Abstract
Breast cancer is one of the most common cancers endangering women's health all over the world. Traditional Chinese medicine is increasingly recognized as a possible complementary and alternative therapy for breast cancer. Chaihu-Shugan-San is a traditional Chinese medicine prescription, which is extensively used in clinical practice. Its therapeutic effect on breast cancer has attracted extensive attention, but its mechanism of action is still unclear. In this study, we explored the molecular mechanism of Chaihu-Shugan-San in the treatment of breast cancer by network pharmacology. The results showed that 157 active ingredients and 8074 potential drug targets were obtained in the TCMSP database according to the screening conditions. 2384 disease targets were collected in the TTD, OMIM, DrugBank, GeneCards disease database. We applied the Bisogenet plug-in in Cytoscape 3.7.1 to obtain 451 core targets. The biological process of gene ontology (GO) involves the mRNA catabolic process, RNA catabolic process, telomere organization, nucleobase-containing compound catabolic process, heterocycle catabolic process, and so on. In cellular component, cytosolic part, focal adhesion, cell-substrate adherens junction, and cell-substrate junction are highly correlated with breast cancer. In the molecular function category, most proteins were addressed to ubiquitin-like protein ligase binding, protein domain specific binding, and Nop56p-associated pre-rRNA complex. Besides, the results of the KEGG pathway analysis showed that the pathways mainly involved in apoptosis, cell cycle, transcriptional dysregulation, endocrine resistance, and viral infection. In conclusion, the treatment of breast cancer by Chaihu-Shugan-San is the result of multicomponent, multitarget, and multipathway interaction. This study provides a certain theoretical basis for the treatment of breast cancer by Chaihu-Shugan-San and has certain reference value for the development and application of new drugs.
1. Introduction
Breast cancer is one of the most common cancers endangering women's health all over the world. The GLOBO-CAN 2018 statistics show alarming results that there are 8.6 million new cases of female cancer and 4.2 million female cancer deaths worldwide. The proportion of breast cancer is 24.2% and 15.0%, respectively, ranking first in female cancer incidence and death [1]. It is predicted that, by the 2050s, the global incidence of breast cancer will reach nearly 3,200,000 new cases of breast cancer each year. These datasets reflect the high incidence of breast cancer and the urgent global need for breast cancer prevention and treatment measures [2].
Traditional Chinese medicine (TCM) has a long history in the etiology, pathogenesis, prevention, and treatment of breast cancer. According to the principle of TCM syndrome differentiation and treatment, the clinical syndrome of breast cancer is mainly “Liver-Qi” stagnation. Chaihu-Shugan-San is one of the classical prescriptions for the treatment of “Liver-Qi” stagnation. It has the effect of soothing “Liver-Qi.” It has a history of 485 years and is widely used in clinical practice [3–5]. Chaihu-Shugan-San includes seven kinds of traditional Chinese medicine such as Bupleurum chinense DC (Chinese name: Chaihu), Radix Paeoniae Alba (Chinese name: Baishao), Citrus reticulata Blanco (Chinese name Chenpi), Cyperus rotundus L (Chinese name: Xiangfu), Glycyrrhiza uralensis Fisch (Chinese name: Gancao), Citrus aurantium L (Chinese name: Zhiqiao), and Ligusticum chuanxiong Hort (Chinese name: Chuanxiong) [6]. Traditional Chinese medicine meridian tropism is one of the core components of the theory of TCM. According to the theory of TCM, the characteristics of the selective distribution of the effective components of traditional Chinese medicine in the body are basically consistent with the relationship between the viscera and viscera of the corresponding meridian tropism. Chaihu, Chenpi, Xiangfu, Chuanxiong, and Baishao in Chaihu-Shugan-San belong to the liver meridian, which can soothe the liver and regulate Qi well. In traditional Chinese medicine theory, the main location of breast cancer is in the liver and often related to the spleen and kidney in the process of disease. Chaihu-Shugan-San meridian attribution is consistent with breast cancer meridian attribution and can better play the therapeutic effect.
As a classical prescription, Chaihu-Shugan-San has been widely studied in pharmacology. Chaihu-Shugan-San has the pharmacological effects of antidepressant [7], regulation of neuro-endocrine-immune network [8, 9], anti-inflammation, antioxidative stress [10], lipid-lowering and glucose-lowering, and antifibrosis [11]. In terms of antitumor, in a study of 86 patients with stage III breast cancer, the control group was given a standardized CAF regimen (cyclophosphamide + doxorubicin + 5-fluorouracil), and the experimental group was treated with Chaihu-Shugan-San on the basis of CAF. The short-term effective rate of the observation group was significantly higher than that of the control group (81.4% vs. 58.14%); the Karnofsky improvement rate of the observation group was significantly higher than that of the control group (48.84% vs. 34.88%) [12]. As a safe complementary alternative therapy, Chaihu-Shugan-San combined with other chemotherapy regimens can improve the therapeutic effect, alleviate the myelosuppression caused by chemotherapeutic drugs, and improve the prognosis of breast cancer patients [13, 14]. However, the mechanism of Chaihu-Shugan-San in the treatment of breast cancer remains to be further explored.
Classical prescriptions are currently the preferred way to treat diseases in TCM clinic, but they lack a scientific basis to reasonably explain the mechanism of TCM prescriptions from the whole to the local level or from the cellular to the molecular level [15]. Network pharmacology is an emerging discipline based on the integration of systems biology, molecular biology, pharmacology, and a variety of network computing platforms in the context of the era of big data, which can more directly explain the association between TCM prescriptions and diseases [16]. Therefore, this study constructed a multidimensional network of “ingredient-target-pathway” through network pharmacology to explore the potential molecular mechanism of Chaihu-Shugan-San in the treatment of breast cancer and to provide a certain theoretical basis for Chaihu-Shugan-San in the treatment of breast cancer.
2. Materials and Methods
2.1. Screening of Active Components and Target Prediction in Chaihu-Shugan-San
In this study, the chemical components of the seven herbs were searched on Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, http://tcmspw.com/tcmsp.php, updated on May 31, 2014) [17]. Search keywords are Chaihu, Baishao, Chenpi, Xiangfu, Gancao, Zhiqiao, and Chuanxiong, and only oral bioavailability (OB) ≥30% and drug-likeness (DL) ≥0.18 were considered in this study. The Canonical SMILES sequence of the compound was searched in the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) [18], and this sequence was used to predict the target of the compound in the Swiss Target Prediction online database (http://www.swisstargetprediction.ch/) [19] and collect target protein gene names and UniProt ID in the prediction results.
2.2. Network Construction of Components and Targets
The chemical composition and potential targets of the above Chaihu-Shugan-San were uploaded to Cytoscape 3.7.1 [20] software to build up the component-target network. In the network, the degree centrality (DC) represents the number of nodes in the network that directly interacts with the node. The greater the degree, the more the biological functions it participates in; the betweenness centrality (BC) refers to the proportion of the number of nodes passing through the shortest path in the network, and the larger the BC is, the more influential the node is. Closeness centrality (CC) reflects the degree of proximity between nodes, and the reciprocal of the shortest path distance from one node to other nodes is CC. The closer the nodes are, the larger the CC is; the average shortest path length (ASPL) is the average of the shortest path length between all points in the network. The smaller the average path of a node, the more crucial this node is in the network.
2.3. Prediction of Breast Cancer Targets
With “breast cancer” or “malignant breast tumors” as keywords, we searched in Online Mendelian Inheritance in Man (OMIM, http://www.omim.org/, updated on May 4, 2018) [21], DrugBank (https://www.drugbank.ca/, version 5.1.6, updated on Apr 22, 2020) [22], Therapeutic Target Database (TTD, http://db.idrblab.net/ttd/, updated on Nov 11, 2019) [23], and GeneCards (https://www.genecards.org/, version 4.14.0) [24] to collect breast cancer-related targets. In the GeneCards database, the higher the score value is, the closer the relationship between the target and disease is, and the score value greater than the median is used as the screening condition to extract the key target. The above retrieval results were combined to remove duplicates and serve as the prediction target library of breast cancer.
2.4. Protein-Protein Interaction (PPI) Network Construction and Selection of Core Targets
The BisoGenet plug-in in Cytoscape 3.7.1 draws the PPI network and maps the Chaihu-Shugan decoction component targets and breast cancer-related disease targets into the protein interaction relationship network, using Cytoscape 3.7.1 merge two protein interaction networks, to extract the intersection of the network. Based on the intersection network, the CytoNCA plug-in [25] is used to screen out the nodes whose degree centrality (DC) is greater than 2 times the median of all nodes. After multiple screening, the core PPI network is finally obtained.
2.5. GO Functional Enrichment Analysis and KEGG Pathway Enrichment Analysis
The core target of PPI network selected above was imported into Metascape (https://metascape.org/gp/index.html, updated on March 20, 2020) [26] database for KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis and GO (Gene Ontology) biological process enrichment analysis. Parameter is set to min overlap >3, p value cutoff <0.01, and min enrichment >1.5. Taking p value as parameter and sorting from small to large as screening condition, KEGG pathway and GO biological process of the top 20 eligible were selected and uploaded to OmicShare (http://www.omicshare.com/tools) platform for data visualization.
2.6. Constructing PPI Network of Ingredient-Disease-KEGG Pathway
The top 20 KEGG pathways, Chaihu-Shugan-San active ingredients, and disease common targets were uploaded to Cytoscape 3.7.1 software to obtain the multidimensional network diagram of component-disease -KEGG pathway.
3. Results
3.1. Active Ingredient and Target of Chaihu-Shugan-San
OB ≥30%, DL ≥0.18 as the screening conditions, after searching TCMSP database, ChaiHu-ShuGan-San obtained a total of 157 chemical components, 13 compounds from Baishao, 17 compounds from Chaihu, 5 compounds from Chenpi, 7 compounds from Chuanxiong, 5 compounds from Zhiqiao, 92 qualified compounds from Gancao, and 18 compounds from Xiangfu (as shown in Table S1 in Supplementary Materials). 8074 targets were obtained by inputting 158 chemical components into Swiss Target Prediction online database.
3.2. Compound-Target Network Construction
The compound-target network consists of 945 nodes and 8200 edges. 29 of 157 compounds were not found in the database and not involved in the network construction (Figure 1). In this network, the average degree value is 15.647, and most of the proteins share common ligands with other proteins, which reflects the mechanism of the joint action between multicomponents and multitargets of Chaihu-Shugan-San, and conform to the characteristics of the traditional Chinese formula. Table 1 shows the detailed topological parameters of the top 10 compounds with high DC.
Figure 1.
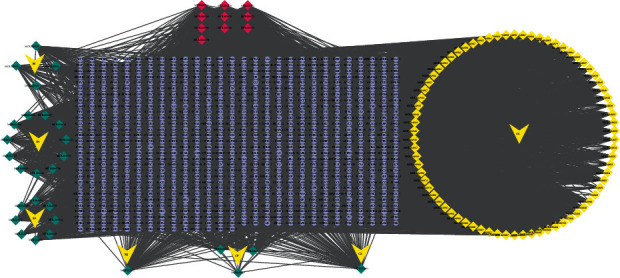
Compound-target network: green diamond nodes represent compounds, red diamond nodes represent common compounds, yellow v nodes represent herb names, and purple circular nodes represent corresponding potential targets of compounds.
Table 1.
Topological parameter of top 10 compounds.
| ID | Molecule name | DC | BC | CC | ASPL |
|---|---|---|---|---|---|
| MOL000422 | Kaempferol | 317 | 0.011 | 0.377 | 2.651 |
| MOL000354 | Isorhamnetin | 303 | 0.008 | 0.373 | 2.681 |
| MOL000359 | Sitosterol | 233 | 0.005 | 0.360 | 2.774 |
| MOL004328 | Naringenin | 199 | 0.058 | 0.397 | 2.520 |
| MOL000358 | Beta-sitosterol | 139 | 0.004 | 0.357 | 2.802 |
| MOL000098 | Quercetin | 116 | 0.009 | 0.377 | 2.654 |
| MOL004609 | Areapillin | 101 | 0.008 | 0.372 | 2.690 |
| MOL003044 | Chrysoeriol | 101 | 0.008 | 0.371 | 2.690 |
| MOL000006 | Luteolin | 101 | 0.008 | 0.371 | 2.690 |
| MOL004071 | Hyndarin | 101 | 0.063 | 0.374 | 2.677 |
3.3. Screening of Breast Cancer Targets
Breast cancer or malignant breast tumors were used as keywords to search in TTD, OMIM, DrugBank, and GeneCards databases. 37 disease targets were obtained from TTD database, 1163 disease targets were screened from OMIM database, 202 disease targets were screened from Drugbank database, and 1286 disease targets with score >13.96 were obtained from GeneCards database. The duplicates were deleted after merging, and 2,384 breast cancer-related targets were finally obtained.
3.4. Construction of PPI Network of Chaihu-Shugan-San and Disease Targets
To further explore the pharmacological mechanism of Chaihu-Shugan-San on breast cancer, Chaihu-Shugan-San and breast cancer protein were input into the BisoGenet plug-in of Cytoscape 7.2.1 software for merging. CytoNCA plug-in performs topological analysis and takes 2 times of the average degree value as the screening condition. In the first screening, a network composed of 2,728 nodes and 109,005 edges was obtained by the median DC >46. Finally, a PPI network with 451 nodes and 17,140 edges was constructed by further screening with the median DC >156. The process is shown in Figure 2. Topological parameters of the top 10 targets with high DC are shown in Table 2, and other detailed results are shown in Table S2.
Figure 2.
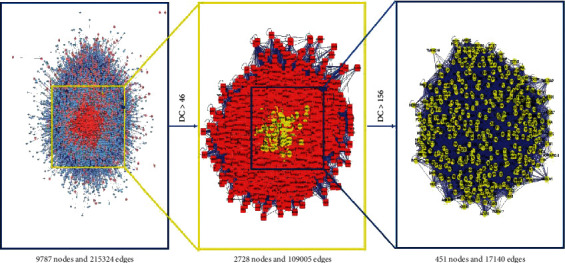
Network topology analysis of PPI.
Table 2.
Topological parameter of top 10 core targets.
| Target | DC | BC | CC | ASPL |
|---|---|---|---|---|
| HDAC1 | 1976 | 0.075 | 0.534 | 1.874 |
| HTT | 1695 | 0.094 | 0.526 | 1.900 |
| AKT1 | 1136 | 0.031 | 0.495 | 2.019 |
| HDGF | 1095 | 0.021 | 0.499 | 2.002 |
| RC3H1 | 930 | 0.026 | 0.501 | 1.996 |
| CBX8 | 836 | 0.013 | 0.488 | 2.050 |
| HDAC2 | 813 | 0.015 | 0.493 | 2.029 |
| SUMO1 | 810 | 0.021 | 0.500 | 1.999 |
| RPSA | 761 | 0.013 | 0.489 | 2.047 |
| RPLP0 | 736 | 0.011 | 0.481 | 2.081 |
3.5. GO Biological Process and Enrichment Analysis of KEGG Pathway
GO biological process consists of molecular function (MF), biological process (BP), and cellular component (CC) to interpret antitumor biological processes at key targets. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis studies the key target of antitumor signaling pathways. The results of GO enrichment analysis showed that there were 2000 biological processes, 274 cell components, and 564 molecular functions. KEGG enrichment has 154 signaling pathways. According to the ranking of p values, the top 20 were selected to plot the bubble chart (Figure 3). The left side of each chart is the top enriched name. The color of bubbles from blue to red represents the p value from large to small. The larger the bubbles, the larger the gene count of the pathway. The horizontal axis represents the ratio of the pathway genes to the total input genes. The top 20 signal pathways of KEGG enrichment are shown in Table S3.
Figure 3.
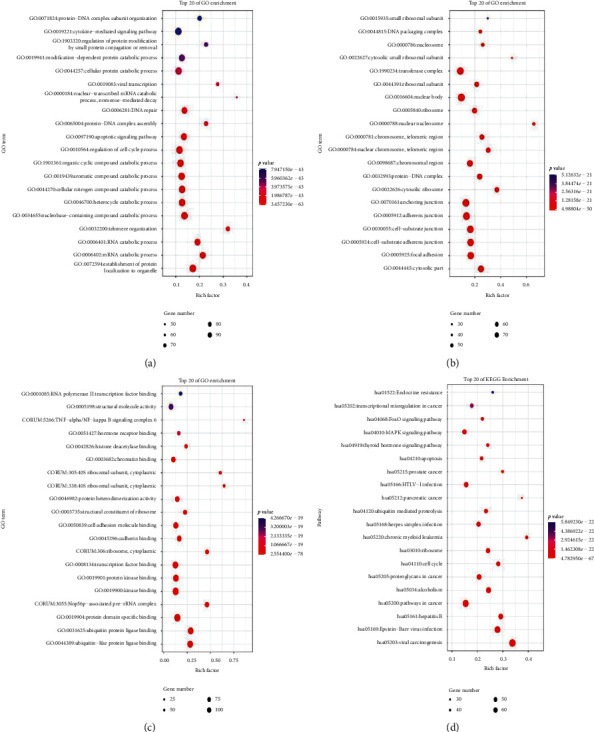
GO function enrichment analysis and enrichment analysis of KEGG signaling pathway (top 20). (a) BP. (b) CC. (c) MF. (d) KEGG.
3.6. The Multidimensional Network of “Component-Target Disease-KEGG Signaling Pathway” Was Constructed
Combining the component-target network and the first 20 KEGG signaling pathway targets, a multidimensional network of “component-disease target-KEGG signaling pathway” was obtained by Cytoscape 7.2.1 software (as shown in Figure 4). The results showed that the effective components of Chaihu-Shugan-San could treat breast cancer by multitarget and multisignal pathways.
Figure 4.
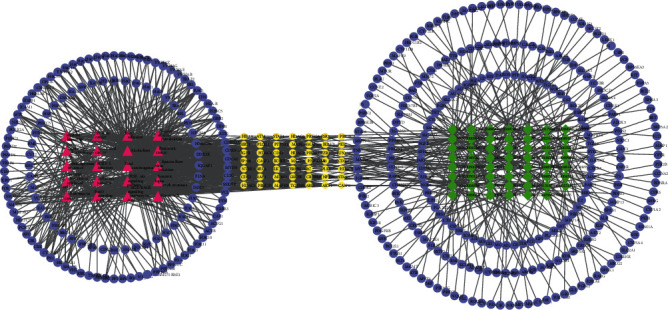
Component-disease target-KEGG signaling pathway. Green represents active ingredients of drugs; purple represents targets; yellow represents common targets; and red represents signaling pathways.
4. Discussion
Traditional Chinese medicine compound acts on diseases through multimolecule, multitarget, and multipathway and plays a certain therapeutic effect. Network pharmacology is a science based on the macroconnection under the background of the big data era. It systematically analyzes the molecular mechanism of action from all levels, which is consistent with the holistic view of TCM and the thought of syndrome differentiation and treatment. Chaihu-Shugan-San prescription, in which Chaihu is the monarch drug, is good at soothing the “Liver-Qi.” Xiangfu and Chuanxiong are the minister drugs, which can relieve “Liver-Qi.” Chenpi and Zhiqiao, regulating “Qi” stagnation and Baishao, nourishing “Blood” and softening the “Liver,” are adjuvants. Gancao is used as a guide medicine to reconcile various drugs. The combination of various drugs can regulate “Liver-Qi” and smooth “Qi.” Chaihu-Shugan-San has a history of more than 480 years. The basic compatibility of clinical medication is Chenpi 6 g, Chaihu 6 g, Chuanxiong 4.5 g, Xiangfu 4.5 g, Zhiqiao 4.5 g, Shaoyao 4.5 g, and Gancao 1.5 g, which is added or subtracted according to the actual situation of patients. The official preparation method is to add 250 ml of water to the herbs and boil them for 30 min. Su [27] and his team identified 33 chemical constituents in Chaihu-Shugan-San. Among them, gallic acid (source: Shaoyao), oxidized paeoniflorin (source: Shaoyao), paeoniflorin (source: Shaoyao), paeoniflorin (source: Shaoyao), glycyrrhizin (source: Gancao), naringin (source: Chenpi, Zhiqiao), hesperidin (source: Chenpi, Zhiqiao), and ferulic acid (source: Chuanxiong) had higher contents, all above 1000 mg/g [28]. Although some studies have comprehensively elucidated the treatment of depression [6, 29], nonalcoholic fatty liver disease [30, 31] and functional dyspepsia [5] by Chaihu-Shugan-San, no studies have comprehensively elucidated the mechanism of Chaihu-Shugan-San in the treatment of breast cancer. Therefore, with the aid of network pharmacology, this study analyzed the specific molecular mechanism of Chaihu-Shugan-San in the treatment of breast cancer from a microscopic perspective.
The results of network analysis showed that the active ingredients in Chaihu-Shugan-San mainly included beta-sitosterol, kaempferol, quercetin, naringenin, isorhamnetin, and nobiletin. Beta-sitosterol can promote the apoptosis of breast cancer cells by activating the Fas signaling pathway and caspase-8 activity [32] and is expected to be an orphan nutrition drug against cancer [33]. Kaempferol has shown a good affinity for PAK4 in molecular docking and is considered to be a potential inhibitor in triple-negative breast cancer [34], and kaempferol can prevent G2/M phase of the cell cycle by downregulating CDK1 in human breast cancer MDA-MB-453 cells [35], and blocking RhoA and Rac1 signaling pathways to inhibit breast cancer cell migration and invasion [36] is a powerful antioxidant inducer and can inhibit oncogene transformation and induce cancer cell apoptosis and DNA damage. Quercetin, naringenin, and isorhamnetin, such as flavonoids, can prevent breast cancer cell migration through inflammatory and apoptotic cell signaling [37, 38], and quercetin can induce autophagy by inhibiting the Akt-mTOR pathway [39].
The PPI network showed that the active components in Chaihu-Shugan-San may function through the core targets such as histone deacetylase 1 (HDAC1), huntingtin (HTT), RAC-alpha serine/threonine-protein kinase (AKT1), hepatoma-derived growth factor (HDGF), roquin-1 (RC3H1), chromobox protein homolog 8 (CBX8), histone deacetylase 2 (HDAC2), small ubiquitin-related modifier 1 (SUMO1), 40 S ribosomal protein SA (RPSA), and 60 S acidic ribosomal protein P0(RPLP0). HDAC1 plays an important role in transcriptional regulation and cell cycle progression [40]. HDAC1 can promote the proliferation and migration of breast cancer cells by activating the Snail/IL-8 signaling pathway [41]. Downregulation of HTT transcription and protein levels is a key factor in poor prognosis and metastasis development of breast cancer [42]. AKT1 is involved in the regulation of many tumor processes, including tumor proliferation, cell survival, metabolism, growth, and angiogenesis. The mutation frequency of AKT1 in Chinese breast cancer patients is 3.2%, and it is considered to be a sensitive target for the treatment of breast cancer. A study involving 313 Chinese breast cancer patients found that the mutation frequency of AKT1 in Chinese breast cancer patients was 3.2%, and it is considered a sensitive target for the treatment of breast cancer [43]. HDAC2 is a poor prognostic factor in patients receiving anthracycline therapy and is positively correlated with breast cancer metastasis, progression, increased Ki-67, multidrug resistance protein, and negatively correlated with overall survival of patients [44]. The occurrence and development of breast cancer are closely related to the core proteins, which fully prove that the treatment of breast cancer by Chaihu-Shugan-San is the result of multimolecular, multitarget, and multipathway interaction.
The biological process of Gene Ontology (GO) involves the mRNA catabolic process, RNA catabolic process, telomere organization, nucleobase-containing compound catabolic process, heterocycle catabolic process, and so on. In cellular component, cytosolic part, focal adhesion, cell-substrate adherens junction, and cell-substrate junction are highly correlated with breast cancer. In the molecular function category, most proteins were addressed to ubiquitin-like protein ligase binding, protein domain specific binding, and Nop56p-associated pre-rRNA complex. In addition, the results of KEGG pathway analysis showed that the pathways mainly involved in apoptosis, cell cycle, transcriptional dysregulation, endocrine resistance, and viral infection. Estrogen receptor (ER) signal transduction pathway plays a central role in the development of breast cancer. ER can not only regulate the expression of certain genes through its mediated signal transduction pathway but also has extensive connections with many other signal transduction pathways, forming a signal transduction regulatory network [45]. ER binds to receptor proteins in the nucleus, and the receptor is activated. Activated ER-α and ER-β form homodimers or heterodimers. Some coregulators form complexes with dimers. The complexes bind to ER response elements to initiate transcription, thereby regulating the function of target genes, leading to abnormal cell proliferation and differentiation, and ultimately leading to tumorigenesis [46]. The increased mutation rate of ER-α in precancerous lesions of breast cancer affects the junction between ER-α zinc finger region and ligand binding domain, resulting in high sensitivity of the body to estrogen. Under the action of low levels of hormones, ER-α is highly bound to TNF-2 co-activator, which leads to the occurrence of tumors. For ER receptor-positive breast cancer patients, quantitative expression of ER receptor is an independent imaging factor to evaluate their prognosis, recurrence, and metastasis [47]. Abnormal activation of MAPK signal transduction pathway can lead to cell loss of apoptosis and differentiation ability, promote malignant transformation, abnormal proliferation, produce tumors, and further promote the proliferation of tumor cells. Therefore, inhibitors of some key kinases in the MAPK signaling pathway have become a hotspot in the treatment of breast cancer in recent years. Studies have found that Kruppel-like factor 4 [48] and pre-mRNA processing factor 4 [49] affect the growth, migration, and apoptosis of breast cancer cells through MAPK and are expected to become new targets for the treatment of breast cancer. Studies found that the activation or loss of FOXO function can inhibit the growth and metastasis of breast tumors [50], and the dysregulation of FOXO transcription factors has also become a key molecule in the endocrine resistance mechanism [51]. More and more attention has been paid to the relationship between viral infection and breast cancer. In particular, human papilloma virus (HPV) has a strong cause-and-effect relationship with breast cancer. Many studies have found that different HPV genotypes are associated with the prevalence of breast cancer and the nuclear prognosis. The relationship between viral infection and breast cancer has been paid more and more attention [52–54]. The relationship between Epstein–Barr Virus (EBV) and breast cancer has also been extensively studied, but the current evidence is less and more controversial [55]. It is proved again that Chaihu-Shugan-San treatment of breast cancer is through a combination of multiple biological pathways and multiple signaling pathways, but this multifeature is not only found in a single disease. Chaihu-Shugan-San is mainly involved in the regulation of neurotransmitters, regulation of inflammatory mediators of TRP channels, calcium signaling pathways, cyclic adenosine monophosphate signaling pathways, and neuroactive ligand-receptor interactions to play an antidepressant role [56]. Chaihu-Shugan-San can improve cognitive impairment in Alzheimer's disease through multitarget action, and its effect is verified by biological experiments [57]. These all embody the principle of “treating different diseases with the same treatment” in TCM.
Acknowledgments
This work was supported by grants from the China National Natural Science Foundation (Grant no. 81973640).
Data Availability
All data generated or analyzed during this study are included in this paper.
Disclosure
Kunmin Xiao and Kexin Li are the first authors.
Conflicts of Interest
All authors state that they have no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Shijie Zhu conceived and designed the experiments. Kunmin Xiao and Kexin Li performed the experiments and wrote the manuscript. Sidan Long and Chenfan Kong contributed to analysis tools.
Supplementary Materials
Table S1: active ingredients of Chaihu-Shugan-San. Table S2: topological parameters of Chaihu-Shugan-San targets. Table S3: the top 20 signal pathways of KEGG enrichment.
References
- 1.Bray F., Ferlay J., Soerjomataram I., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Benson J. R., Jatoi I. The global breast cancer burden. Future Oncology. 2012;8(6):697–702. doi: 10.2217/fon.12.61. [DOI] [PubMed] [Google Scholar]
- 3.Yang N., Jiang X., Qiu X., Hu Z., Wang L., Song M. Modified Chaihu shugan powder for functional dyspepsia: meta-analysis for randomized controlled trial. Evidence-Based Complementary and Alternative Medicine. 2013;2013:10. doi: 10.1155/2013/791724.791724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Fan R., Huang X. Meta-analysis of the clinical effectiveness of traditional Chinese medicine formula Chaihu-Shugan-San in depression. Journal of Ethnopharmacology. 2012;141(2):571–577. doi: 10.1016/j.jep.2011.08.079. [DOI] [PubMed] [Google Scholar]
- 5.Qin F., Liu J. Y., Yuan J. H. Chaihu-Shugan-San, an oriental herbal preparation, for the treatment of chronic gastritis: a meta-analysis of randomized controlled trials. Journal of Ethnopharmacology. 2013;146(2):433–439. doi: 10.1016/j.jep.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 6.He Z., Fan R., Zhang C., et al. Chaihu-Shugan-San reinforces CYP3A4 expression via pregnane X receptor in depressive treatment of liver-Qi stagnation syndrome. Evidence-Based Complementary and Alternative Medicine. 2019;2019:11. doi: 10.1155/2019/9781675.9781675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia K. K., Pan S. M., Ding H., et al. Chaihu-shugan san inhibits inflammatory response to improve insulin signaling in liver and prefrontal cortex of CUMS rats with glucose intolerance. Biomedicine & Pharmacotherapy. 2018;103:1415–1428. doi: 10.1016/j.biopha.2018.04.171. [DOI] [PubMed] [Google Scholar]
- 8.Huang X., Guo Y., Huang W. H., et al. Searching the cytochrome P450 enzymes for the metabolism of meranzin hydrate: a prospective antidepressant originating from Chaihu-Shugan-San. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0113819.e113819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su Z. H., Jia H. M., Zhang H. W., Feng Y. F., An L., Zou Z. M. Hippocampus and serum metabolomic studies to explore the regulation of Chaihu-Shu-Gan-San on metabolic network disturbances of rats exposed to chronic variable stress. Molecular BioSystems. 2014;10(3):549–561. doi: 10.1039/c3mb70377k. [DOI] [PubMed] [Google Scholar]
- 10.Liang Y., Zhang Y., Deng Y., et al. Chaihu-Shugan-San decoction modulates intestinal microbe dysbiosis and alleviates chronic metabolic inflammation in NAFLD rats via the NLRP3 inflammasome pathway. Evidence-Based Complementary and Alternative Medicine. 2018;2018:11. doi: 10.1155/2018/9390786.9390786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Q. H., Xu Y. J., Feng G. H., et al. p38 MAPK signal pathway involved in anti-inflammatory effect of Chaihu-Shugan-San and Shen-ling-Bai-zhu-San on hepatocyte in non-alcoholic steatohepatitis rats. African Journal of Traditional, Complementary and Alternative Medicines. 2013;11(1):213–221. doi: 10.4314/ajtcam.v11i1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L., Ni Y., Yin F. Effect of Chaihu Shugan powder combined with CAF in the treatment of stage III breast cancer and its effect on serum MMPs, estrogen level and tumor markers. Journal of Chinese Medicinal Materials. 2020;43(1):220–221. [Google Scholar]
- 13.Quan F., Shi B., Yu M. Clinical effects of Chaihu Shugan powder combined with Paclitaxel and cyclophosphamide in treatment of breast cancer. Grassroots Medical Forum. 2019;23(26):3703–3704. [Google Scholar]
- 14.He W. Advanced breast cancer treated with Bupleurum powder for soothing the liver combined with chemotherapy randomized parallel group study. Journal of Practical Traditional Chinese Internal Medicine. 2014;28(8):83–86. [Google Scholar]
- 15.LI S., Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chinese Journal of Natural Medicines. 2014;11(2):110–120. doi: 10.3724/sp.j.1009.2013.00110. [DOI] [PubMed] [Google Scholar]
- 16.Hopkins A. L. Network pharmacology: the next paradigm in drug discovery. Nature Chemical Biology. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 17.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6(1) doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihlenfeldt W.-D. A virtual file system for the PubChem chemical structure and bioassay database. Chemistry Central Journal. 2008;2(1) doi: 10.1186/1752-153x-2-s1-p26. [DOI] [Google Scholar]
- 19.Gfeller D., Michielin O., Zoete V. Shaping the interaction landscape of bioactive molecules. Bioinformatics. 2013;29(23):3073–3079. doi: 10.1093/bioinformatics/btt540. [DOI] [PubMed] [Google Scholar]
- 20.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamosh A., Scott A. F., Amberger J. S., Bocchini C. A., McKusick V. A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Research. 2004;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wishart D. S., Feunang Y. D., Guo A. C., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Research. 2018;46(1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., Zhang S., Li F., et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Research. 2020;48(1):D1031–D1041. doi: 10.1093/nar/gkz981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stelzer G., Rosen N., Plaschkes I., et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Current Protocols in Bioinformatics. 2016;54(1) doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 25.Tang Y., Li M., Wang J., Pan Y., Wu F. X. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y., Zhou B., Pache L., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications. 2019;10(1):p. 1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su Z. H., Zou G. A., Preiss A., Zhang H. W., Zou Z. M. Online identification of the antioxidant constituents of traditional Chinese medicine formula Chaihu-Shu-Gan-San by LC-LTQ-Orbitrap mass spectrometry and microplate spectrophotometer. Journal of Pharmaceutical and Biomedical Analysis. 2010;53(3):454–461. doi: 10.1016/j.jpba.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 28.Ni X., Cao M., Wu Z., Li L. Research progress on chemical components and pharmacological effects of Chaihu Shugan powder. Shanghai Journal of Traditional Chinese Medicine. 2017;51(9):109–113. [Google Scholar]
- 29.Liu Q., Sun N. N., Wu Z. Z., Fan D. H., Cao M. Q. Chaihu-Shugan-San exerts an antidepressive effect by downregulating miR-124 and releasing inhibition of the MAPK14 and Gria3 signaling pathways. Neural Regeneration Research. 2018;13(5):837–845. doi: 10.4103/1673-5374.232478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nie H., Deng Y., Zheng C., et al. A network pharmacology-based approach to explore the effects of Chaihu Shugan powder on a non-alcoholic fatty liver rat model through nuclear receptors. Journal of Cellular and Molecular Medicine. 2020;24(9):5168–5184. doi: 10.1111/jcmm.15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang W. N., Li D., Jiang T., et al. Protective effects of chaihu shugan san () on nonalcoholic fatty liver disease in rats with insulin resistance. Chinese Journal of Integrative Medicine. 2018;24(2):125–132. doi: 10.1007/s11655-016-2252-4. [DOI] [PubMed] [Google Scholar]
- 32.Awad A. B., Chinnam M., Fink C. S., Bradford P. G. Beta-Sitosterol activates Fas signaling in human breast cancer cells. Phytomedicine. 2007;14(11):747–754. doi: 10.1016/j.phymed.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Sayeed M. S. B., Ameen S. S. Beta-sitosterol: a promising but orphan nutraceutical to fight against cancer. Nutrition and Cancer. 2015;67(8):1216–1222. doi: 10.1080/01635581.2015.1087042. [DOI] [PubMed] [Google Scholar]
- 34.Arowosegbe M. A., Amusan O. T., Adeola S. A., et al. Kaempferol as a potential PAK4 inhibitor in triple negative breast cancer: extra precision glide docking and free energy calculation. Current Drug Discovery Technologies. 2019;16 doi: 10.2174/1570163816666190823135948. [DOI] [PubMed] [Google Scholar]
- 35.Zhu L., Xue L. Kaempferol suppresses proliferation and induces cell cycle arrest, apoptosis, and DNA damage in breast cancer cells. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics. 2019;27(6):629–634. doi: 10.3727/096504018x15228018559434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S., Yan T., Deng R., et al. Low dose of kaempferol suppresses the migration and invasion of triple-negative breast cancer cells by downregulating the activities of RhoA and Rac1. Onco Targets and Therapy. 2017;10:4809–4819. doi: 10.2147/ott.s140886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Z., Jin G., Ge Y., Guo Z. Naringenin inhibits migration of breast cancer cells via inflammatory and apoptosis cell signaling pathways. Inflammopharmacology. 2019;27(5):1021–1036. doi: 10.1007/s10787-018-00556-3. [DOI] [PubMed] [Google Scholar]
- 38.Hu J., Zhang Y., Jiang X., et al. ROS-mediated activation and mitochondrial translocation of CaMKII contributes to Drp1-dependent mitochondrial fission and apoptosis in triple-negative breast cancer cells by isorhamnetin and chloroquine. Journal of Experimental & Clinical Cancer Research. 2019;38(1):p. 225. doi: 10.1186/s13046-019-1201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia L., Huang S., Yin X., Zan Y., Guo Y., Han L. Quercetin suppresses the mobility of breast cancer by suppressing glycolysis through Akt-mTOR pathway mediated autophagy induction. Life Sciences. 2018;208:123–130. doi: 10.1016/j.lfs.2018.07.027. [DOI] [PubMed] [Google Scholar]
- 40.Zhang N., Zhang H., Liu Y., et al. SREBP1, targeted by miR-18a-5p, modulates epithelial-mesenchymal transition in breast cancer via forming a co-repressor complex with Snail and HDAC1/2. Cell Death & Differentiation. 2019;26(5):843–859. doi: 10.1038/s41418-018-0158-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang Z., Ding S., Huang H., et al. HDAC1 triggers the proliferation and migration of breast cancer cells via upregulation of interleukin-8. Biological Chemistry. 2017;398(12):1347–1356. doi: 10.1515/hsz-2017-0155. [DOI] [PubMed] [Google Scholar]
- 42.Thion M. S., McGuire J. R., Sousa C. M., et al. Unraveling the role of Huntingtin in breast cancer metastasis. Journal of the National Cancer Institute. 2015;107(10):djv208. doi: 10.1093/jnci/djv208. [DOI] [PubMed] [Google Scholar]
- 43.Li G., Guo X., Chen M., et al. Prevalence and spectrum of AKT1, PIK3CA, PTEN and TP53 somatic mutations in Chinese breast cancer patients. PLoS One. 2018;13(9) doi: 10.1371/journal.pone.0203495.e0203495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao H., Yu Z., Zhao L., et al. HDAC2 overexpression is a poor prognostic factor of breast cancer patients with increased multidrug resistance-associated protein expression who received anthracyclines therapy. Japanese Journal of Clinical Oncology. 2016;46(10):893–902. doi: 10.1093/jjco/hyw096. [DOI] [PubMed] [Google Scholar]
- 45.Hayashi S. I., Yamaguchi Y. Estrogen signaling and prediction of endocrine therapy. Cancer Chemotherapy & Pharmacology. 2005;56(1):27–31. doi: 10.1007/s00280-005-0096-2. [DOI] [PubMed] [Google Scholar]
- 46.Knox A. J., Meegan M. J., Lloyd D. G. Estrogen receptors: molecular interactions, virtual screening and future prospects. Current Topics in Medicinal Chemistry. 2006;6(3):217–243. doi: 10.2174/156802606776173438. [DOI] [PubMed] [Google Scholar]
- 47.Dieci M. V., Piacentini F., Dominici M., et al. Quantitative expression of estrogen receptor on relapse biopsy for ER-positive breast cancer: prognostic impact. Anticancer research. 2014;34(7):3657–3662. [PubMed] [Google Scholar]
- 48.Jia Y., Zhou J., Luo X., et al. KLF4 overcomes tamoxifen resistance by suppressing MAPK signaling pathway and predicts good prognosis in breast cancer. Cellular Signalling. 2018;42:165–175. doi: 10.1016/j.cellsig.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 49.Park S., Han S. H., Kim H. G., et al. PRPF4 is a novel therapeutic target for the treatment of breast cancer by influencing growth, migration, invasion, and apoptosis of breast cancer cells via p38 MAPK signaling pathway. Molecular and Cellular Probes. 2019;47 doi: 10.1016/j.mcp.2019.101440.101440 [DOI] [PubMed] [Google Scholar]
- 50.Hornsveld M., Smits L. M., Meerlo M., et al. FOXO transcription factors both suppress and support breast cancer progression. Progression. Cancer Research. 2018;78(9):2356–2369. doi: 10.1158/0008-5472.can-17-2511. [DOI] [PubMed] [Google Scholar]
- 51.Bullock M. FOXO factors and breast cancer: outfoxing endocrine resistance. Endocrine-Related Cancer. 2016;23(2):R113–R130. doi: 10.1530/erc-15-0461. [DOI] [PubMed] [Google Scholar]
- 52.Petry K. U. HPV and cervical cancer. Scandinavian Journal of Clinical and Laboratory Investigation. 2014;74:59–62. doi: 10.3109/00365513.2014.936683. [DOI] [PubMed] [Google Scholar]
- 53.Kan C. Y., Iacopetta B. J., Lawson J. S., Whitaker N. J. Identification of human papillomavirus DNA gene sequences in human breast cancer. British Journal of Cancer. 2005;93(8):946–948. doi: 10.1038/sj.bjc.6602778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khodabandehlou N., Mostafaei S., Etemadi A., et al. Human papilloma virus and breast cancer: the role of inflammation and viral expressed proteins. BMC Cancer. 2019;19(1):61. doi: 10.1186/s12885-019-5286-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glaser S. L., Hsu J. L., Gulley M. L. Epstein-barr virus and breast cancer: state of the evidence for viral carcinogenesis. Cancer Epidemiology, Biomarkers & Prevention. 2004;13(5):688–697. [PubMed] [Google Scholar]
- 56.Liu Y. Y., Hu D., Fan Q. Q., et al. Study on mechanism of chaihu shugan powder () for treating depression based on network pharmacology. Chinese Journal of Integrative Medicine. 2019 doi: 10.1007/s11655-019-3172-x. [DOI] [PubMed] [Google Scholar]
- 57.Zeng Q., Li L., Siu W., et al. A combined molecular biology and network pharmacology approach to investigate the multi-target mechanisms of Chaihu Shugan San on Alzheimer’s disease. Biomedicine & Pharmacotherapy. 2019;120 doi: 10.1016/j.biopha.2019.109370.109370 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: active ingredients of Chaihu-Shugan-San. Table S2: topological parameters of Chaihu-Shugan-San targets. Table S3: the top 20 signal pathways of KEGG enrichment.
Data Availability Statement
All data generated or analyzed during this study are included in this paper.


