Abstract
Corals are the ecosystem engineers of coral reefs, one of the most biodiverse marine ecosystems. The ability of corals to form reefs depends on the precipitation of calcium carbonate (CaCO3) under biological control. However, several mechanisms underlying coral biomineralization remain elusive, for example, whether corals employ different molecular machineries to deposit different CaCO3 polymorphs (i.e., aragonite or calcite). Here, we used tandem mass spectrometry (MS/MS) to compare the proteins occluded in the skeleton of three octocoral and one scleractinian species: Tubipora musica and Sinularia cf. cruciata (calcite sclerites), the blue coral Heliopora coerulea (aragonitic skeleton), and the scleractinian aragonitic Montipora digitata. Reciprocal Blast analysis revealed extremely low overlap between aragonitic and calcitic species, while a core set of proteins is shared between octocorals producing calcite sclerites. However, the carbonic anhydrase CruCA4 is present in the skeletons of both polymorphs. Phylogenetic analysis highlighted several possible instances of protein co-option in octocorals. These include acidic proteins and scleritin, which appear to have been secondarily recruited for calcification and likely derive from proteins playing different functions. Similarities between octocorals and scleractinians included presence of a galaxin-related protein, carbonic anhydrases, and one hephaestin-like protein. Although the first two appear to have been independently recruited, the third appear to share a common origin. This work represents the first attempt to identify and compare proteins associated with coral skeleton polymorph diversity, providing several new research targets and enabling both future functional and evolutionary studies aimed at elucidating the origin and evolution of coral biomineralization.
Keywords: biomineralization, corals, proteomics, Octocorallia
Significance
In corals (Cnidaria: Anthozoa), skeletons play important ecological functions and services as they form the structural foundation of coral reefs. Several genes have been linked to the ability of corals to calcify, but to date, information for subclass Octocorallia is extremely limited. Notably, skeleton proteomes (i.e., proteins occluded within the mineral) have not been characterized yet. Here, we employed proteomics and phylogenetics to characterize the skeletal proteome for three octocoral species exhibiting different skeleton mineralogy (aragonite or calcite). The extremely low proteome overlap observed here shows that aragonite and calcite-forming octocorals have evolved very different proteins repertoires to form their skeletons. On the other hand, some octocoral skeletogenic proteins appear homologous to proteins found in scleractinian skeletons, pointing to common calcification-related functions in different coral groups.
Introduction
The capacity of animals to actively control the deposition of mineral skeletons has been a long debated topic, with different models of calcification being proposed over the years. In the “organic matrix mediated” (Lowenstam 1981) or “biologically controlled” (Mann 1983) scenario, an animal employs sets of macromolecules to guide the deposition of its mineral skeletal structures. In line with this, several biomineralization-related processes including crystal nucleation and growth (Mitterer 1978; Wheeler et al. 1981; Liu et al. 2012; Von Euw et al. 2017) or the induction of a given calcium carbonate (CaCO3) polymorph (i.e., aragonite and calcite) (Falini et al. 1996; Amos et al. 2010; Goffredo et al. 2011; Rahman et al. 2011) appear to be regulated by proteins included in the skeleton organic matrix (OM). This consists of a diverse array of proteins, polysaccharides (Goldberg 2001; Marin et al. 2016; Naggi et al. 2018), and lipids (Farre and Dauphin 2009; Farre et al. 2010) occluded within the mineral fraction of the skeleton. Over the last few years, advances in coupled proteomic and transcriptomic research have enabled the simultaneous characterization of several OM proteins in different groups of marine calcifying invertebrates, including mollusks (Marie et al. 2010, 2013; Mann and Jackson 2014), scleractinian corals (Drake et al. 2013; Ramos-Silva et al. 2013; Takeuchi et al. 2016), brachiopods (Jackson et al. 2015), and echinoderms (Mann et al. 2008; Flores et al. 2016; Flores and Livingston 2017). These studies showed that invertebrate skeletal proteomes include varying fractions of novel proteins—producing no significant matches against predicted proteins in genome sequence databases—and do exhibit contrasting rates of conservation between and within lineages. For instance, about 40% of the skeletal proteome is shared among echinoderms (Flores and Livingston 2017), whereas in mollusks the fraction of shared proteins is only about 10% (Kocot et al. 2016). Although a core set of proteins appears to be conserved across mollusks, irrespective of the morphological features of the shell (Arivalagan et al. 2017), the occurrence of both aragonite and calcite layers within the shell allowed Marie et al. (2012) to identify shell matrix proteins associated with different CaCO3 polymorphs. The different proteins specifically associated with the aragonitic or calcitic shell layers suggest that mollusks may use different molecular mechanisms for the deposition of these structures (Marie et al. 2012).
In corals (class Anthozoa, phylum Cnidaria), putative relationships between structural characteristics of the skeleton, like its CaCO3 polymorph, and the molecular machinery employed for its formation, have hitherto only been marginally addressed, although some skeletal coral proteins have been suggested to drive the in vitro crystallization of specific calcium carbonate polymorphs (Goffredo et al. 2011; Rahman et al. 2011). However, no study to date has leveraged mass spectrometry-based protein discovery methods to characterize and compare skeletal proteomes across corals that exhibit different biomineralization strategies, that is, those that produce aragonite versus those that produce calcite. CaCO3 skeleton-producing anthozoan corals belong to two different clades, namely the order Scleractinia (stony corals; subclass Hexacorallia) and the subclass Octocorallia (soft corals). As major contributors to CaCO3 deposition, scleractinians have been the focus of extensive biomineralization-related research, and skeletal proteomes have been characterized for different scleractinian species (Drake et al. 2013; Ramos-Silva et al. 2013; Takeuchi et al. 2016). However, the uniformity in biomineralization strategies (i.e., aragonitic exoskeleton) present among scleractinians makes this group inappropriate to investigate the biological regulation of skeletal polymorph deposition and its evolution. On the contrary, both calcite and aragonite-forming corals are present within Octocorallia (class Anthozoa). Thus, octocoral skeletons offer a unique opportunity to compare the skeletal repertoires associated with different skeletal structures and CaCO3 polymorphs. However, information on octocoral biomineralization-related proteins is extremely limited (but see Debreuil et al. 2012; Rahman et al. 2011). For instance, the lack of comparative developmental information and phylogenetic resolution (McFadden et al. 2006, 2010) renders evolutionary relationships between aragonite and calcite skeletons elusive.
To shed new light on aragonite and calcite formation in octocorals, we used tandem mass spectrometry (MS/MS) to characterize the skeletal proteome of three species exhibiting different skeleton morphologies and mineralogies: the leather coral Sinularia cf. cruciata and the pipe organ coral Tubipora musica, both characterized by the production of calcite sclerites, and the massive, aragonitic blue coral Heliopora coerulea. To compare skeletal repertoires between scleractinians and aragonitic octocorals, we additionally examined the proteome of the stony scleractinian coral Montipora digitata. Our work represents the first study of skeletome diversity across and within anthozoan corals providing 1) the identification of several new coral biomineralization-related proteins and 2) a comparative analysis examining putative relations between polymorph and type of skeletal structure, and the molecular machinery employed by corals for its formation.
Materials and Methods
Extraction of OM Proteins
Samples of T. musica, H. coerulea, S. cf. cruciata, and M. digitata cultured in research aquaria (closed artificial seawater systems) were bleached in 5% NaOCl (Sigma-Aldrich) for 72 h to remove the tissue and other potential contaminants. They were subsequently rinsed several times with ultrapure water and oven-dried at 37 °C. Clean skeletons were ground to powder with a mortar and pestle, and again bleached (5% NaClO solution for 5 h), washed with ultrapure water, and oven-dried at 37 °C. Four and seven grams of clean skeleton powder were used (per extraction) for calcite and aragonite-forming corals, respectively. The skeleton powder was decalcified with 10% acetic acid for 24 h at room temperature on an orbital shaker. The decalcification solution was centrifuged (14,000 × g, 30 min, 25 °C) to separate the acid soluble (ASM) and insoluble (AIM) fractions. The obtained insoluble pellets were washed several times with ultrapure water, dried and stored at −80 °C until further analysis. The supernatants (ASM) were desalted and concentrated using Amicon Ultrafiltration devices (15 ml, 3 kDa cutoff), and the ASM proteins were precipitated following the method described in Wessel and Flügge (1984). Briefly, four volumes of methanol, one chloroform, and three volumes of water were added to one volume of sample and the solution was centrifuged at 14,000 × g for 20 min at 25 °C. After centrifugation, the supernatant was discarded. Three volumes of methanol were added and the solution was centrifuged again. The resulting protein pellets were air-dried and stored at −80 °C. For each species, two skeleton samples from the same colony were independently processed.
SDS-PAGE Analysis
ASM and AIM proteins were dissolved in 2× Laemmli buffer (95% buffer and 5% beta-mercaptoethanol) (BioRad). As observed by Ramos-Silva et al. (2013), AIM pellets were only partly dissolved. Samples were denatured for 2 min at 95 °C and loaded onto 12.5% polyacrylamide gels. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was run on a BioRad MiniProtean Tetra Cell at constant voltage for about 70 min. Proteins were visualized after staining with ProteoSilver Silver Stain Kit (Sigma-Aldrich) (supplementary fig. 1, Supplementary Material online) with the Precision Plus Protein Dual Color Standards (BioRad) as a size marker.
Proteomic Analysis
For mass spectrometry analysis, sample aliquots were loaded onto a 12.5% acrylamide gel and run as described above. In an effort to reduce potential variability due to technical factors, we included replicates within the experimental design. For each species, two protein extractions were performed. From each extraction, three ASM and AIM triplicates were loaded on gels and analyzed with mass spectrometry. Information on numbers of detected proteins between replicates and extractions is available in supplementary figures 6 and 7, Supplementary Material online, respectively. However, the presence of technical causes underlying the nondetection of skeleton organic matrix proteins (SOMPs) cannot be completely excluded. After trypsin digestion proteins were analyzed on a Bruker Impact II Q-Tof mass spectrometer (Bruker Corp., Billerica, MA) coupled with an Ultimate 3000 RSLC nano liquid chromatography (Thermo Fisher, Waltham, MA). Peptide separation was performed using an Acclaim PepMap RSLC column with 75 μm diameter, 25 cm length, C18 particles of 2 μm diameter and 100 Å pore size (Thermo Fisher). Data were analyzed using MaxQuant 1.5.2.8 (Cox and Mann 2008). The following parameters were used: mass error = max 10 ppm, two allowable cleavages, and false discovery rate = 0.01%. Common contaminants, potential symbiont, and bacterial sequences were filtered. Peptides were mapped against respective octocoral (Conci et al. 2019) and M. digitata (González-Pech et al. 2017) transcriptomes.
Assemblies are available online at https://gitlab.lrz.de/palmuc/concietal_octoskeletomes and https://github.com/PalMuc/Montipora_digitata_resources, last accessed August 11, 2020). Sample processing and mass spectrometry were performed by the MSBioLMU Unit at the Biology Department I of the Ludwig-Maximilians University in Munich (Germany). Sequences with at least two unique matching peptides were considered for downstream analysis (supplementary material 1, Supplementary Material online).
Bioinformatic Analysis of OM Proteins
Identified OM proteins were annotated by BlastP (cutoff e-value: 1e−10) against the NCBI nonredundant database. To determine skeletal proteins shared between species and putative orthology (best reciprocal hits), a bidirectional Blast analysis was performed (supplementary material 8, Supplementary Material online). Distribution of homologs (supplementary material 2, Supplementary Material online) of octocoral and M. digitata SOMPs was subsequently assessed within a set of cnidarian genomes and transcriptomes (Shinzato et al. 2011; Pratlong et al. 2015; Voolstra et al. 2015, 2017; Liew et al. 2016; Jeon et al. 2019) with BlastP applying the same search criteria described above. Presence of signal peptide, transmembrane regions, and GPI-anchor was predicted with SignalP 4.0 (Petersen et al. 2011), TMHMM 2.0 (Krogh et al. 2001), and PredGPI (Pierleoni et al. 2008), respectively. Protein isoelectric point was determined with ProtParam (Gasteiger et al. 2005). The amino acid composition of acidic proteins (i.e., isoelectric point <4.5) (Marin and Luquet 2007) detected was computed with a custom script available at https://gitlab.lrz.de/palmuc/Concietal_proteomics_skeletomes (last accessed August 11, 2020). Relative amino acid frequencies within acidic proteins were determined on sequences predicted as complete after removal of the signal peptide sequence. To incorporate information on aspartic acid residue distribution within protein sequences, the median distance between consecutive aspartic acids was calculated. Median distance values and amino acid frequencies were then used to perform principal component analysis (PCA) of acidic proteins.
Phylogenetic Analyses
For phylogenetic inference, protein queries were blasted against a database of cnidarian sequences (supplementary material 3, Supplementary Material online). The following e-value cutoffs were used: 1e−05 (scleritin), 1e−20 (carbonic anhydrase [CA]), and 1e−50 (hephaestin-like). Sequences predicted as “internal” (i.e., lacking both 3′ and 5′ ends) by TransDecoder (Galaxy version 3.0.1) were discarded and a minimum length filter of 250 and 600 residues was applied to significant hits for CA and hephaestin-like, respectively. For the former, sponge and human sequences used in Voigt et al. (2014) were added to the data set. CAs from the green algae Chlamydomonas reinhaardtii (P20507) and Desmodesmus sp. (AOL92959.1) were used as the outgroup. Sequences were aligned with both MUSCLE (Edgar 2004) and MAFFT v.7 (Katoh and Standley 2013), and best-fit models were estimated with Prottest 3.4 (Darriba et al. 2011). Maximum-likelihood analysis was performed in Seaview 4 (Gouy et al. 2010) using PhyML 3.1 (Guindon and Gascuel 2003), whereas MrBayes v.3.2 was used for Bayesian inferences. Trees were sampled every 100th generation (nruns = 2) and burn-in fraction for each analysis was determined after visual inspection of the trace files using Trace v1.6 (available at http://tree.bio.ed.ac.uk/software/tracer, last accessed August 11, 2020). All alignments, trees, and protein sequences used for phylogenetic analyses are available at https://gitlab.lrz.de/palmuc/Concietal_proteomics_skeletomes (last accessed August 11, 2020).
Results
Shared and Species-Specific Components of the Anthozoan Skeletome
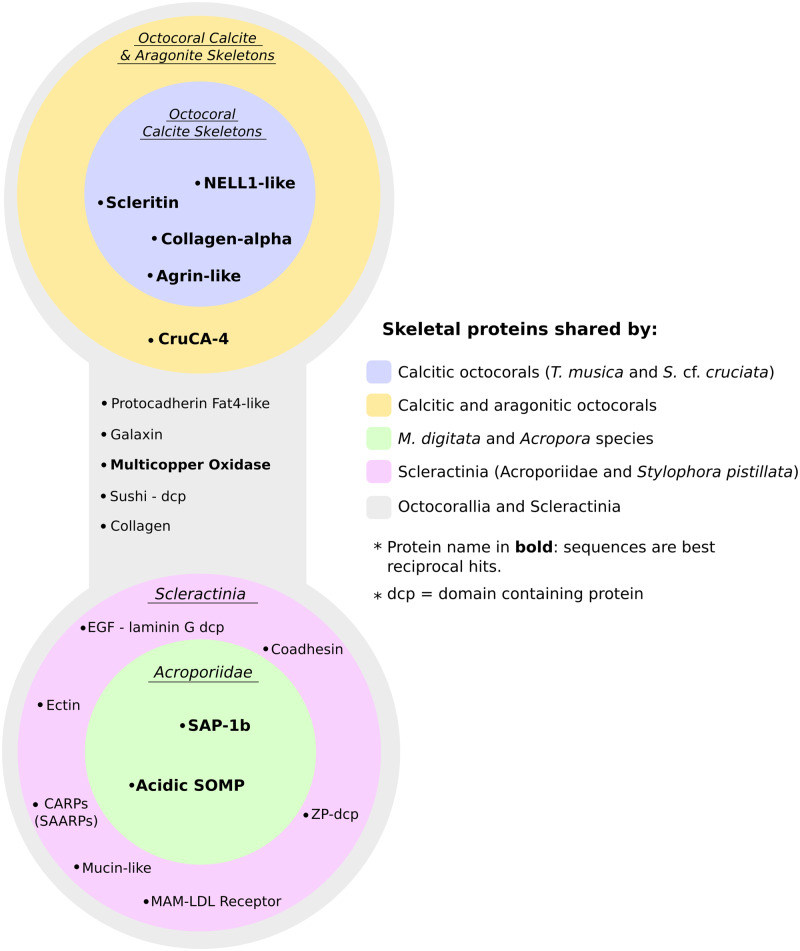
The discovery and subsequent annotation of anthozoan SOMPs retrieved between 12 and 54 proteins (supplementary materials 4–7, Supplementary Material online) with low protein numbers shared between octocoral species. However, simultaneously small sets of skeletal proteins shared between organisms at different taxonomic levels were identified (fig. 1). These included instances of proteins being secreted in the skeleton of both scleractinians and octocorals. The aragonitic octocoral H. coerulea did not exhibit higher similarity to aragonitic scleractinians compared with calcitic soft corals. Thus, proteins shared between Octocorallia and Scleractinia do not appear to be linked to the skeleton polymorph.
Fig. 1.
Overview of shared skeletome proteins across Anthozoa. Alcyonacea includes the proteomes of Tubipora musica (this study) and Sinularia cf. cruciata (this study). Calcitic octocorals: Alcyonacea + involvement of scleritin in Corallium rubrum (Debreuil et al. 2012). Octocorallia: Alcyonacea + Heliopora coerulea (this study) + involvement of CruCA4 in C. rubrum (Le Goff et al. 2016). Acroporiidae: proteomes of Montipora digitata (this study) + Acropora digitifera (Takeuchi et al. 2016) + Acropora millepora (Ramos-Silva et al. 2013). Scleractinia: Acroporidae + proteome of Stylophora pistillata (Drake et al. 2013).
Common to all four species analyzed is a Sushi-domain containing protein also containing NIDO (IPR003886), AMOP (IPR005533), and Von Willebrand factor D (IPR001846) domains. This same arrangement is present in the mucin-like B3EWY9 found in the Acropora millepora SOMP, and the proteins identified in this study also share local similarity with protein P13 from the skeleton of Stylophora pistillata. Other proteins secreted in both scleractinian and octocoral skeletons include a putative homolog of galaxin, a hephaestin-like multicopper oxidase (MCO), one cell adhesion protocadherin Fat 4-like protein, and CAs. The first one, galaxin, was detected in the sclerites of T. musica (TR44621|c0_g1_i1) and we named it octogalaxin-1. In addition to the characteristic presence of multiple di-cysteine motifs, this protein is, as in scleractinians, predicted to be secreted and the signal peptide is followed by a R-X-R-R endoprotease target motif (Fukuda et al. 2003). The MCO was found in S. cf. cruciata (TR42435|c0_g1_i1) and the protocadherin-like proteins were identified in the skeleton of H. coerulea (DN66065_c0_g1_i4). Although our comparative analysis of octocoral skeletal proteomes did not find evidence of a conserved octocoral biomineralization toolkit, we found homologs of Corallium rubrum’s CA CruCA4 (Le Goff et al. 2016) in both aragonitic and calcitic species. One CruCA4 homolog was found in S. cruciata, whereas in the aragonitic blue coral two homologs of CruCA4 were detected. As reported for C. rubrum (Le Goff et al. 2016; Del Prete et al. 2017), the histidine residue involved in proton transfer is not conserved in all other homologs of the protein (supplementary fig. 2, Supplementary Material online). Mutation of the His64 residues has been linked to decreases in catalytic efficiency (Vullo et al. 2008). Additionally, one H. coerulea CAs (DN64689_c5_g1_i4) is predicted to be an acatalytic CA-related protein, as it lacks one of the three zinc-binding histidines (supplementary fig. 2, Supplementary Material online) (Supuran 2008; Picaud et al. 2009). Four additional proteins are present in the sclerites of both calcitic octocorals analyzed and they represent best reciprocal hits between the two species. This sclerite “toolkit” includes 1) scleritin, 2) an agrin-like protein consisting of repeated Kazal domains (IPR036058) and one C-terminal WAP (IPR008197) domain, 3) a kinase C-binding, NELL1-like protein, and 4) one collagen alpha-chain like protein. Two scleritin-like sequences (TR40200|c16_g1_i1 and TR42410|c0_g2_i1) were detected in T. musica, whereas only one match was produced in Sinularia. Agrin is a glycoprotein which in humans participates in cell–matrix interactions (Groffen et al. 1998), and agrin-like and agrin-like protease inhibitors have been recently found in the skeleton of the seastar Patiria miniata (Flores and Livingston 2017). NELL-1 is, on the other hand, involved in bone formation in vertebrates (Aghaloo et al. 2007; Zou et al. 2011). The NELL1-like protein identified here also exhibits local similarity to P32 (kielin-like), a secreted protein found in the skeleton of St. pistillata (Drake et al. 2013).
In the scleractinian M. digitata, with the exception of the secreted acidic protein SAP-1a, we retrieved the entire acidic protein repertoire previously isolated from the skeletons of A. millepora and Acropora digitifera (Ramos-Silva et al. 2013; Takeuchi et al. 2016). This includes both secreted aspartic acid-rich proteins SAARP-1 and SAARP-2, the acidic secreted OM protein B3EWY7 and the secreted acidic protein SAP-1b. In addition, putative orthologs for A. millepora mucin-like (B3EWY9), coadhesin-like (B3EWZ3.1), and CA (B8V7P3.1) were also present. Of note is the presence in M. digitata of a lithostatine-like protein containing a c-type lectin domain. The presence of this domain is a common feature for skeletal proteins in several marine invertebrates, such as mollusks (Mann et al. 2000; Weiss et al. 2000; Matsubara et al. 2008; see Sarashina and Endo 2006 for a review) and birds (Mann and Siedler 2004, 2006). SOMPs with sequence similarity to lithostatin and c-type lectin-like proteins have been characterized from the skeletons of echinoderms (Wilt 2002) but have, to our knowledge, not been reported in corals to date.
Similarity between Scleractinian and Octocoral Acidic Proteins
Two acidic proteins (DN60904_c0_g1_i1 and DN65627_c8_g3_i2) were detected in the OM of H. coerulea and one in T. musica (TR43768|c0_g2_i1). For both proteins, significant matches in public databases (BlastP e-value cutoff: 1e−05) were limited to Octocorallia. As for scleractinians acidic SOMPs, homologs of H. coerulea acidic protein 1 exhibit higher isoelectric points which are related to lower aspartic acid contents. In an effort to investigate sequence similarities between octocoral and scleractinian acidic SOMPs and their nonacidic homologs in a phylogenetic independent way, we conducted a PCA based on sequence amino acid composition and the distribution (i.e., median distance) of aspartate residues along the sequence (fig. 2).
Fig. 2.
PCA of acidic SOMPs found in scleractinians (Drake et al. 2013; Ramos-Silva et al. 2013) and Heliopora coerulea, and their putative nonacidic homologs found in other cnidarian species (Conci et al. 2019). Black arrow: H. coerulea acidic protein 1 (DN60904_c0_g1_i1). The four octocoral sequences are represented by a yellow star symbol (independent of protein acidity). The acidic protein found in the sclerites of Tubipora musica was not included as its partial sequence does not allow to calculate amino acid frequencies accurately. D, aspartic acid; E, glutamic acid; G, glycine; K, lysine; R, arginine.
Despite not having significant similarity to scleractinian sequences, H. coerulea acidic protein-1 grouped together with the scleractinian acidic protein ASOMP-P27 (Drake et al. 2013; Ramos-Silva et al. 2013; Conci et al. 2019). Main sequence features contributing to the clustering patterns observed are similarities in aspartic acid content and distribution within the protein sequence. Nonacidic homologs of H. coerulea acidic protein-1 clustered with other nonacidic proteins. Apart from exhibiting lower aspartate contents, proteins within this group are characterized by a higher lysine and glycine content compared with their acidic putative homologs.
Evolutionary History of Octocoral and Scleractinian SOMPs
To further explore the evolutionary history of octocoral and scleractinian SOMPs, we estimated phylogenies for selected proteins derived from the skeletomes. Here, we focused on 1) scleritin, to examine phylogenetic relationships between the proteins found in calcite sclerites and scleritin expressed by H. coerulea and 2) the hephaestin-like MCO, as it represents the only detected instance of a potentially orthologous protein found in both octocoral and scleractinian skeletons. Finally, we surveyed a taxonomically comprehensive data set of cnidarian transcriptomes, with the aim to further resolve evolutionary relationships of (octo)coral skeletal CAs,
Information on scleritin secretion into octocoral skeletons was integrated with previously estimated scleritin presence–absence data (fig. 3a). Data-mining of octocoral transcriptomes showed the presence of multiple scleritin encoding transcripts within a single species (e.g., C. rubrum). Phylogenetic analysis split scleritin homologs into two distinct and well supported clades (fig. 3b and supplementary fig. 3, Supplementary Material online). The three sequences identified in T. musica and S. cf. cruciata grouped together with the scleritin originally described in C. rubrum by Debreuil et al. (2012), alongside all other scleritin homologs found in octocoral species characterized by the presence of calcitic sclerites (fig. 3a).
Fig. 3.
(a) Presence–absence of scleritin in octocoral skeletomes in relation to skeletal structures. (b) Phylogenetic analysis of scleritin. Arrows point to sequences secreted into calcite sclerites. *Protein identified in Debreuil et al. (2012). Protein sequences were aligned with MUSCLE and maximum-likelihood analysis (400 replicated) was done with Seaview 4. Bayesian analysis was performed with MrBayes 3.2. Black dot on node indicates full support (100% bootstrap value and 1.0 posterior probability). Bootstrap values >50 and posterior probabilities >0.95 given at each node. Posterior probabilities are not shown regardless of their value if bootstrap support is <50, if bootstrap was below 50. Bootstrap support is <50, posterior. Involvement of scleritin in Corallium rubrum biomineralization based on Debreuil et al. (2012). Phylogeny based on MAFFT aligning algorithm in supplementary figure 3, Supplementary Material online. All alignments available online at https://gitlab.lrz.de/palmuc/Concietal_proteomics_skeletomes (last accessed August 11, 2020).
We, therefore, referred to this clade as “skeletal” because all the scleritin sequences implicated to date in octocoral biomineralization are included in it. The H. coerulea sequence grouped on the other hand with scleritin-like sequences from C. rubrum.
To infer a phylogenetic tree for hephaestin-like proteins, putative homologs were searched across Cnidaria using the three MCOs included in Takeuchi et al (2016) as a search query. Each query protein occupied a different clade populated by scleractinian and corallimorph sequences (fig. 4 and supplementary fig. 4, Supplementary Material online). Proteins present in scleractinian skeletons all grouped within clade 1.
Fig. 4.
(a) Phylogenetic analysis (400 bootstrap replicates) of cnidarian MCOs. Arrows point to proteins found in coral skeletons. Bold text: sequences determined from genome data, others from transcriptomes. Aligning algorithm: MAFFT. Best-fit model: WAG + G + I. Number on nodes: bootstrap support and posterior probability values. Support values in bold: node supported (>50) in MUSCLE-based phylogeny (supplementary fig. 4, Supplementary Material online). (b) Section of multiple consensus (60%) sequences alignment for the three “corallimorph + scleractinian” clades. Blue box highlights the absence of the type-I copper-binding histidine in clades 2 and 3. Histidine classification based on Takeuchi et al. (2016). All alignments available at https://gitlab.lrz.de/palmuc/Concietal_proteomics_skeletomes (last accessed August 11, 2020).
Homologs identified in black corals (Antipatharia) grouped within clade 1 but with low support. Analysis of the consensus sequence alignment shows that one of the histidines involved in copper binding is not present in hephaestin-like proteins from clades 2 and 3. Contrarily, all copper-binding residues listed in Takeuchi et al (2016) are conserved across clade 1 and all other cnidarian groups, including the protein secreted in the sclerites of S. cf. cruciata.
Cytosolic and mitochondrial CAs formed a single clade, whereas secreted and membrane-bound CAs were found to be polyphyletic. All homologs of the CA CruCA4 occupied the same clade (fig. 5 and supplementary fig. 5, Supplementary Material online). This group also included the CA-related protein present in the skeleton of H. coerulea. Scleractinian biomineralization-related CAs did, on the other hand, split into three distinct groups.
Fig. 5.
Maximum-likelihood analysis (400 bootstrap replicates) with focus on cnidarian carbonic anhydrases and carbonic anhydrases-related proteins. Sequences aligned with MAFFT. MUSCLE-based phylogeny in supplementary figure 5, Supplementary Material online. Best-fit model: WAG + G + I. Involvement of CruCA4, STPCA, STPCA-2, and B8V7P3 based on Le Goff et al. (2016), Moya et al. (2008), Bertucci et al. (2011), and Ramos-Silva et al. (2013), respectively. Other taxa include: Homo sapiens, Porifera, Cubozoa, Hydrozoa, Staurozoa, Scyphozoa, Ceriantharia, Actiniaria, and Corallimorpharia. Outgroup: Chlamydomonas reinhaardtii (P20507) and Desmodesmus sp. (AOL92959.1).
The CA sequence detected in M. digitata grouped together with A. millepora B8V7P3, but not with STPCA-2 (Bertucci et al. 2011) which is present in the skeleton of St. pistillata (Drake et al. 2013). This suggests that scleractinian families might secrete different CAs as part of their skeleton OM. Finally, the third group comprises scleractinian homologs of St. pistillata STPCA (Moya et al. 2008).
Discussion
Determining which morphomineralogical features of coral skeletons are biologically controlled, and which result from environmental effects, remains a key unresolved aspect of coral biomineralization. Here, we capitalized on the copresence of aragonite and calcite-forming species within Octocorallia, a unique feature among Anthozoa. We provide the first insight into the diversity of proteins occluded within the coral’s CaCO3 skeleton, that is, the skeletome, of species employing different calcification strategies. The identification of several octocoral SOMPs, in addition to providing new targets for follow up research, also allowed to perform comparative analyses with previously published and the newly characterized M. digitata proteome. Our work represents the first examination of the diversity of skeletal toolkits across Anthozoa and its relation to the variety of biomineralization strategies displayed by this group.
We have reported low overall proteome overlap between octocoral skeletons of different polymorphs, while simultaneously highlighting instances of skeletal proteins shared both within and between scleractinians and both aragonitic and calcitic octocorals. It has to be noted that protein presence in the skeleton does not automatically constitute evidence for involvement in biomineralization. For instance, random incorporation within the mineral fraction cannot be excluded. Furthermore, multiple factors linked to sample characteristics, processing, and amount of material analyzed (Chandramouli and Qian 2009; Michalski et al. 2011; Feist and Hummon 2015) can alter the detectability of peptides during mass spectrometry and can potentially have contributed to the low proteome similarities observed. However, comparisons between the different replicates (see supplementary figs. 6 and 7, Supplementary Material online) indicate that the results presented here are robust to potential variability between extractions and across samples. For other proteins, presence in the skeleton may not be directly linked to mineral formation, while still necessary for biomineralization or other functions played by the skeleton. For instance, protease inhibitors such as agrin-like proteins are common components of skeleton matrices, where they might prevent protease-driven matrix degradation (Marie et al. 2010; Flores and Livingston 2017). Presence of MCOs (hephaestin-like proteins), on the other hand, could be linked to octocoral sclerites serving as “deposits” for toxic metals, as proposed for scleractinian skeletons (Ramos-Silva et al. 2013). Alternatively, this could also be linked to ultraviolet radiation absorbance of Fe3+ and the capacity of the skeleton to serve as an anti-UV defense structure (Reef et al. 2009). Hephaestin-like proteins have been isolated from other mineralized structures including scleractinian skeletons (Ramos-Silva et al. 2013), and brachiopod shells (Luo et al. 2015). More recently, these proteins have also been implicated in radula formation (Hilgers et al. 2018). Here, we have shown that the MCO found in octocoral and scleractinian skeletons is found across Cnidaria. Thus, the protein might have played a different ancestral role and secondarily acquired a biomineralization-related function within certain lineages.
Co-option of proteins for biomineralization appears to be a common evolutionary mechanism among different marine calcifiers (Ramos-Silva et al. 2013; Aguilera et al. 2017; Shimizu et al. 2017). But it has been unknown whether this also applies to octocorals. Phylogenetic information is an essential requirement to identify and study co-option events (McLennan 2008). However for octocorals, phylogenetic relationships, particularly between major clades, remain largely unresolved. Efforts to resolve deep divergences are currently hampered by several factors including rapid radiation (McFadden et al. 2006), slow mitochondrial evolution, and inconsistent results between nuclear and mitochondrial markers (see McFadden et al. 2010 for review). Nonetheless, by combining transcriptomic–proteomic information with phylogenetic reconstructions, we detected several cases of possible co-option events during the evolution of octocoral skeletomes. In addition to MCOs, these include scleritin (Debreuil et al. 2012), galaxins (Reyes-Bermudez et al. 2009), CAs (Tambutté et al. 2007; Le Goff et al. 2016), and aspartic acid-rich proteins, on which extensive research has been conducted (Mass et al. 2013, 2016; Von Euw et al. 2017). The latter are of particular interest as they have been implicated in several processes, including regulation of CaCO3 polymorph. For example, scleractinian coral acid-rich proteins (CARPs) have been shown to select for different polymorphs at different magnesium concentrations (Laipnik et al. 2020). Similarly, one extracellular matrix protein (ECMP-67) from the octocoral Lobophytum crassum drived the in vitro formation of calcite under aragonite-favoring conditions (Rahman et al. 2006, 2011). Acidic proteins found in H. coerulea and T. musica could thus be key to the formation of octocoral skeletons and represent interesting targets for future investigations. In vivo functional studies, assessing phenotypic changes linked to acidic proteins function, are also necessary to fully determine the degree of control that these proteins might exert on the skeleton polymorph.
Reciprocal Blast analysis did not identify homology between the acidic proteins in T. musica and H. coerulea, or between octocoral and scleractinian sequences. Also, no homology to known scleractinian acidic proteins could be determined. As for scleractinian corals, however, octocoral acidic proteins possess nonacidic homologs in other species. PCA based on amino acid frequencies and aspartic acid distribution showed that the H. coerulea acidic protein 1, and its nonacidic homologs, exhibits similar amino acidic compositions to scleractinian sequences. As proposed for scleractinian SAARPs (Bhattacharya et al. 2016; Takeuchi et al. 2016), acidic protein 1 in H. coerulea appears to have arisen through co-option from a nonacidic protein.
The last comprehensive phylogenetic analysis of Octocorallia splits the group into three major clades (McFadden et al. 2006). Species from the genera Sinularia and Tubipora grouped within the same group, whereas Corallium and Heliopora species were included in two other clades. When data-mining available transcriptomes, both scleritin- and galaxin-related proteins possess homologs in representatives of all three major octocoral clades (Conci et al. 2019). Thus, they were available to octocorals early on in their evolutionary history and possibly in the octocoral common ancestor. At the skeletome level however, their distribution is restricted to certain species. As for scleractinian galaxin, which supposedly originated via co-option of an extracellular protein playing a different ancestral function (Forêt et al. 2010), (octo)galaxin and scleritin could also represent instances of existing proteins acquiring novel functions. For galaxin-related proteins, phylogenetic analyses (Bhattacharya et al. 2016; Conci et al. 2019) have shown that these proteins are polyphyletic in anthozoans, pointing to different independent recruitments in scleractinians and octocorals. Nevertheless, the presence of the scleractinian-characteristic endoprotease target motif (Fukuda et al. 2003) in the (octo)galaxin of T. musica’s, and the fact that polyphyly within galaxin phylogenies is likely affected by inclusion of false homologs and sensitivity to analytical parameters (Conci et al. 2019), makes it difficult to rule out the hypothesis of an ancient, biomineralization-related recruitment of (octo)galaxins prior to the divergence of octocorals from the remaining Anthozoa. Similarity in protein, features between galaxins sensu stricto (see Conci et al. 2019) and galaxin-like proteins, combined with the current lack of support for the deep phylogenetic relationships among these proteins, make understanding the evolution of galaxins difficult. Future studies should attempt to provide robust phylogenies for these proteins in order to assess whether their recruitment for biomineralization in Anthozoa is ancient or convergent. Different aspects of scleritin evolution also remain elusive. In C. rubrum, scleritin expression appears limited to scleroblasts (Debreuil et al. 2012), the cells responsible for sclerites formation. Although histological information is limited, scleroblasts in H. coerulea have not been described yet, while the presence of a calcifying epithelium closely associated with the skeleton has been reported (Le Tissier 1991). Furthermore, no information on scleritin presence in octocorals lacking sclerites (e.g., Clavularidae, Alderslade and Mcfadden 2011) is to our knowledge available. The expression of scleritin in H. coerulea suggests that the protein might therefore be produced by different cell types in different octocorals. Determining which H. coerulea cell population(s) express scleritin could help to determine whether the proteins 1) play a different biomineralization-related role or 2) have retained their ancestral nonrelated function. Additionally, more new genomic data are required to assess whether the presence of multiple scleritin homologs in some octocorals represents a true gene expansion event within calcite-forming species.
On the other hand, the ubiquitous expression of the CA CruCA4 across Octocorallia is matched by its presence in both aragonite and calcite skeletal structures. This protein holds particular interest as it represents the only (and possibly orthologous) skeletal protein found in aragonite and calcite skeletons. Notably, this pattern has also been observed in mollusks, where a CA (nacrein) was among the very few proteins conserved in the aragonitic (nacre) and calcitic (prism) shell layers (Marie et al. 2012). The presence of the same CAs within aragonite and calcite structures is consistent with experimental observations that have excluded a role of these enzymes in polymorph regulation (Rodriguez-Navarro et al. 2019). Our phylogenetic analysis of cnidarian CAs pointed to the polyphyly of secreted CAs (as previously observed for the Metazoa [Le Roy et al. 2014]) while mitochondrial and cytosolic CAs grouped in the same clade. All CruCA4 homologs occupied a single moderately supported clade. Within this group, some species were represented by multiple sequences, suggesting that recent duplication events might have occurred. One of these appears to have led to the appearance of an acatalytic CA-related protein—also secreted into the skeleton—in H. coerulea. Phylogenetic analysis of CAs did not support a common origin of octocoral and scleractinian skeletal CAs. CAs found in the skeleton of St. pistillata and acroporids occupied two separate clades, pointing to independent origins. Independent acquisitions of different biomineralization-related functions are known from several metazoan lineages, and currently render phylogenetic reconstructions problematic (for review see Le Roy et al. 2014). As previously discussed for scleritin, genomic information on CAs numbers across lineages could increase phylogenetic resolution and help determine CAs evolution.
Finally, several skeletal proteins were found in common between M. digitata and Acropora species. These include almost the entire “acidic toolkit” (both SARPs, acidic SOMPs and SAP1-b), and other putative extracellular components like mucin- and coadhesin-like proteins. Although the shared fraction accounts for <25% of the total Acropora skeleton proteome, it included the most abundant components (based on emPAI values; Ishihama et al. 2005; Ramos-Silva et al. 2013).
The discovery and description of skeletal proteins represents one of the first essential steps to study biological control in animal biomineralization. Here, we have applied a proteomics-based approach to identify and characterize the skeletal proteome of coral species that exhibit different calcification strategies. In addition to new evolutionary insights on coral biomineralization, this work provides several new targets for future functional investigations. The new data availability for both calcite and aragonite-forming octocoral species is of particular interest, as it opens up the possibility for in vivo investigations on biological control over CaCO3 polymorph in corals.
Supplementary Material
Acknowledgments
S.V. was supported by the German Research Foundation (DFG) (grant Va1146-2/1 “MINORCA”). G.W. was supported by LMU Munich’s Institutional Strategy LMUexcellent (10.13039/501100005722) within the framework of the German Excellence Initiative and the German Research Foundation (DFG) (grant Wo896/18-1 “MINORCA”) and from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie Early Research Training Network grant agreement no. 764840 (ITN IGNITE). We thank Dr Peter Naumann for technical assistance and maintenance of the aquaria facilities and coral culturing, as well as Simone Schätzle and Gabriele Büttner for assistance in the laboratory. S.V. is indebted to N. Villalobos Trigueros, M. Vargas Villalobos, S. Vargas Villalobos, and S. Vargas Villalobos for their constant support.
Literature Cited
- Aghaloo T, et al. 2007. A study of the role of nell-1 gene modified goat bone marrow stromal cells in promoting new bone formation. Mol Ther. 15(10):1872–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera F, McDougall C, Degnan BM. 2017. Co-option and de novo gene evolution underlie molluscan shell diversity. Mol Biol Evol. 34(4):779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderslade P, Mcfadden CS. 2011. A new sclerite-free genus and species of Clavulariidae (Coelenterata: Octocorallia). Zootaxa 3104(1):64–68. [Google Scholar]
- Amos FF, Destine E, Ponce CB, Evans JS. 2010. The N- and C-terminal regions of the pearl-associated EF hand protein, PFMG1, promote the formation of the aragonite polymorph in vitro. Cryst Growth Des. 10(10):4211–4216. [Google Scholar]
- Arivalagan J, et al. 2017. Insights from the shell proteome: biomineralization to adaptation. Mol Biol Evol. 34(1):66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci A, Tambutté S, Supuran CT, Allemand D, Zoccola D. 2011. A new coral carbonic anhydrase in Stylophora pistillata. Mar Biotechnol. 13(5):992–1002. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, et al. 2016. Comparative genomics explains the evolutionary success of reef-forming corals. Elife 5:e13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli K, Qian P-Y. 2009. Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum Genomics Proteomics. 2009: 239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conci N, Wörheide G, Vargas S. 2019. New non-bilaterian transcriptomes provide novel insights into the evolution of coral skeletomes. Genome Biol Evol. 11:3068–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debreuil J, et al. 2012. Molecular cloning and characterization of first organic matrix protein from sclerites of red coral, Corallium rubrum. J Biol Chem. 287(23):19367–19376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prete S, et al. 2017. Kinetic properties and affinities for sulfonamide inhibitors of an α-carbonic anhydrase (CruCA4) involved in coral biomineralization in the Mediterranean red coral Corallium rubrum. Bioorg Med Chem. 25(13):3525–3530. [DOI] [PubMed] [Google Scholar]
- Drake JL, et al. 2013. Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc Natl Acad Sci U S A. 110(10):3788–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falini G, Albeck S, Weiner S, Addadi L. 1996. Control of aragonite or calcite polymorphism by mollusk shell macromolecules. Science 271(5245):67–69. [Google Scholar]
- Farre B, Cuif J-P, Dauphin Y. 2010. Occurrence and diversity of lipids in modern coral skeletons. Zoology 113(4):250–257. [DOI] [PubMed] [Google Scholar]
- Farre B, Dauphin Y. 2009. Lipids from the nacreous and prismatic layers of two Pteriomorpha mollusc shells. Comp Biochem Physiol B Biochem Mol Biol. 152(2):103–109. [DOI] [PubMed] [Google Scholar]
- Feist P, Hummon AB. 2015. Proteomic challenges: sample preparation techniques for microgram-quantity protein analysis from biological samples. Int J Mol Sci. 16(2):3537–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores RL, Gonzales K, Seaver RW, Livingston BT. 2016. The skeletal proteome of the brittle star Ophiothrix spiculata identifies C-type lectins and other proteins conserved in echinoderm skeleton formation. Aims Mol Sci. 3(3):357–367. [Google Scholar]
- Flores RL, Livingston BT. 2017. The skeletal proteome of the sea star Patiria miniata and evolution of biomineralization in echinoderms. BMC Evol Biol. 17(1):125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forêt S, et al. 2010. New tricks with old genes: the genetic bases of novel cnidarian traits. Trends Genet. 26(4):154–158. [DOI] [PubMed] [Google Scholar]
- Fukuda I, et al. 2003. Molecular cloning of a cDNA encoding a soluble protein in the coral exoskeleton. Biochem Biophys Res Commun. 304(1):11–17. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, et al. 2005. Protein identification and analysis tools on the ExPASy server In: Walker JM, editor. The proteomics protocols handbook. Totowa, NJ: Humana Press; p. 571–607. [Google Scholar]
- Goffredo S, et al. 2011. The skeletal organic matrix from Mediterranean coral Balanophyllia europaea influences calcium carbonate precipitation. PLoS One 6(7):e22338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg WM. 2001. Acid polysaccharides in the skeletal matrix and calicoblastic epithelium of the stony coral Mycetophyllia reesi. Tissue Cell 33(4):376–387. [DOI] [PubMed] [Google Scholar]
- González-Pech RA, Vargas S, Francis WR, Wörheide G. 2017. Transcriptomic resilience of the Montipora digitata holobiont to low pH. Front Mar Sci. 4:403. [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27(2):221–224. [DOI] [PubMed] [Google Scholar]
- Groffen AJ, et al. 1998. Agrin is a major heparan sulfate proteoglycan in the human glomerular basement membrane. J Histochem Cytochem. 46(1):19–27. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Hilgers L, Hartmann S, Hofreiter M, von Rintelen T. 2018. Novel genes, ancient genes, and gene co-option contributed to the genetic basis of the radula, a molluscan innovation. Mol Biol Evol. 35(7):1638–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, et al. 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4(9):1265–1272. [DOI] [PubMed] [Google Scholar]
- Jackson DJ, et al. 2015. The Magellania venosa biomineralizing proteome: a window into brachiopod shell evolution. Genome Biol Evol. 7(5):1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, et al. 2019. The draft genome of an octocoral, Dendronephthya gigantea. Genome Biol Evol. 11(3):949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocot KM, Aguilera F, McDougall C, Jackson DJ, Degnan BM. 2016. Sea shell diversity and rapidly evolving secretomes: insights into the evolution of biomineralization. Front Zool. 13:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Laipnik R, et al. 2020. Coral acid rich protein selects vaterite polymorph in vitro. J Struct Biol. 209(2):107431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff C, et al. 2016. Carbonic anhydrases in cnidarians: novel perspectives from the octocorallian Corallium rubrum. PLoS One 11(8):e0160368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy N, Jackson DJ, Marie B, Ramos-Silva P, Marin F. 2014. The evolution of metazoan α-carbonic anhydrases and their roles in calcium carbonate biomineralization. Front Zool. 11:75. [Google Scholar]
- Le Tissier MD. 1991. The nature of the skeleton and skeletogenic tissues in the Cnidaria Hydrobiologia 216:397–402. [Google Scholar]
- Liew YJ, Aranda M, Voolstra CR. 2016. Reefgenomics.Org—a repository for marine genomics data. Database 2016:baw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, et al. 2012. The role of matrix proteins in the control of nacreous layer deposition during pearl formation. Proc Biol Sci. 279(1730):1000–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstam HA. 1981. Minerals formed by organisms. Science 211(4487):1126–1131. [DOI] [PubMed] [Google Scholar]
- Luo Y-J, et al. 2015. The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization. Nat Commun. 6:8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Jackson DJ. 2014. Characterization of the pigmented shell-forming proteome of the common grove snail Cepaea nemoralis. BMC Genomics. 15(1):249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Poustka AJ, Mann M. 2008. The sea urchin (Strongylocentrotus purpuratus) test and spine proteomes. Proteome Sci. 6(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Siedler F. 2004. Ostrich (Struthio camelus) eggshell matrix contains two different C-type lectin-like proteins. Isolation, amino acid sequence, and posttranslational modifications. Biochim Biophys Acta 1696(1):41–50. [DOI] [PubMed] [Google Scholar]
- Mann K, Siedler F. 2006. Amino acid sequences and phosphorylation sites of emu and rhea eggshell C-type lectin-like proteins. Comp Biochem Physiol B Biochem Mol Biol. 143(2):160–170. [DOI] [PubMed] [Google Scholar]
- Mann K, Weiss IM, André S, Gabius H-J, Fritz M. 2000. The amino-acid sequence of the abalone (Haliotis laevigata) nacre protein perlucin: detection of a functional C-type lectin domain with galactose/mannose specificity. Eur J Biochem. 267(16):5257–5264. [DOI] [PubMed] [Google Scholar]
- Mann S. 1983. Mineralization in biological systems In: Inorganic elements in biochemistry. Structure and bonding. Berlin/Heidelberg (Germany: ): Springer; p. 125–174. [Google Scholar]
- Marie B, et al. 2010. Proteomic analysis of the organic matrix of the abalone Haliotis asinina calcified shell. Proteome Sci. 8(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, et al. 2012. Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc Natl Acad Sci U S A. 109(51):20986–20991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, et al. 2013. The shell‐forming proteome of Lottia gigantea reveals both deep conservations and lineage‐specific novelties. FEBS J. 280(1):214–232. [DOI] [PubMed] [Google Scholar]
- Marin F, Bundeleva I, Takeuchi T, Immel F, Medakovic D. 2016. Organic matrices in metazoan calcium carbonate skeletons: composition, functions, evolution. J Struct Biol. 196(2):98–106. [DOI] [PubMed] [Google Scholar]
- Marin F, Luquet G. 2007. Unusually acidic proteins in biomineralization In: Buerlein E, editor. Handbook of biomineralization. Weinheim (Germany: ): Wiley-VCH Verlag GmbH; p. 273–290. [Google Scholar]
- Mass T, et al. 2013. Cloning and characterization of four novel coral acid-rich proteins that precipitate carbonates in vitro. Curr Biol. 23(12):1126–1131. [DOI] [PubMed] [Google Scholar]
- Mass T, et al. 2016. Temporal and spatial expression patterns of biomineralization proteins during early development in the stony coral Pocillopora damicornis. Proc Biol Sci. 283: 20160322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara H, et al. 2008. Modulating effect of acorn barnacle C-type lectins on the crystallization of calcium carbonate. Fish Sci. 74(2):418–424. [Google Scholar]
- McFadden CS, France SC, Sánchez JA, Alderslade P. 2006. A molecular phylogenetic analysis of the Octocorallia (Cnidaria: Anthozoa) based on mitochondrial protein-coding sequences. Mol Phylogenet Evol. 41(3):513–527. [DOI] [PubMed] [Google Scholar]
- McFadden CS, Sánchez JA, France SC. 2010. Molecular phylogenetic insights into the evolution of Octocorallia: a review. Integr Comp Biol. 50(3):389–410. [DOI] [PubMed] [Google Scholar]
- McLennan DA. 2008. The concept of co-option: why evolution often looks miraculous. Evol Educ Outreach 1(3):247–258. [Google Scholar]
- Michalski A, Cox J, Mann M. 2011. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J Proteome Res. 10(4):1785–1793. [DOI] [PubMed] [Google Scholar]
- Mitterer RM. 1978. Amino acid composition and metal binding capability of the skeletal protein of corals. Bull Mar Sci. 28:173–180. [Google Scholar]
- Moya A, et al. 2008. Carbonic anhydrase in the scleractinian coral Stylophora pistillata: characterization, localization, and role in biomineralization. J Biol Chem. 283(37):25475–25484. [DOI] [PubMed] [Google Scholar]
- Naggi A, et al. 2018. Structure and function of stony coral intraskeletal polysaccharides. ACS Omega 3(3):2895–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8(10):785–786. [DOI] [PubMed] [Google Scholar]
- Picaud SS, et al. 2009. Crystal structure of human carbonic anhydrase-related protein VIII reveals the basis for catalytic silencing. Proteins 76(2):507–511. [DOI] [PubMed] [Google Scholar]
- Pierleoni A, Martelli P, Casadio R. 2008. PredGPI: a GPI-anchor predictor. BMC Bioinf. 9(1):392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratlong M, et al. 2015. The red coral (Corallium rubrum) transcriptome: a new resource for population genetics and local adaptation studies. Mol Ecol Resour. 15(5):1205–1215. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Isa Y, Uehara T. 2006. Studies on two closely related species of octocorallians: biochemical and molecular characteristics of the organic matrices of endoskeletal sclerites. Mar Biotechnol. 8(4):415–424. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Oomori T, Wörheide G. 2011. Calcite formation in soft coral sclerites is determined by a single reactive extracellular protein. J Biol Chem. 286(36):31638–31649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Silva P, et al. 2013. The skeletal proteome of the coral Acropora millepora: the evolution of calcification by co-option and domain shuffling. Mol Biol Evol. 30(9):2099–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reef R, Kaniewska P, Hoegh-Guldberg O. 2009. Coral skeletons defend against ultraviolet radiation. PLoS One 4(11):e7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Bermudez A, Lin Z, Hayward DC, Miller DJ, Ball EE. 2009. Differential expression of three galaxin-related genes during settlement and metamorphosis in the scleractinian coral Acropora millepora. BMC Evol Biol. 9(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro C, et al. 2019. The multiple roles of carbonic anhydrase in calcium carbonate mineralization. CrystEngComm 21(48):7407–7423. [Google Scholar]
- Sarashina I, Endo K. 2006. Skeletal matrix proteins of invertebrate animals: comparative analysis of their amino acid sequences. Paleontol Res. 10(4):311–336. [Google Scholar]
- Shimizu K, Luo Y-J, Satoh N, Endo K. 2017. Possible co-option of engrailed during brachiopod and mollusc shell development. Biol Lett. 13(8):20170254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476(7360):320–323. [DOI] [PubMed] [Google Scholar]
- Supuran CT. 2008. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov. 7(2):168–181. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Yamada L, Shinzato C, Sawada H, Satoh N. 2016. Stepwise evolution of coral biomineralization revealed with genome-wide proteomics and transcriptomics. PLoS One 11(6):e0156424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambutté S, et al. 2007. Characterization and role of carbonic anhydrase in the calcification process of the azooxanthellate coral Tubastrea aurea. Mar Biol. 151(1):71–83. [Google Scholar]
- Voigt O, Adamski M, Sluzek K, Adamska M. 2014. Calcareous sponge genomes reveal complex evolution of α-carbonic anhydrases and two key biomineralization enzymes. BMC Evol Biol. 14:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Euw S, et al. 2017. Biological control of aragonite formation in stony corals. Science 356(6341):933–938. [DOI] [PubMed] [Google Scholar]
- Voolstra C, et al. 2015. The ReFuGe 2020 Consortium—using ‘omics’ approaches to explore the adaptability and resilience of coral holobionts to environmental change. Front Mar Sci. 2:68. [Google Scholar]
- Voolstra CR, et al. 2017. Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci Rep. 7(1):17583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vullo D, Nishimori I, Scozzafava A, Supuran CT. 2008. Carbonic anhydrase activators: activation of the human cytosolic isozyme III and membrane-associated isoform IV with amino acids and amines. Bioorg Med Chem Lett. 18(15):4303–4307. [DOI] [PubMed] [Google Scholar]
- Weiss IM, Kaufmann S, Mann K, Fritz M. 2000. Purification and characterization of perlucin and perlustrin, two new proteins from the shell of the mollusc Haliotis laevigata. Biochem Biophys Res Commun. 267(1):17–21. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 138(1):141–143. [DOI] [PubMed] [Google Scholar]
- Wheeler AP, George JW, Evans CA. 1981. Control of calcium carbonate nucleation and crystal growth by soluble matrx of oyster shell. Science 212(4501):1397–1398. [DOI] [PubMed] [Google Scholar]
- Wilt FH. 2002. Biomineralization of the spicules of sea urchin embryos. Zool Sci. 19(3):253–261. [DOI] [PubMed] [Google Scholar]
- Zou X, et al. 2011. NELL-1 binds to APR3 affecting human osteoblast proliferation and differentiation. FEBS Lett. 585(15):2410–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.