8161-HAS3 EVs”. EVs isolated from parental melanoma MV3 cells are called as “MV3 parental EVs”. EVs isolated from cells after siRNA treatments for IHH are called as “IHHi EVs” and the same with dox induction are labelled as “IHHi-HAS3 EVs”. Similarly, EVs isolated from cells with the siRNA cocktail against HAS2 and HAS3 are called as “HAS2 + 3i EVs”. EVs isolated from MV3 cells with transient transfection of GFP-ΔHAS3 are called as “ΔHAS3 EVs”. The nomenclature is explained in the table (Table 1).
Table 1.
Nomenclature of different types of EVs and cell lines used in this study
| Cell line | Treatment | Collected EVs and their nomenclature |
|---|---|---|
| MV3-GFP-HAS3 | No dox induction | MV3 EVs |
| MV3-GFP-HAS3 | dox induction | HAS3 EVs |
| C8161-GFP-HAS3 | No dox induction | C8161 EVs |
| C8161-GFP-HAS3 | dox induction | C8161 HAS3 EVs |
| MV3 parental | No treatment | MV3 (P) EVs |
| MV3 parental | GFP-ΔHAS3 transfection | ΔHAS3 EVs |
| MV3 parental | HAS2 + 3i transfection | HAS2 + 3i EVs |
| MV3-GFP-HAS3 | IHHi transfection | IHHi EVs |
| MV3-GFP-HAS3 | IHHi transfection and dox induction | IHHi-HAS3 EVs |
Nanoparticle tracking analysis
The concentration and size distribution of EVs collected from MV3 cells were analyzed using a Nanoparticle Tracking analyzer (NTA) (Malvern Instruments Ltd., Malvern, UK) with an NS300 view unit. The settings used for data acquisition were as follows: camera level 13, acquisition time 30 s and detection threshold 3, kept constant during measurements. Analysis was done using the NTA 3.1 software (Nanosight, Amesbury, UK).
Transfection
For transfections, cells were seeded in a 12 well plate (HaCaT: 11 × 104 cells/well, WM115: 16 × 104 cells/well, MV3: 1.1 × 104), 6 cm dishes (HaCaT: 7 × 105 cells/dish, WM115: 1.1 × 106 cells/dish) and 15 cm dishes (MV3: 3 × 106 cells/dish, MV3-GFP HAS3: 3.2 × 106 cells/dish). Once they reached 40–50% confluency, they were transfected with plasmid DNA or siRNAs.
Plasmid transfection was done with Lipofectamine 3000 (Invitrogen) and siRNA transfections were done with Lipofectamine RNAiMAX (Invitrogen), according to the manufacturers’ instructions. c-Myc T58A (Addgene, USA), EGFP-C1 (Clontech, USA), pcDNA, GFP-mHAS3 D216A [46] and GFP-ΔHAS3 plasmids were used in 12 well, 6 cm and 15 cm plate formats. Predesigned siRNAs targeting claspin: siRNA-1 (60 nM) (ID: s34330), siRNA-2 (60 nM) (ID: s34332), c-Myc: siRNA-1 (HaCaT: 30 nM, WM115: 60 nM) (ID: s9130), siRNA-2 (HaCaT: 30 nM, WM115: 60 nM) (ID: s9131), IHH: siRNA-1(MV3-GFP HAS3: 50 nM) (ID: s7257), siRNA-2 (MV3-GFP HAS3: 50 nM) (ID: s7258), HAS2: siRNA-1 (MV3: 50 nM) (ID: s6457), siRNA-2 (MV3: 50 nM) (ID: s6459) and HAS3: siRNA-1 (MV3-GFP HAS3: 50 nM) (ID: s194495), siRNA-2 (MV3-GFP HAS3: 50 nM) (ID: s6460) were purchased from Ambion, Thermo Fisher Scientific Waltham, MA, USA. Scrambled (control) siRNA (30 nM) from Origene (ref no: SR30004) was used. The cell media containing the transfection mix was removed after 24 h and the cells were either treated with EVs for 24 h or re-plated for proliferation or treated for 48 h with EV-free serum media for collecting EVs.
Treatments
Both MV3- and HAS3-EVs were diluted to a standard 200 µg/ml protein concentration in PBS. MV3- and HAS3-EVs were added to the target cells for treatment using a calculation of 120 µg protein to a seeding density of 0.3 × 106 cells, for example in a 6-well growth area. For culture plates with higher growth area, the amount of EVs were calculated according to the corresponding seeding density i.e. 320 µg for 6 cm dish with 0.8 × 106 seeded cells and 0.8 mg for 10 cm dish with 2.0 × 106 seeded cells. The final concentration for Gli inhibitor, GANT58 (Sigma-Aldrich, USA) was 10 µM in HaCaT cells, 1 µM in WM115 cells and 5 µM in MV3 EGFP HAS3 cells. In some experiments, 1 mM Glucosamine (Sigma), 20 mM Mannose (Sigma,) and 0.5 mM 4MU (Sigma) were used. HaCaT cells were treated with hyaluronan oligosaccharides of various lengths (HA6, HA8 and HA10 (Seikagaku Kogyo Co., Tokyo, Japan) at a final concentration of 0.1, 0.2 and 0.5 mg/ml.
Transmission electron microscopy (TEM)
The EV preparations (10 µl) isolated from MV3 cell culture media were layered onto carbon-coated glow-discharged nickel grids (Agar Scientific Ltd, Sanstedt, UK). Grids were fixed in 2% paraformaldehyde for 10 min, contrasted using 2% neutral uranyl acetate (UA) for 10–15 min in the dark and embedded in 1.8% methyl cellulose (25 Ctp)/0.4% UA. Samples were imaged using a JEOL JEM 2100F transmission electron microscope (Jeol Ltd, Tokyo, Japan) operated at 200 kV.
Confocal microscopy
Fluorescent images were taken with 40 × (NA 1.3) and 63 × (NA 1.4) oil objectives on a Zeiss Axio Observer equipped with a Zeiss LSM 700 confocal module (Carl Zeiss Microimaging GmBH, Jena, Germany) and a Zeiss XL-LSM S1 incubator with temperature and CO2 control. ZEN software (Carl Zeiss Microimaging GmBH) was used for image processing and 3D rendering.
Cell lysis and Western blotting
Cells (HaCaT and WM115) grown in 6 cm dishes were treated with EVs for 24 h and lysed using NE-PER™ kit (Thermo Scientific) to isolate nuclear and cytoplasmic fractions. Protein content was determined using a Pierce BCA protein assay kit (Thermo Scientific, Epsom, UK). The proteins in the cell fractions were separated using a 10% SDS-PAGE and transferred to nitrocellulose membranes (Protran, Whatman), blocked with 5% bovine serum albumin (BSA) in Tris buffered saline containing 0.1% Tween 20 (TBST) or 5% skimmed milk-TBST for 1 h at room temperature and then incubated overnight with anti-claspin (1:2000, 5% BSA-TBST) (Proteintech, USA), anti-c-Myc (1:500, 5% BSA-TBST) (Santa Cruz, USA), anti-CD63 (1:1000, 5% BSA-TBST) (Abcam, UK), anti-Actin (1:2000, 5% milk-TBST) (Santa Cruz), anti-lamin β1 (1:2000, 5% milk-TBST) (Abcam), anti-Gli1 (1:1000, 5% BSA-TBST) (Abcam), anti-Gli2 (1:1000, 5% BSA-TBST) (Proteintech), anti-DHH (1:1000, 5% BSA-TBST) (Abcam), anti-IHH (1:1000, 5% BSA-TBST) (Abcam), anti-MAPK p42/44 (1:1000, 2% BSA-TBST) (Cell Signaling), anti-NFκB p65 (1:200, 1% BSA-TBST) (Santa Cruz), anti-EGFR (1:1000, 5% BSA-TBST) (Cell Signaling), anti-CD44 (1:2000, 1% BSA-TBST) (Chemicon, Temecula, CA), anti-EGF (1:1000, 5% BSA-TBST) (Abcam), anti-GFP (1:2000, 5% BSA-TBST) (Invitrogen) and anti-Calnexin (1:2000, 5% BSA-TBST) (Cell Signaling). Alexa-Fluor plus 555 anti-mouse or Alexa-Fluor plus 680 anti-rabbit secondary antibodies in 2% milk-TBST (1:10,000 dilution) (Thermo Scientific) were used. The membranes were washed three times for 5 min with TBST after antibody incubations and visualized using a BioRad ChemiDoc scanner.
RNA isolation and q-PCR
The cells were lysed using NucleoZOL (Macherey–Nagel, Germany) and RNA extraction was done according to the manufacturer’s instructions. For q-PCR, the primer sequences used are mentioned in supplementary Table 2. Roche FastStart Universal SYBR Master mix was used. Relative mRNA levels were calculated according to 2^(− ΔΔCT) method with GAPDH used as the housekeeping gene.
Hyaluronan assay
MV3-GFP-HAS3 cells grown in six-well plates (2 × 105 cells/well) were induced with doxycycline (0.25 µg/ml). 24 h post seeding, the cells were treated with mannose or GANT58 for 6 h. The growth medium was then collected and stored at − 20 °C. Culture medium was also collected from 1 × 105 HaCaT cells grown in 12 well plate, treated with MV3- or HAS3-EVs for 24 h. The cells were then lysed using NucleoZOL (Macherey–Nagel, Germany) and RNA extraction was carried out. In some cases, cell numbers were also counted. The HA content in the culture medium was analyzed using a previously established enzyme-linked sorbent assay [94]. The HA content was normalized to RNA concentration.
RNA sequencing and data analysis
HaCaT cells were grown in six well plates (4.5 × 105 cells/well) and treated with MV3- and HAS3-EVs for 24 h. Cells were lysed, and RNA was extracted using a high pure RNA isolation kit (Roche). The isolated RNA samples (N = 3 per group) were then sent for RNA-seq library generation (Illumina TruSeq stranded mRNA library preparation kit) and single-end sequencing using Illumina Hi seq (Illumina HiSeq 3000 instrument) at the Finnish Functional Genomics Centre, Turku center for biotechnology (Finland).
The 50 nt RNA-seq reads were quality controlled using FastQC (version 0.10.1) and trimmed with Trimmomatic (version 0.33) [95] using long TruSeq 3-SE.v2 adapters with the essential settings: ILLUMINACLIP:0:30:10, LEADING:3, TRAILING:3, MINLEN:35. Trimmed reads originating from mitochondrial DNA or ribosomal RNA, or composed of a single nucleotide, were then removed using bowtie2 (version 2.2.3) [96]. The remaining reads were aligned to the Gencode human transcriptome version 24 basic (for genome version hg38) using Tophap2 (version 2.0.13) [97] with essential non-default settings: read-gap-length 1, segment-mismatches 1, library-type fr-first strand, coverage-search, prefilter-multihits. The initial, unprocessed read counts ranged from 16.1 to 21.7 million per sample, out of which 11.6–17.1 million survived preprocessing and were aligned. All samples had very high mean Sanger 1.9 encoded sequence quality scores (> 39). Gene-wise read counts were collected in R (version 3.3.2) using GenomicAlignments:summarizeOverlaps (version 1.10.1) and, after normalization using variance stabilizing transformation (vst), they were subjected to sample-level quality control: no batch effects were identified. To identify differentially expressed genes between experimental groups, gene-wise read counts were analyzed in R (version 3.4.1) using DESeq 2 (version 1.16.1), employing Wald statistics and FC shrinkage [98].
Sample preparation for proteomics
HaCaT cells grown in 6 cm dishes were treated with (7x105 cells/dish) MV3- and HAS3-EVs for 24 h. Along with HAS3-EVs, c-Myc inhibitor was also added to the cells with appropriate control samples. The cells were lysed using RIPA buffer (with protease inhibitor cocktail) (Sigma-Aldrich). 10 µg of total protein amounts were digested using a modified FASP protocol, as previously reported [99] .
Liquid chromatography high-definition tandem mass spectrometry
The samples were analyzed in a randomized order. 300 ng of digested proteins/replicate (three technical replicates per sample) were used in nano-LC-HD-MSE analysis. The nano-LC-HD-MSE analyses were executed as described [100, 101]. Database searches were carried out against UniProtKB/Swiss-Prot reviewed human database (release 2017.10.03 with 20,239 entries) with the Ion Accounting algorithm and using the following parameters: peptide and fragment tolerance, automatic; maximum protein mass, 750 kDa; minimum fragment ions matches per protein, ≥ 7; minimum fragment ions matches per peptide, ≥ 3; minimum unique peptide matches per protein, ≥ 2; primary digest reagent as trypsin; missed cleavages allowed, 2; fixed modification, carbamidomethylation C; variable modifications, deamidation (N, Q) and oxidation of methionine (M); and false discovery rate < 4%.
Bioinformatic analyses
For differential expression analysis (defining differentially expressed proteins, DEPs), the list was limited to those quantified with a fold change (FC), FC > 1.5 and P ≤ 0.05 by analysis of variance for all comparisons. The list of up/downregulated protein changes with their corresponding unique UniProtKB/Swiss-Prot identifiers served as inputs into Ingenuity® Pathway Analysis (IPA) bioinformatics analyses (Qiagen Bioinformatics, Redwood City, CA, USA) and screened for associated canonical pathways. The identified/quantified proteins with their respective relative abundance protein ratios are reported in supplementary Table 1.
Cell proliferation
4 × 104 cells/well of HaCaT and 6 × 104 cells/well of WM115 were added to a 12 well plate and treated with EVs. During the EV treatment, cells were grown in 5% EV free serum media. The cells were then harvested and counted for a period of 4 days. During treatment with siRNAs, plasmids and inhibitors, the cells were incubated with EVs for an additional 2 days. At the end of 2 days, they were trypsinized and counted.
Type I collagen 96 well plate invasion assay
Cells were treated with EVs for 24 h and plated as 5 × 104/well on a pre-coated 96 well plate with type I collagen, as per manufacturer’s instructions (#3457-096-K, R&D Systems, MN 55413, USA). 24 h later, invading cells were stained with Calcein-AM in cell dissociation buffer and the resulting fluorescence was measured at 488 nm in a plate reader.
Cell cycle array
A high throughput ELISA based antibody array (Full moon biosystems, California, USA) was used to check the expression levels of cell cycle proteins (six replicates each) after treatment with EVs. HaCaT cells grown in 6 cm dishes were lysed using RIPA buffer, treated and the resulting lysates were added to the antibody arrays according to the manufacturer’s protocol. The protein expression levels were quantified by Full moon biosystems services.
Tissue staining
The specimens including 14 benign nevi, 15 dysplastic nevi, 12 in situ melanomas and 16 invasive melanomas were obtained from the archives of the Kuopio University Hospital under the ethical approval of Kuopio University Hospital and The Finnish National Supervisory Authority for Welfare and Health (VALVIRA). For claspin immunohistochemical staining, 5 µm thick formalin-fixed and paraffin-embedded samples were dewaxed and rehydrated. For antigen retrieval, specimens were incubated in 10 mM citrate buffer, pH 6.0 for 10 min at 120 °C in a pressure cooker and then cooled for 15 min. Endogenous peroxidase was blocked with 1% hydrogen peroxide and unspecific staining with 2% milk powder in 0.1 M phosphate buffer. Thereafter, the sections were incubated with claspin polyclonal antibody (Proteintech Antibodies, 23206-1-AP) in a humidity chamber at + 4 °C overnight. The primary antibody was used at 1:600. Thereafter, the samples were incubated with biotinylated anti-rabbit secondary antibody (1:300, Vector laboratories, Burlingame, CA, USA) at room temperature for 1 h, followed by incubation with the ABC reagent. 3, 3′-diaminobenzidine (DAB) was used as a chromogen to visualize the staining and the nuclei were counterstained with Mayer’s hematoxylin. Finally, the samples were dehydrated and mounted in Depex. The stainings were viewed with an Axio Lab.A1 microscope (Carl Zeiss, Microscopy GmbH, Jena, Germany) and the intensities of the claspin staining’s were estimated as follows: negative (0), weak (1), moderate (2) or strong (3).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 3. Supplementary Fig. 1: a Western blots showing expression of CD44, c-Myc, MAPK p42/44, EGFR, NFkB-p65, E-cad, and slug in HaCaT cells treated with MV3- and HAS3-EVs. Actin was used as the loading control. b–i Immunostaining of slug in HaCaT cells treated with MV3 and HAS3-EVs. Panels b–d, f–h show HaCaT cells immunostained for slug with MV3- and HAS3-EVs treatment. e, i shows enlarged image of the area indicated by a white box in (c, g). Effect of various concentrations of short HA oligosaccharides HA6 (j) and HA8 (k) on the binding of EVs originating from HAS3-induced MV3 cells to HaCaT cells. Data represent mean ± S.E. of four independent experiments. *P value < 0.05, **P value < 0.01, one-way ANOVA (Tukey’s test). Scale bars represent 20 µm in B, C, D, F, G, H and 5 µm in E, I. E-Cad E-cadherin. (JPEG 1856 kb)
Supplementary material 4. Supplementary Fig. 2: a Nanoparticle tracking analysis of EV levels and b size distribution secreted from uninduced (C8161-EVs) and induced (C8161-HAS3 EVs) C8161-GFP-HAS3 cells. c Effect of melanoma-derived EVs (C8161 cells) on HaCaT cells proliferation. d Effect of doxycycline on EV secretion in MV3 (P) and c MV3-GFP cells. e HA secretion analysis in MV3 cells treated with or without doxycycline, GFP and GFP-mHAS3 D216A transfection. f HA secretion analysis in MV3-GFP cells with or without doxycycline. Data represent mean ± S.E. of three independent experiments. (JPEG 1128 kb)
Supplementary material 5. Supplementary Fig. 3: a Detailed view of a differential proteomics network demonstrating the involvement of claspin, CLSPN in the cell cycle process. The fold changes with respective P values (by ANOVA) and the number of unique peptides identified in proteomics analysis are given by each node. b Western blot presenting truncated GFP-ΔHAS3 bands at ~ 35 kDa in MV3 cells; mock = pcDNA3 vector. c Effect of HAS2 + 3 siRNA cocktail on HA secretion in MV3 cells. Knocking down endogenous HAS2 and HAS3 in MV3 cells decreased HA secretion significantly while cells transiently transfected with a truncated version of GFP-HAS3 plasmid (GFP-ΔHAS3) did not affect HA secretion. d HA secretion in GFP-HAS3 transfected MV3 cells. **P value < 0.01, one-way ANOVA (Tukey’s test). (JPEG 1125 kb)
Acknowledgements
Open access funding provided by University of Eastern Finland (UEF) including Kuopio University Hospital. Sarah Wojciechowski, Jouni Härkönen, Eija Rahunen, Virpi Miettinen and Taija Hukkanen are acknowledged for their expert technical assistance. The authors thank Prof. Markku Tammi and Prof. Raija Tammi for critical reading of the manuscript. We also wish to acknowledge Dr. Rabah Soliymani and Prof. Marc Baumann from Meilahti Clinical Proteomics Facility, HiLIFE, supported by Biocenter Finland. We are thankful for the opportunity to use the facilities of the SIB Labs. This project is funded by the Academy of Finland (K.R. Grants #276426, #284520 and #312519), Jane and Aatos Erkko Foundation and grant from the Sigrid Jusélius Foundation (S.P.-S.). Personal grants to UTA from Centre for International Mobility (CIMO), Matti and Vappu Maukonen Foundation, K. Albin Johanssons stiftelse Foundation, Paavo Koistinen Foundation, Kuopio University Foundation, Northern Savo Cancer Foundation and Otto A. Malm Foundation. Project grants to Anna-Liisa Levonen from Sigrid Jusélius Foundation and Cancer Foundation Finland. This research is also supported by the EATRIS, the European Infrastructure for Translational Medicine.
Abbreviations
- HAS
Hyaluronan synthase
- HA
Hyaluronan
- HH
Hedgehog signaling
- EVs
Extracellular vesicles
- CLSPN
Claspin
- EMT
Epithelial-to-mesenchymal transition
- IHH
Indian hedgehog
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer—implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15:617–638. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 2.D’Souza-Schorey Crislyn C, Clancy JW. Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev. 2012;26:1287–1299. doi: 10.1101/gad.192351.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briscoe J, Thérond PP. The mechanisms of hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 4.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 5.Qian Z, Shen Q, Yang X, Qiu Y, Zhang W. The role of extracellular vesicles: an epigenetic view of the cancer microenvironment. Biomed Res Int. 2015 doi: 10.1155/2015/649161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poste G, Nicolson GL. Arrest and metastasis of blood-borne tumor cells are modified by fusion of plasma membrane vesicles from highly metastatic cells. Proc Natl Acad Sci. 2006;77:399–403. doi: 10.1073/pnas.77.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kultti A, Rilla K, Tiihonen R, Spicer AP, Tammi RH, Tammi MI. Hyaluronan synthesis induces microvillus-like cell surface protrusions. J Biol Chem. 2006;281:15821–15828. doi: 10.1074/jbc.M512840200. [DOI] [PubMed] [Google Scholar]
- 10.Rilla K, Oikari S, Jokela TA, Hyttinen JMT, Kärnä R, Tammi RH, Tammi MI. Hyaluronan synthase 1 (HAS1) requires higher cellular UDP-GlcNAc concentration than HAS2 and HAS3. J Biol Chem. 2013;288:5973–5983. doi: 10.1074/jbc.M112.443879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tammi RH, Passi AG, Rilla K, Karousou E, Vigetti D, Makkonen K, Tammi MI. Transcriptional and post-translational regulation of hyaluronan synthesis. FEBS J. 2011;278:1419–1428. doi: 10.1111/j.1742-4658.2011.08070.x. [DOI] [PubMed] [Google Scholar]
- 12.Deen AJ, Rilla K, Oikari S, Kärna R, Bart G, Häyrinen J, Bathina AR, Ropponen A, Makkonen K, Tammi RH, Tammi MI. Rab10-mediated endocytosis of the hyaluronan synthase HAS3 regulates hyaluronan synthesis and cell adhesion to collagen. J Biol Chem. 2014;289:8375–8389. doi: 10.1074/jbc.M114.552133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porsch H, Bernert B, Mehić M, Theocharis AD, Heldin CH, Heldin P. Efficient TGFβ-induced epithelial-mesenchymal transition depends on hyaluronan synthase HAS2. Oncogene. 2013;32:4355–4365. doi: 10.1038/onc.2012.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo Y-Z, Fang W-Y, Huang C-C, Tsai S-T, Wang Y-C, Yang C-L, Wu L-W. Hyaluronan synthase 3 mediated oncogenic action through forming inter-regulation loop with tumor necrosis factor alpha in oral cancer. Oncotarget. 2017;8:15563–15583. doi: 10.18632/oncotarget.14697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kultti A, Zhao C, Singha NC, Zimmerman S, Osgood RJ, Symons R, Jiang P, Li X, Thompson CB, Infante JR, Jacobetz MA, Tuveson DA, Frost GI, Shepard HM, Huang Z. Accumulation of extracellular hyaluronan by hyaluronan synthase 3 promotes tumor growth and modulates the pancreatic cancer microenvironment. Biomed Res Int. 2014 doi: 10.1155/2014/817613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feitelson MA, Arzumanyan A, Kulathinal RJ, Blain SW, Holcombe RF, Mahajna J, Marino M, Martinez-Chantar ML, Nawroth R, Sanchez-Garcia I, Sharma D, Saxena NK, Singh N, Vlachostergios PJ, Guo S, Honoki K, Fujii H, Georgakilas AG, Bilsland A, Amedei A, Niccolai E, Amin A, Ashraf SS, Boosani CS, Guha G, Ciriolo MR, Aquilano K, Chen S, Mohammed SI, Azmi AS, Bhakta D, Halicka D, Keith WN, Nowsheen S. Sustained proliferation in cancer: mechanisms and novel therapeutic targets. Semin Cancer Biol. 2015;35:S25–S54. doi: 10.1016/j.semcancer.2015.02.006.Sustained. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta Rev Cancer. 2002;1602:73–87. doi: 10.1016/S0304-419X(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 18.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O’Connell BC, Mateyak MK, Tam W, Kohlhuber F, Dang CV, Sedivy JM, Eick D, Vogelstein B, Kinzler KW. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci USA. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miliani de Marval PL, Macias E, Rounbehler R, Sicinski P, Kiyokawa H, Johnson DG, Conti CJ, Rodriguez-Puebla ML. Lack of cyclin-dependent kinase 4 inhibits c-Myc tumorigenic activities in epithelial tissues. Mol Cell Biol. 2004;24:7538–7547. doi: 10.1128/MCB.24.17.7538-7547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 21.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Mannava S, Grachtchouk V, Zhuang D, Soengas MS, Gudkov AV, Prochownik EV, Nikiforov MA. c-Myc depletion inhibits proliferation of human tumor cells at various stages of the cell cycle. Oncogene. 2008;27:1905–1915. doi: 10.1038/sj.onc.1210823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sar F, Lindsey-Boltz LA, Subramanian D, Croteau DL, Hutsell SQ, Griffith JD, Sancar A. Human claspin is a ring-shaped DNA-binding protein with high affinity to branched DNA structures. J Biol Chem. 2004;279:39289–39295. doi: 10.1074/jbc.M405793200. [DOI] [PubMed] [Google Scholar]
- 24.Azenha D, Lopes MC, Martins TC. Claspin functions in cell homeostasis—A link to cancer? DNA Repair. 2017;59:27–33. doi: 10.1016/j.dnarep.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Yang CC, Suzuki M, Yamakawa S, Uno S, Ishii A, Yamazaki S, Fukatsu R, Fujisawa R, Sakimura K, Tsurimoto T, Masai H. Claspin recruits Cdc7 kinase for initiation of DNA replication in human cells. Nat Commun. 2016;7:12135. doi: 10.1038/ncomms12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.i Altaba AR, Sánchez P, Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 27.McMillan R, Matsui W. Molecular pathways: the hedgehog signaling pathway in cancer. Clin Cancer Res. 2012;18:4883–4888. doi: 10.1158/1078-0432.CCR-11-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pak E, Segal RA. Hedgehog signal transduction: key players, oncogenic drivers, and cancer therapy. Dev Cell. 2016;38:333–344. doi: 10.1016/j.devcel.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pietrobono S, Santini R, Gagliardi S, Dapporto F, Colecchia D, Chiariello M, Leone C, Valoti M, Manetti F, Petricci E, Taddei M, Stecca B. Targeted inhibition of hedgehog-gli signaling by novel acylguanidine derivatives inhibits melanoma cell growth by inducing replication stress and mitotic catastrophe. Cell Death Dis. 2018 doi: 10.1038/s41419-017-0142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soleti R, Benameur T, Porro C, Panaro MA, Andriantsitohaina R, Martínez MC. Microparticles harboring sonic hedgehog promote angiogenesis through the upregulation of adhesion proteins and proangiogenic factors. Carcinogenesis. 2009;30:580–588. doi: 10.1093/carcin/bgp030. [DOI] [PubMed] [Google Scholar]
- 31.MacKie AR, Klyachko E, Thorne T, Schultz KM, Millay M, Ito A, Kamide CE, Liu T, Gupta R, Sahoo S, Misener S, Kishore R, Losordo DW. Sonic hedgehog-modified human CD34+ cells preserve cardiac function after acute myocardial infarction. Circ Res. 2012;111:312–321. doi: 10.1161/CIRCRESAHA.112.266015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takabe P, Bart G, Ropponen A, Rilla K, Tammi M, Tammi R, Pasonen-Seppänen S. Hyaluronan synthase 3 (HAS3) overexpression downregulates MV3 melanoma cell proliferation, migration and adhesion. Exp Cell Res. 2015;337:1–15. doi: 10.1016/j.yexcr.2015.07.026. [DOI] [PubMed] [Google Scholar]
- 33.Deen AJ, Arasu UT, Pasonen-Seppänen S, Hassinen A, Takabe P, Wojciechowski S, Kärnä R, Rilla K, Kellokumpu S, Tammi R, Tammi M, Oikari S. UDP-sugar substrates of HAS3 regulate its O-GlcNAcylation, intracellular traffic, extracellular shedding and correlate with melanoma progression. Cell Mol Life Sci. 2016;73:3183–3204. doi: 10.1007/s00018-016-2158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lesley J, Hascall VC, Tammi M, Hyman R. Hyaluronan binding by cell surface CD44. J Biol Chem. 2000;275:26967–26975. doi: 10.1074/jbc.M002527200. [DOI] [PubMed] [Google Scholar]
- 35.Moon S-O, Lee J-H, Kim T-J. Changes in the expression of c-Myc, RB and tyrosine-phosphorylated proteins during proliferation of NIH 3T3 cells induced by hyaluronic acid. Exp Mol Med. 1998;30:29–33. doi: 10.1038/emm.1998.4. [DOI] [PubMed] [Google Scholar]
- 36.Ohkawa T, Ueki N, Taguchi T, Shindo Y, Adachi M, Amuro Y, Hada T, Higashino K. Stimulation of hyaluronan synthesis by tumor necrosis factor-α is mediated by the p50/p65 NF-κB complex in MRC-5 myofibroblasts. Biochim Biophys Acta Mol Cell Res. 1999;1448:416–424. doi: 10.1016/S0167-4889(98)00155-4. [DOI] [PubMed] [Google Scholar]
- 37.Karjalainen JM, Tammi RH, Tammi MI, Eskelinen MJ, Ågren UM, Parkkinen JJ, Alhava EM, Kosma VM. Reduced level of CD44 and hyaluronan associated with unfavorable prognosis in clinical stage I cutaneous melanoma. Am J Pathol. 2000;157:957–965. doi: 10.1016/S0002-9440(10)64608-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang SJ, Bourguignon LYW. Hyaluronan and the interaction between CD44 and epidermal growth factor receptor in oncogenic signaling and chemotherapy resistance in head and neck cancer. Arch Otolaryngol Head Neck Surg. 2006;132:771–778. doi: 10.1001/archotol.132.7.771. [DOI] [PubMed] [Google Scholar]
- 39.Bin Park G, Ko HS, Kim D. Sorafenib controls the epithelial-mesenchymal transition of ovarian cancer cells via EGF and the CD44-HA signaling pathway in a cell type-dependent manner. Mol Med Rep. 2017;16:1826–1836. doi: 10.3892/mmr.2017.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouchard C, Staller P, Eilers M. Control of cell proliferation by Myc. Trends Cell Biol. 1998;8:202–206. doi: 10.1016/S0962-8924(98)01251-3. [DOI] [PubMed] [Google Scholar]
- 41.Cannonier SA, Gonzales CB, Ely K, Guelcher SA, Sterling JA. Hedgehog and TGFβ signaling converge on Gli2 to control bony invasion and bone destruction in oral squamous cell carcinoma. Oncotarget. 2016;7:76062–76075. doi: 10.18632/oncotarget.12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, Cleveland JL, Tansey WP, Lowe SW. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conzen SD, Gottlob K, Kandel ES, Khanduri P, Wagner AJ, O’Leary M, Hay N. Induction of cell cycle progression and acceleration of apoptosis are two separable functions of c-Myc: transrepression correlates with acceleration of apoptosis. Mol Cell Biol. 2000;20:6008–6018. doi: 10.1128/mcb.20.16.6008-6018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Cunningham M, Zhang X, Tokarz S, Laraway B, Troxell M, Sears RC. Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res. 2011;71:925–936. doi: 10.1158/0008-5472.CAN-10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bart G, Vico NO, Hassinen A, Pujol FM, Deen AJ, Ruusala A, Tammi RH, Squire A, Heldin P, Kellokumpu S, Tammi MI. Fluorescence resonance energy transfer (FRET) and proximity ligation assays reveal functionally relevant homo-and heteromeric complexes among hyaluronan synthases HAS1 HAS2, and HAS3. J Biol Chem. 2015;290:11479–11490. doi: 10.1074/jbc.M115.640581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rilla K, Siiskonen H, Spicer AP, Hyttinen JMT, Tammi MI, Tammi RH. Plasma membrane residence of hyaluronan synthase is coupled to its enzymatic activity. J Biol Chem. 2005;280:31890–31897. doi: 10.1074/jbc.M504736200. [DOI] [PubMed] [Google Scholar]
- 47.Spicer AP, Seldin MF, Olsen AS, Brown N, Wells DE, Doggett NA, Itano N, Kimata K, Inazawa J, McDonald JA. Chromosomal localization of the human and mouse hyaluronan synthase genes. Genomics. 1997;41:493–497. doi: 10.1006/geno.1997.4696. [DOI] [PubMed] [Google Scholar]
- 48.Vigetti D, Deleonibus S, Moretto P, Karousou E, Viola M, Bartolini B, Hascall VC, Tammi M, De Luca G, Passi A. Role of UDP-N-acetylglucosamine (GlcNAc) and O-GlcNacylation of hyaluronan synthase 2 in the control of chondroitin sulfate and hyaluronan synthesis. J Biol Chem. 2012;287:35544–35555. doi: 10.1074/jbc.M112.402347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siiskonen H, Poukka M, Tyynelä-Korhonen K, Sironen R, Pasonen-Seppänen S. Inverse expression of hyaluronidase 2 and hyaluronan synthases 1–3 is associated with reduced hyaluronan content in malignant cutaneous melanoma. BMC Cancer. 2013;13:1. doi: 10.1186/1471-2407-13-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qu X, Shen L, Zheng Y, Cui Y, Feng Z, Liu F, Liu J. A signal transduction pathway from TGF-β1 to SKP2 via Akt1 and c-Myc and its correlation with progression in human melanoma. J Investig Dermatol. 2014;134:159–167. doi: 10.1038/jid.2013.281. [DOI] [PubMed] [Google Scholar]
- 51.Van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 52.Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, Lucci A, Ivan C, Calin GA, Kalluri R. Cancer exosomes perform cell-independent MicroRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, García-Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, Lothe IMB, Kure EH, Hernandez J, Doussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallya K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J, Lyden D. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watt FM, Frye M, Benitah SA. MYC in mammalian epidermis: how can an oncogene stimulate differentiation? Nat Rev Cancer. 2008;8:316. doi: 10.1038/nrc2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leung JY, Ehmann GL, Giangrande PH, Nevins JR. A role for Myc in facilitating transcription activation by E2F1. Oncogene. 2008;27:4172–4179. doi: 10.1038/onc.2008.55. [DOI] [PubMed] [Google Scholar]
- 56.Hallmann A. Key elements of the retinoblastoma tumor suppressor pathway in Volvox carteri. Commun Integr Biol. 2009;2:396–399. doi: 10.4161/cib.2.5.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife. 2014 doi: 10.7554/eLife.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Müller H, Helin K. The E2F transcription factors: key regulators of cell proliferation. Biochim Biophys Acta Rev Cancer. 2000;1470:M1–M12. doi: 10.1016/S0304-419X(99)00030-X. [DOI] [PubMed] [Google Scholar]
- 60.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/S0959-437X(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 61.Bertoli C, Skotheim JM, De Bruin RAM. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verlinden L, Vanden Bempt I, Eelen G, Drijkoningen M, Verlinden I, Marchal K, De Wolf-Peeters C, Christiaens MR, Michiels L, Bouillon R, Verstuyf A. The E2F-regulated Gene Chk1 is highly expressed in triple-negative estrogen receptor-/progesterone receptor-/HER-2-breast carcinomas. Cancer Res. 2007;67:6574–6581. doi: 10.1158/0008-5472.can-06-3545. [DOI] [PubMed] [Google Scholar]
- 63.Bertoli C, Herlihy AE, Pennycook BR, Kriston-Vizi J, De Bruin RAM. Sustained E2F-dependent transcription is a key mechanism to prevent replication-stress-induced DNA damage. Cell Rep. 2016;15:1412–1422. doi: 10.1016/j.celrep.2016.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smits VAJ, Cabrera E, Freire R, Gillespie DA. Claspin -checkpoint adaptor and DNA replication factor. FEBS J. 2018;286:441–455. doi: 10.1111/febs.14594. [DOI] [PubMed] [Google Scholar]
- 65.Rovida E, Stecca B. Mitogen-activated protein kinases and hedgehog-gli signaling in cancer: a crosstalk providing therapeutic opportunities? Semin Cancer Biol. 2015;35:154–167. doi: 10.1016/j.semcancer.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 66.Mas C, Correa R, i Altaba AR, Clement V, Zbinden M, Beermann F, Stecca B, Piguet V. Melanomas require hedgehog-gli signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci. 2007;104:5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O’Reilly KE, de Miera EVS, Segura MF, Friedman E, Poliseno L, Han SW, Zhong J, Zavadil J, Pavlick A, Hernando E, Osman I. Hedgehog pathway blockade inhibits melanoma cell growth in vitro and in vivo. Pharmaceuticals. 2013;6:1429–1450. doi: 10.3390/ph6111429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Faião-Flores F, Alves-Fernandes DK, Pennacchi PC, Sandri S, Vicente ALSA, Scapulatempo-Neto C, Vazquez VL, Reis RM, Chauhan J, Goding CR, Smalley KS, Maria-Engler SS. Targeting the hedgehog transcription factors GLI1 and GLI2 restores sensitivity to vemurafenib-resistant human melanoma cells. Oncogene. 2017;36:1849–1861. doi: 10.1038/onc.2016.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pelillo C, Bergamo A, Mollica H, Bestagno M, Sava G. Colorectal cancer metastases settle in the hepatic microenvironment through α5β1 integrin. J Cell Biochem. 2015;116:2385–2396. doi: 10.1002/jcb.25189. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Hu C. WIF-1 and Ihh expression and clinical significance in patients with lung squamous cell carcinoma and adenocarcinoma. Appl Immunohistochem Mol Morphol. 2018;26:454–461. doi: 10.1097/PAI.0000000000000449. [DOI] [PubMed] [Google Scholar]
- 71.Mohelnikova-Duchonova B, Kocik M, Duchonova B, Brynychova V, Oliverius M, Hlavsa J, Honsova E, Mazanec J, Kala Z, Ojima I, Hughes DJ, Doherty JE, Murray HA, Crockard MA, Lemstrova R, Soucek P. Hedgehog pathway overexpression in pancreatic cancer is abrogated by new-generation taxoid SB-T-1216. Pharmacogenomics J. 2017;17:452–460. doi: 10.1038/tpj.2016.55. [DOI] [PubMed] [Google Scholar]
- 72.Fukaya M, Isohata N, Ohta H, Aoyagi K, Ochiya T, Saeki N, Yanagihara K, Nakanishi Y, Taniguchi H, Sakamoto H, Shimoda T, Nimura Y, Yoshida T, Sasaki H. Hedgehog signal activation in gastric pit cell and in diffuse-type gastric cancer. Gastroenterology. 2006;131:14–29. doi: 10.1053/j.gastro.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 73.Dagklis A, Demeyer S, DeBie J, Radaelli E, Pauwels D, Degryse S, Gielen O, Vicente C, Vandepoel R, Geerdens E, Uyttebroeck A, Boeckx N, De Bock CE, Cools J. Hedgehog pathway activation in T cell acute lymphoblastic leukemia predicts response to SMO and GLI1 inhibitors. Blood. 2016;128:2642–2654. doi: 10.1182/blood-2016-03-703454. [DOI] [PubMed] [Google Scholar]
- 74.Hollern DP, Yuwanita I, Andrechek ER. A mouse model with T58A mutations in Myc reduces the dependence on KRas mutations and has similarities to claudin-low human breast cancer. Oncogene. 2013;32:1296–1304. doi: 10.1038/onc.2012.142. [DOI] [PubMed] [Google Scholar]
- 75.Yang Z, Zhang C, Qi W, Cui Y, Xuan Y. GLI1 promotes cancer stemness through intracellular signaling pathway PI3K/Akt/NFκB in colorectal adenocarcinoma. Exp Cell Res. 2018;373:145–154. doi: 10.1016/j.yexcr.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 76.Kim J, Hyun J, Wang S, Lee C, Jung Y. MicroRNA-378 is involved in hedgehog-driven epithelial-to-mesenchymal transition in hepatocytes of regenerating liver article. Cell Death Dis. 2018 doi: 10.1038/s41419-018-0762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jo JH, Park SB, Park S, Lee HS, Kim C, Jung DE, Song SY. Novel gastric cancer stem cell-related marker LINGO2 is associated with cancer cell phenotype and patient outcome. Int J Mol Sci. 2019;20:1–17. doi: 10.3390/ijms20030555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tian L, Deng Z, Xu L, Yang T, Yao W, Ji L, Lu Y, Zhang J, Liu Y, Wang J. Downregulation of ASPP2 promotes gallbladder cancer metastasis and macrophage recruitment via aPKC-ι/GLI1 pathway. Cell Death Dis. 2018 doi: 10.1038/s41419-018-1145-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jian W, Bai Y, Li X, Kang J, Lei Y, Xue Y. Phosphatidylethanolamine-binding protein 4 promotes the epithelial-to-mesenchymal transition in non–small cell lung cancer cells by activating the sonic hedgehog signaling pathway. J Cell Biochem. 2018;120:5386–5395. doi: 10.1002/jcb.27817. [DOI] [PubMed] [Google Scholar]
- 80.Adolphe C, Hetherington R, Ellis T, Wainwright B. Patched1 functions as a gatekeeper by promoting cell cycle progression. Cancer Res. 2006;66:2081–2088. doi: 10.1158/0008-5472.CAN-05-2146. [DOI] [PubMed] [Google Scholar]
- 81.Maurer J, Brabletz S, Orian-Rousseau V, Mock K, Preca B-T, Brabletz T, Bajdak K, Lehmann W, Sundararajan V, Bronsert P, Stemmler MP, Matzge-Ogi A. A novel ZEB1/HAS2 positive feedback loop promotes EMT in breast cancer. Oncotarget. 2017;8:11530–11543. doi: 10.18632/oncotarget.14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu S, Cheng C. Akt signaling is sustained by a CD44 splice isoform–mediated positive feedback loop. Cancer Res. 2017;77:3791–3801. doi: 10.1158/0008-5472.CAN-16-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu J, Li Q, Kuehn MR, Litingtung Y, Vokes SA, Chiang C. Sonic hedgehog signaling directly targets hyaluronic acid synthase 2, an essential regulator of phalangeal joint patterning. Dev Biol. 2013;375:160–171. doi: 10.1016/j.ydbio.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Colombino M, Capone M, Lissia A, Cossu A, Rubino C, De Giorgi V, Massi D, Fonsatti E, Staibano S, Nappi O, Pagani E, Casula M, Manca A, Sini MC, Franco R, Botti G, Caracò C, Mozzillo N, Ascierto PA, Palmieri G. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol. 2012;30:2522–2529. doi: 10.1200/JCO.2011.41.2452. [DOI] [PubMed] [Google Scholar]
- 85.Hogan SA, Levesque MP, Cheng PF. Melanoma immunotherapy: next-generation biomarkers. Front Oncol. 2018;8:1–10. doi: 10.3389/fonc.2018.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Franklin C, Livingstone E, Roesch A, Schilling B, Schadendorf D. Immunotherapy in melanoma: recent advances and future directions. Eur J Surg Oncol. 2017;43:604–611. doi: 10.1016/j.ejso.2016.07.145. [DOI] [PubMed] [Google Scholar]
- 87.Rodríguez-Cerdeira C, Gregorio MC, López-Barcenas A, Sánchez-Blanco E, Sánchez-Blanco B, Fabbrocini G, Bardhi B, Sinani A, Guzman RA. Advances in immunotherapy for melanoma: a comprehensive review. Mediat Inflamm. 2017 doi: 10.1155/2017/3264217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Friedlander P, Wassmann K, Christenfeld AM, Fisher D, Kyi C, Kirkwood JM, Bhardwaj N, Oh WK. Whole-blood RNA transcript-based models can predict clinical response in two large independent clinical studies of patients with advanced melanoma treated with the checkpoint inhibitor, tremelimumab. J Immunother Cancer. 2017 doi: 10.1186/s40425-017-0272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sharma P, Ludwig S, Muller L, Hong CS, Kirkwood JM, Ferrone S, Whiteside TL. Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma. J Extracell Vesicles. 2018;7:1435138. doi: 10.1080/20013078.2018.1435138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Broggi MAS, Maillat L, Clement CC, Bordry N, Corthésy P, Auger A, Matter M, Hamelin R, Potin L, Demurtas D, Romano E, Harari A, Speiser DE, Santambrogio L, Swartz MA. Tumor-associated factors are enriched in lymphatic exudate compared to plasma in metastatic melanoma patients. J Exp Med. 2019;216:1091–1107. doi: 10.1084/jem.20181618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.García-Silva S, Benito-Martín A, Sánchez-Redondo S, Hernández-Barranco A, Ximénez-Embún P, Nogués L, Mazariegos MS, Brinkmann K, Amor López A, Meyer L, Rodríguez C, García-Martín C, Boskovic J, Letón R, Montero C, Robledo M, Santambrogio L, Sue Brady M, Szumera-Ciećkiewicz A, Kalinowska I, Skog J, Noerholm M, Muñoz J, Ortiz-Romero PL, Ruano Y, Rodríguez-Peralto JL, Rutkowski P, Peinado H. Use of extracellular vesicles from lymphatic drainage as surrogate markers of melanoma progression and BRAF V600E mutation. J Exp Med. 2019;216:1061–1070. doi: 10.1084/jem.20181522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koistinen V, Härkönen K, Kärnä R, Arasu UT, Oikari S, Rilla K. EMT induced by EGF and wounding activates hyaluronan synthesis machinery and EV shedding in rat primary mesothelial cells. Matrix Biol. 2017;63:38–54. doi: 10.1016/j.matbio.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 93.Arasu UT, Kärnä R, Härkönen K, Oikari S, Koistinen A, Kröger H, Qu C, Lammi MJ, Rilla K. Human mesenchymal stem cells secrete hyaluronan-coated extracellular vesicles. Matrix Biol. 2017;64:54–68. doi: 10.1016/j.matbio.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 94.Hiltunen ELJ, Anttila M, Kultti A, Ropponen K, Penttinen J, Yliskoski M, Kuronen AT, Juhola M, Tammi R, Tammi M, Kosma VM. Elevated hyaluronan concentration without hyaluronidase activation in malignant epithelial ovarian tumors. Cancer Res. 2002;62:6410–6413. doi: 10.1177/000842989902800304. [DOI] [PubMed] [Google Scholar]
- 95.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scifo E, Szwajda A, Soliymani R, Pezzini F, Bianchi M, Dapkunas A, Debski J, Uusi-Rauva K, Dadlez M, Gingras AC, Tyynelä J, Simonati A, Jalanko A, Baumann MH, Lalowski M. Proteomic analysis of the palmitoyl protein thioesterase 1 interactome in SH-SY5Y human neuroblastoma cells. J Proteomics. 2015;123:42–53. doi: 10.1016/j.jprot.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 100.Laakkonen EK, Soliymani R, Karvinen S, Kaprio J, Kujala UM, Baumann M, Sipilä S, Kovanen V, Lalowski M. Estrogenic regulation of skeletal muscle proteome: a study of premenopausal women and postmenopausal MZ cotwins discordant for hormonal therapy. Aging Cell. 2017;16:1276–1287. doi: 10.1111/acel.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lalowski MM, Björk S, Finckenberg P, Soliymani R, Tarkia M, Calza G, Blokhina D, Tulokas S, Kankainen M, Lakkisto P, Baumann M, Kankuri E, Mervaala E. Characterizing the key metabolic pathways of the neonatal mouse heart using a quantitative combinatorial omics approach. Front Physiol. 2018 doi: 10.3389/fphys.2018.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 3. Supplementary Fig. 1: a Western blots showing expression of CD44, c-Myc, MAPK p42/44, EGFR, NFkB-p65, E-cad, and slug in HaCaT cells treated with MV3- and HAS3-EVs. Actin was used as the loading control. b–i Immunostaining of slug in HaCaT cells treated with MV3 and HAS3-EVs. Panels b–d, f–h show HaCaT cells immunostained for slug with MV3- and HAS3-EVs treatment. e, i shows enlarged image of the area indicated by a white box in (c, g). Effect of various concentrations of short HA oligosaccharides HA6 (j) and HA8 (k) on the binding of EVs originating from HAS3-induced MV3 cells to HaCaT cells. Data represent mean ± S.E. of four independent experiments. *P value < 0.05, **P value < 0.01, one-way ANOVA (Tukey’s test). Scale bars represent 20 µm in B, C, D, F, G, H and 5 µm in E, I. E-Cad E-cadherin. (JPEG 1856 kb)
Supplementary material 4. Supplementary Fig. 2: a Nanoparticle tracking analysis of EV levels and b size distribution secreted from uninduced (C8161-EVs) and induced (C8161-HAS3 EVs) C8161-GFP-HAS3 cells. c Effect of melanoma-derived EVs (C8161 cells) on HaCaT cells proliferation. d Effect of doxycycline on EV secretion in MV3 (P) and c MV3-GFP cells. e HA secretion analysis in MV3 cells treated with or without doxycycline, GFP and GFP-mHAS3 D216A transfection. f HA secretion analysis in MV3-GFP cells with or without doxycycline. Data represent mean ± S.E. of three independent experiments. (JPEG 1128 kb)
Supplementary material 5. Supplementary Fig. 3: a Detailed view of a differential proteomics network demonstrating the involvement of claspin, CLSPN in the cell cycle process. The fold changes with respective P values (by ANOVA) and the number of unique peptides identified in proteomics analysis are given by each node. b Western blot presenting truncated GFP-ΔHAS3 bands at ~ 35 kDa in MV3 cells; mock = pcDNA3 vector. c Effect of HAS2 + 3 siRNA cocktail on HA secretion in MV3 cells. Knocking down endogenous HAS2 and HAS3 in MV3 cells decreased HA secretion significantly while cells transiently transfected with a truncated version of GFP-HAS3 plasmid (GFP-ΔHAS3) did not affect HA secretion. d HA secretion in GFP-HAS3 transfected MV3 cells. **P value < 0.01, one-way ANOVA (Tukey’s test). (JPEG 1125 kb)