Abstract
Bacteria of the Planctomycetes phylum have many unique cellular features, such as extensive membrane invaginations and the ability to import macromolecules. These features raise intriguing questions about the composition of their cell envelopes. In this study, we have used microscopy, phylogenomics, and proteomics to examine the composition and evolution of cell envelope proteins in Tuwongella immobilis and other members of the Planctomycetes. Cryo-electron tomography data indicated a distance of 45 nm between the inner and outer membranes in T. immobilis. Consistent with the wide periplasmic space, our bioinformatics studies showed that the periplasmic segments of outer-membrane proteins in type II secretion systems are extended in bacteria of the order Planctomycetales. Homologs of two highly abundant cysteine-rich cell wall proteins in T. immobilis were identified in all members of the Planctomycetales, whereas genes for peptidoglycan biosynthesis and cell elongation have been lost in many members of this bacterial group. The cell wall proteins contain multiple copies of the YTV motif, which is the only domain that is conserved and unique to the Planctomycetales. Earlier diverging taxa in the Planctomycetes phylum contain genes for peptidoglycan biosynthesis but no homologs to the YTV cell wall proteins. The major remodeling of the cell envelope in the ancestor of the Planctomycetales coincided with the emergence of budding and other unique cellular phenotypes. The results have implications for hypotheses about the process whereby complex cellular features evolve in bacteria.
Keywords: cell envelope, Planctomycetes, phylogeny, cellular complexity, bacterial evolution
Significance
The Planctomycetes are bacteria with complex cellular architectures that resemble those of eukaryotic cells, but how and why this complexity arose is not known. The findings presented in this article suggests that the remodeling of the bacterial cell wall was a key event that enabled invaginations of the cytoplasmic membrane and import of macromolecules and thereby laid the basis for the evolution of their complex cell structures.
Introduction
The Planctomycetes have been an enigma ever since their discovery. Over the years, they have been described as fungi (Gimesi 1924), bacteria (Hirsch 1972), or as a missing link between prokaryotes and eukaryotes (Devos and Reynaud 2010). Based on 16S rRNA sequence comparisons, the Planctomycetes are now described as bacteria related to the phyla Verrumicrobia and Chlamydiae in the PVC superphylum (Wagner and Horn 2006). Cell biology features such as elaborate intracellular membrane networks and FtsZ-independent cell proliferation distinguishes the Planctomycetes from the prototype bacterial cell. Knowledge about the cell wall structure in the Planctomycetes is essential for a deeper understanding of their complex cell biology features. However, the nature of the cell wall and the suggested loss of cell wall components, which is the topic of this study, has been a matter of much controversy over the years.
In bacteria, the cell wall consists of peptidoglycan, which is a polymer made up of glycan strands that are cross-linked by short peptides to form a mesh-like structure (Vollmer 2008; Vollmer et al. 2008). Genes coding for proteins involved in peptidoglycan synthesis are located in the highly conserved “division and cell wall” (dcw) gene cluster (Tamames et al. 2001). In Escherichia coli, the cluster contains 15 genes with the order: mraZW–ftsLI–murEF–mraY–murD–ftsW–murGC–ddlB–ftsQAZ. The cytoplasmic enzymes MurCDEF catalyze the sequential addition of amino acids to N-acetyl-glucosamine (GlcNAc). The membrane protein MraY links the newly synthesized precursor molecule to a transport lipid in the inner membrane to generate a disaccharide–pentapeptide–lipid subunit, which is flipped across the inner membrane to the periplasm with the help of the MurJ flippase (Sham et al. 2014). Once inside the periplasmic space, glycosyltransferases add the precursor molecule to the nascent peptidoglycan chain, after which the pentapeptides in the growing chain are cross-linked to the sacculus with the aid of transpeptidases, also referred to as penicillin-binding proteins or PBPs.
In rod-shaped bacteria that divide by binary fission, cell wall synthesis is normally symmetric and coupled to cell elongation, after which the elongated cell divides into two daughter cells of equal sizes (Typas et al. 2012). A key component in this process is the MreB elongasome complex, which is located on the inner side of the cytoplasmic membrane. The MreB protein is a homolog of actin that forms filaments and controls the position of the penicillin-binding proteins PBP1 and PBP2, which are attached to the growing peptidoglycan chain (Jones et al. 2001; Typas et al. 2012; Strahl et al. 2014). The two systems are interconnected such that peptidoglycan synthesis drives the movement of the MreB filaments via the PBPs, and vice versa MreB activity is required for peptidoglycan synthesis (Dominguez-Escobar et al. 2011; Typas et al. 2012).
The Mollicutes are currently the only known group of bacteria that lacks a peptidoglycan cell wall, however, these bacteria are host-associated and rely on their eukaryotic host cells for maintenance of turgor pressure and osmotic stability. There are also exceptions to the prototype model of bacterial cell division by binary fission. For example, the Alphaproteobacteria display a wide spectrum of cell division processes, from symmetric division and tip extension to asymmetric binary fission and budding (Randich and Brun 2015). Cell wall architectures in Agrobacterium tumefaciens, which divides by binary fission, and Hyphomonas neptunius, which divides by budding, differ in both the amino acid content of the pentapeptide side chains and the degree of crosslinking in the peptidoglycan layer. Furthermore, the manner in which the spatiotemporal activities of peptidoglycan synthetases and hydrolases are regulated in relation to the cell cycle differs in bacteria that divide by binary fission versus budding (Cserti et al. 2017). Thus, regardless of whether the cells proliferate by binary fission or budding, peptidoglycan biosynthesis is directly coupled to cell elongation and division in bacteria.
The Planctomycetes are unique in their ability to multiply without the otherwise essential FtsZ protein. Cell proliferation processes range from binary fission in Phycisphaerae and Candidatus Brocadiae to polar budding in Planctomycetaceae and round cell budding in Isosphaeraceae and Gemmataceae (reviewed in Wiegand et al. [2020]). The cell cycle of Gemmata obscuriglobus, which has been most extensively studied, starts with the formation of a daughter cell bud into which the chromosomal DNA is transported (Lee et al. 2009).
However, the composition of the cell wall has been more elusive. Early studies suggested that the Planctomycetes do not contain a peptidoglycan cell wall, but rather a stable proline- and cysteine-rich protein envelope (König et al. 1984; Liesack et al. 1986). Consistently, cysteine-rich proteins with YTV-domain repeats were identified in cell envelope preparations of Rhodopirellula baltica (Hieu et al. 2008) and subsequently also in Gem. obscuriglobus (Sagulenko et al. 2017). The YTV domain (Pfam ID: PF07639) is 43 amino acids in length and characterized by repeats of the motif YTVxRPVxET, but the function of this domain is unknown. Genes for peptidoglycan biosynthesis could not be identified in the Planctomycetes (Pilhofer et al. 2008), adding support to the hypothesis that these bacteria have a highly unique cell wall structure.
More recently, it was discovered that members of the class Ca. Brocadiae (also called anammox bacteria), which represent an early-diverging lineage in the phylum, are sensitive to antibiotics that target the peptidoglycan and contain genes for peptidoglycan biosynthesis (van Teeseling et al. 2015). Based on these findings, it was proposed that the anammox bacteria have a conventional bacterial peptidoglycan cell wall layer (van Teeseling et al. 2015). Furthermore, experimental studies provided indications of a peptidoglycan cell wall also in Planctopirus limnophila and Gem. obscuriglobus, and based on the results from sensitive sequence-based methods it was argued that genes for peptidoglycan biosynthesis are present in all members of the Planctomycetes (Jeske et al. 2015; Rivas-Marin et al. 2016; Wiegand et al. 2018). The identification of bacterial outer-membrane proteins and a lipopolysaccharide in Gem. obscuriglobus (Kerger et al. 1988; Speth et al. 2012; Mahat et al. 2016) also indicated that these bacteria contain a conventional Gram-negative bacterial cell envelope. Taken together, these results led to a paradigm shift in that the Planctomycetes were regarded similar to other Gram-negative bacteria, with the major difference being their mode of cell division (Wiegand et al. 2018).
In the light of these results, it was puzzling that a renewed bioinformatics analysis using more stringent criteria concluded that genes for peptidoglycan biosynthesis are missing from several Planctomycetes species (Wiegand et al. 2020). Furthermore, knockout mutants in major cell division genes in Pl. limnophila yielded no obvious phenotype, as presented in both Wiegand et al. (2020) and Rivas-Marin et al. (2020).
At about the same time, novel Planctomycetes strains were isolated with unique phenotypes that have never been observed previously in bacteria. Prime among these were Saltatorellota, which in addition to being able to change the shape of its cells, can divide by both budding and binary fission (Wiegand et al. 2019, 2020). Genes for peptidoglycan biosynthesis were identified in the Saltatorellota genomes, however, it was suggested that the peptidoglycan layer is remodeled during the cell-shifting and budding growth phases, respectively.
Even more remarkable was the isolation of Candidatus Uab amorphum, another early-diverging strain related to the anammox bacteria (Shiratori et al. 2019). This was the first description of a bacterial cell that is able to engulf other bacterial cells through a phagocytosis-like process. Genes for peptidoglycan biosynthesis were identified also in this genome, although it was argued that the cell wall needs to be degraded under the engulfment process (Shiratori et al. 2019). The study highlighted an actin-like protein that may form fibers involved in the engulfment of bacterial cells (Shiratori et al. 2019). These results raise many new questions and challenge the view that the Planctomycetales have a conventional bacterial cell envelope.
Given the many contradictory results and hypotheses regarding cell wall biosynthesis and cell elongation in the Planctomycetes, we have here examined the cell envelope structure in Tuwongella immobilis, a recently described genus and species of the Gemmataceae family (Seeger et al. 2017). We have also performed a broad bioinformatics study of cell envelope components in all members of the Planctomycetes for which genome sequence data are available. Our evolutionary analyses reveal dramatic changes in cell envelope, cell elongation, and cell division proteins in the Planctomycetales. We discuss the results in view of different models for the evolution of cellular complexity in bacteria.
Results
Evolution of Transenvelope Spanning Protein Complexes
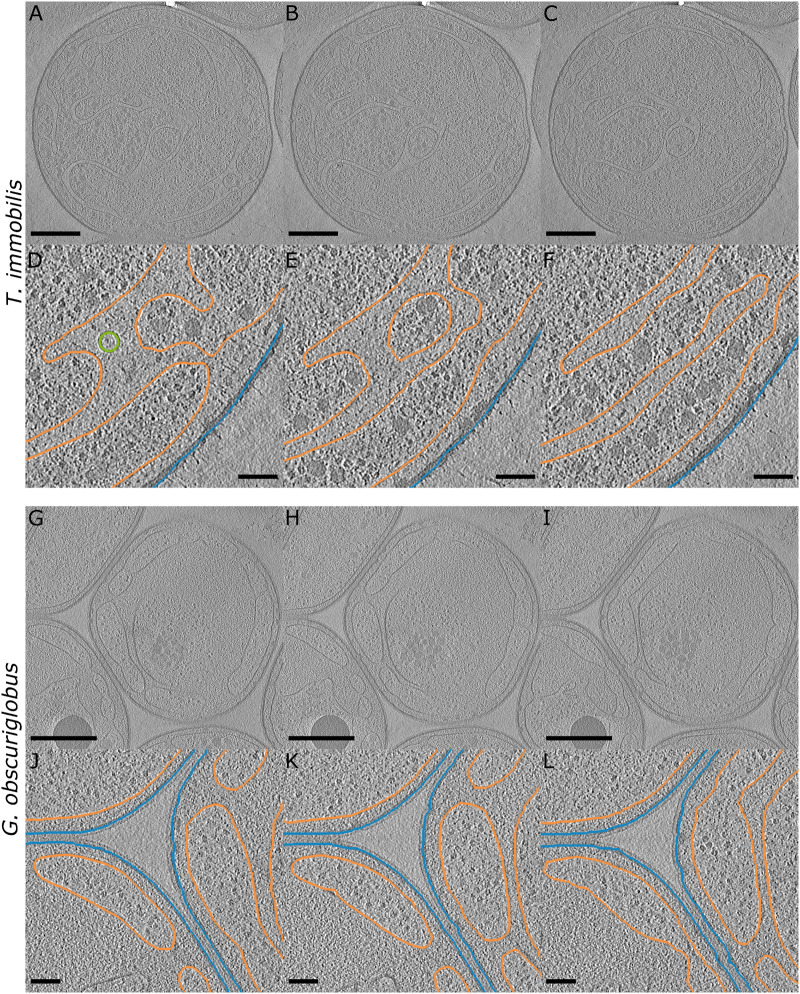
We acquired cryo-electron tomography (cryo-ET) data after cryo-focused ion beam (FIB) milling from both T. immobilis and Gem. obscuriglobus, resulting in the reconstruction of two tomograms for each species at a pixel size of 7.37 Å (fig. 1 and supplementary movies 1–4, Supplementary Material online). Cells from both species were well preserved by the plunge-freezing and FIB-milling processes. The complex inner membrane network, typical for members of the Gemmataceae family, was clearly visible and the ribosome-rich cytoplasm was separated from the periplasmic space. The inner membrane appeared to form enclosed compartment-like structures in some tomographs but the fully reconstructed tomograms showed that those structures were not enclosed within the volume of the prepared specimen. In Gem. obscuriglobus, we also observed a fibrillar structure over a length of >200 nm and a width of 45 nm (supplementary movie 3, Supplementary Material online). Additionally, we observed large spherical particles of high contrast in both T. immobilis (supplementary movie 2, Supplementary Material online) and Gem. obscuriglobus (supplementary movies 3 and 4, Supplementary Material online). Those particles, which resembled polyphosphate granules observed in Pseudomonas aeruginosa (Racki et al. 2017), varied in diameter between 140 nm in T. immobilis and 290–380 nm in Gem. obscuriglobus.
Fig. 1.
Cryo-electron tomographs of Tuwongella immobilis and Gemmata obscuriglobus cells. (A–C and G–I) Tomographs of T. immobilis and Gem. obscuriglobus with interslice distances of 50 voxel (37 nm); scale bar: 500 nm, thickness: 1 voxel; for corresponding full tomogram, see supplementary movies 1 and 4, Supplementary Material online. (D–F and J–L) Partial segmentation of inner membrane (orange), outer membrane (blue), and membrane vesicle (green) of the corresponding tomographs shown in (A–C) and (G–I). Scale bar: 100 nm.
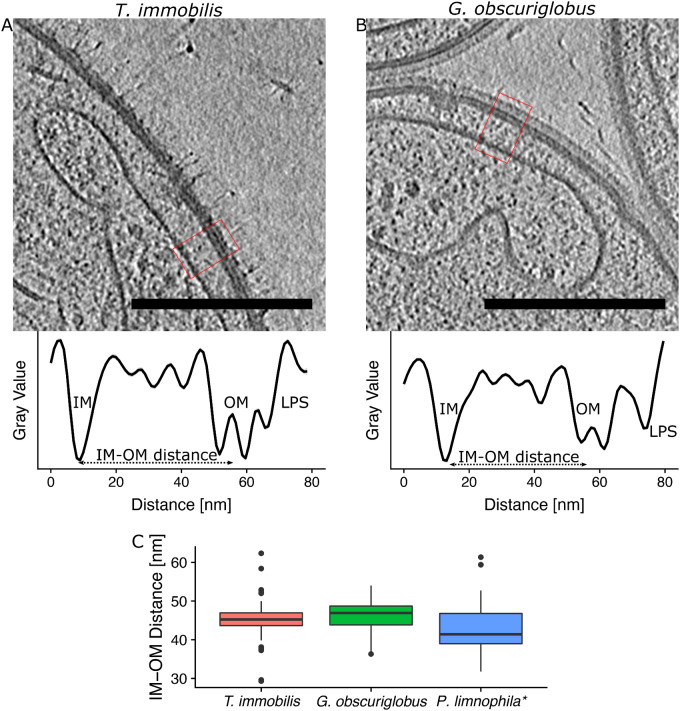
Both the inner and the outer-membrane layers were clearly visible in density profiles of T. immobilis and Gem. obscuriglobus, although the inner membrane looked like a single layer (fig. 2). However, at larger magnification, it was confirmed that the inner layer is composed of a lipid bilayer with a thickness of ∼3–4 nm. The outer membrane consists of three density layers, of which the two inner layers are separated by 8–10 nm. The extracellular layer is more clearly visible in Gem. obscuriglobus, and was 13 nm away from the outer-membrane layers (fig. 2B) as compared with only 7 nm in T. immobilis (fig. 2A). At sites where the inner membrane aligns in proximity to the outer membrane, the average distance between the inner membrane and the outer-membrane layers was estimated to be 45 nm in T. immobilis and 47 nm in Gem. obscuriglobus. These values are similar to an average distance of 41 nm in Pl. limnophila (fig. 2C), as estimated from previously published cryo-ET data of this species (Boedeker et al. 2017).
Fig. 2.
Cryo-electron tomography analyses of the cell envelopes of Tuwongella immobilis and Gemmata obscuriglobus. Tomograms of (A) T. immobilis and (B) Gem. obscuriglobus; scale bar: 250 nm. Corresponding density profiles were recorded within the highlighted area (red square) across the cell envelope (from inside to outside: IM, inner membrane; OM, outer membrane; LPS, putative lipopolysaccharide); scale bar: 250 nm, thickness: 25 voxels. (C) Boxplots of the measured IM–OM distances of T. immobilis (n = 60) and of Gem. obscuriglobus (n = 60). *Measurements for Planctopirus limnophila (n = 60) are based on previously published cryo-ET data (Boedeker et al. 2017).
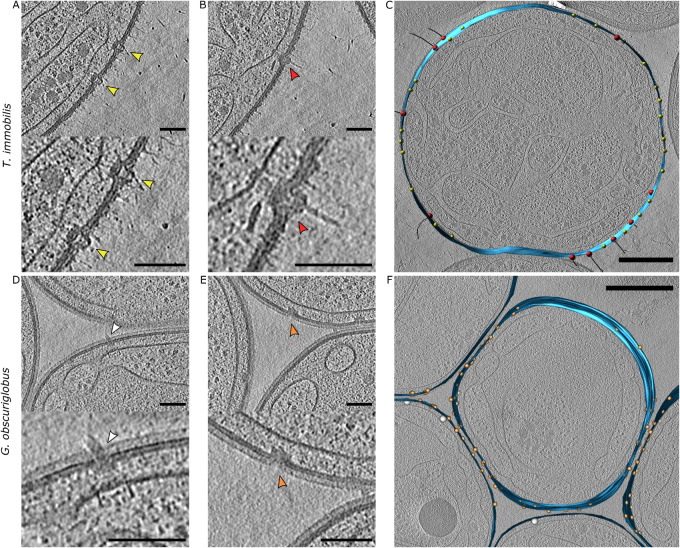
The resolution of the tilt series also allowed us to identify outer-membrane and transenvelope spanning protein complexes (fig. 3). In T. immobilis, we observed a “hat-like” outer-membrane protein complex with a diameter of 25 nm on the cell surface and 35 nm on the periplasmic side (fig. 3A). Likewise, we identified an outer-membrane protein complex in Gem. obscuriglobus with an estimated diameter of 25 nm on the cell surface and 39 nm on the periplasmic side (fig. 3E). Despite different structures as inferred by visual inspection, both outer-membrane protein complexes were highly abundant on the cell surfaces of T. immobilis (fig. 3C) and Gem. obscuriglobus (fig. 3F).
Fig. 3.
Cryo-electron tomography analyses of protein complexes that span across the cell envelopes in Tuwongella immobilis and Gemmata obscuriglobus. Outer-membrane embedded protein complexes in tomograms of (A) T. immobilis and (E) Gem. obscuriglobus without visible connection to the cytoplasmic membrane. Outer membrane embedded protein complexes with clear connection to cytoplasmic membrane in tomograms of (B) T. immobilis and (D) Gem. obscuriglobus; scale bar: 100 nm, thickness: 25 voxels. Segmentation of embedded protein complexes on the cell surface (indicated by spheres) in tomographs (thickness: 1 voxel) of (C) T. immobilis and (F) Gem. obscuriglobus. Colors of spheres correspond to arrow colors in (A and B) and (D and E); scale bar: 500 nm, thickness: 1 voxel.
The transenvelope spanning protein complexes included a flagellum-like complex in Gem. obscuriglobus with a diameter of ∼40 nm on the cytoplasmic side of the membranes (fig. 3D). The distance between the membrane layers at the site of the flagellum was estimated to only 33 nm and the diameter of the tip-like structure of the flagellum was measured to ∼14 nm. No flagellum-like complexes were identified in the T. immobilis cells, consistent with the lack of genes for a flagellum in the genome. Instead, we observed pili-like structures in T. immobilis (fig. 3B). The diameter of the pilus-like complexes was estimated to 25 nm on the cell surface and 32 nm on the cytoplasmic side of the membranes. The diameter of the pilus was ∼8 nm. Such pili-like structures were highly abundant on the cell surface of T. immobilis (fig. 3C).
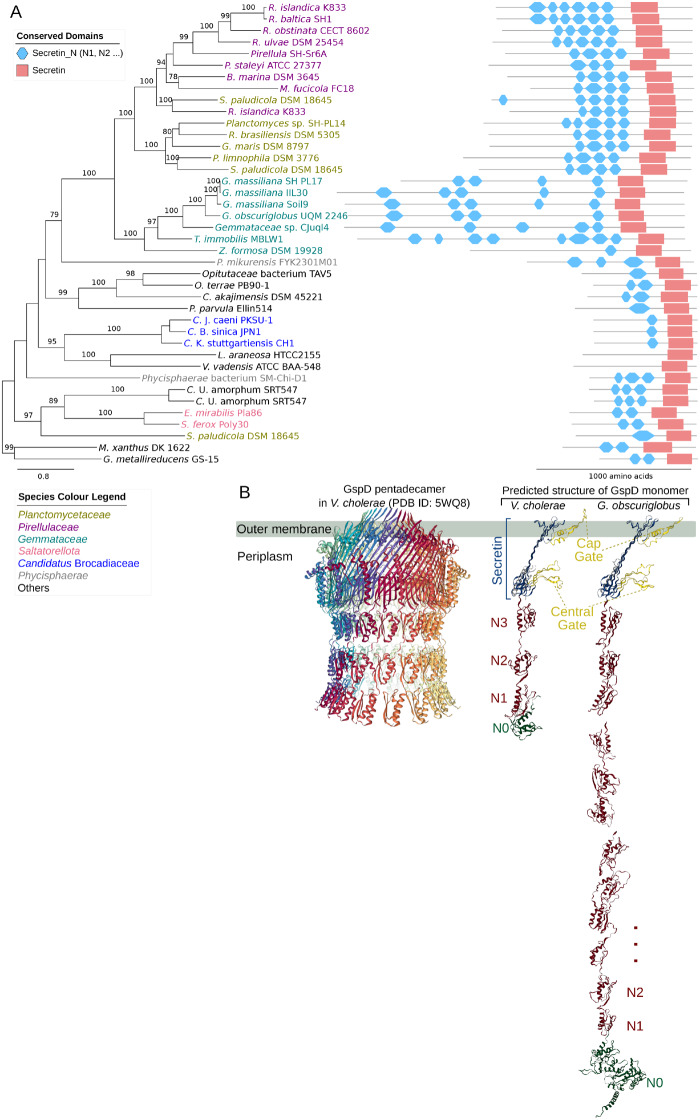
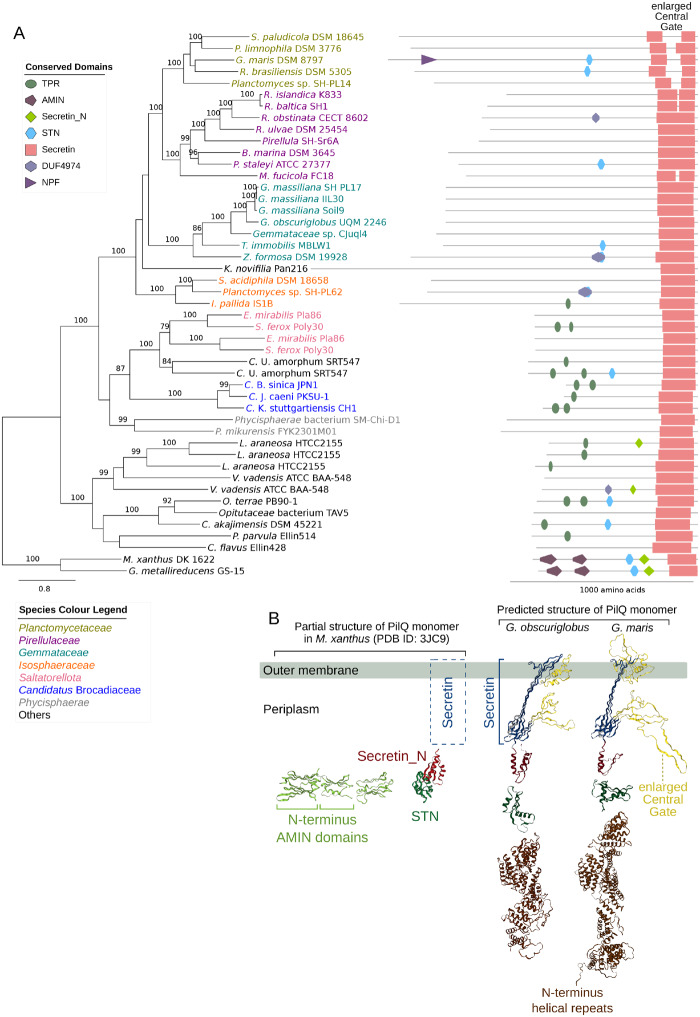
Consistent with the visual identification of the pili-like structures, we identified gene clusters for type IV pili (T4P) and type II secretion systems (T2SS) in the genomes of the Planctomycetes species (supplementary figs. S1 and S2, Supplementary Material online). The outer-membrane components GspD and PilQ of T2SS and T4P complexes, respectively, are normally ∼700–1,000 amino acids long. Interestingly, our analysis showed that the GspD and PilQ proteins are more than twice as long in members of the Planctomycetales than the homologous proteins in the early-diverging Planctomycetes species and outgroups (figs. 4A and 5A).
Fig. 4.
Phylogeny and structure of the outer-membrane protein GspD. (A) Phylogeny and domain architecture of the outer-membrane channel protein, GspD, in the type II secretion system. The phylogeny was inferred with the maximum likelihood method with 100 bootstraps. Only bootstrap support values ≥70 are shown. (B) Structure of the GspD pentadecamer complex (pdb ID: 5WQ8) (Yan et al. 2017) and the right face of the GspD monomer in Vibrio cholera are shown next to the predicted structure of the GspD monomer in Gemmata obscuriglobus.
Each monomer of the GspD pentadecamer normally consists of a C-terminal secretin domain embedded in the outer membrane and 4–6 Secretin_N domains (referred to as N1, N2, and so on) that extend into the periplasmic space, as exemplified in Vibrio cholerae (fig. 4B). Tertiary structure predictions of the GspD protein in Gem. obscuriglobus identified the secretin domain and an extended periplasmic tail with up to 12 N-domains (fig. 4B). Likewise, the PilQ proteins also consists of a C-terminal secretin domain, however, a large insertion that consists of ∼50% glycine was found in the central gate of the secretin channel in Gimesia maris and other members of the Planctomycetaceae. Furthermore, the N-terminus of the PilQ monomer consists helical repeats in all members of the Planctomycetales, instead of the AMIN domains that have been described to be spread inside the peptidoglycan, as exemplified in Myxococcus xanthus (fig. 5B). Thus, the periplasmic segments of the GspD and PilQ proteins have undergone extensive modifications in the Planctomycetales.
Fig. 5.
Phylogeny and structure of the outer-membrane protein PilQ. (A) Phylogeny and domain architecture of the outer-membrane channel protein, PilQ, in the type IV pili system. The phylogeny was inferred with the maximum likelihood method with 100 bootstraps. Only bootstrap support values ≥70 are shown. (B) The partial structure of the PilQ monomer in Myxococcus xanthus (pdb ID: 3JC9) (Chang et al. 2016) is shown beside the predicted structure of the PilQ monomer in Gemmata obscuriglobus and Gimesia maris.
Identification and Evolution of Cell Wall Proteins with YTV Domains
Next, we investigated the sensitivity of T. immobilis cells to mutanolysin that target the peptidoglycan, using Gem. obscuriglobus and Pl. limnophila as controls. Mutanolysin had no or only a slow lytic effect on EDTA-treated T. immobilis cells, as monitored by UV–VIS spectroscopy, phase contrast microscopy, and transmission electron microscopy (TEM) (supplementary figs. S3–S5, Supplementary Material online). Nor did the addition of mutanolysin to Gem. obscuriglobus indicate lysis as the UV–VIS traces were within the same levels as the negative control, although the addition of DTT indicated some lytic effect in T. immobilis and Pl. limnophila (supplementary figs. S3 and S5, Supplementary Material online). Thus, the cell wall of T. immobilis cannot be effectively lysed by mutanolysin under conditions that degrade the peptidoglycan-containing cell walls of EDTA-treated E. coli cells within minutes (supplementary fig. S3, Supplementary Material online).
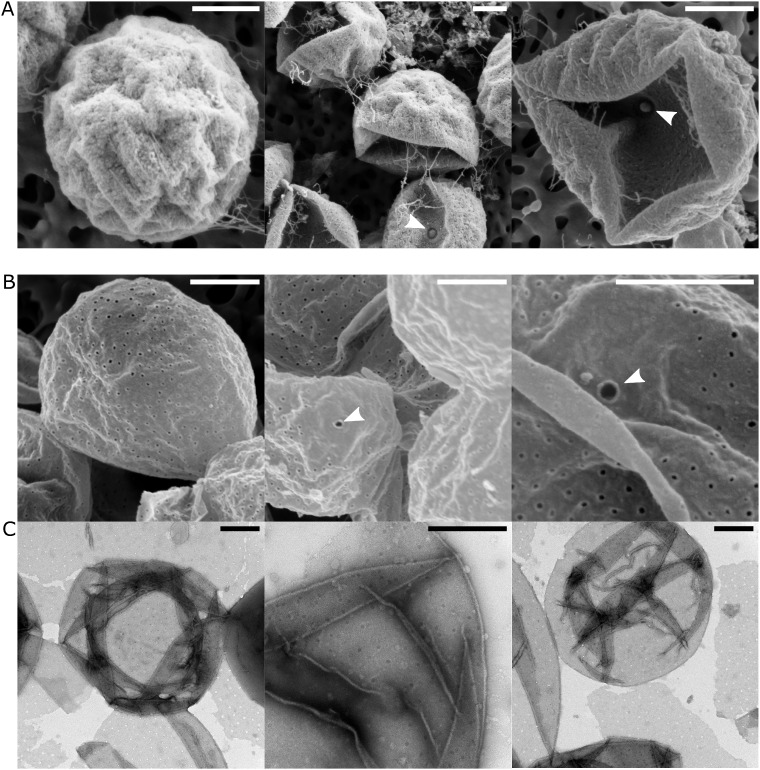
However, a strong lytic effect was observed on the three tested Planctomycetes species upon application of 5% SDS (supplementary fig. S3, Supplementary Material online). After treatments with 5% SDS at 70 °C only an empty, single-layered cell wall structure remained (supplementary figs. S4 and S5, Supplementary Material online), which was investigated in more detail by scanning electron microscopy (SEM) and negative stain TEM (nsTEM) (fig. 6). Notably, the isolated cell wall in T. immobilis contained uniformly distributed small pores of ∼25 nm in diameter (fig. 6B), with a distribution pattern resembling the appearance of pores on the cell surfaces of intact cells (fig. 6A). We also observed a single, large pore in intact cells and the isolated cell wall with an inner diameter of 55 nm and an outer diameter of 85 nm, which may correspond to the budding pore (fig. 6A and B). The surrounding area of the large pore appeared to contain fewer small pores. In areas of the nsTEM micrographs where the smaller pores were uniformly distributed, the density was estimated to 50 pores per µm2 (fig. 6C).
Fig. 6.
Electron microscopy (EM) analysis of the isolated cell wall of Tuwongella immobilis. (A) Scanning EM micrographs of untreated T. immobilis cells. White arrowheads point toward single-pore structures in the bacterial cell envelope. Scale bar: 500 nm. (B) Scanning EM micrographs of the isolated cell wall of T. immobilis. In addition to uniformly distributed smaller pores, a large single pore can is identified in the cell wall (white arrowhead). Scale bar: 500 nm. (C) Negative stain TEM micrographs of T. immobilis cell wall preparations served to estimate the surface density of the pores to ∼50 pores per µm2. Scale bar: 500 nm.
The purified cell wall remained intact after 3-h treatment with mutanolysin and/or DTT, but was no longer visible after incubation with proteinase K for 45 min (supplementary fig. S6, Supplementary Material online). This result suggests that proteins represent major components of the cell wall.
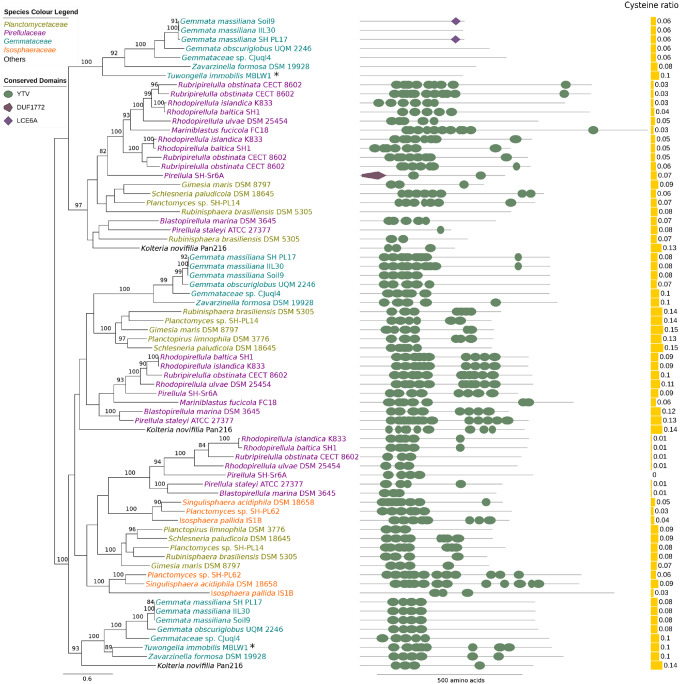
A proteomics analysis of the isolated cell wall from T. immobilis by LC-MS/MS identified a total of 130 proteins, including many inner and outer-membrane proteins in low quantity that may have copurified with the cell wall (supplementary tables S1 and S2, Supplementary Material online). Two proteins (encoded by GMBLW1_35240, GMBLW1_26360) accounted for almost 50% of the total score of the MS-spectrum and were thus highly abundant in the cell wall preparation. The same two proteins were top-ranked in both Sample 1 taken from the top of the gel and Sample 2 taken from the rest of the gel (supplementary fig. S7, Supplementary Material online), but the protein scores in Sample 1 were 90–95% compared with the scores in Sample 2 (supplementary tables S1 and S2, Supplementary Material online). The two proteins have an unusually high proportion of cysteine and valine residues (C: 10%; V: 15–17%). Based on Pfam predictions, it was predicted that both proteins contain a signal peptide and a noncytoplasmic domain. Additionally, GMBLW1_26360 was predicted to contain several YTV-domain repeats (Pfam Acc: PF07639).
Homologs to the two cysteine-rich cell wall proteins of T. immobilis were identified in all members of the Planctomycetales (supplementary table S3, Supplementary Material online). A phylogeny inferred from the homologous proteins revealed several distinct clades, indicative of at least three duplication events in the common ancestor of the Planctomycetales (fig. 7). Species diversification patterns within each clade were mostly consistent with vertical inheritance, although the cysteine content and the predicted number of YTV domains varied extensively. Many paralogous proteins with YTV domains were broadly present in all members of the Planctomycetales, whereas homologs to the cysteine-rich cell wall protein without YTV domains in T. immobilis were solely found in members of the Gemmataceae. Not even weak hits to any of these proteins were detected in Phycisphaerae, Saltatorellota, Ca. U. amorphum, or Ca. Brocadiae. Nor could any other proteins with YTV domains be identified in these earlier diverging lineages or in any other bacteria. Thus, this is the only domain that is uniquely present and conserved in all members of the Planctomycetales and as such may represent one of the few true innovations in the Planctomycetes phylum.
Fig. 7.
Phylogeny and domain architecture of YTV proteins identified in the cell wall of Tuwongella immobilis. A phylogeny of the proteins identified in the cell wall of T. immobilis (GMBLW1_35240 and GMBLW1_26360), and their homologs in other Planctomycetes species is shown alongside the domain architecture of each protein. The phylogeny was inferred with the maximum likelihood method with 100 bootstraps. Only bootstrap support values ≥70 are shown. The relative fraction of cysteine residues in each protein is shown next to the protein domain architecture.
Phyletic Distribution Patterns of Peptidoglycan Biosynthesis Genes
We also reinvestigated the phyletic distribution patterns of peptidoglycan biosynthesis genes in the Planctomycetes. We selected for this analysis genes that in other bacteria are known to be involved in peptidoglycan biosynthesis and cell division, such as those located in the dcw and mreB gene clusters. The analysis included a representative set of 32 Planctomycetes genomes, plus an additional ten genomes from other members of the PVC superphylum and two outgroup taxa (supplementary table S4, Supplementary Material online). The search for homologs was done in a stepwise manner as detailed in Materials and Methods section and included the use of the already identified genes in the dcw gene cluster for peptidoglycan biosynthesis and cell division in Pl. limnophila as queries in BLAST searches (supplementary tables S5 and S6, Supplementary Material online).
The results showed that the large majority of genes in the dcw gene cluster were absent from Pirellulaceae, Gemmataceae, and Isosphaeraceae and several genes in the cluster could also not be identified in Ca. U. amorphum (fig. 8). Furthermore, the genes identified in Planctomycetaceae and Phycisphaerae were spread across multiple genomic loci in contrast to those identified in the genomes of Saltatorellota, Ca. Brocadiae, and the outgroup species. We also observed that the murABCDE genes in Kolteria novifilia were located at the same genomic site, but had a different gene order structure compared with the dcw cluster genes in the outgroup species.
Fig. 8.
Phyletic distribution profiles of genes for peptidoglycan biosynthesis and cell division in the Planctomycetes. The presence of genes involved in cell elongation, division, and peptidoglycan synthesis is shown by gray squares. The genes belonging to the division and cell wall (dcw) gene cluster, and the cell elongation (mreB–rodA) gene cluster are marked. The colocalization of genes is indicated by dotted lines. Connecting rectangles between the squares indicate gene fusion. A stripped version of the 16S rRNA phylogeny (shown in supplementary fig. S8, Supplementary Material online) has been used to arrange the species in taxonomical order. Branches with 100% bootstrap support in the 16S rRNA tree have been indicated with a star (*). The mur genes in Kolteria novifilia have an atypical gene order, which is murBAEDC, hence their gene order could not be shown by connecting lines in this figure, indicated with two stars (**).
To place the inferred presence/absence patterns into an evolutionary framework, we mapped the phyletic distribution profiles of the genes involved in cell elongation, division, and peptidoglycan synthesis onto a 16S rRNA phylogenetic tree of the Planctomycetes (fig. 8 and supplementary fig. S8, Supplementary Material online). The topology of the tree confirmed that all members of the Planctomycetales formed a monophyletic clade with 100% bootstrap support to the exclusion of the earlier diverging lineages Ca. Brocadiae, Saltatorellota, and the single-lineage Ca. U. amorphum although the divergence order of the latter lineages was not well resolved in the 16S rRNA tree. Thus, based on the tree topology, it is suggested that deletions of genes in the dcw gene cluster in Pirellulaceae, Gemmataceae, and Isosphaeraceae in the Planctomycetales have occurred independently from the loss of these genes in Ca. U. amorphum.
The results further showed that the phyletic distribution pattern of the dcw gene cluster in the Planctomycetes matched the phyletic distribution pattern of mreBCD, pbp2, rodA, mviN, and murA (fig. 8). This correlation may not be surprising since cell elongation is linked with the biosynthesis of the peptidoglycan in all other bacteria. Additionally, ftsZ, ftsA, rodZ, and amiA, involved in the final septation step, were missing from the Planctomycetes including the early-diverging lineages Phycisphaerae, Saltatorellota, Ca. U. amorphum, and Ca. Brocadiae. Moreover, the ftsQ gene was uniquely missing from Saltatorellota and Ca. U. amorphum. Thus, our analysis confirmed that major cell division genes were lost already in the common ancestor of the Planctomycetes, whereas genes for peptidoglycan biosynthesis were lost subsequently and at least twice independently.
To ensure that the presence/absence patterns obtained from the careful investigation of the smaller genome data set were representative of other species from the same clades, we searched an extended data set that included 121 Planctomycetes genomes, using the genes identified in the smaller data set as queries. The heatmap showing the resulting e-values and bit scores (supplementary fig. S8, Supplementary Material online) confirmed the observed phyletic distribution patterns (fig. 8).
Evolution of Peptidoglycan Biosynthesis and Cell Division Proteins
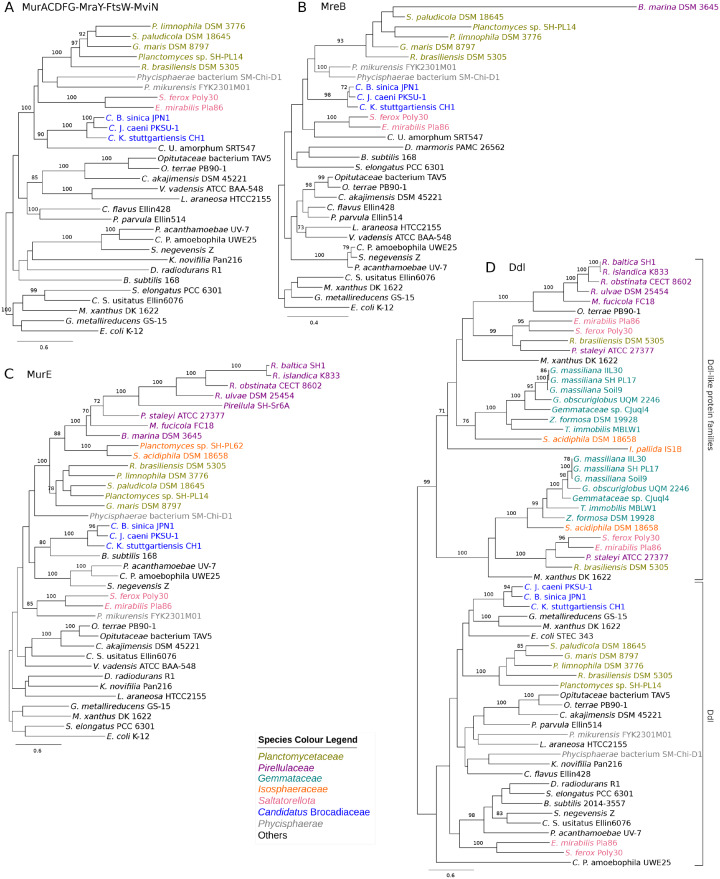
To study the evolution of genes coding for cell wall and cell division proteins in the few Planctomycetes species where they could be identified, we inferred a phylogenetic tree based on a concatenated alignment of the MurACDFG, MviN, FtsW, and MraY proteins (fig. 9A). The phylogeny based on concatenated alignment of MurACDFG, MviN, FtsW, and MraY supported the hypothesis that the genes in the dcw cluster have been vertically inherited from the common ancestor of all Planctomycetes, although the diversification pattern of the early-diverging lineages was not well resolved. The single-protein trees supported the clustering of genomes into clades, but the resolution was in most cases not high enough to resolve the diversification order of these clades (supplementary fig. S9, Supplementary Material online). However, none of the single-protein trees was incongruent with the concatenated tree topology with significant support.
Fig. 9.
Phylogenies of proteins involved in cell wall biosynthesis. Phylogenetic trees were inferred based on (A) a concatenated alignment of MurACDFG, MraY, FtsW, and MviN proteins (B) MreB proteins, (C) MurE proteins, and (D) proteins from Ddl and 2 Ddl-like protein families. Taxa in which these genes were identified as present (fig. 8) were included in addition to five outgroup species Escherichia coli, Candidatus Solibacter usitatus, Synechococcus elongatus, Bacillus subtilis, and Deinococcus radiodurans. The phylogeny was inferred with the maximum likelihood method with 100 bootstraps. Only bootstrap support values ≥70 are shown.
A phylogeny was also inferred based on an alignment of the MreB proteins (fig. 9B), which was largely congruent with the topology of the tree inferred from the concatenated alignment. Surprisingly, a gene with homology to mreB was also identified in Blastopirellula marina, although this species lacks most other genes in the dcw gene cluster. However, the branch to this lineage was exceptionally long. Likewise, a phylogeny inferred from the MurE proteins revealed exceptionally long branches to Rhodopirellula clade (fig. 9C). Thus, the few retained homologs to peptidoglycan biosynthesis genes in the Rhodopirellula clade are likely to have been adopted for other functions.
We also noted dramatic changes to the ddl genes that code for d-alanine–d-alanine ligase that synthesizes di-alanine, a substrate for the synthesis of short peptides of the peptidoglycan. In all species that contain the dcw gene cluster, the ddl gene is located upstream of the ftsQ gene, but no such gene was identified near the ftsQ gene in the species that lack the dcw gene cluster. However, paralogous ddl genes were identified elsewhere in these genomes. A phylogeny confirmed that the Ddl-like paralogs belong to a clade that is distinct from the Planctomycetaceae clade that contains the Ddl proteins encoded by genes located in the dcw gene cluster (fig. 9D). The two Saltatorellota species contain both the Ddl and the Ddl-like proteins, possibly suggesting ancestral duplication and differential retention. Two paralogous Ddl-like proteins are present in the Gemmataceae. Moreover, the FtsQ protein in the Gemmataceae lacked some of the highly conserved amino acids at the C-terminal end (supplementary fig. S10, Supplementary Material online), indicative of subfunctionalization.
The loss and modification of genes for cell wall and cell elongasome components in the Planctomycetales was preceded by loss of genes for the cell division proteins FtsA, FtsZ, and RodZ and AmiA already in the common ancestor of the Planctomycetes phylum. Furthermore, the ftsZ gene is absent from Methylacidimicrobium tartarophylax, which is a member of the Verrumicrobia and has an enlarged periplasm (Dunfield et al. 2007). Interestingly, a phylogeny inferred from an FtsZ protein alignment revealed dramatically extended branches to Lentisphaera and Methylacidiphilae (supplementary fig. S11, Supplementary Material online), suggesting functional modifications of the FtsZ protein also in sister lineages in which the protein is retained.
Discussion
The Planctomycetes are thought to represent a deeply branching lineage of bacteria that have secondarily acquired cellular traits that resemble those of eukaryotes, but how this transition to a higher level of cellular complexity has occurred is still a mystery. Rather than the simplistic idea that the eukaryotic-like traits result from the acquisition of eukaryotic genes, our results suggest that the rebuilding and loss of the bacterial peptidoglycan cell wall was a key process in the evolution toward cellular complexity. The cell envelope structure of the Planctomycetes has been debated for decades and there is still no consensus regarding the source and composition of the various membrane layers and the cell wall. The different hypotheses are mainly due to different interpretations of similar results and therefore merit a deeper discussion. Below, we discuss the composition and evolution of cell envelope components in more detail, as inferred from our results and those of previous studies.
The most visually striking feature of these bacteria is the invagination of the inner membrane, which dramatically increases the volume of the periplasmic space and raises questions about the structures of protein complexes that normally span across the inner and outer membranes. Even at sites where the inner and outer-membrane layers are in close proximity to each other the width between them is >40 nm, despite of which we observed typical bacterial-like transenvelope spanning protein complexes, such as pili and flagella. However, the outer-membrane components of the T2SS/T4P were predicted to be twice as long in members of the Planctomycetales compared with homologs in earlier diverging taxa and the outgroup species. Length extensions were identified in the periplasmic segments of the proteins, indicating that the length increase is an adaptation to a wider periplasmic space.
Another highly atypical feature is the large number of pore-like structures on the cell surface, referred to in the literature as crateriform structures (Fuerst and Webb 1991). These pore-like structures were clearly visible in intact cells as well as in the isolated cell wall structure of T. immobilis, and Gem. obscuriglobus contains similar perforations. The diameter of the uniformly distributed small pores was ∼25 nm, with an estimated abundance of ∼50 pores per µm2 of membrane sheet. We also identified highly abundant outer-membrane protein complexes with diameters of ∼25 nm on the surfaces of the T. immobilis and Gem. obscuriglobus cells. Thus, we hypothesize that the identified small pores in the cell wall correspond to the sites at which the outer-membrane protein complexes are normally situated. Pore-like structures of ∼30 nm with an estimated abundance of ∼200 pores per µm2 of membrane sheet were identified in Gem. obscuriglobus on one of three membrane types (Sagulenko et al. 2017). It has been argued that the pore-containing membrane surrounds the nucleoid (Sagulenko et al. 2017), although others have suggested that it represents the outer membrane (Jogler et al. 2019). Although we have shown in this study that T. immobilis has a pore-containing cell wall, future studies are needed to resolve the intracellular locations of the pore-containing membranes in Gem. obscuriglobus.
A third atypical feature concerns the composition of the cell wall. Our results showed that two cysteine-rich proteins accounted for >50% of the total protein content of the isolated cell wall. Cysteine-rich proteins with YTV domains have been identified previously in cell wall preparations of R. baltica (Hieu et al. 2008) and Gem. obscuriglobus (Sagulenko et al. 2017), adding support to the hypothesis of a proteinaceous cell wall (König et al. 1984; Liesack et al. 1986). Our bioinformatics analysis confirmed that homologs to these proteins are present in all members of the Planctomycetales but not in Phycisphaerae or any of the earlier diverging bacteria. Thus, our analysis suggests that the cysteine-rich cell wall proteins with YTV domains emerged and duplicated in the common ancestor of the Planctomycetales.
Interestingly, the presence of cysteine-rich cell wall layer is not unique to the Planctomycetales, but has also been identified in the intracellular human pathogen Chlamydia trachomatis, another member of the PVC superphylum (Aistleitner et al. 2015). In this species, two other cell wall proteins were identified, OmcA and OmcB. The cell wall protein OmcA is associated with the outer membrane through a lipid anchor and has a cysteine content of 15%, whereas OmcB is an outer-membrane spanning protein with a cysteine-content of 4%. The cysteine residues in these two proteins are located within the periplasm, and thus available for disulfide bond formation (Pilhofer et al. 2013; Liechti et al. 2014). This arrangement means that the stability of the cell wall can be modulated by changes in the redox potential, such that the cell wall is less rigid under reduced conditions. This has important consequences for the life cycle of Ch. trachomatis; in the extracellular stage, the disulfide bridges contribute to the formation of a rigid cell wall, whereas the cysteine bonds are reduced during the replicative stage in the protective host cell cytoplasm, resulting in a less rigid cell envelope (Christensen et al. 2019).
Homologs to OmcA and OmcB are present in all members of the Protochlamydia, whereas Waddlia and Parachlamydia species only contain OmcA and Simkania none of the two (Aistleitner et al. 2015). We identified homologs to OmcB also in the Planctomycetes (supplementary fig. S12, Supplementary Material online), although these proteins are not as cysteine-rich as in Chlamydia. Nevertheless, it may be hypothesized that cysteine-rich cell wall proteins control the rigidity of the cell wall also in the Planctomycetales in response to changes in the oxidation–reduction potential, in analogy to the role of such proteins in Ch. trachomatis. Future studies of the expression patterns and oxidation stages of the cysteine-rich proteins associated with the cell wall in the Planctomycetales are needed to test this hypothesis.
Importantly, our results suggest that several members of the Planctomycetales do not contain a conventional peptidoglycan, whereas Ch. trachomatis in addition to the cysteine-rich outer-membrane proteins also contains a peptidoglycan (Liechti et al. 2014). Our result contradicts an earlier suggestion that genes for peptidoglycan biosynthesis are present in all species of the Planctomycetales (Jeske et al. 2015). A comparison of the results from the two studies showed that the genes that we classified as present were marked as significant hits in Jeske et al. (2015), whereas those that we classified as absent were marked as nonsignificant hits. Thus, the inferred presence/absence pattern of genes for peptidoglycan biosynthesis depends on how to judge weak sequence similarities. In the previous study (Jeske et al. 2015), the proteins required for peptidoglycan biosynthesis in E. coli were used as queries in PSI-BLAST searches at five iterations to identify homologs in the Planctomycetes species. Some hits showed BLAST identities >30% and E values <1e-6; these were marked with (+) (supplementary table 2 in Jeske et al. [2015]). However, other hits were not statistically significant (e.g., murCD and murF were identified at E values >1e-6 and protein identities were <30%). These nonsignificant hits were marked with (+) (supplementary table 2 in Jeske et al. [2015]).
A re-examination of the analyses described in Jeske et al. (2015) showed that the weak PSI-BLAST hits (+) obtained for the MurCDF proteins in Pirellulaceae, Isosphaeraceae, and Gemmataceae were in most cases to a single protein, FolC, which is a distant homolog of the Mur ligase proteins involved in folate rather than peptidoglycan biosynthesis, and in some cases to CphA, cyanophycin synthetase. Phylogenies inferred from MurC and FolC/CphA confirmed the absence of MurCDF in Pirellulaceae, Isosphaeraceae, and Gemmataceae, whereas the paralogous protein FolC is present in all members of the Planctomycetes. Likewise, the weak hits in the PSI-BLAST search for the MurG protein were to glycosyltransferases, which consist of many paralogs broadly grouped into large protein families.
Furthermore, since the species associated with weak hits are not more distantly related to Pl. limnophila than the species with strong hits, there is no reason a priori to expect such large differences in sequence similarity for orthologs that code for the same functions. Phylogenies confirmed that the weak hits in the PSI-BLAST search for the Mur proteins were to paralogous proteins with inferred other functions. Likewise, a recent study (Wiegand et al. 2020) reinvestigated the analyses in Jeske et al. (2015) using more stringent criteria, and also came to the conclusion that these genes are absent. Thus, it can be concluded that genes for peptidoglycan biosynthesis are absent from the genomes of Pirellulaceae, Isosphaeraceae, and Gemmataceae.
Despite the lack of such genes, it has been argued that all members of the Planctomycetales possess a peptidoglycan cell wall (Wiegand et al. 2020) based on the identification of GlcNAc and MurNAc after digestion with mutanolysin in Pl. limnophila and Gem. obscuriglobus (Jeske et al. 2015). However, we found that the cell wall structures purified from T. immobilis were resistant to muralytic enzymes but rapidly digested by proteinase K. These results support earlier findings reporting that proteins are a major component of the cell wall, but does not rule out the possibility that the cell wall also contains a simple or different form of glycan strand.
Another argument in support of the hypothesis that these bacteria do not contain a conventional peptidoglycan cell wall is that the loss of genes for peptidoglycan biosynthesis in Gemmataceae, Isosphaerae, and Pirellulaceae coincides with the loss of genes in the mreBCD–pbp2–rodA gene cluster. The 16S rRNA phylogeny suggests that Gemmataceae groups with Isosphaeraceae, whereas Pirellulaceae forms a clade with Planctomycetaceae. Based on this clustering pattern, we infer two independent losses of genes for peptidoglycan biosynthesis and cell elongation. Although the diversification of these clades are not well supported in the 16S rRNA tree, the exact same topology was obtained in a tree inferred from a concatenated alignment of 76 proteins encoded by single-copy genes with bootstrap support values of >98% at the critical nodes (Mahajan et al. 2020). We therefore consider two independent losses to be a likely scenario, although we recognize that if Gemmataceae, Isosphaeraceae, and Pirellulaceae would instead belong to the same clade, a single loss would suffice to explain the phyletic distribution pattern of these genes in the Planctomycetales.
The standard set of genes for peptidoglycan biosynthesis could also not be detected the early-diverging lineage Ca. U. amorphum and thus, a case can be made for multiple, independent losses. These losses make sense since it has been shown that Gem. obscuriglobus is able to import proteins in an “endocytosis-like” process (Fuerst and Sagulenko 2010, 2011; Lonhienne et al. 2010) and more stunningly that Ca. U. amorphum is capable of phagocytosis (Shiratori et al. 2019).
Genes for peptidoglycan biosynthesis and the MreB protein complex are retained in the Planctomycetaceae, but some of these may have slightly altered functions. For example, an mreB knock-out mutant in Pl. limnophila showed no phenotypic differences compared with the wild-type strain, suggesting that MreB is not essential for cell elongation in this species (Rivas-Marin et al. 2020; Wiegand et al. 2020). Likewise, FtsI and FtsW, which are required during the elongation and septation stages of binary cell division, were found to be nonessential for cell division in Pl. limnophila (Rivas-Marin et al. 2020; Wiegand et al. 2020). Furthermore, a phylogenetic tree inferred from the MurE proteins revealed atypically long branches to several species in the Rhodopirellula clade, which testify to the hypothesis of altered selective constraints on these genes.
We have shown previously that the common ancestor of the Planctomycetales acquired >1,000 novel protein families by duplications and domain rearrangements, including novel signal transduction and regulatory systems (Mahajan et al. 2020). In this study, we have shown that the ancestor of the Planctomycetales gained novel cysteine-rich cell wall proteins not found in other groups of bacteria. Additionally, we have shown that the periplasmic segment of the outer-membrane proteins has been extended in size, and that the peptidoglycan cell wall and cell elongasome protein complex have been lost or modified. We argue that the rebuilding of the cell wall and the transition from binary fission to budding enabled the uptake of macromolecules (including proteins) and facilitated massive invaginations of the cytoplasmic membrane, thereby increasing the periplasmic volume for storage and degradation of the imported macromolecules. The altered cell envelope structures and cell proliferation processes in the Planctomycetales should thus be viewed as adaptive processes underlying the evolution of cellular complexity.
Materials and Methods
Cultivation and Isolation of the Cell Wall
Tuwongella immobilis, Gem. obscuriglobus, and Pl. limnophila were cultured on M1 medium agar plates (Schlesner 1994) at 32 °C for a period of 4 days prior to harvesting. Cells were harvested into 10 mM Tris, 1 mM EDTA, pH 7.4 (TE), pelleted by centrifugation (4,500×g, 5 min, room temperature), and the supernatant was removed. Escherichia coli K12 MG1655 was grown in LB medium at 37 °C in a Biosan TS-1 bioreactor (2,000 rpm, 1 s−1 reverse spinning rate) for 2.5 h. Cells were harvested by centrifugation (4,500×g, 10 min, room temperature) and the supernatant was removed. Bacterial cell pellets were resuspended in TE buffer and the absorbance at λ = 600 nm (OD600) was measured. Aliquots corresponding to a cell density of OD600 ≈ 0.5 were transferred to fresh tubes and the pellets were washed two more times in TE buffer.
Isolation of Cell Wall
Tuwongella immobilis was cultivated in 100 ml M1 medium at 32 °C until the early stationary phase (OD600 ≈ 1.0, 6 days). The cell suspension was split into aliquots of 50 ml and centrifuged at 4,500×g for 20 min at 4 °C. The supernatant was discarded and the pellet was washed twice in 1× TE buffer. The washed pellet was stored at −20 °C until further processing. To isolate the cell wall, the frozen pellet from 50-ml cell suspension was thawed at room temperature and resuspended in 10 ml 5% SDS (v/v) in 1× TE buffer. After overnight incubation at 70 °C under gentle orbital shaking, the SDS-insoluble cell wall was pelleted by ultracentrifugation (100,000×g, 40 min, 4 °C, Beckman Optima XPN-100, 90Ti rotor). The pellet was washed twice by resuspension in 4 ml 5% SDS in 1× TE buffer, followed by ultracentrifugation (100,000×g, 30 min, 4 °C, Beckman Optima XPN-100, 90Ti rotor). The washed pellet was resuspended in 500 µl 1× TE buffer and dissolved by mild tip-sonication (2 mm probe, 30% amplitude, 5×10 s, on ice, Vibra-Cell VCX 130 sonicator).
Real-Time Spectroscopy
The effects of enzymatic and chemical treatments were monitored in real-time using a Bioscreen C plate reader (Oy Growth Curves Ab Ltd., Finland) operated at 37 °C. Washed cell pellets with a cell density adjusted to OD600 of 0.5 were resuspended in the following buffers: 1) 10 mM Tris, 10 mM EDTA, pH 7.4 (“EDTA”), 2) 10 mM Tris, 10 mM EDTA, 10 mM DTT, pH 7.4 (“EDTA/DTT”), 3) 10 mM Tris, 10 mM EDTA, 10 mM DTT, 100 µg/ml mutanolysin, pH 7.4 (“EDTA/DTT/Mut”), 4) 10 mM Tris, 10 mM EDTA, 100 µg/ml mutanolysin, pH 7.4 (“EDTA/Mut”), and 5) 10 mM Tris, 10 mM EDTA, 5% SDS (“EDTA/SDS”). Samples were analyzed in technical triplicates using a volume of 300 µl per well. Absorbance at λ = 600 nm was measured every 5 min after shaking the plate for 60 s over the timecourse of 16 h. A cell-free sample, only containing 10 mM Tris, 10 mM EDTA, pH 7.4 was used as a blank. The raw data and a corresponding smoothed line (geom_smooth function of R ggplot, span=0.1) were plotted as a function of time.
Microscopy Analyses
Phase Contrast Microscopy
After treating the cells under the different conditions as described earlier, the triplicate samples were pooled and centrifuged at 17,000×g for 10 min at room temperature. The supernatant was removed leaving ∼50 µl buffer remaining, cells were resuspended, and immobilized on 1% agarose slabs for phase contrast microscopy on an Olympus CX41 microscope using a 100× objective.
Transmission Electron Microscopy
The pooled and concentrated cell suspensions used for phase-contrast microscopy were centrifuged at 17,000×g for 10 min at room temperature, the supernatant was discarded, and remaining pellets were fixed in 10 mM phosphate buffer, 2.5% glutaraldehyde, 1% formaldehyde, pH 7.4. After fixation for 15 min at room temperature, the fixed samples were stored at +4 °C. Further processing was performed by the electron microscopy unit, EMiL, at Karolinska Institutet, Huddinge, Sweden as previously described (Seeger et al. 2017).
Scanning Electron Microscopy
Isolated cell wall sacculi of T. immobilis were centrifuged for 20 min at 17,000×g and the supernatant was removed. The pellet was fixed in 10 mM phosphate buffer, 2.5% glutaraldehyde, 1% formaldehyde, pH 7.4 for 15 min at room temperature before storage at 4 °C. As a control, untreated T. immobilis cells, grown on M1 agar plates for 5 days at 32 °C, were transferred into fixation solution and fixed as the cell wall samples. The fixed samples were adhered onto a pore membrane and washed with 0.01 M phosphate buffer, pH 7.4, and MilliQ water prior to stepwise ethanol dehydration and critical-point drying using carbon dioxide (Leica EM CPD 030). The pore membranes were mounted on specimen stubs using carbon adhesive tabs and sputter-coated with platinum (Quorum Q150T ES). SEM images were acquired using an Ultra 55 field emission scanning electron microscope (Zeiss, Oberkochen, Germany) at 3 kV and the SE2 detector.
Negative-Staining TEM
About 3 µl of the isolated T. immobilis cell wall were applied on glow-discharged carbon-coated and formvar-stabilized 400 mesh copper grids (Ted Pella) and incubated for ∼30 s. Excess of sample was blotted off and the grid was washed with MilliQ water prior to negative staining using 2% uranyl acetate. TEM imaging was done using a Hitachi HT7700 (Hitachi High-technologies) transmission electron microscope operated at 100 kV equipped with a 2kx2k Veleta CCD camera (Olympus Soft Imaging System).
Cryo-EM Tomography
Tuwongella immobilis and Gem. obscuriglobus were grown on M1 agar plates for 3 days at 32 °C and for additional 2 days at 20 °C, harvested into 10 mM phosphate buffer, pH 7.4, and the cell concentration was adjusted to OD600 ≈ 8. The cell suspension was plunge-frozen on Quantifoil R2/1 holey carbon films on 200 gold mesh (Jena Bioscience) using a FEI Vitrobot (Thermo Fisher Scientific, blot time: 6 s, blot force: −5, wait time: 15 s, 20 °C, 100% humidity). Cryo-FIB milling with gallium ion source was performed on a FEI Dual beam Scios (Thermo Fisher Scientific, Netherlands). Autogrids with vitrified cells were mounted onto a dedicated cryo-holder and transferred onto a cooled (−180 °C) cryo-stage in the dual-beam microscope. The lamellae were prepared with two parallel rectangular patterns at both sides (top and bottom) of the cells, at milling angle of 10–15°, accelerating voltage of 30 kV, and ion currents between 30 and 300 pA. The final thickness of the lamella (milled at the lowest ion current) was ∼150–300 nm. After cryo-FIB milling, the autogrids were transferred into an autoloader cassette for cryo-ET using a FEI Titan Krios Cryo-transmission electron microscope (Thermo Fisher Scientific, Netherlands). Tilt series were acquired using a K2 camera (C2 aperture: 70, objective aperture: 100, magnification: 19,500×, pixel size: 7.37 Å, spot size: 9, illuminated area: 5.3 µm, energy filter slit width: 20) starting bidirectionally from 20° (−60°/+60°, 1° increment for T. immobilis and −50°/+50°, 1° increment for Gem. obscuriglobus) with a defocus of −6 µm. The total electron dose per tilt series was 99e−/Å for T. immobilis and 95e−/Å for Gem. obscuriglobus.
Additional tilt series at higher magnification were acquired for T. immobilis (magnification: 33,000×, pixel size: 4.37 Å, spot size: 8, illuminated area: 3.2 µm, energy filter slit width: 20) starting bidirectionally from 20° (−60°/+60°, increment 2°) with a defocus of −4 µm. The total electron dose was 95e−/Å2. Motion correction for the acquired tilt series was performed by using motioncor2. Tomograms were reconstructed using the Etomo package of IMOD 4.9.10 using the workflow based on patch-tracking. Nonlinear anisotropic diffusion (NAD) filtering was performed on the reconstructed tomogram (K = 30, iterations=20).
Measurements of the Distance between the Inner and the Outer Membranes
The distance between the inner (IM) and outer (OM) membranes was measured using ImageJ 1.52e (Schneider et al. 2012) as part of the Fiji distribution (Schindelin et al. 2012). At sites where the inner membrane aligned in proximit to the outer membrane, the average distance between the inner and outer-membrane layers was estimated. For this, six slices were extracted using the IMOD Slicer function (interslice distance ≈22 nm, thickness: 25) both from the tomograms of T. immobilis and Gem. obscuriglobus. The OM–IM distances were measured at several sections across the cell envelope in the x–y plane. Ten average density profiles (gray value as function of distance in [nm]) were recorded per slice using a linewidth corresponding to ∼20 nm. The OM–IM distances were then determined between the gray value minima (middle) of the outer membrane and the inner membrane. OM–IM distances in Pl. limnophila were measured similarly on six previously published cryo-EM tomographs (Boedeker et al. 2017).
Proteomics Analysis
LC-MS/MS Analysis of Isolated T. immobilis Cell Wall
A small aliquot of the isolated cell wall preparation was mixed to a final concentration of 1× NuPAGE LDS Sample Buffer, 1× NuPAGE Sample Reducing Agent, and incubated at 70 °C for 10 min. About 20 µl sample and 5 µl Novex Sharp Unstained Protein Standard were loaded on a NuPAGE 4–12% Bis–Tris protein gels (1.0 mm) and electrophoresis was performed in 1× MOPS SDS Running buffer supplemented with NuPage Antioxidant for 55 min at 200 V. The gel was washed, stained, and destained using SimplyBlue SafeStain based on the manufacturer’s microwave protocol and imaged on a ChemiDoc MP imaging system (Bio-Rad). For LC-MS/MS analysis, the top of the gel, expected to contain the large cell wall sacculi, was cut and stored at 4 °C (1). The rest of the gel lane was cut and stored at 4 °C until processing (2). The proteins from both samples were in-gel digested by trypsin as previously described (Shevchenko et al. 2000). Briefly, gel pieces were destained, reduced with 10 mM DTT, and alkylated with 50 mM iodoacetoamide. Incubation with 12.5 ng/ml trypsin (Promega) in 25 mM NaHCO3 was performed overnight, at 37 °C in the dark. Tryptic peptides were extracted using sonication and by adding 60% ACN in 5% formic acid. Peptides were dried in a Speedvac system. Thereafter, the samples were resolved in 30 µl 0.1% FA (formic acid). The resulting peptides were separated in reversed-phase on a C18-column, applying a 90-min long gradient, and electrosprayed on-line to a QExactive Plus mass spectrometer (Thermo Finnigan). Tandem mass spectrometry was performed applying HCD. Database searches were performed using the Sequest algorithm, embedded in Proteome Discoverer 1.4 (Thermo Fisher Scientific) against the genome of T. immobilis isolate MBLW1 (GenBank accession number: LR593887.1). The search parameters were set to enzyme: trypsin (fixed modification was Carbamidomethyl [C], and variable modifications were Oxidation [M] and Deamidated [NQ]). The search criteria for protein identification were set to at least two matching peptides of 95% confidence level per protein (supplementary tables S1 and S2, Supplementary Material online).
Bioinformatics Analyses
Genome Data
Genome sequence data of 44 bacterial species (supplementary table S4, Supplementary Material online) were downloaded from the NCBI public databases. The data set included 32 genomes from a representative set of species from the Planctomycetes (Ludwig et al. 2010) for which whole-genome sequence data are available (although only assembled genome sequence data from culture enrichments experiments are available for Ca. Brocadiae). The data set included genomes from the four major clades in the order Planctomycetales (represented by Planctomycetaceae, Pirellulaceae, Gemmataceae, and Isosphaeraceae), as well as genomes from the classes Phycisphaerae and Ca. Brocadiae. We also included the recently sequenced genomes from the newly described genus Saltarotellota. The genomes have been color-coded according to the classification of taxa into lineages in all figures and supplementary material.
The data set also included an additional ten completed genomes from species of Lentisphaera, Verrumicrobia, and Chlamydia in the PVC superphylum (Ludwig et al. 2010) for which whole-genome sequence data were available. The genome data available from the new phyla Kiritimatiellaeota and Omnitrophia in the PVC superphylum are mostly single-cell genomes or metagenomes and thus less suitable for studies of phyletic gene presence/absence patterns. Three of the four whole genomes available for Kiritimatiellaeota were included in the extended analysis of peptidoglycan biosynthesis genes and in the absence of whole-genome data, we also included two single-cell genomes from Omnitrophica.
For most analyses, we used the Deltaproteobacteria Myxococcus xanthus and Geobacter metallireducens as the outgroups, although some analyses also included other outgroup taxa, as detailed in supplementary table S4b, Supplementary Material online. M. xanthus was selected as the outgroup because of its complex lifestyle, long genes, and large genome of 9.1 Mb. As such it is one of the few bacterial species from an unrelated phylum with similar characteristics as the Planctomycetes. We added Geo. metallireducens to get one additional species from the Deltaproteobacteria.
For genomes downloaded after June 2016, we used the Gene IDs and annotations made available through NCBI, which are based on the NCBI prokaryotic genome annotation pipeline (Tatusova et al. 2016), whereas most other genomes were reanalyzed to make the annotations of the genomes in the data set comparable (supplementary table S4, Supplementary Material online). For the latter genomes, gene predictions were made using Prodigal v2.60 (Hyatt et al. 2010) and annotations were based on searches against the UniprotKB protein database (UniProt 2015) and the NCBI nonredundant protein database (nr) using BlastP (Altschul et al. 1990).
Assignment of Conserved Protein Domains
Conserved domains were assigned to the proteins using the pfam_scan.pl script and the Pfam 32.0 database (Punta et al. 2012). However, some of the domains in the outer-membrane proteins and the cell wall proteins are highly repeated and some of copies of these repeat domains have diverged significantly from the HMM models that are described in Pfam database. Hence, a subsidized e-value cut-off (parameters for pfam_scan.pl: -e_dom 0.1 -e_seq 0.01) was used to search for Pfam domains with weaker homology in the GspD, PilQ, and OmcB proteins and in proteins containing the YTV domains.
Clustering of Proteins into Families
All-against-all homology search was performed using proteins from all the genomes and blastall v2.2.26 (Altschul et al. 1990). The homology search results were filtered based on the aligned length of the proteins, as described previously (Mahajan et al. 2020), and clustered into protein families using OrthoMCL v2 (Dongen 2000; Li et al. 2003).
Identification of Genes for Peptidoglycan Biosynthesis and Cell Division
Genes for peptidoglycan biosynthesis and cell division functions were identified by a three-step procedure. First, we identified protein families containing proteins encoded by genes annotated to be implicated in the biosynthesis and assembly of the peptidoglycan. In the second step, neighbors of the genes thus identified in the individual genomes were examined for sequence similarity, domain architecture similarity, and phylogenetic relationships to the positional homologs in related species. Homologs of the neighbor genes thus identified were used as queries in searches against the 44 reference proteomes, using blastall v2.2.26, and putative hits were examined for domain architecture similarity and phylogenetic relationships. The second step was repeated until no new orthologs could be detected. Thirdly, to verify the absence of these genes in some lineages, proteins encoded by genes for peptidoglycan biosynthesis and cell division in Pl. limnophila were used as the queries in protein-based searches with blastall v2.2.26 against the other 43 proteomes. The details of the BLAST hits to genes in Pl. limnophila are shown in supplementary table S5, Supplementary Material online and the list of identified genes is shown in supplementary table S6, Supplementary Material online.
The proteins identified in the representative set of 44 Planctomycetes genomes were used as queries in sequence similarity searches against an expanded set of 121 Planctomycetes genomes using blastall v2.2.26. Proteins involved in peptidoglycan biosynthesis in unrelated species such as E. coli and Bacillus subtilis were also included as queries in these searches, to ensure that no divergent homologs putatively acquired by horizontal gene transfer were missed. We also performed an hmmscan search of all proteomes using HMMER version 3.1b (Eddy 2011) against an hmm database built with known peptidoglycan biosynthesis proteins from three species in the clade containing Candidatus Kuenenia stuttgartiensis CH1 and five species in the clade containing Pl. limnophila. However, also this search did not recover any additional orthologs but yielded only hits to very distant paralogs with other functions.
Identification of Genes for YTV Domain Proteins
The YTV domain proteins were identified using the annotations from the Pfam database and protein family clusters (supplementary table S3, Supplementary Material online). The proteins thus identified were aligned using mafft-linsi algorithm (Katoh et al. 2005), and three different segments of the alignment that consisted of the YTV motif were manually curated and used to build three different HMM models with the aid of hmmbuild from HMMER version 3.1b (Eddy 2011). Using these HMM models, we searched for distant homologs of YTV domain proteins in all the 44 proteomes in our main data set, but no divergent homologs were detected outside the Planctomycetales.
Phylogenetic Inference
Proteins were aligned using the mafft-linsi alignment algorithm in MAFFT v7.310 (Katoh et al. 2005), thereafter, the poorly conserved edges and long gaps were trimmed from the alignments using Gblocks 0.91b (settings: −b3 = 50 −b5 = a) (Talavera and Castresana 2007). The alignments of MurABCDFG, MviN, MraY, and FtsW were concatenated together and the missing proteins (four in K. novifilia and two in Ca. U. amorphum) were replaced with gaps “-.” Maximum likelihood phylogenies were inferred with the PROTCATLG (MurB, MreB, and multiprotein alignment). For all other proteins, their sequences were aligned using the mafft-linsi alignment algorithm in MAFFT v7.310, and the phylogeny was inferred using PROTGAMMALG models and 100 bootstraps in RAxML 8.0.26 (Stamatakis 2014). The number of taxa and the number of positions in the alignment for each phylogeny are summarized in supplementary table S7, Supplementary Material online.
The 16S rRNA sequences from an extended data set of 146 genomes were aligned using SINA aligner v1.2.11 (Pruesse et al. 2012) and the maximum likelihood phylogeny was inferred using CATGTR model with 100 bootstraps in RAxML 8.0.26. Only bootstrap support values of 70 or above are shown in the figures.
Protein Tertiary Structure Modeling
The structures of the PilQ, GspD monomers were modeled using online structure prediction server RaptorX (Kallberg et al. 2012). The C-terminal, which includes the Secretin and Secretin_N domains, of the GspD protein in Gem. obscuriglobus, and the PilQ proteins in Gem. obscuriglobus and Gi. maris was modeled using the GspD protein in E. coli (PDB ID: 5wq7). The N0 domain of the GspD protein in Gem. obscuriglobus was modeled using the same in Ps. aeruginosa (pdb ID: 5wln). The N-terminus helical repeats of the PilQ protein in Gem. obscuriglobus and Gi. maris were modeled using structure of a TPR repeat domain in the Ski2 protein in Sacharomyces cerevisiae (pdb ID: 4buj).
Data Availability
The metadata and repositories that hold the data are summarized in supplementary table S8, Supplementary Material online.
Supplementary Material
Acknowledgments
We thank Cornelius Wijaya for initial studies of the effects of biochemical treatments on lysis of studied Planctomycetes species. We thank the National Microscopy Infrastructure, NMI (VR-RFI 2016-00968) at the Umeå Core facility for Electron Microscopy, the SciLifeLab cryo-EM node in Umeå for cryo-ET and the mass spectrometry-based proteomic facility in Uppsala for mass spectrometry analysis. This work was supported by grants to S.G.E.A. from the Swedish Research Council (2014-4460, 2018-4135) and the Knut and Alice Wallenberg Foundation (2011.0148, 2017.0322).
Data deposition: This project has been deposited at BioStudies under the accession S-BSST390.
Literature Cited
- Aistleitner K, et al. 2015. Conserved features and major differences in the outer membrane protein composition of Chlamydiae. Environ Microbiol. 17(4):1397–1413. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Boedeker C, et al. 2017. Determining the bacterial cell biology of Planctomycetes. Nat Commun. 8(1):14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YW, et al. 2016. Architecture of the type IVa pilus machine. Science 351(6278):aad2001–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S, McMahon RM, Martin JL, Huston WM. 2019. Life inside and out: making and breaking protein disulfide bonds in Chlamydia. Crit Rev Microbiol. 45(1):33–50. [DOI] [PubMed] [Google Scholar]
- Cserti E, et al. 2017. Dynamics of the peptidoglycan biosynthetic machinery in the stalked budding bacterium Hyphomonas neptunium. Mol Microbiol. 103(5):875–895. [DOI] [PubMed] [Google Scholar]
- Devos DP, Reynaud EG. 2010. Intermediate steps. Science 330(6008):1187–1188. [DOI] [PubMed] [Google Scholar]
- Dominguez-Escobar J, et al. 2011. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333(6039):225–228. [DOI] [PubMed] [Google Scholar]
- Dongen S. 2000. A cluster algorithm for graphs. Amsterdam: (The Netherlands; ): CWI (Centre for Mathematics and Computer Science). [Google Scholar]
- Dunfield PF, et al. 2007. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450(7171):879–882. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst JA, Sagulenko E. 2010. Protein uptake by bacteria: en endocytosis-like process in the planctomycete Gemmata obscuriglobus. Commun Integr Biol. 3(6):572–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst JA, Sagulenko E. 2011. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol. 9(6):403–413. [DOI] [PubMed] [Google Scholar]
- Fuerst JA, Webb RI. 1991. Membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus. Proc Natl Acad Sci U S A. 88(18):8184–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimesi N. 1924. Hydrobiologiai tanulmányok (Hydrobiologische Studien) I. Planctomyces bekefii Gim. nov. gen. et sp. Budapest: Ciszterci Rend Kiad; p. 1–8. [Google Scholar]
- Hieu CX, et al. 2008. Detailed proteome analysis of growing cells of the planctomycete Rhodopirellula baltica SH1T. Proteomics 8(8):1608–1623. [DOI] [PubMed] [Google Scholar]
- Hirsch P. 1972. Two identical genera of budding and stalked bacteria: Planctomyces Gimesi 1924 and Blastocaulis Henrici and Johnson 1935. Int J Syst Evol Microbiol. 22(2):107–111. [Google Scholar]
- Hyatt D. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske O, et al. 2015. Planctomycetes do possess a peptidoglycan cell wall. Nat Commun. 6(1):7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogler C, Wiegand S, Devos DP. 2019. Commentary: manifold routes to a nucleus. Front Microbiol. 10:1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJF, Carballido-Lopez R, Errington J. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104(6):913–922. [DOI] [PubMed] [Google Scholar]
- Kallberg M, et al. 2012. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 7:1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerger B, et al. 1988. The budding bacteria, Pirellula and Planctomyces, with atypical 16S rRNA and absence of peptidoglycan, show eubacterial phospholipids and uniquely high proportions of long chain beta-hydroxy fatty acids in the lipopolysaccharide lipid A. Arch Microbiol. 149(3):255–260. [Google Scholar]
- König E, Schlesner H, Hirsch P. 1984. Cell-wall studies on budding bacteria of the Planctomyces/Pasteuria Group and on a Prosthecomicrobium sp. Arch Microbiol. 138(3):200–205. [Google Scholar]
- Lee K-C, Webb RI, Fuerst JA. 2009. The cell cycle of the planctomycete Gemmata obscuriglobus with respect to cell compartmentalization. BMC Cell Biol. 10(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13(9):2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti GW, et al. 2014. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506(7489):507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesack W, Konig H, Schlesner H, Hirsch P. 1986. Chemical-composition of the peptidoglycan-free cell envelopes of budding bacteria of the Pirella/Planctomyces group. Arch Microbiol. 145(4):361–366. [Google Scholar]
- Lonhienne TG, et al. 2010. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci U S A. 107(29):12883–12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Euzéby J, Whitman WB. 2010. Road map of the phyla Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes In: Bergey's Manual® of Systematic Bacteriology. New York: Springer. p. 1–19. [Google Scholar]
- Mahajan M, et al. 2020. Paralogization and new protein architectures in Planctomycetes bacteria with complex cell structures. Mol Biol Evol. 37(4):1020–1040. [DOI] [PubMed] [Google Scholar]
- Mahat R, Seebart C, Basile F, Ward NL. 2016. Global and targeted lipid analysis of Gemmata obscuriglobus reveals the presence of lipopolysaccharide, a signature of the classical Gram-negative outer membrane. J Bacteriol. 198(2):221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M, et al. 2008. Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J Bacteriol. 190(9):3192–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M, et al. 2013. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun. 4(1):2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glockner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28(14):1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, et al. 2012. The Pfam protein families database. Nucleic Acids Res. 40(D1):D290–D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racki LR, et al. 2017. Polyphosphate granule biogenesis is temporally and functionally tied to cell cycle exit during starvation in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 114(12):E2440–E2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randich AM, Brun YV. 2015. Molecular mechanisms for the evolution of bacterial morphologies and growth modes. Front Microbiol. 6:580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Marin E, Canosa I, Devos DP. 2016. Evolutionary cell biology of division mode in the bacterial Planctomycetes-Verrucomicrobia-Chlamydiae superphylum. Front Microbiol. 7:1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Marin E, et al. 2020. Non-essentiality of canonical cell division genes in the planctomycete Planctopirus limnophila. Sci Rep. 10(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko E, et al. 2017. Nuclear pore-like structures in a compartmentalized bacterium. PLoS One 12(2):e0169432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9(7):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesner H. 1994. The development of media suitable for the microorganisms morphologically resembling Planctomyces spp, Pirellula spp, and other Planctomycetales from various aquatic habitats using dilute media. Syst Appl Microbiol. 17(1):135–145. [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, et al. 2017. Tuwongella immobilis gen. nov., sp. nov., a novel non-motile bacterium within the phylum Planctomycetes. Int J Syst Evol Microbiol. 67(12):4923–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham LT, et al. 2014. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345(6193):220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Chernushevich I, Wilm M, Mann M. 2000. De novo peptide sequencing by nanoelectrospray tandem mass spectrometry using triple quadrupole and quadrupole/time-of-flight instruments. In: Chapman JR, editors: Mass spectrometry of proteins and peptides. Methods in Molecular Biology TM, vol. 146 Totowa (NJ): Humana Press; p. 1. [DOI] [PubMed] [Google Scholar]
- Shiratori T, Suzuki S, Kakizawa Y, Ishida KI. 2019. Phagocytosis-like cell engulfment by a planctomycete bacterium. Nat Commun. 10(1):5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth DR, van Teeseling MCF, Jetten MSM. 2012. Genomic analysis indicates the presence of an asymmetric bilayer outer membrane in Planctomycetes and Verrucomicrobia. Front Microbiol. 3:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl H, Burmann F, Hamoen LW. 2014. The actin homologue MreB organizes the bacterial cell membrane. Nat Commun. 5(1):3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Tamames J, González-Moreno M, Mingorance J, Valencia A, Vicente M. 2001. Bringing gene order into bacterial shape. Trends Genet. 17(3):124–126. [DOI] [PubMed] [Google Scholar]
- Tatusova T, et al. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44(14):6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 10(2):123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C. 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeseling MCF, et al. 2015. Anammox Planctomycetes have a peptidoglycan cell wall. Nat Commun. 6(1):6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W. 2008. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev. 32(2):287–306. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 32(2):149–167. [DOI] [PubMed] [Google Scholar]
- Wagner M, Horn M. 2006. The Planctomycetes, Verrumicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol. 17(3):241–249. [DOI] [PubMed] [Google Scholar]
- Wiegand S, et al. 2019. The novel shapeshifting bacterial phylum Saltatorellota. bioRxiv. doi: 10.1101/817700. [Google Scholar]
- Wiegand S, et al. 2020. Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat Microbiol. 5(1):126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand S, Jogler M, Jogler C. 2018. On the maverick Planctomycetes. FEMS Microbiol Rev. 42(6):739–760. [DOI] [PubMed] [Google Scholar]
- Yan ZF, Yin M, Xu DD, Zhu YQ, Li XM. 2017. Structural insights into the secretin translocation channel in the type II secretion system. Nat Struct Mol Biol. 24(2):177–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metadata and repositories that hold the data are summarized in supplementary table S8, Supplementary Material online.