Abstract
Phylogenomic analyses are recovering previously hidden histories of hybridization, revealing the genomic consequences of these events on the architecture of extant genomes. We applied phylogenomic techniques and several complementary statistical tests to show that introgressive hybridization appears to have occurred between close relatives of Arabidopsis, resulting in cytonuclear discordance and impacting our understanding of species relationships in the group. The composition of introgressed and retained genes indicates that selection against incompatible cytonuclear and nuclear–nuclear interactions likely acted during introgression, whereas linkage also contributed to genome composition through the retention of ancient haplotype blocks. We also applied divergence-based tests to determine the species branching order and distinguish donor from recipient lineages. Surprisingly, these analyses suggest that cytonuclear discordance arose via extensive nuclear, rather than cytoplasmic, introgression. If true, this would mean that most of the nuclear genome was displaced during introgression whereas only a small proportion of native alleles were retained.
Keywords: introgression, Arabidopsis, phylogenomics, cytonuclear interactions
Significance
The Brassicaceae (mustard family) is an agriculturally and scientifically important group of plants, yet phylogenetic relationships and major evolutionary events in the group have not been fully resolved. We show that hybridization and introgression occurred, impacting the genomes of plants in this group. Our findings will inform future molecular biology and evolutionary analyses that utilize Brassicaceae species.
Introduction
Hybridization is a driving force in plant evolution (Stebbins 1969), occurring naturally in ∼10% of all plants, including 22 of the world’s 25 most important crops (Yakimowski and Rieseberg 2014). Botanists have long realized that through backcrossing to parents, hybrids can serve as bridges for the transfer of genes between species, a process known as introgression. As more genome sequences become available, comparative analyses have revealed the watermarks of historical introgression events in plant and animal genomes (Rieseberg et al. 1996; Green et al. 2010; Dasmahapatra et al. 2012; Novikova et al. 2016). Cytonuclear discordance is a hallmark of many introgression events, occurring, in part, because nuclear and cytoplasmic DNAs differ in their mode of inheritance. In plants, this discord is often referred to as “chloroplast capture,” which has been observed in cases where introgression of the chloroplast genome occurs in the near absence of nuclear introgression or via nuclear introgression to a maternal recipient (Rieseberg and Soltis 1991). Moreover, discordant nuclear and cytoplasmic introgression creates an opportunity for independently evolved nuclear and cytoplasmic alleles to interact, either of which may have accumulated mutations that result in incompatibilities with deleterious effects when they are united in hybrids. Such incompatibilities could exert a selective pressure that influences which hybrid genotypes are permissible thereby favoring the cointrogression or coretention of alleles for interacting genes (Sloan et al. 2017).
Disentangling introgression from speciation is particularly important because introgression may facilitate the transfer of adaptive traits. Robust statistical techniques (Huson et al. 2005; Than et al. 2008; Joly et al. 2009; Green et al. 2010; Durand et al. 2011; Stolzer et al. 2012; Hufford et al. 2013; Pease and Hahn 2015; Stenz et al. 2015; Rosenzweig et al. 2016) have been developed to detect the signatures of historical introgression in extant and extinct genomes. Although existing techniques are able to identify the taxa that exchanged genes during introgression using a four-taxon system, most methods do not explicitly distinguish which taxon served as donor and which as recipient during introgression (i.e., polarization of introgression directionality), an important distinction considering that introgression impacts the evolution of the recipient lineage (Rieseberg and Soltis 1991; Dasmahapatra et al. 2012). Most methods that polarize introgression generally do so only when a fifth taxon is available. Moreover, this species needs to have diverged from its sister taxon involved in introgression prior to the proposed introgression event (Eaton and Ree 2013; Eaton et al. 2015; Pease and Hahn 2015). Recently, however, divergence-based four-taxon tests have been developed to permit polarization in cases where a fifth taxon cannot be sampled (Hibbins and Hahn 2019; Forsythe et al. 2020).
The wealth of genomic and functional data in Arabidopsis (Lamesch et al. 2012), combined with publicly available genome sequence for 26 species, makes the plant family Brassicaceae an ideal group for comparative genomics. Phylogeny of the group has been the focus of numerous studies (Bailey et al. 2006; Beilstein et al. 2006, 2008, 2010; Oyama et al. 2008; Couvreur et al. 2010; Huang et al. 2016; Nikolov et al. 2019), providing a robust estimate of its evolutionary history. Although the genus Arabidopsis is well circumscribed (Al-Shehbaz and O’Kane 2002; Beilstein et al. 2010), the identity of its closest relatives remains an open question. Phylogenetic studies to date recover three monophyletic groups: clade A, including the sequenced genomes of Arabidopsis thaliana (Arabidopsis Genome Initiative 2000) and Arabidopsis lyrata (Hu et al. 2011); clade B, including the Boechera stricta genome (Lee et al. 2017); and clade C, including the genomes of Capsella rubella, Capsella grandiflora (Slotte et al. 2013), and Camelina sativa (Kagale et al. 2014) (supplementary table S1, Supplementary Material online). Analyses using nuclear markers strongly support the topology A(BC), which is most often cited as the species tree (Bailey et al. 2006; Beilstein et al. 2008; Oyama et al. 2008; Couvreur et al. 2010; Huang et al. 2016). Organellar markers strongly support the topology B(AC) (Koch et al. 2001; Beilstein et al. 2006, 2008; Franzke et al. 2009) (fig. 1a and b and supplementary table S1, Supplementary Material online). The processes underlying this incongruence remain unclear.
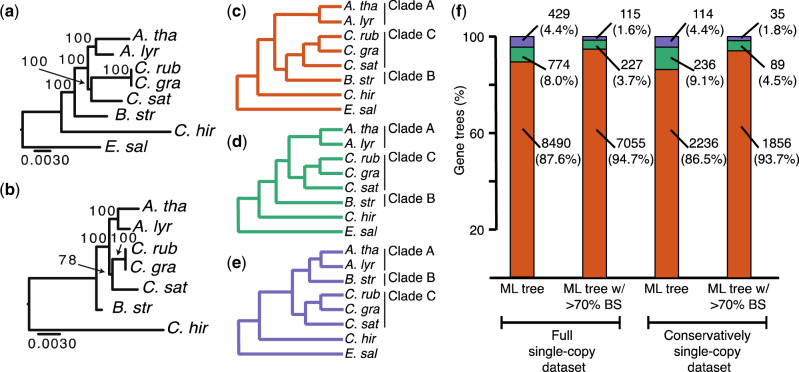
Fig. 1.
Incongruent gene tree topologies are observed within and between nuclear and organellar genomes. (a) Chloroplast and (b) mitochondria ML trees with branch support from 100 bootstrap replicates. Scale bars represent mean substitutions/site. (c–f) ML gene tree topologies inferred from nuclear single-copy genes rooted by Eutrema salsugineum. (c) A(BC), (d) B(AC), and (e) C(AB) topologies. (f) Numbers and frequencies of gene trees displaying A(BC) (orange), B(AC) (green), and C(AB) (purple). Single-copy genes are shown categorized by data set and by level of BS.
Here, we exploit a suite of genomic resources to build on previous single-gene phylogenetic analyses suggesting a putative chloroplast capture event involving Arabidopsis and its closest relatives. We infer gene trees for markers in all three cellular genomes from six available whole-genome sequences. We document cytonuclear discordance and ask if it arose through introgression of organelles or nuclear genes. Further, using a divergence-based approach (Forsythe et al. 2020), we ask which lineage was the recipient of introgressed alleles? Finally, we explore the extent to which physical linkage as well as selection against incompatible alleles at interacting loci, shaped the recipient genome.
Materials and Methods
Experimental Design
Our approach employs publicly available whole genome sequences to infer historical processes that affect the composition and architecture of extant plant genomes. The focus is on Arabidopsis and its closest relatives because there is preliminary evidence of cytonuclear discordance (Koch et al. 2001; Bailey et al. 2006; Beilstein et al. 2006, 2008; Oyama et al. 2008; Franzke et al. 2009; Couvreur et al. 2010; Huang et al. 2016). The objectives of the study were to 1) identify ortholog groups for protein-coding genes from the nuclear and organellar genomes of eight species in Brassicaceae; 2) determine the extent to which these genes have incongruent histories; 3) evaluate evolutionary scenarios that could have produced incongruent histories by determining the relative timing of branching events for different histories; and 4) explore the relative roles of selection and linkage in governing which genes exhibit incongruent histories.
We adopted a phylogenomic approach to identify genes with incongruent histories both from within and among nuclear and organellar genomes in representative species from each of the three monophyletic clades described. In addition, we included Cardamine hirsuta (Gan et al. 2016) and Eutrema salsugineum (Yang et al. 2013) as outgroup genomes. In order to analyze markers spanning nuclear, chloroplast, and mitochondria genomes, we developed a phylogenomic pipeline (supplementary fig. S1a, Supplementary Material online), using CyVerse Atmosphere (Merchant et al. 2016) cyberinfrastructure. The inputs to the pipeline were coding sequences (CDSs) from whole proteomes from each of the eight species used in the study. The workflow of the pipeline is 1) gene family clustering, 2) single-copy gene family filtering, 3) multiple sequence alignment of CDS, 4) inference of maximum likelihood (ML) gene trees, 5) sorting of gene tree topologies, and 6) statistical analyses of topology results. Custom perl, shell, and R scripts used to parse and format files, implement software in high-throughput, and perform downstream analyses are available at https://github.com/EvanForsythe/Brassicaceae_phylogenomics (last accessed 7-29-20).
Phylogenomic Pipeline
Clustering of Putative Orthologs
CDSs for A. thaliana, A. lyrata, C. rubella, C. grandiflora, B. stricta, and E. salsugineum were obtained from Phytozome (Hu et al. 2011; Goodstein et al. 2012; Lamesch et al. 2012; Slotte et al. 2013; Yang et al. 2013; Lee et al. 2017); Cam. sativa and Car. hirsuta were obtained from NCBI (Kagale et al. 2014; Gan et al. 2016). We filtered CDS data sets to contain only the longest gene model when multiple splice-variants were annotated per locus. CDSs were translated into amino acid (AA) sequences using the standard codon table. The resulting whole proteome AA sequences for the eight species were used as input to cluster orthologs via OrthoFinder (version 1.1.4) (Emms and Kelly 2015) under default parameters (supplementary fig. S1a, Supplementary Material online). Two different filtering strategies with varying stringency were applied to the resulting clusters to yield two data set partitions referred to as “full single-copy data set” and “conservatively single-copy data set.” Both filtering strategies are described below.
Full Single-Copy Data Set Filtering
The full single-copy data set was identified by sorting OrthoFinder results to include only clusters that contained exactly one sequence per species, except in the case of Cam. sativa, as it is a hexaploid of relatively recent origin. Thus, clusters with up to three Cam. sativa paralogs (i.e., homeologs) were retained, and we expected these homeologs to form a clade under phylogenetic analysis (supplementary fig. S1b, Supplementary Material online). Gene clusters that yielded trees deviating from this expectation were omitted from further analysis. The full single-copy data set also contains groups classified as retained duplicates (supplementary fig. S1c, Supplementary Material online). Retained duplicate clusters contain exactly two sequences per species (three to six in Cam. sativa). The A. thaliana retained duplicate sequences in each cluster represent known homeologs from the α whole-genome duplication that occurred at the base of Brassicaceae (Bowers et al. 2003), and thus is shared by all sampled species in this study. We retained only those gene clusters that produced trees in which the paralogs formed reciprocally monophyletic clades (supplementary fig. S1c, Supplementary Material online).
Conservative Single-Copy Data Set Filtering
We also used a more stringent set of criteria to develop a conservatively single-copy data set. For this data set, we compared the results obtained from OrthoFinder with results from previously published assessments of plant single-copy or low copy gene families (Duarte et al. 2010; De Smet et al. 2013). The criteria and taxon sampling of our OrthoFinder filtering and the filtering strategies of the two previous analyses differed, meaning each analysis provides its own level of stringency. Moreover, both previous analyses included A. thaliana, allowing for direct comparison with our results. We filtered our clusters to include only those genes recovered by both OrthoFinder and by at least one published analysis. We refer to these as conservatively single-copy. Conservatively, single-copy genes plus the retained duplicates described above constitute the conservatively single-copy data set. Chloroplast and mitochondrial gene data sets were filtered using the same criteria used to filter the full single-copy data set.
Multiple Sequence Alignment and Gene Tree Inference of Nuclear Genes
For single-copy genes, we generated AA-guided multiple sequence alignment of CDS using the MAFFT algorithm (version 6.850) (Katoh and Standley 2013), implemented using ParaAT (version 1.0) (Zhang et al. 2012), under the default settings for both. A multiple sequence alignment of CDS for each gene cluster was used to infer a ML gene tree using RAxML (version 8) (Stamatakis 2014) under the general time reversible model with gamma distributed rate heterogeneity. Support values for nodes were calculated from 100 bootstrap replicates using rapid bootstrapping.
Assembly and Annotation of Mitochondria and Chloroplast Genomes
Whole-genome sequence reads for A. lyrata, B. stricta, C. rubella, C. grandiflora, and Cam. sativa were acquired from NCBI’s Sequence Read Archive (SRA). The run IDs of SRA files used to assemble organelle genomes for each species were A. lyrata (DRR013373, DRR013372); B. stricta (SRR3926938, SRR3926939); C. rubella (SRR065739, SRR065740); C. grandiflora (ERR1769954, ERR1769955); and Cam. sativa (SRR1171872, SRR1171873). Both SRAs for each species were independently aligned to the A. thaliana mitochondrial genome (Ensembl 19) using RMTA (Peri et al. 2020) with default settings for paired-end reads within CyVerse’s Discovery Environment (Merchant et al. 2016). A 15–30× coverage was recovered for each alignment. Mapped read alignment files were converted from BAM to SAM using SAMtools (Li et al. 2009). Mitochondrial consensus sequences were generated (base pair call agreement with 75% of all reads) from each alignment within Geneious (version 7.0; Biomatters) (Kearse et al. 2012). Each mitochondrial consensus sequence was annotated based on the A. thaliana mitochondrial genome annotation (Ensembl 19). CDSs were then extracted using gffread from the Cufflinks package (Trapnell et al. 2010). The same method was used to assemble the B. stricta chloroplast genome. All other chloroplast genome sequences were publicly available.
Multiple Sequence Alignment and Tree Inference from Chloroplast and Mitochondria Markers
Single-copy chloroplast and mitochondrial genes were identified, aligned, and used to infer phylogeny as described previously for nuclear genes. It should be noted that mitochondrial reads were not available for E. salsugineum, leading us to use Car. hirsuta as the sole outgroup for the mitochondria analysis. Summary of individual gene tree results is presented in supplementary fig. S2d and e, Supplementary Material online. We also generated concatenated alignments for both the chloroplast and mitochondrial genes using SequenceMatrix (Vaidya et al. 2011). We inferred trees (fig. 1a and b) from both concatenated alignments using RAxML with the same parameters described above.
Downstream Analyses
Gene Tree Topology Analysis
Tree sorting was performed in batch using the R packages, Ape (Paradis et al. 2004), Phangorn (Schliep 2011), and Phytools (Revell 2012). Gene trees from the retained duplicates were midpoint rooted and split at the root into two subtrees, each of which contained a sequence from all eight analyzed species. Subtrees were analyzed as individual trees alongside all other single-copy gene families as described below. First, each gene tree was rooted at E. salsugineum. Next trees were sorted by considering the topological arrangement of the A, B, and C lineages. For example, a tree was categorized A(BC) if B. stricta, C. rubella, C. grandiflora, and Cam. sativa formed a monophyletic clade. Thus, the branch in the tree leading to the monophyletic clade (the branch uniting B. stricta, C. rubella, C. grandiflora, and Cam. sativa in the above example) was considered the topology-defining branch. Statistical support for any given tree was summarized as the bootstrap value along the topology-defining branch.
Because the focus of our analysis was on topological incongruence of A, B, and C clades, our topology assessment was not designed to detect topological arrangements within A, B, and C clades or in other parts of the trees. If a gene cluster failed to form either a monophyletic A or C clade following phylogenetic analysis, it was marked as “other topology” and removed from further downstream analysis. Exact topologies of all trees, including those recorded as “other topology,” are provided in supplementary table S2, Supplementary Material online.
Applying D-, F-, and DGT-Statistics to Assess the Effects of Incomplete Lineage Sorting and Introgression
To determine whether the observed gene tree incongruences could have been caused primarily by incomplete lineage sorting (ILS), we calculated Patterson’s D-statistic (D) (also known as the ABBA–BABA or four-taxon test) (Green et al. 2010; Durand et al. 2011). D is typically applied to whole-genome alignments of three ingroup taxa and one outgroup taxon. It is calculated by scanning the alignment to identify site patterns consistent with two possible resolutions of ILS (ABBA and BABA). Due to the relatively deep divergence and numerous chromosomal rearrangements between genomes used here, it was not feasible to construct accurate whole-genome alignments. Instead, we identified ABBA and BABA site patterns within single-gene multiple sequence alignments used to infer gene trees. We calculated D and F using the total number ABBA and BABA sites from all nuclear gene alignments (or subsets of nuclear genes corresponding to individual chromosomes or conservatively single-copy genes). We excluded Cam. sativa sequences from this analysis due to the presence of multiple Cam. sativa paralogs in some trees. We considered only biallelic sites in which the two outgroups, E. salsugineum and Car. hirsuta, have the same allele. We also required individual species within each clade to have the same allele. For example, an ABBA site would be one in which E. salsugineum, Car. hirsuta, A. thaliana, A. lyrata, C. rubella, C. grandiflora, and B. stricta display T, T, G, G, G, G, and T, respectively. Note that all members of clade A and C share the derived allele. An example of a BABA site would be T, T, G, G, T, T, and G, respectively. In this case, members of clades A and B share the derived allele. We also tallied AABB sites (e.g., T, T, T, T, G, G, and G, respectively), in which clades B and C share the derived allele, although AABB sites are not a component of D or F. In addition, we calculated D and F using the methodology above but without the requirement for the two outgroups, E. salsugineum and Car. hirsuta, to share an allele. We calculated D and F according to the equations from Zheng and Janke (2018). All site counts and statistics are shown in supplementary table S3, Supplementary Material online.
We also applied the rationale of D to gene tree topology counts by calculating a related statistic, DGT. We used gene tree topologies as proxies for site patterns. Because B(AC) and C(AB) trees were closest in frequency in the nuclear genome, we asked whether their frequencies were statistically significantly different using DGT. B(AC) trees and C(AB) trees were treated as ABBA and BABA sites, respectively, whereas A(BC) was treated as AABB. DGT was then calculated as follows:
We calculated DGT for the set of all nuclear genes as well as for subsets of genes present on each of C. rubella’s nuclear chromosomes, as C. rubella serves as an estimate of the ancestral karyotype for the included species (Schranz et al. 2007). Results from all DGT calculations are given in supplementary table S4, Supplementary Material online.
Phylogenetic Network Reconstruction and Introgression Analysis
To evaluate the likelihood that the observed incongruence was caused by introgression, we also reconstructed ML phylogenetic networks using InferNetwork_ML in PhyloNet (version 3.6.1) (Than et al. 2008). We input all nuclear gene trees (supplementary fig. S1d, Supplementary Material online, Full single-copy genes data set) and implemented InferNetwork_ML using the command “InferNetwork_ML (all) h –n 100 –di –o –pl 8,” where h is the number of reticulations allowed in a given network. The method ignores gene tree branch lengths, utilizing gene tree topologies alone to infer reticulation events. We performed separate analyses using h = 0 (a tree), h = 1, and h = 2, outputting the 100 most likely trees/networks (designated with –n) from each analysis. We followed the analysis strategies of Wen et al. (2016a), manually inspecting networks to identify those with edges consistent with both the major nuclear topology [A(BC)] as well as the major chloroplast and mitochondrial topology [B(A, C)] (supplementary fig. S2l–o, Supplementary Material online). Additionally, we reported the most likely tree/network from each analysis (supplementary fig. S2k, p, and q, Supplementary Material online). As an additional means of asking whether ILS alone adequately explains incongruence, we performed Tree Incongruence Checking in R (TICR) (Stenz et al. 2015). We used a population tree inferred from PhyloNet (h = 0) (supplementary fig. S2j, Supplementary Material online) with a table of concordance factors for all quartets. We performed the TICR test as implemented in the R package, phylolm (Tung Ho and Ané 2014), according to the methods outlined in https://github.com/crsl4/PhyloNetworks.jl/wiki/TICR-test:-tree-versus-network%3F (last accessed 7-29-20).
Identification of Introgressed Topology and Species Branching Order
In order to identify the topology most likely to represent introgression, we measured node depths on trees displaying either A(BC) or B(AC). As above, Cam. sativa sequences were not considered in order to avoid complications associated with paralogous sequences. For each nuclear gene tree, we calculated pairwise synonymous divergence (dS) between taxa on the tree using PAML (version 4.8) (Yang 2007). To infer the pairwise distance between two clades on the tree, we took the average dS score between each combination of taxa present in the two clades. For example, the depth of the node uniting clades A and C on B(AC) trees would be the average of dS(A. thaliana, C. rubella), dS(A. lyrata, C. rubella), dS(A. thaliana, C. grandiflora), and dS(A. lyrata, C. grandiflora). To calculate normalized dS, each dS node depth (as described above) was divided by the average pairwise dS of each ingroup species versus the outgroup, Car. hirsuta.
We also calculated node depths from ultrametric gene trees. Before measuring node depths, gene trees were smoothed to ultrametric trees using semiparametric penalized likelihood rate smoothing (Sanderson 2002). We implemented the rate smoothing algorithm designated by the chronopl function in the Ape package. We tested six values of the smoothing parameter (λ), which controls the tradeoff between parametric and nonparametric formulation of rate smoothing, to assess the sensitivity of node depths to different values of λ. We calculated node depth on ultrametric trees for nodes representing T1 and T2 on each given topology (supplementary fig. S3a, Supplementary Material online). We plotted the frequency distributions of node depths (supplementary fig. S3b, Supplementary Material online) as well as descriptive statistics (supplementary fig. S3c–t, Supplementary Material online).
In order to account for intragenic recombination, we split each gene alignment into 200-nt alignments, the goal being to reduce the probability of recombination occurring in the middle of our alignment. For each window, we calculated a distance matrix and inferred a neighbor joining “window tree” using Ape in R (Paradis et al. 2004). We calculated the depth of the T1 node for each window displaying either A(BC) or B(AC) from the distance matrix by averaging the pairwise distance values similar to our treatment of dS node depths above. We documented the number of discordant windows in alignments for A(BC) (supplementary fig. S4a, Supplementary Material online) and B(AC) (supplementary fig. S4b, Supplementary Material online) trees and used boxplots to compare distributions of A(BC) and B(AC) node depths (fig. 2g and supplementary fig. S4c, Supplementary Material online).
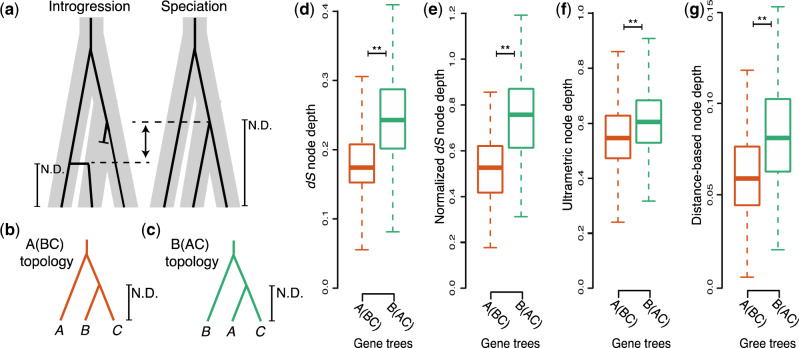
Fig. 2.
Node depths indicate extensive introgression led to transfer of nuclear genes. (a) Model depicting expected node depths (N.D.) for genes undergoing introgression (left) or speciation (right). Speciation history is represented by thick gray bars. Individual gene histories are represented by black branches. Blunt ended branches represent a native allele that was replaced by an introgression allele. Vertical arrow indicates expected difference in node depth. (b, c) The informative node depths on A(BC) (b) and B(AC) (c) trees. (d–f) Boxplots depicting observed median and quartile node depths measured from dS (d), normalized dS (e), ultrametric gene trees (f), and concordant windows within gene alignments (g).
Polarization of Introgression Directionality with Divergence-Based Introgression Polarization
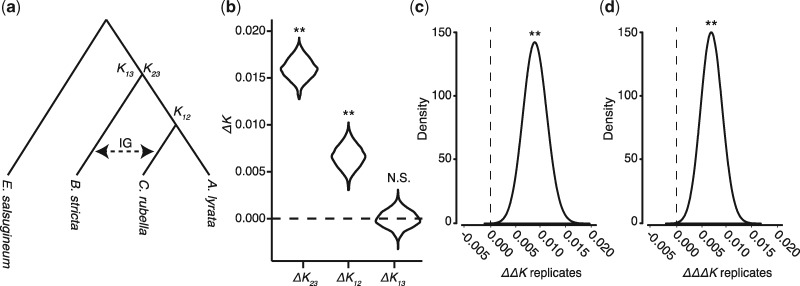
To search for evidence of asymmetry in introgression directionality, we applied Divergence-based Introgression Polarization (DIP) (Forsythe et al. 2020) to the full single-copy data set. Scripts and more information on running DIP are available at https://github.com/EvanForsythe/DIP (last accessed 7-29-20). Following the nomenclature of Forsythe et al. 2020, we used A. lyrata, C. rubella, B. stricta, and E. salsugineum as P1, P2, P3, and O, respectively. We treated gene alignments as separate windows, pruning the alignments down to just the representative species described above. We performed all three versions of DIP described by Forsythe et al. (2020). With the above taxon sampling scheme, a 1× DIP profile of nonzero ΔK23, nonzero ΔK12, and ΔK13 equal to zero would indicate introgression from clade B to clade C. A profile of nonzero ΔK23, ΔK12 equal to zero, and nonzero ΔK13 would indicate introgression from clade C to clade B. For 2× and 3× DIP, positive ΔΔK and ΔΔΔK values would indicate clade B to clade C introgression, whereas negative values would indicate introgression in the opposite direction.
Cytonuclear Function Enrichment Analysis
We used the Cytonuclear Molecular Interaction Reference for Arabidopsis data set (Forsythe et al. 2019) to identify nuclear-encoded genes in our data set that are both organelle localized and involved in interactions with organelle-encoded genes/gene products. The data set is available at http://cymira.colostate.edu/ (last accessed 7-29-20). We performed this analysis on our full single-copy data set. For each category, the percentage of B(AC) trees annotated with that category was compared with the percentage of A(BC) trees with the category. Comparisons were quantified with an enrichment score (E). For example, we used the following equation to ask if B(AC) or A(BC) topology genes are enriched for chloroplast interaction:
Positive E indicates enrichment for a category among B(AC) trees, whereas negative E indicates enrichment among A(BC) trees (supplementary table S6, Supplementary Material online).
Network Analysis of Protein–Protein Interactions
Experimentally curated protein–protein interaction data for Arabidopsis were downloaded from Arabidopsis thaliana Protein Interaction Network (version 2.6.70) (Brandão et al. 2009). Interaction data were filtered to contain only genes included in the full single-copy data set. An undirected interaction network was visualized and analyzed using the igraph package (http://igraph.org [last accessed 7-29-20]) in R. Each node in the graph represents a single-copy nuclear gene family, whereas each edge in the graph indicates a physical interaction in Arabidopsis. Nodes were colored by gene tree topology and diameter of nodes are proportional to bootstrap support (BS) values for the gene tree (see supplementary fig. S2a–c, Supplementary Material online).
We asked if genes displaying the same topology are clustered with each other in the network by calculating nominal assortativity (Newman 2003). Assortative mixing/clustering of gene tree topology results across the network was quantified by the assortativity coefficient (A) of the network. Positive A indicates clustering of genes with the same topology, whereas negative A indicates overdispersal. We calculated the observed A for our network as well as a null distribution of A generated by randomly assigning a topology to nodes in 10,000 replicates of our network.
Mapping of Gene Coordinates to the C. rubella Nuclear Genome
Topology results were mapped to the nuclear genome of C. rubella using the gene coordinates from the GFF file associated with the genome assembly. Genome maps were visualized using the R package, Sushi (Phanstiel et al. 2014), made available through Bioconductor (Gentleman et al. 2004). Colored horizontal lines indicate genes displaying each topology. The length of each line represents the BS value found at the topology-defining branch in the gene tree (see supplementary fig. S2a–c, Supplementary Material online).
Detection of Linkage Disequilibrium
Topology results mapped to the C. rubella genome were used to ask if genes displaying the same topology are clustered together linearly along chromosomes. We assessed the physical clustering of A(BC), B(AC), and C(AB) genes with two measures: 1) number of instances in which genes with the same topology are located within 10 kb of each other (supplementary fig. S6a, Supplementary Material online) and 2) number of instances in which neighboring genes share topology, regardless of distance (supplementary fig. S6b, Supplementary Material online). We established a null distribution for both measurements by generating 10,000 maps of the C. rubella genome in which observed location of single-copy genes and the overall gene tree frequencies were maintained, but the assignment of topologies to genes was randomized across chromosomes. Measure 1 and measure 2 were calculated for each of the 10,000 replicates to obtain null distributions.
Statistical Analyses
Statistical tests were performed in R (version 3.4). Below, we describe methods used to assess the significance of our results. Our general strategy was to provide sufficient information to enable readers to make their own interpretations of the data; toward that goal, we have included Bonferroni corrected and uncorrected (raw) P values for each experiment where corrections could be applied (supplementary tables S4–S6, Supplementary Material online, or within supplementary text, Supplementary Material online).
D-, F-, and DGT-Statistics
We calculated D, F, and DGT for both the full single-copy and conservatively single-copy data sets. Confidence intervals were obtained by resampling either data set to generate 10,000 bootstrap replicates, recalculating D/F/DGT for each replicate. The resulting distributions were compared using the Z-test. To account for potential autocorrelation bias caused by nonindependence of linked genes, D/F/DGT were also calculated using block bootstrapping. For D and F, block bootstrapping was achieved by simply bootstrap resampling from the available gene alignments and recalculating D/F with each replicate. For DGT, block bootstrapping was accomplished by splitting the data set into 100 equal size blocks of neighboring genes based on position along C. rubella chromosomes. Blocks were then bootstrap resampled 10,000 times and DGT was recalculated with each replicate to obtain a distribution. P values from analyses of the whole genome were Bonferroni adjusted for four comparisons for DGT. Raw P values are reported in the main body and adjusted P values are shown in supplementary table S4, Supplementary Material online.
Phylogenetic Network Reconstruction and Introgression Analysis
PhyloNet models were statistically compared by calculating AIC and BIC scores for each tree/network with the following expressions:
and
where k is the number of free parameters in the model, n is the number of input gene trees, and L is the ML value of the model. We compared hypotheses by calculated difference in AIC and BIC scores for each given tree/network relative to the most likely network (ΔAIC and ΔBIC).
Node-Depth-Based Test of Species Branching Order
Frequency distributions of node depths were plotted. Two-tailed T-tests and Wilcoxon rank sum tests were performed to assess differences in distribution means and medians, respectively. P values were Bonferroni corrected for six comparisons. Raw P values are reported in the main body and adjusted P values are shown in supplementary table S5, Supplementary Material online.
Divergence-Based Test of Introgression Directionality
Statistics were calculated for 1×, 2×, and 3× DIP as described by Forsythe et al. (2020).
Functional Category Enrichment
Enrichment of functional categories was assessed by comparing categories of A(BC) genes versus B(AC) genes. For each category, two-by-two contingency tables were constructed and used to perform two-tailed Fisher’s exact tests. Reported P values were Bonferroni corrected for five comparisons.
Protein–Protein Interaction Network
Clustering in the interaction network was quantified with an assortativity coefficient (A) (Newman 2003). To assess significance of the observed A, we randomly assigned one of the three topologies (keeping the frequency of each topology the same as in the original data set) to genes in 10,000 copies of the network. We computed A for each of the 10,000 networks to obtain a null distribution of A and used the null distribution to perform a two-tailed Z-test.
Haplotype Block Linear Clustering
We quantified linear clustering of topologies by counting the number of occurrences of proximal and neighboring genes in the observed data. We assessed the significance of the observed values by generating null distributions from 10,000 data sets in which the topologies were randomized. We used the null distributions to perform two-tailed Z-tests. P values were Bonferroni corrected for six comparisons.
Results
Summary of Previous Studies of the Species Branching Order
Considerable efforts have been made to develop Brassicaceae as a comparative genetic and genomic system. Despite alternative estimates of the branching order for Arabidopsis and its relatives, all trees from these studies share three distinct clades. The genus Arabidopsis as outlined by Al-Shehbaz and O’Kane (2002) is monophyletic and comprises nine species. We refer to this group as clade A, represented by the genomes of Arabidopsis (Lamesch et al. 2012) and A. lyrata (Hu et al. 2011). Boechereae is a diverse tribe containing eight genera, including Boechera, which comprises more than 70 species (Alexander et al. 2010). Boechereae is sister to Halimolobeae, which contains five genera and 39 species (Bailey et al. 2007). We refer to species in Boechereae and Halimolobeae as clade B, represented by the recently sequenced genome of B. stricta (Lee et al. 2017). A third monophyletic clade is composed of 15 species in Capsella, Camelina, and Catolobus (Slotte et al. 2006; Galasso et al. 2015). Genome sequence in this group includes C. rubella, C. grandiflora (Slotte et al. 2013), and the paleohexaploid oil-seed crop, Cam. sativa (Kagale et al. 2014).
Although clades A, B, and C are well resolved in the literature, their relationship to one another differs by marker. To understand incongruence implied by previous analyses, we reviewed eight phylogenetic studies, paying particular attention to the relative relationships of species from clade A, B, and C (supplementary table S1, Supplementary Material online). We find that phylogenies inferred from organellar markers (Koch et al. 2001; Beilstein et al. 2006, 2008; Franzke et al. 2009) are incongruent with those inferred from nuclear markers, or concatenation of nuclear and organellar markers (Bailey et al. 2006; Beilstein et al. 2008; Oyama et al. 2008; Couvreur et al. 2010; Huang et al. 2016). We find that all studies of individual chloroplast and mitochondria markers yield B(AC). On the other hand, all studies that include nuclear markers yield A(BC). The statistical support for both of these topologies varies by study but each topology is well supported in at least some of the studies, indicating the phylogenetic incongruence is not likely to be caused by a lack of phylogenetic resolution or error in phylogenetic reconstruction. The observation of phylogenetic incongruence motivated our current phylogenomic analysis.
Gene Tree Incongruence within and between Organelle and Nuclear Genomes
We searched for incongruent histories present within and among nuclear and organellar genomes in representative species from each clade. We included Car. hirsuta (Gan et al. 2016) and E. salsugineum (Yang et al. 2013) as outgroups. We considered three biological processes capable of producing incongruent genealogical histories: gene duplication and loss, ILS, and introgression. In addition, we assessed the possible contribution of phylogenetic error or “noise.”
Chloroplast assemblies and annotations were available for all analyzed species except for B. stricta. We assembled the Chloroplast genome from B. stricta from whole-genome sequencing reads. We annotated the genome and extracted CDS from 85 protein-coding genes. Ortholog clustering revealed 77 orthologous gene clusters, 32 of which passed our filters as single-copy. We performed multiple sequence alignment for the 32 single-copy families and concatenated the alignments into an alignment with a total length of 30,645 nt that produced B(AC) as a well-supported most likely tree. This result is consistent with the trees previously inferred from chloroplast markers (see supplementary table S1, Supplementary Material online). We also analyzed each gene separately. Of the 32 genes, 13 were B(AC), zero were A(BC), and one was C(AB). Eighteen of the gene trees lacked statistically supported resolution. The chloroplast gene trees displaying B(AC) show variable BS but 7 of 13 are supported by at least 70% BS at the topology-defining branch (supplementary fig. S2f, Supplementary Material online; green bars). The one chloroplast gene tree indicating the C(AB) topology lacked BS (<50%) at its topology-defining branch (supplementary fig. S2f, Supplementary Material online; purple bar). Regardless of whether chloroplast genes are analyzed individually or are concatenated they strongly support B(AC) as the chloroplast branching order.
Mitochondria assemblies and annotations were unavailable for A. lyrata, B. stricta, C. rubella, C. grandiflora, Cam. sativa, and E. salsugineum. We assembled mitochondrial genomes for these five species from raw sequencing reads. We were unable to assemble the E. salsugineum mitochondrial genome so we included only Car. hirsuta as an outgroup for mitochondrial analyses. We annotated the genomes and extracted CDS from 85 protein-coding genes. Ortholog clustering revealed 24 orthologous gene clusters, 21 of which passed our filter as single-copy. We performed multiple sequence alignment for the 21 single-copy families and concatenated the alignments into an alignment with a total length of 7,014 nt that yielded a well-supported B(AC). Of the 21 individual mitochondrial gene trees, four displayed B(AC), zero displayed A(BC), and one displayed C(AB). Sixteen of the gene trees lacked statistically supported resolution. Three of the four B(AC) gene trees have at least 70% BS at the topology-defining branch (supplementary fig. S2g, Supplementary Material online; green bars). The one C(AB) tree lacked BS at its topology-defining branch (supplementary fig. S2g, Supplementary Material online; purple bar). In sum, regardless of whether mitochondrial genes are analyzed individually or are concatenated, they support B(AC) as the mitochondrial topology.
Given the well-known history of whole-genome duplication of the nuclear genome in Brassicaceae, we took extensive measures to minimize the possibility that duplication and loss biased our inferences. We identified single-copy nuclear genes as well as genes that were retained in all species postduplication (see Discussion). We identified 10,193 single-copy nuclear genes using OrthoFinder (Emms and Kelly 2015) (denoted as “full single-copy data set”) (supplementary fig. S1a–c, Supplementary Material online). The full single-copy data set comprises 37.17% and 35.83% of the Arabidopsis and C. rubella genomes, respectively. These genes were indicated as single-copy by OrthoFinder because they form clusters that include exactly one locus from each species (with the exception of the polyploid Cam. sativa, see Materials and Methods). These single-copy genes span the eight chromosomes of C. rubella (supplementary fig. S1d, Supplementary Material online), whose karyotype serves as an estimate of the ancestral karyotype for these species (Schranz et al. 2007). ML analyses yielded 8,490 (87.6%) A(BC), 774 (8.0%) B(AC), and 429 (4.4%) C(AB) nuclear gene trees (fig. 1c–f). A complete list of the gene tree topologies resulting from phylogenetic analyses of these markers is included in supplementary table S2, Supplementary Material online.
The most parsimonious explanation for our single-copy genes is that they were either not duplicated in our focal species or, if duplicated, were returned to single-copy before a speciation occurred, and thus behave as unduplicated in a phylogenetic context, meaning that any observed incongruent topologies resulted from a process other than duplication. However, although not parsimonious, it is important to consider the possibility that ancestral duplication, paralog retention through two speciation events, and lineage-specific loss events led to hidden out-paralogs in our data set. To further reduce the probability that this series of events contributed to incongruent gene trees, we performed a reanalysis after further filtering our data set to include only genes that were previously indicated as reliable single-copy markers in angiosperms (Duarte et al. 2010; De Smet et al. 2013). This filter reduced our single-copy data set to 2,098 genes (supplementary fig. S1e and f, Supplementary Material online). We combined this data set with genes that were duplicated during whole-genome duplication (Bowers et al. 2003) but did not undergo loss in focal species to yield a data set of 2,747 genes, which we denote as “conservatively single-copy,” so named because they are the genes that are least likely to contain hidden out-paralogs. The conservatively single-copy data set comprises 10.02% and 9.66% of the Arabidopsis and C. rubella genomes, respectively. ML analyses of these genes yielded 2,236 (86.5%) A(BC), 236 (9.1%) B(AC), and 114 (4.4%) C(AB) trees (fig. 1b–f), consistent with our results from the full single-copy data set.
To ask whether phylogenetic noise contributed to incongruent nuclear gene tree topologies, we also filtered our single-copy nuclear gene tree results to contain only trees in which the observed topology was supported by at least 70% BS at the topology-defining branch (see supplementary fig. 2Sa–c, Supplementary Material online) and found that B(AC) and C(AB) trees were still present (fig. 1f). Together, these analyses consistently support the incongruent histories present in the organellar and nuclear genomes and indicate that incongruence cannot be fully explained by gene duplication and loss or by phylogenetic noise.
Contribution of Introgression to Incongruent Gene Trees
A number of approaches have been developed to determine the relative contributions of ILS and introgression to gene tree incongruence. Site-based comparative approaches such as the D-statistic (Green et al. 2010; Durand et al. 2011) are typically applied to whole-genome alignments and calculated by determining the frequency of site patterns. Given the relatively deep divergence of our study taxa, it was not feasible to construct accurate whole-genome alignments among them, and thus we used multiple sequence alignments from single-copy genes to calculate D- and F-statistics. We applied D- and F-statistics to compare the frequencies of the two site patterns consistent with the B(AC) and C(AB) gene tree topologies, which have the closest frequency to each other in our phylogenetic analyses above. The deep scale of divergence among our taxa raised concerns of homoplastic mutations obscuring site patterns. The outgroup typically indicates the ancestral character state in site-based comparative introgression analyses (Green et al. 2010; Durand et al. 2011). Our inclusion of two graded outgroups allowed us to filter for sites in which the two outgroups share an allele, thus reducing the number of potential homoplastic sites in the data. Applying this approach, we found significantly positive D and F across the whole nuclear genome for all data sets and resampling techniques (supplementary table S3, Supplementary Material online), thereby rejecting the null hypothesis that ILS alone was responsible for the observed incongruence of markers across the nuclear genome. This result is consistent with B(AC) occurring in a larger proportion of nuclear gene trees than C(AB).
We also calculated D and F using E. salsugineum as the sole outgroup. Interestingly, these analyses returned significantly negative D and F across the whole nuclear genome for all data sets and resampling techniques (supplementary table S3, Supplementary Material online), contradicting results calculated from sites in which both outgroup species share an allele. In sum, D and F values calculated with E. salsugineum as outgroup indicate that introgression occurred, but the introgression event is inferred to occur between different taxa than those inferred when both outgroup species share an allele (see Discussion). To explore this contradictory result, we partitioned the data set by gene tree topology and recalculated D and F. Regardless of outgroup, we observe extremely high (positive) D and F for genes that display the B(AC) topology and extremely low (negative) D and F for genes that display the C(AB) topology, indicating that B(AC) topology genes are highly enriched for ABBA sites, whereas C(AB) topology genes are highly enriched for BABA sites. This result informs our rationale for treating B(AC) trees and C(AB) trees as proxies for ABBA and BABA sites, respectively, in performing DGT analyses below. In sum, our site-based introgression analyses consistently indicate that introgression occurred; however, these statistics differ in their inference of the species involved in introgression depending on whether sites are filtered to avoid putative homoplastic sites. The potential impact of homoplasy on these site-based statistics, which largely rely on parsimony logic, lead us to employ additional analyses that make use of ML trees.
Given that calculated D and F appear to be strongly correlated with gene tree topology, we used gene tree topologies as proxies for site patterns to calculate a related statistic, referred to here as DGT (Huson et al. 2005). We found positive DGT across all chromosomes, however for chromosomes 2, 4, and 7, the significance of DGT depended on the data set and whether we resampled the data by bootstrap or block-bootstrap. When a significant DGT was detected, it was reflected by both raw and Bonferroni adjusted P values. Loss of power is expected with this method because it involves condensing multiple ABBA/BABA sites into a single-gene tree, thus the sample size is much smaller. These results indicate a significant excess of B(AC) genes in comparison to C(AB) genes in the nuclear genome, consistent with our observed gene trees frequencies (fig. 1f). DGT results are consistent with positive D and F results calculated using the outgroup filter but contradicts the negative D and F results obtained using E. salsugineum as sole outgroup, meaning we cannot fully resolve the species involved in introgression using the above methods, leading us to explore additional analytical frameworks in an effort to arrive at a consensus signal.
Phylogenetic Network Reconstruction and Coalescent-Based Introgression Analyses
Coalescent-based approaches (Than et al. 2008; Stenz et al. 2015) use gene trees to distinguish between organismal histories that are tree-like (incongruencies among trees arise from ILS) and network-like (incongruencies result from ILS + introgression). Phylogenetic networks are emerging as natural means of capturing reticulate evolutionary histories in the presence of ILS (Wen et al. 2016a; Copetti et al. 2017). We used PhyloNet to reconstruct the most likely species tree (0 reticulations) and networks (one or two reticulations) from nuclear gene trees. We show the first and second most likely species trees, which are consistent with the A(BC) and B(AC), respectively (supplementary fig. S2j and k, Supplementary Material online). We also present the most likely networks containing edges that incorporate A(BC) and B(AC) (supplementary fig. S2l–o, Supplementary Material online). Finally, we show the unconstrained most likely networks (supplementary fig. S2p and q, Supplementary Material online). For each reticulation inferred by PhyloNet, two reticulation edges are inferred (supplementary fig. S2l–q, Supplementary Material online; blue branches). Inheritance probabilities (i.e., the proportion of gene trees displaying an edge) are shown next to each edge. The analysis is agnostic to which of the two edges represents introgression and which represents speciation (Wen et al. 2016a).
All network models shown are substantially more likely than models that yield bifurcating trees (supplementary fig. S2j–q, Supplementary Material online; ΔAIC ≥ 87.80 and ΔBIC ≥ 73.50), providing an additional line of evidence that introgression played a role in generating incongruent gene trees, consistent with our D-, F-, and DGT-statistic results. We find that the overall most likely reticulation events involve introgression from clade A to Car. hirsuta (supplementary fig. S2p and q, Supplementary Material online), which was initially included as an outgroup. This was unexpected, as the major nuclear, chloroplast, and mitochondria topologies do not show evidence of clade A and Car. hirsuta forming a clade. Given that the goal of this study is to investigate processes leading to cytonuclear discordance, we focus on reticulation events involving clades A, B, and C.
Among the set of networks that address potential introgression between clades A, B, and C, the networks shown in supplementary figure S2m and n, Supplementary Material online, indicate that clade C was the recipient of introgressed alleles from either clade B or clade A. The networks shown in supplementary figure S2l and o, Supplementary Material online, indicate an alternative scenario, in which clade B was either the recipient of introgressed alleles from clade C or from a more distantly related “ghost lineage” that is either not sampled or extinct. The highest likelihood network in this set displays an alternative history in which clade A was the recipient of introgressed alleles from either clade C or a more distant ghost lineage (supplementary fig. S2q, Supplementary Material online). Although several alternative introgression scenarios are represented among the most likely networks, none of these indicates introgression between clade A and B, thus phylogenetic network analyses are consistent with positive D, F, and DGT.
To test the robustness of the network-based analyses above, we also performed a quartet-based analysis, TICR (Stenz et al. 2015), using the population tree from PhyloNet, which displays A(BC) with branch lengths in coalescence units (supplementary fig. S2r–u, Supplementary Material online). The TICR test indicates that the population tree does not fit the quartet concordance factors adequately (P = 0.00058; χ2). These results suggest that the observed pattern does not have a simple evolutionary explanation, thereby indicating a complex evolutionary history in the group. In sum, both comparative genomic and coalescent-based approaches support an evolutionary history that includes introgression. Inconsistency across different analytical methods prevents the confident resolution of a specific introgression event; however, only one approach indicated that C(AB) trees resulted from introgression, whereas the remaining analyses indicated that either A(BC) or B(AC) trees resulted from introgression. Based on this weight of evidence, we proceed under the working hypothesis that A(BC) and B(AC) trees are indicative of speciation/introgression histories, whereas C(AB) trees are largely the result of ILS, although the uncertainty in this model should be weighed as it pertains to downstream analyses (see Discussion).
Recovery of the Species Branching Order and Introgression Events
To uncover which lineages were affected by introgression, we determined the relative timing of the B(AC) and A(BC) branching events by calculating node depths (fig. 2) (Fontaine et al. 2015). Introgression nodes are expected to be younger than speciation nodes (Fontaine et al. 2015; Rosenzweig et al. 2016; Lee-Yaw et al. 2019) because introgression produces incongruent trees when it occurs between nonsister species subsequent to speciation (Green et al. 2010; Durand et al. 2011; Dasmahapatra et al. 2012) (illustrated by fig. 2a). Therefore, we calculated the depth of the node uniting clade A with clade C in nuclear B(AC) trees and compared it with the depth of the node uniting the B and C clades in nuclear A(BC) trees (fig. 2a–c, N.D.). We calculated node depths using four separate measures to account for potential biases (fig. 2d–g). To account for selection on AAs, we used synonymous divergence (dS) (fig. 2d and supplementary fig. S5, Supplementary Material online). To account for potential differing rates of evolution across the genome, we normalized dS using the divergence between the clade of interest and an outgroup (i.e., “relative node depth”) (Rosenzweig et al. 2016) (fig. 2e). To account for potential differences in rates of evolution between lineages, we also calculated node depths from ultrametric trees in which the rates of evolution had been smoothed across the tree using a penalized likelihood approach (Sanderson 2002) (fig. 2f and supplementary fig. S3 and table S5, Supplementary Material online). Because our ultrametric approach required the user-defined λ parameter, we explored the effect of different λ values on the calculation of node depths and found that node depths on A(BC) trees were consistently significantly shallower than node depths on B(AC) trees. Additionally, we accounted for potential intragene discordance due to recombination within a gene, by divided each gene alignment into 200-bp windows, inferred a neighbor joining tree for each window, and only calculated node depth from windows that were concordant with the ML tree for the gene, thus minimizing the probability of recombination within the loci from which node depth is calculated (fig. 2g and supplementary fig. S4, Supplementary Material online). For all four node depth measures, the node depth for A(BC) was significantly shallower than for B(AC) (fig. 2d–g and supplementary figs. S3 and S4 and table S6, Supplementary Material online; P < 2.2e-16, Wilcoxon).
Recognizing that there are likely deep coalescing genes within our A(BC) and B(AC) bins, we removed the genes with the deepest nodes in both A(BC) and B(AC) bins and still found the same significant pattern (supplementary fig. S3o–t, Supplementary Material online; P < 2.2e-16, Wilcoxon). Hence, node depth data are most consistent with a scenario in which A and C diverged from each other prior to the exchange of genes between clade B and C via introgression. This surprising result stands in opposition to previously published trees inferred from single or concatenated nuclear genes, which strongly favor A(BC) (Al-Shehbaz and O’Kane 2002; Oyama et al. 2008; Beilstein et al. 2010; Huang et al. 2016). However, it bolsters the argument that B(AC) best represents the species branching order despite the low frequency of these genes in the nucleus (similar to Fontaine et al. [2015]) and further suggests that the vast majority of nuclear genes in either B or C arrived there via introgression. We discuss the implications of this finding on the concept of the species branching order (see Discussion). It should be noted that our downstream analyses of selection and linkage (fig. 4 and supplementary fig. S6 and table S6, Supplementary Material online) are framed in the context of nuclear introgression but would remain equally valid if cytonuclear discordance arose via organellar introgression.
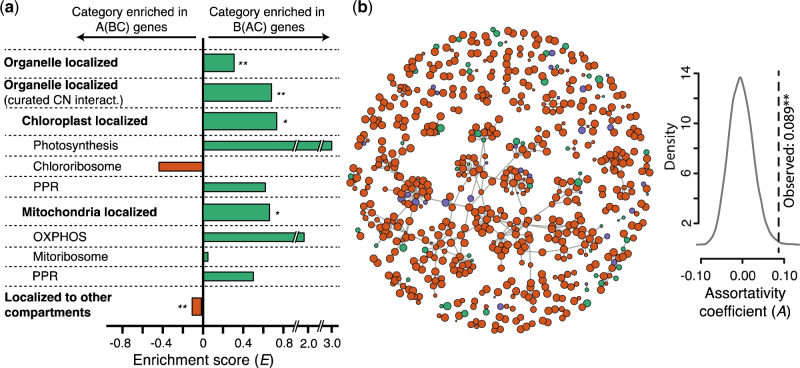
Fig. 4.
Selection for cytonuclear and nuclear–nuclear interactions influenced introgression. (a) Enrichment (E) for GO terms = (% B(AC) genes – % A(BC) genes)/(% B(AC) + A(BC) genes). (b) (Left) Protein–protein interaction network for Arabidopsis protein complexes. Node fill, gene tree topology; node diameters proportional to BS (supplementary fig. S2a–c, Supplementary Material online). (c) (Right) Assortativity coefficient (A) of the network. Null distribution of A (gray curve); dotted line, observed A. Significance levels (**P < 0.01 and * P < 0.05) are based on Bonferroni corrected P values.
Identification of Introgression Donor and Recipient Linages
We next asked whether transfer of genetic material during introgression was directionally asymmetric and, if so, which of the two clade ancestors was the donor and which was the recipient of introgressed alleles. We applied DIP (Forsythe et al. 2020), which is calculated from pairwise sequence divergence between taxa involved in introgression and a sister taxon by comparing divergence values obtained from introgressed loci versus nonintrogressed loci (see Materials and Methods) (fig. 3a). We applied three variations of DIP, 1×, 2×, and 3× DIP, designed to increase sensitivity to bidirectional introgression and minimize bias. 1× DIP yielded a positive ΔK23 (P < 2.2e-16), positive ΔK12 (P < 2.2e-16), and ΔK13 not significantly different from zero (P = 0.66) (fig. 3b). This pattern matches our expectations for asymmetric introgression from clade B to clade C. Likewise, 2× and 3× DIP yielded positive ΔK (P < 2.2e-16) and ΔΔK (P = 0.002), respectively, also indicative of introgression from clade B to clade C. Taken together, DIP analyses indicate predominant introgression from clade B to clade C.
Fig. 3.
Asymmetric introgression led to transfer of nuclear genes from clade B to clade C. (a) Model depicting the taxon sampling and design of DIP analyses. (b–d) Results from 1× DIP (b), 2× DIP (c), and 3× DIP (d) analyses. Distributions represent bootstrap resampling replicates. See Forsythe et al. (2020) for detailed explanation of DIP.
The Role of Cytonuclear Interactions during Introgression
According to our leading model, the introgression that occurred during the evolution of clade C resulted in a genome in which the majority of nuclear alleles were displaced by alleles from clade B whereas native organellar genomes were maintained. We asked whether we could detect patterns within the set of nuclear genes that were also maintained alongside organelles during introgression. We hypothesized that during the period of exchange, selection would favor the retention of alleles that maintain cytonuclear interactions, especially when replacement with the paternal allele is deleterious (Sloan et al. 2017). Using Cytonuclear Molecular Interaction Reference for Arabidopsis, a curated set of Arabidopsis genes involved in cytonuclear interactions (Forsythe et al. 2019), we asked if B(AC) nuclear genes were significantly enriched for interactions in the chloroplast and mitochondria, indicating that these genes are more likely to be retained than are other nuclear genes. We calculated enrichment (E) for each category by comparing the percentage of B(AC) nuclear genes in a given category to the percentage of A(BC) genes in that category (see Materials and Methods). Positive E indicates enrichment among B(AC) genes; negative E indicates enrichment among A(BC) genes (fig. 4a and supplementary table S6, Supplementary Material online). B(AC) nuclear genes are significantly enriched for organellar localized genes (P = 1.00e-3, Fisher’s) as well as genes that are both organelle localized and known to be involved in cytonuclear interactions (P = 1.23e-3). Enrichment was also detected for the chloroplast and mitochondria individually (P = 3.12e-2 and 2.07e-2, respectively). We saw a general trend of enrichment in the same direction at the level of organellar functional categories but did not perform statistical tests on these due to their small number. We observed the opposite enrichment pattern for genes targeted to other parts of the cell (P = 1.17e-3) (fig. 4a and supplementary table S6, Supplementary Material online). In sum, these results suggest a role for selection in shaping which genes were displaced during introgression.
The Role of Nuclear–Nuclear Interactions during Introgression
We also asked if interactions between/among nuclear genes influenced the likelihood of replacement by foreign alleles. Using Arabidopsis protein–protein interaction data (Brandão et al. 2009), we constructed an interaction network of the full set of single-copy nuclear genes (fig. 4b, left). To assess whether genes with shared history are clustered in the network, we calculated its assortativity coefficient (A). We assessed significance by generating a null distribution for A using 10,000 networks of the same size and shape with randomized topology assignments. In our empirical network, A was significantly positive (A = 0.0885, P = 0.00189, Z-test), and hence topologies are nonrandomly clustered (fig. 4b, right), indicating that selection acted against genotypes containing interactions between introgressed and nonintrogressed alleles.
The Role of Physical Linkage during Introgression
Although it appears gene function exerted influence on nuclear introgression, we also asked whether blocks of genes with similar histories were physically clustered on chromosomes. We looked for evidence of haplotype blocks using the C. rubella genome map (fig. 5). Previous studies in this group estimate linkage disequilibrium to decay within 10 kb (Kim et al. 2007; Song et al. 2009), creating blocks of paternal or maternal genes around that size. We assessed the physical clustering of genes with shared history by two measures: 1) number of instances in which genes with the same topology are located within 10 kb of each other (supplementary fig. S6a, Supplementary Material online) and 2) number of instances in which neighboring genes share topology, regardless of distance (supplementary fig. S6b, Supplementary Material online). The second measure provides a simple measure of clustering without requiring an estimate of ancestral linkage. We compared both measures with a null distribution generated from 10,000 replicated chromosome maps in which the topology assignments were randomized across the marker genes. By both measures, we found significant clustering of A(BC) (measure 1: P = 3.022e-8; measure 2: P = 1.41364e-10, Z-test) and B(AC) (measure 1: P = 0.003645; measure 2: P = 1.7169e-11) genes (supplementary fig. S6c–h, Supplementary Material online). The observed clustering indicates that haplotype blocks of cotransferred and nontransferred genes are detectable in extant genomes, pointing to physical linkage as a factor influencing whether genes are transferred or retained.
Fig. 5.
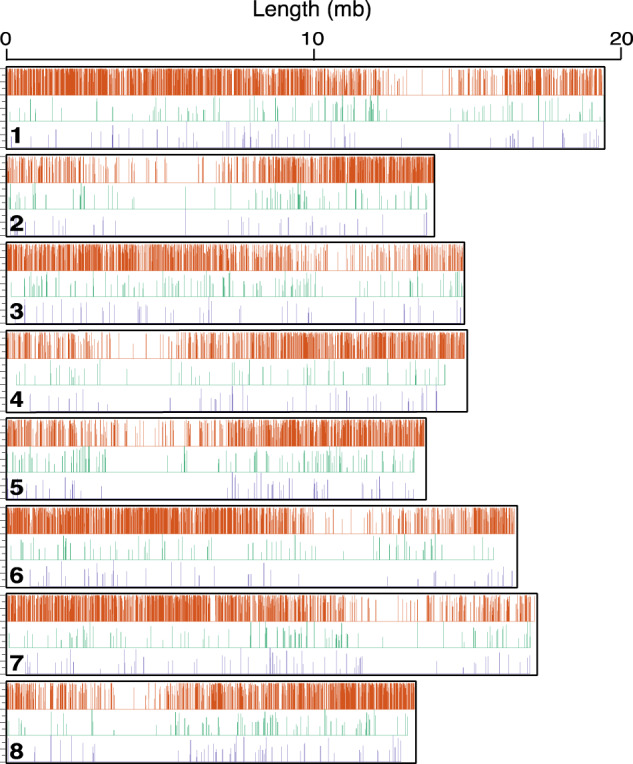
Introgressed and retained haplotype blocks are detectable. Nuclear genes mapped to Capsella rubella. Vertical lines, genes (colored by topology). Line heights proportional to BS (supplementary fig. S2a–c, Supplementary Material online).
Discussion
Phylogenomic studies in plants face unique challenges. The prevalence of gene and genome duplication complicates the detection of orthologs, and thus choosing markers that minimize duplication is extremely important when applying tests of introgression originally developed for animals (Green et al. 2010). Because duplication history cannot be definitively known, we can never be sure that cryptic duplication has not introduced phylogenetic incongruence into our data set; this is a risk in any phylogenetic study, especially in plants. We acknowledge that all nuclear genes have undergone duplication at some point in Brassicaceae (Bowers et al. 2003) and address this challenge by specifically targeting genes least likely to have undergone duplication during the speciation and introgression events we detected. If duplication was biasing the results we obtained from our full single-copy data set, we expected that the proportion of B(AC) trees would have decreased in our conservatively single-copy data set. However, the proportions we observed were not substantially impacted by our conservative single-copy filter. In fact, the proportion of B(AC) genes was slightly higher in the conservatively single-copy genes, the opposite of what we would expect if duplication was creating incongruent trees. Moreover, results of the D-, F-, and DGT-statistics from both data sets significantly favored introgression (supplementary tables S3 and S4, Supplementary Material online), another indication that biases associated with cryptic duplication and loss are not driving our conclusions of introgression.
We applied several methods to distinguish between introgression and ILS. Like all applications of D and related statistics, it is important to acknowledge that ancestral population structure may produce signatures that mimic introgression (Eriksson and Manica 2012). However, when this possibility was thoroughly explored in the case of Neanderthals, introgression remained the favored hypothesis (Lohse and Frantz 2014). Advanced simulation beyond the scope of this study would be required to definitively rule out ancestral population structure in our Brassicaceae system. It is worth noting that, regardless of the measure or approach employed, our results (supplementary fig. S2 and tables S3 and S4, Supplementary Material online), were consistent with an explanation of introgression rather than ILS or duplication and loss.
On the other hand, our analyses were not consistent in their indication of the taxa involved in introgression. D and F were sensitive to the methodology used in filtering the sites included in ABBA/BABA site counts. The more conservative filter, which requires sites to be monoallelic in the two outgroups, leads to fewer total sites being tallied. When only a single outgroup taxon is used to root the tree, more sites meet the necessary criterion to be included in the calculation of D and F. These additional sites appear to be heavily biased toward BABA, causing D and F to shift from positive to negative when these sites are included. It should be noted that application of this filter is nonstandard in D and F analyses and the effects of such a filter have not been thoroughly explored. Inconsistency in D and F analyses led us to explore numerous analytical methods. Although each method significantly supported histories that include introgression, results were not cohesive enough to confidently indicate a single clear introgression scenario. Future studies that employ whole-genomes from additional taxa will likely add resolution to this question. Given the genomes and analytical techniques currently at our disposal, our best interpretation is that C(AB) trees resulted largely from ILS. This interpretation is based on the fact that, although B(AC) is the organelle topology and A(BC) is the major topology in the nucleus, C(AB) is not well supported by the organelles or the nucleus, making it unlikely to represent the signature of the cytonuclear discordance we set out to study. We rely on this interpretation to perform downstream analyses of divergence and functional enrichment but acknowledge that further work is needed to confidently sort out the full series of evolutionary events underlying phylogenetic incongruence in the group. Future analysis of genome-scale data that include denser sampling of representative taxa (Nikolov et al. 2019) may hold the key to resolving some of the questions about complex evolution raised by this study.
One of the major implications of cytonuclear discordance is the potential for cytonuclear incompatibility to arise. We searched for evidence of such incompatibility by using a curated cytonuclear data set (Forsythe et al. 2019) to ask if cytonuclear genes shared the evolutionary history of the organelles more than expected by chance, which would be expected if selection acted to maintain coadapted nuclear and cytoplasmic alleles. We found that nuclear genes encoding organelle-localized and organelle-interacting proteins were enriched for B(AC), the organelle topology (fig. 4a and supplementary table S6, Supplementary Material online). This nonrandom distribution of cytonuclear functions in genes displaying B(AC) versus A(BC) suggests that selection against cytonuclear incompatibility acted. The genes displaying this pattern may constitute a core set whose replacement by introgressed alleles is deleterious. We also find evidence that selection acted to maintain nuclear–nuclear interactions (fig. 4b). In general, our results suggest that epistatic interactions between genes exerted selective pressure that influenced which genes were displaced and which were retained.
We document the presence of statistically detectable genomic blocks of cointrogressed/coretained genes (fig. 5 and supplementary fig. S6, Supplementary Material online). Given observations of nonrandom gene order in eukaryotes (Hurst et al. 2004; Nützmann et al. 2016), it is difficult to fully disentangle functional versus physical linkage, meaning it is possible that chromosomal proximity of interacting genes may contribute to the shared history we documented among interacting genes. However, the dearth of proven functional clusters in plant genomes (Wisecaver et al. 2017) suggests this phenomenon, alone, is unlikely to explain the signatures of selection we describe above. It is also possible that selection drove the displacement or retention of entire haplotype blocks via hitchhiking. Disentangling the interplay of physical linkage versus selection during introgression remains an area of future work.
Our initial interpretation of the observed phylogenetic incongruence was that A(BC) resulted from simple speciation events and B(AC) resulted from introgression between clades A and C, a pattern we referred to as cytoplasmic introgression. However, in light of recent findings from mosquitos (Fontaine et al. 2015; Wen et al. 2016b), we thought it important to consider alternative hypotheses. Using the same approach that revealed introgression in mosquitos, we calculated the mean node depth for each of the alternative topologies we recovered for nuclear genes. In addition, we employed several strategies to account for the effects of selection (fig. 2d), effective population size variation across the genome (fig. 2e), lineage-specific effects (fig. 2f), intragenic recombination (fig. 2g), and mixed distributions caused by the presence of ILS loci in B(AC) and A(BC) trees (supplementary fig. S3o–t, Supplementary Material online) on our node depth calculations. In all cases, our node depth comparisons rejected the hypothesis that the node uniting clades A and C on B(AC) trees resulted from an introgression event, and instead indicated that the node uniting clades B and C on A(BC) trees resulted from an introgression between clades B and C. Thus, given currently available genomic data, our results suggest that the “true” species branching order is B(AC), despite this topology being found for only a small minority of nuclear genes.
There is growing debate about the efficacy of bifurcating phylogenies in describing organismal evolution, prompting the development of powerful network frameworks that highlight reticulation in species relationships (Than et al. 2008; Nakhleh 2013; Hahn and Nakhleh 2016). Although our analysis reinforces the importance of considering reticulation, we also show that bifurcating trees should not be entirely abandoned in the face of reticulation. The presence of reticulation does not preclude the occurrence of simple bifurcating speciation events, it simply means some bifurcations result from speciation, whereas others result from introgression. Therefore, some gene trees will have nodes representing speciation events, whereas other genes trees will have a node or nodes that represent introgression. We define the species branching order as the topology of the gene tree in which all nodes represent speciation events, even if this history does not represent the majority of the genome. Our finding of massive nuclear introgression leads to a dilemma regarding which branching order should be used in future comparative studies in this group. For many (if not most) practical purposes, it is reasonable to continue to use A(BC) because it represents the history of most of the genome. However, we would argue that studies using this topology should bear in mind that the true history is more complicated than simple speciation and consider the potential implications. Integrating all available information into a useful model for studying trait evolution represents a future goal in systematics.
In summary, our comparative genomic analyses suggest that the original observation of “chloroplast capture” is the result of introgression among the ancestors of the extant genera Arabidopsis, Boechera, and Capsella. Moreover, selection and linkage influenced the genes that were ultimately introgressed and retained. To our surprise, we found evidence that the species branching order in this group is more accurately reflected by B(AC), and thus similar to the findings of Fontaine et al. (2015), it appears that nuclear introgression obscured speciation such that the latter was only recoverable from extensive genomic data. What makes introgression here particularly interesting is that its impact on the genome is evident despite the fact that it must have occurred prior to the radiation of clade A 13–9 Ma (Beilstein et al. 2010; Huang et al. 2016). Hence, it is likely that, as additional high-quality genomes become available, comparative analyses will reveal histories that include nuclear introgression, even when the genomes considered are more distantly related. In short, our findings explore the genomic battle underlying chloroplast capture to reveal an onslaught of alleles via directional introgression. A core set of nuclear genes resisted displacement by exogenous alleles; purifying selection removed genotypes with chimeric epistatic combinations that were deleterious, just as Bateson–Dobzhansky–Muller first described (Orr 1996; Sloan et al. 2017). Will other introgression events reveal similar selective constraints as those we detail? If so, it could point us toward key interactions between cytoplasmic and nuclear genomes that lead to successful introgression, thereby refining our understanding of the factors governing the movement of genes among species.
Data Availability
Gene tree data are linked to the online version of the paper. Scripts and input files used to perform analyses are available at https://github.com/EvanForsythe/Brassicaceae_phylogenomics.
Supplementary Material
Acknowledgments
The data reported in this article are provided in the Supplementary Material online. Scripts used to perform analyses are available at https://github.com/EvanForsythe/Brassicaceae_phylogenomics. This work was funded by the National Science Foundation (Grant Nos. 1409251, 1444490, and 1546825 to M.A.B.) and NSF-IOS (1758532 to A.D.L.N.). We thank M.J. Sanderson, M.M. McMahon, E. Lyons, D.B. Sloan, M.P. Simmons, R.N. Gutenkunst, A.E. Baniaga, and S.M. Lambert for helpful discussions and M.T. Torabi, M.C. Borgstrom, and D.S. Clausen for statistical consultation. Finally, this work benefited greatly from input of the PaBeBaMo research group in the School of Plant Sciences, University of Arizona.
Author Contributions
E.S.F. and M.A.B. conceived the study. A.D.L.N. performed organellar genome assembly. E.S.F. performed all other analyses. E.S.F. and M.A.B. wrote the manuscript with input from A.D.L.N. All authors approved of manuscript before submission.
Literature Cited
- Alexander PJ, Windham MD, Govindarajulu R, Al-Shehbaz IA, Bailey CD. 2010. Molecular phylogenetics and taxonomy of the genus Boechera and related genera (Brassicaceae: Boechereae). Syst Bot. 35(3):559–577. [Google Scholar]
- Al-Shehbaz IA, O’Kane SL. 2002. Taxonomy and phylogeny of Arabidopsis (Brassicaceae). Arab B 6:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815. [DOI] [PubMed] [Google Scholar]
- Bailey CD, Al-Shehbaz IA, Rajanikanth G. 2007. Generic limits in tribe Halimolobeae and description of the new genus Exhalimolobos (Brassicaceae). Syst Bot. 32(1):140–156. [Google Scholar]
- Bailey CD, et al. 2006. Toward a global phylogeny of the Brassicaceae. Mol Biol Evol. 23(11):2142–2160. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Kellogg EA. 2006. Brassicaceae phylogeny and trichome evolution. Am J Bot. 93(4):607–619. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Mathews S, Kellogg EA. 2008. Brassicaceae phylogeny inferred from phytochrome A and ndhF sequence data: tribes and trichomes revisited. Am J Bot. 95(10):1307–1327. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. 2010. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci U S A. 107(43):18724–18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JL, Chapman BA, Rong J, Paterson AH. 2003. Unraveling angiosperms genome evolution by phylogenetic analysis of chromosomal duplications events. Nature 422(6930):433–438. [DOI] [PubMed] [Google Scholar]
- Brandão MM, Dantas LL, Silva-Filho MC. 2009. AtPIN: Arabidopsis thaliana protein interaction network. BMC Bioinf. 10(1):454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copetti D, et al. 2017. Extensive gene tree discordance and hemiplasy shaped the genomes of North American columnar cacti. Proc Natl Acad Sci U S A. 114:201706367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur TLP, et al. 2010. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol Biol Evol. 27(1):55–71. [DOI] [PubMed] [Google Scholar]
- Dasmahapatra KK, et al. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R, et al. 2013. Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc Natl Acad Sci U S A. 110(8):2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte JM, et al. 2010. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol Biol. 10:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 28(8):2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton DAR, Hipp AL, González-Rodríguez A, Cavender-Bares J. 2015. Historical introgression among the American live oaks and the comparative nature of tests for introgression. Evolution (N Y). 69(10):2587–2601. [DOI] [PubMed] [Google Scholar]
- Eaton DAR, Ree RH. 2013. Inferring phylogeny and introgression using RADseq data: an example from flowering plants (Pedicularis: Orobanchaceae). Syst Biol. 62(5):689–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson A, Manica A. 2012. Effect of ancient population structure on the degree of polymorphism shared between modern human populations and ancient hominins. Proc Natl Acad Sci U S A. 109(35):13956–13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine MC, et al. 2015. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347(6217):1258524–1258526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe ES, Sharbrough J, Havird JC, Warren JM, Sloan DB. 2019. CyMIRA: the Cytonuclear Molecular Interactions Reference for Arabidopsis. Genome Biol Evol. 11(8):2194–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe ES, Sloan DB, Beilstein MA. 2020. Divergence-based introgression polarization. Genome Biol Evol. 12(4):463–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzke A, German D, Al-Shehbaz IA, Mummenhoff K. 2009. Arabidopsis family ties: molecular phylogeny and age estimates in Brassicaceae. Taxon 58(2):425–427. [Google Scholar]
- Galasso I, Manca A, Braglia L, Ponzoni E, Breviario D. 2015. Genomic fingerprinting of Camelina species using cTBP as molecular marker. Am J Plant Sci. 06(08):1184–1200. [Google Scholar]
- Gan X, et al. 2016. The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat Plants 2(11):16167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5(10):R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40:1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, et al. 2010. A draft sequence of the Neandertal genome. Science 328(5979):710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Nakhleh L. 2016. Irrational exuberance for resolved species trees. Soc Study Evol. 70(1):7–17. [DOI] [PubMed] [Google Scholar]
- Hibbins MS, Hahn MW. 2019. The timing and direction of introgression under the multispecies network coalescent. Genetics 211(3):1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 43(5):476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-H, et al. 2016. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol Biol Evol. 33(2):394–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford MB, et al. 2013. The genomic signature of crop-wild introgression in maize. PLoS Genet. 9(9):e1003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pál C, Lercher MJ. 2004. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 5:299–310. [DOI] [PubMed] [Google Scholar]
- Huson DH, et al. 2005. Reconstruction of reticulate networks from gene trees. Res Comput Mol Biol Proc. 3500:233–249. [Google Scholar]
- Joly S, McLenachan PA, Lockhart PJ. 2009. A statistical approach for distinguishing hybridization and incomplete lineage sorting. Am Nat. 174(2):E54–E70. [DOI] [PubMed] [Google Scholar]
- Kagale S, et al. 2014. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat Commun. 5:3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, et al. 2007. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 39(9):1151–1155. [DOI] [PubMed] [Google Scholar]
- Koch M, Haubold B, Mitchell-Olds T. 2001. Molecular systematics of the Brassicaceae: evidence from coding plastidic matK and nuclear Chs sequences. Am J Bot. 88(3):534–544. [PubMed] [Google Scholar]
- Lamesch P, et al. 2012. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40:1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-R, et al. 2017. Young inversion with multiple linked QTLs under selection in a hybrid zone. Nat Ecol Evol. 1:0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Yaw JA, Grassa CJ, Joly S, Andrew RL, Rieseberg LH. 2019. An evaluation of alternative explanations for widespread cytonuclear discordance in annual sunflowers (Helianthus). New Phytol. 221(1):515–526. [DOI] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse K, Frantz L. 2014. Neandertal admixture in Eurasia confirmed by maximum-likelihood analysis of three genomes. Genetics 196(4):1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant N, et al. 2016. The iPlant collaborative: cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol. 14(1):e1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhleh L. 2013. Computational approaches to species phylogeny inference and gene tree reconciliation. Trends Ecol Evol. 31:1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M. 2003. Mixing patterns in networks. Phys Rev E 67(2):026126. [DOI] [PubMed] [Google Scholar]
- Nikolov LA, et al. 2019. Resolving the backbone of the Brassicaceae phylogeny for investigating trait diversity. New Phytol. 222(3):1638–1651. [DOI] [PubMed] [Google Scholar]
- Novikova PY, et al. 2016. Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat Genet. 48(9):1077–1082. [DOI] [PubMed] [Google Scholar]
- Nützmann H-W, Huang A, Osbourn A. 2016. Plant metabolic clusters—from genetics to genomics. New Phytol. 211(3):771–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ΘOrr HA. 1996. Dobzhansky, Bateson, and the genetics of speciation. Genetics 144(4):1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama RK, et al. 2008. The shrunken genome of Arabidopsis thaliana. Plant Syst Evol. 273(3–4):257–271. [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20(2):289–290. [DOI] [PubMed] [Google Scholar]
- Pease JB, Hahn MW. 2015. Detection and polarization of introgression in a five-taxon phylogeny. Syst Biol. 64(4):651–662. [DOI] [PubMed] [Google Scholar]
- Peri S, et al. 2020. Read mapping and transcript assembly: a scalable and high-throughput workflow for the processing and analysis of ribonucleic acid sequencing data. Front Genet. 10:1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel DH, Boyle AP, Araya CL, Snyder MP. 2014. Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30(19):2808–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3(2):217–223. [Google Scholar]
- Rieseberg LH, Soltis DE. 1991. Phylogenetic consequences of cytoplasmic gene flow in plants. Evol. Trends Plants 5:65–84. [Google Scholar]
- Rieseberg LH, Whitton J, Linder CR. 1996. Molecular marker incongruence in plant hybrid zones and phylogenetic trees. Acta Bot Neerl. 45(3):243–262. [Google Scholar]
- Rosenzweig BK, Pease JB, Besansky NJ, Hahn MW. 2016. Powerful methods for detecting introgressed regions from population genomic data. Mol Ecol. 25(11):2387–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol Biol Evol. 19(1):101–109. [DOI] [PubMed] [Google Scholar]
- Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27(4):592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz ME, Windsor AJ, Song B-H, Lawton-Rauh A, Mitchell-Olds T. 2007. Comparative genetic mapping in Boechera stricta, a close relative of Arabidopsis. Plant Physiol. 144(1):286–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Havird JC, Sharbrough J. 2017. The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol Ecol. 26(8):2212–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T, Ceplitis A, Neuffer B, Hurka H, Lascoux M. 2006. Intrageneric phylogeny of Capsella (Brassicaceae) and the origin of the tetraploid C. bursa-pastoris based on chloroplast and nuclear DNA sequences. Am J Bot. 93(11):1714–1724. [DOI] [PubMed] [Google Scholar]
- Slotte T, et al. 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet. 45(7):831–835. [DOI] [PubMed] [Google Scholar]
- Song BH, et al. 2009. Multilocus patterns of nucleotide diversity, population structure and linkage disequilibrium in Boechera stricta, a wild relative of Arabidopsis. Genetics 181(3):1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. 1969. The significance of hybridization for plant taxonomy and evolution. Taxon 18(1):26–35. [Google Scholar]
- Stenz NWM, Larget B, Baum DA, Ané C. 2015. Exploring tree-like and non-tree-like patterns using genome sequences: an example using the inbreeding plant species Arabidopsis thaliana (L.) Heynh. Syst Biol. 64(5):809–823. [DOI] [PubMed] [Google Scholar]
- Stolzer M, et al. 2012. Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics 28(18):i409–i415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than C, Ruths D, Nakhleh L. 2008. PhyloNet: a software package for analyzing and reconstructing reticulate evolutionary relationships. BMC Bioinf. 9(1):322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28(5):511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung Ho LS, Ané C. 2014. A linear-time algorithm for Gaussian and non-Gaussian trait evolution models. Syst Biol. 63(3):397–408. [DOI] [PubMed] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27(2):171–180. [DOI] [PubMed] [Google Scholar]
- Wen D, Yu Y, Hahn MW, Nakhleh L. 2016. a. SOM: reticulate evolutionary history and extensive introgression in mosquito species revealed by phylogenetic network analysis. Mol Ecol. 25(11):2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D, Yu Y, Hahn MW, Nakhleh L. 2016. b. Reticulate evolutionary history and extensive introgression in mosquito species revealed by phylogenetic network analysis. Mol Ecol. 25(11):2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver JH, et al. 2017. A global coexpression network approach for connecting genes to specialized metabolic pathways in plants. Plant Cell 29(5):944–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimowski SB, Rieseberg LH. 2014. The role of homoploid hybridization in evolution: a century of studies synthesizing genetics and ecology. Am J Bot. 101(8):1247–1258. [DOI] [PubMed] [Google Scholar]
- Yang R, et al. 2013. The reference genome of the halophytic plant Eutrema salsugineum. Front Plant Sci. 4:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. 2012. ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun. 419(4):779–781. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Janke A. 2018. Gene flow analysis method, the D-statistic, is robust in a wide parameter space. BMC Bioinf. 19(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene tree data are linked to the online version of the paper. Scripts and input files used to perform analyses are available at https://github.com/EvanForsythe/Brassicaceae_phylogenomics.