Abstract
Perfluorooctanoic acid (PFOA) is a persistent organic pollutant prevalent in the environment and implicated in damage to the liver leading to a fatty liver phenotype called hepatocellular steatosis. Our goal is to provide a basis for PFOA-induced hepatocellular steatosis in relation to epigenetic alterations and mRNA splicing. Young adult female mice exposed to different concentrations of PFOA showed an increase in liver weight with decreased global DNA methylation (5-mC). At higher concentrations, the expression of DNA methyltransferase 3A (Dnmt3a) was significantly reduced and the expression of tet methycytosine dioxygenase 1 (Tet1) was significantly increased. There was no significant change in the other Dnmts and Tets. PFOA exposure significantly increased the expression of cell cycle regulators and anti-apoptotic genes. The expression of multiple genes involved in mTOR (mammalian target of rapamycin) signaling pathway were altered significantly with reduction in Pten (phosphatase and tensin homolog, primary inhibitor of mTOR pathway) expression. Multiple splicing factors whose protein but not mRNA levels affected by PFOA exposure were identified. The changes in protein abundance of the splicing factors was also reflected in altered splicing pattern of their target genes, which provided new insights on the previously unexplored mechanisms of PFOA-mediated hepatotoxicity and pathogenesis.
Keywords: PFOA, epigenetics, splicing factors, liver, mouse studies
1. Introduction
Perfluorooctanoic acid (PFOA) is a synthetic compound that belongs to a group of perfluoroalkyl substance (PFAS). PFOA consists a long hydrophobic chain with eight carbons, saturated with fluorine atoms and a hydrophilic polar functional group (Banks et al., 1994; Kissa et al., 2001; Taylor, 1999). PFOA is one of the most frequently used perfluoroalkyl compounds since the 1940s; and is abundant in the environment, with deleterious consequences due exposure through food and drinking water (US EPA, 2001; Martin et al., 2003; Ericson et al., 2008; Vestergren et al. 2008). PFOA has been detected in human cord serum and breast milk (Apelberg et al., 2007; Liu et al., 2010; Mondal et al., 2012). In the human system, PFOA is known to be not easily metabolized due to its long half-life (mean estimated half-life is 2.7 years) as noted in past works (Li et al., 2018; Burris et al., 2002; Olsen et al., 2007). Experiments in animal model shows that PFOA predominantly accumulates in the liver, kidney and serum (Ylinen et al., 1990; Vanden Heuvel et al., 1991; Cui et al., 2009), resulting in hepatotoxicity, developmental toxicity, immunotoxicity, and neurotoxicity (DeWitt et al., 2008; Johansson et al., 2008; Gallo et al., 2012; Christopher and Martin, 1977; Metrick and Marias, 1977; Yang et al., 2000; Yang et al., 2001; Sibinski et al., 1987; Johansson et al., 2008).
Past in vitro and in vivo studies show a lack of genotoxicity associated with PFOA (Fernández Freire et al., 2008; Eriksen et al., 2010; Florentin et al. 2011; Stefani et al., 2014). Thus, one plausible way by which PFOA can exert its effect is through epigenetic programming. DNA methylation is one of the most important and well-established epigenetic indicators that especially plays a role in embryonic development and cellular differentiation (Jahner et al., 1982; Razin et al., 1984). Alteration at regions where cytosine-guanine dinucleotide (CpG sites) are frequent (CpG islands) could typically lead to the repression of gene expression (Hackett and Surani, 2013). CpG methylation is regulated by a group of enzymes known as DNA methyltransferases (Dnmts). Three isoforms of Dnmts, namely DNA methyltransferase 1 (Dnmt1), DNA methyltransferase 3 alpha (Dnmt3a), and DNA methyltransferase 3 beta (Dnmt3b), all have unique functions to perform (Lyko et al., 1999; Tucker et al., 1996). Apart from Dnmts, ten-eleven translocation methylcytosines (Tets), including Tet1, Tet2, and Tet3, also play major roles in DNA methylation status maintenance. These Tet enzymes catalyze the oxidation of 5-methylcytosine (5-mC) to generate 5-hydroxymethylcytosine (5-hmC) causing demethylation in CpG islands (Tahiliani et al., 2009; Ito et al., 2011); thus activating gene transcription. They can also subsequently catalyze the oxidation of 5-mC to 5-formylcytosine (5-fC) and 5-fC to 5-carboxylcytosine (5-caC) (Ito et al. 2011).
The effect of PFOA exposure on methylation has been noted in some in vitro and in vivo studies (Wen et al., 2020; Wan et al., 2010; Rashid el at., 2020; Tian et al., 2012). Prenatal PFOA exposure has been associated with reduced Insulin Line Growth Factor 2 (IGF2) methylation in cord blood (Kobayashi et al., 2017), lower global DNA cytosine methylation in neonates (Guerrero-Preston et al., 2010), and increased methylation of Long Interspersed Nuclear Element-1 (LINE-1) (Watkins et al., 2014). Other studies have reported changed expression in cholesterol metabolism (Fletcher et al., 2013) and lipid metabolism (Wen et al., 2020). However, none have explored genome-wide methylation alterations and its impact in liver where PFOA accumulation can reach high levels.
PFOA-induced liver enlargement is an evident consequence as noted in past studies (Christopher and Martin, 1977; Metrick and Marias, 1977). Hepatocellular hypertrophy/ cytomegaly and increased liver mass were observed in male rats exposed to PFOA (Goldenthal, 1978; Perkins et al., 2004), which are notable features of non-alcoholic fatty liver disease (NAFLD) (Liang et al., 2014). Several animal studies have linked PFOA exposure to fatty liver disease, also known as hepatic steatosis (Sibinski et al. 1987). In chronic studies with Sprague-Dawley rats, PFOA exposure was known to induce hepatocellular adenomas, with increasing liver weight and hepatic β-oxidation activity (Biegel et al., 2001). Study has also reported on epigenetic alterations induced by PFOA in relation to lipid metabolism genes in liver cell model (Wen et al., 2020).
Besides epigenetic changes, gene or protein expression could also be influenced by changes in mRNA splicing and translational factors upon exposure to PFOA. Transcriptome-wide profiling studies have been previously carried out on human livers with NAFLD (Lake et al., 2011; Moylan et al., 2014); but as with most studies they only examined changes in overall mRNA abundance and did not attempt to monitor changes in splice isoforms (Shackel et al., 2006). Nonetheless, when mRNA level changes were profiled in the liver samples from insulin-resistant obese population, the key pathways downregulated in obese liver samples were related to alternative mRNA processing and splicing (Pihlajamäki et al., 2011). Alternative splicing decisions are determined by splice site strength, cis-acting regulatory elements within pre-mRNAs that promote or inhibit exon recognition, and expression/activity of trans-acting factors that bind to these cis elements and regulate the accessibility of the spliceosome to splice sites (Chen and Manley, 2009; Kalsotra and Cooper, 2011; Lee and Rio, 2015). Notably, several new studies have shown that proper expression of alternative splicing factors is important for hepatocyte differentiation and function, implicating its major role in maintaining normal liver physiology (Pihlajamaki et al., 2011; Sen et al., 2013; Elizalde et al., 2014; Sen et al., 2015; Bhate et al., 2015, Cheng et al., 2016; Bangru et al., 2018; Kumar et al., 2019).
Although numerous biochemical studies have been performed on PFOA exposed liver samples, none of the studies have investigated the mechanisms of epigenetic alterations or alternative splicing variations in PFOA exposed liver. Specifically, our study tested the hypothesis that PFOA induces hepatic hypertrophy and steatosis through specific alterations of DNA methylation patterns and changes in alternative splicing factors.
2. Materials and Methods
2.1. Chemicals
PFOA (99% purity) was purchased from Sigma-Aldrich (St. Louis, MO). Stock solutions of PFOA were prepared by diluting PFOA in 0.5% v/v Tween 20 (MP Biomedicals, Solon, OH). Stock solutions were diluted to create doses of 1 mg/kg/day, 5 mg/kg/day, 10 mg/kg/day, and 20 mg/kg/day of PFOA. PFOA concentrations were chosen based on previous studies and their relevance. The highest concentration of serum PFOA following occupational exposure was 92.03 μg/mL and the arithmetic mean was 2.21 μg/mL (Olsen & Zobel, 2007). In mice studies, a serum PFOA level of 171 μg/mL was reached after 17 days of 20 mg/kg/day oral gavage (Lau et al., 2006). Therefore, to consider both community and occupational exposure, we chose to expose mice at low to high concentrations (1, 5, 10 and 20 mg/kg/day) in our study.
2.2. Animals and Dosing Paradigm
Adult female CD-1 mice (Charles River, USA) were housed at 25 °C in conventional ventilated polysulfone cages on 12 hr Light: 12 hr Dark cycles. Mice were fed Harlan Teklad Rodent Diet 8604 and given reverse osmosis filtered water in polysulfone water bottles ad libitum. The animal experimental methods and protocols were approved by the University of Illinois Institutional Animal Care and Use Committee and conform to the guidelines set forth by the National Institute of Health for the Care and Use of Laboratory Animals. 30-day old CD1 female mice were orally dosed with either vehicle control (water) or PFOA dissolved in water (1, 5, 10, or 20 mg/kg/day) consecutively for 10 days. Mice were euthanized during diestrus cycle after 10 days of dosing and liver samples were collected for further studies.
2.3. Oil Red O staining
Lipid droplets with neutral and hydrophobic lipids can be stained red with Oil Red O (ORO). Livers from mice treated with 1mg/kg/day, 10 mg/kg/day PFOA for 10 days and control mice were frozen immediately with liquid nitrogen after sacrifice. Liver samples were then unfrozen and incubated in 10% neutral buffered formalin solution (HT501128, Millipore Sigma; St. Louis, MO, USA) at room temperature for 24 hours. Samples were subsequently freeze-fixed with Tissue-Tek O.C.T. compound (4583, Sakura Finetek; Torrance, CA, USA) and sectioned into 8 p,m thick slices. Sections were then stained with Oil Red O in isopropanol (MAK194, Sigma-Aldrich; St. Louis, MO, USA) per instructions per protocol. Slides were also counterstained with Hematoxylin.
2.4. TUNEL assay
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay is used to detect DNA break that occurs during the last phase of cell apoptosis, as an indicator of apoptotic cells. Liver samples from mice treated with 1mg/kg/day, 10 mg/kg/day PFOA and control mice were frozen immediately with liquid nitrogen after sacrifice. Samples were then unfrozen and incubated in 10% neutral buffered formalin solution (HT501128, Millipore Sigma; St. Louis, MO, USA) at room temperature for 24 hours. Subsequently, liver samples were fixed in paraffin and 10 μm paraffin embedded sections were sliced. TUNEL assay (ab206386, abcam; Cambridge, MA, USA) was then performed per protocol. At the location of DNA fragmentation, diaminobenzidine (DAB) reacted with HRP-labeled samples would generate a brown substrate.
2.5. Quantification of DNA methylation
Genomic DNA of the mice liver samples were extracted and purified with Purelink genomic DNA mini kit (Invitrogen, Thermofisher; Waltham, MA, USA) per kit’s protocol. Additional RNase A treatment was followed per specifications. The concentrations of extracted DNA were measured by a NanoDrop spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Genomic DNA (100 ng) methylation (5-mC) level in both the control and PFOA treated liver samples were determined with MethylFlash Global DNA methylation (5-mC) ELISA easy Kit (Epigentek Group; Farmingdale, NY, USA) following manufacturer’s protocol. The methylated DNA fraction was detected by an ELISA-like reaction with capture and detection antibodies based on absorbance (at 450 nm) with a BioTek microplate reader (BioTek Instruments, Winooski, VT, USA).
2.6. Protein isolation and western blot analysis
Proteins were isolated by homogenization of the frozen tissues (liver) in a bullet blender with appropriate homogenization buffer, 400 μL per 100 mg liver tissue (pH7.5, HEPES-KOH 10 mM, Sucrose 0.32 M, EDTA 5 mM, Proteinase inhibitor (1 tablet per 20 ml buffer)). 20% Sodium Dodecyl sulfate (SDS) was added to a final concentration of 1% (v/v) prior to sonication. Samples were sonicated and clarified by centrifugation. Protein content was measured using BCA protein assay kit (Thermo®). A total of 40 to 60 μg proteins were resolved on a 10% SDS-PAGE electrophoresis gel and transferred onto a PVDF membrane (Immobilon, Millipore). Membranes were visualized for equal loading using Ponceau staining solution (0.5% w/v PonS, 1% Acetic acid). Membranes were blocked with 5% (w/v) milk powder in TBST (Tris-buffered saline, 0.1% Tween 20) for 2 hours at room temperature. Blots were incubated in primary antibodies overnight at 4 °C at pre-determined concentrations (see Supplementary Table S2), washed in TBST, and incubated in HRP conjugated secondary antibodies for 1 hour at room temperature. The immunoreactivity was visualized in ChemiDoc XRS+ using the Clarity Western ECL kit (BioRad®).
2.7. Gene expression and splice isoform analysis
Total RNA was isolated from mouse livers using TRIzol reagent (Life Tech® or Ambion, Thermofisher, Waltham, MA, USA). 5 μg of RNA was reverse transcribed to cDNA using random primer mix (NEB; Ipswich, MA, USA) and Maxima Reverse Transcriptase kit (Thermofisher; Waltham, MA, USA ) or with high capacity cDNA synthesis kit (Applied Biosystems, Thermofisher; Waltham, MA, USA). For the results from Figure 1 to 3, the cDNA was diluted to 25 ng/μl to be used downstream for qRT-PCR and alternative splicing assays. Primers used are listed in Supplementary Tables S1 and S3. The qRT-PCR assays were performed using GoTaq® Green Master Mix (Promega; Madison, WI, USA) with an initial step at 95 °C for 5 min followed by 32 cycles of 95°C for 30 sec, 58°C for 30 sec, and 72°C for 40 sec. Then it was ended with a final extension of 72°C for 3 min. PSI values for the alternatively spliced region were calculated with ImageLab software (BioRad; Hercules, CA, USA) as [(exon inclusion band intensity)/(exon inclusion band intensity + exon exclusion band intensity) × 100]. For the transcripts of splicing factors, qRT-PCR was performed in triplicate with 50 ng of cDNA per reaction on an Eco Real-Time PCR unit (Illumina; San Diego, CA, USA) utilizing PerfeCTa SYBR Green FastMix (Quantabio; Beverly, MA, USA) with an initial activation step at 95°C for 10 min and followed by 40 cycles at 95°C for 10 sec and 60°C for 30 sec.
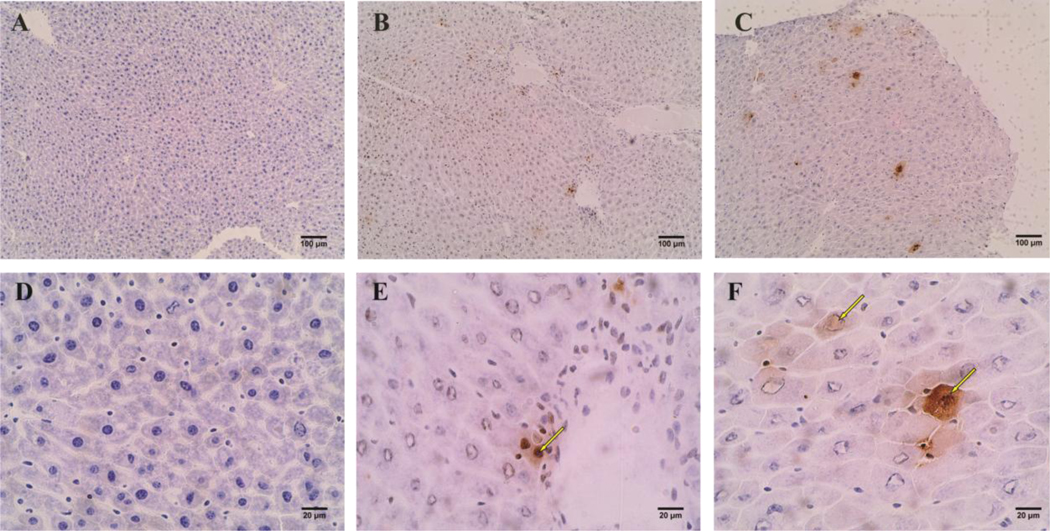
Figure 1. Oil Red O stained mouse liver sections.
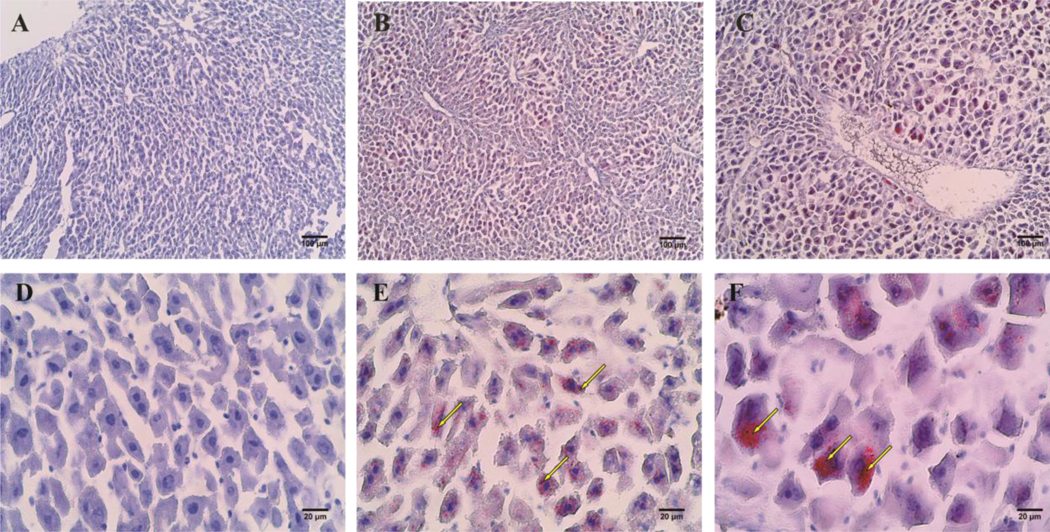
All images were photographed with brightfield microscope with white light source. (A), (B), and (C) were imaged with 10x objective with scale bar of 100 μm. While (D), (E), (F) were imaged with 50x objective with scale bar of 20 μm. (A) and (D) are from control mice. (B) and (E) are from mice treated with 1 mg/kg/day PFOA. (C) and (F) are from mice treated with 10 mg/kg/day PFOA. Red spots exampled with arrows are lipid droplets stained red with Oil Red O.
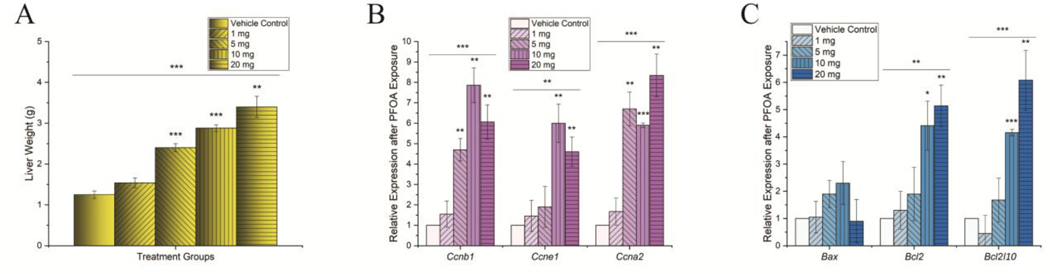
Figure 3. PFOA induced hepatocellular hypertrophy and changes in expression of cell proliferation and apoptosis related genes.
(A) Histograms plot shows liver weight of animals treated with different concentrations of PFOA treatments. Increased PFOA concentration increases liver weight (B) Histograms quantifying the relative mRNA expression of cell cycle genes Ccnb1, Ccne1 and Ccna2. (C) Histograms quantifying the relative mRNA expression of pro and anti-apoptotic genes Bax, Bcl2 and Bcl2l10. The expression levels were normalized with GAPDH. n = 3 for all experimental data. PFOA treatment concentrations are shown as the mass in legends per kg mouse per day. Significance codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1.
2.8. Statistical analyses
Data analyses were conducted with the R statistics (R 3.6.1; R Core Team, 2019). R function “aov” was used to perform the one-way analysis of variance (one-way ANOVA) within each group of data. Two sample t-test with equal variances assumption was conducted between every treatment group’s result and its corresponding control group to evaluate the difference significance between their means. Standard error of the mean was shown in figures as error bar. Significance levels were presented as marks right above each column. Except Figure 6, “.” denotes 0.05≤P<0.1, “*” 0.01≤P<0.05, “**” 0.001≤P<0.01, “***” P≤0.001.
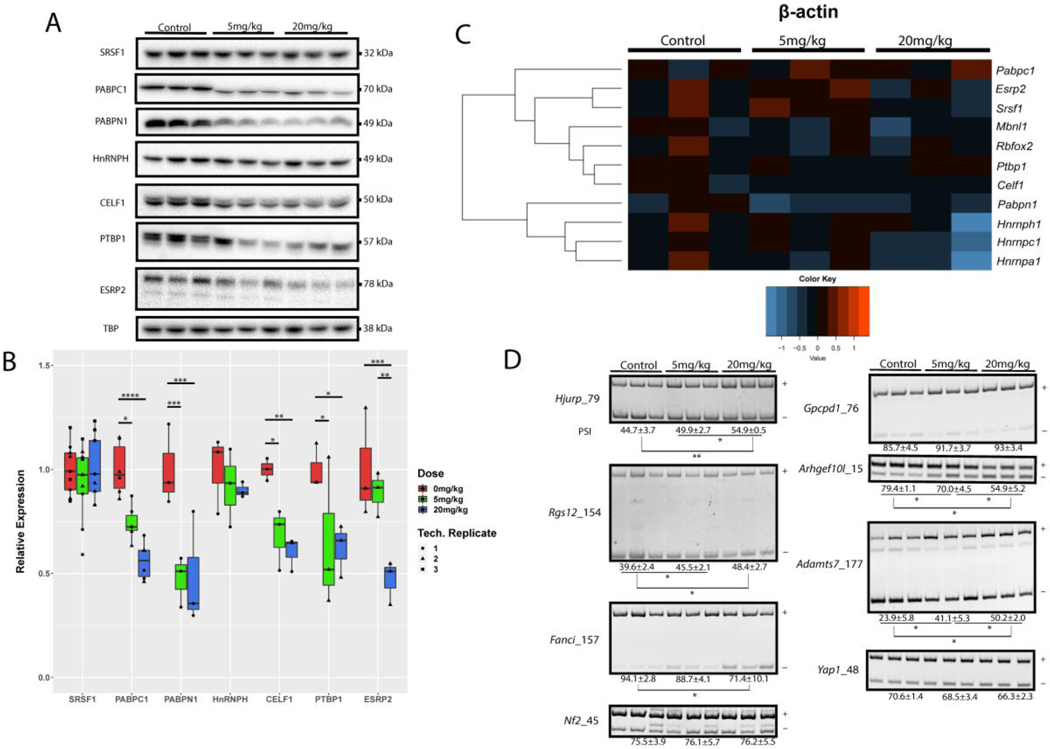
Figure 6. Changes in tissue-specific RNA binding proteins (RBPs) and alternative splicing following PFOA exposure.
(A) Western blot showing dose-dependent changes in abundance of select RNA binding protein levels in mouse livers following PFOA exposure. (B) Quantification of Western Blots shown in A. Data are represented as box plots from at least 2 independent experiments. (C) Heat map representation of the qPCR data showing relative transcript levels of RNA binding proteins following PFOA exposure. The raw Ct values were first normalized to β-actin, and then converted to expression levels using the formula relative expression = 2^(ΔCt(ctl)- ΔCt(treatment)). Z scores of the relative expressions were used to generate the heat map using “heatmap()” function in R. (D) RT-PCR gel images showing alternative splicing changes in developmentally regulated splicing events following PFOA exposure. (+) and (−) bands show inclusion and exclusion of alternative exon, respectively. Percent spliced in (PSI) values were calculated by dividing (+) band intensities with the sum of (+) and (−) band intensities. Significant comparisons are denoted in the figure. * denotes p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001. (n=3 biological replicates for each condition).
3. Results
3.1. Hepatocellular hypertrophy, increased hepatocellular lipid deposit and DNA damage
From the Oil Red O stained liver slides, we saw increased lipid storage in liver cells of mice treated with 1 mg/kg/day or 10 mg/kg/day PFOA (Fig 1B, C, E, F) compared to liver cells of mice treated with control (Fig 1A, D). Larger lipid droplets occurred in liver cells from mouse treated with 10 mg/kg/day PFOA, indicating a dose-dependent increase in lipid storage in liver cells. Another observation from ORO stained cryosections was that cells were enlarged significantly after PFOA treatment, especially at 10 mg/kg/day (Fig 1). Control mouse liver cells had an average diameter of around 15 μm. While the size of liver cells almost doubled in mouse treated with 10 mg/kg/day PFOA, with an overall average diameter of ~30 μm. The same phenomenon of cytomegaly can be observed in TUNEL assay paraffin embedded sections (Fig 2). All control liver tissue looked healthy and had no DNA break as detected by TUNEL assay (Fig 2A, D). Small brown spots were observed in liver tissue from mice treated with 1 mg/kg/day PFOA (Figure 2 B, E). Larger and more frequent brown spots were observed in liver tissue in mice treated with 10 mg/kg/day PFOA, indicating higher DNA damage after PFOA treatment and a possible dose-dependent correlation between liver cell DNA damage or apoptosis upon PFOA exposure. To see if increased PFOA exposure indeed increased the PFOA accumulation in mouse liver, we tested the amount of PFOA in liver sample from mouse treated with vehicle control, 1 and 10 mg/kg/day PFOA through liquid chromatography-mass spectrometry (LC-MS) method. The results show a PFOA accumulation of 6.5 μg and 29.9 μg of accumulation in the liver corresponding to the 1 and 10 mg/kg/day exposed dose in the mouse.
Figure 2. TUNEL assay of mouse liver sections.
All images were photographed with brightfield microscope with white light source. (A), (B), and (C) were imaged with 10x objective with scale bar of 100 μm. While (D), (E), (F) were imaged with 50x objective with scale bar of 20 μm. (A) and (D) are from control mice. (B) and (E) are from mice treated with 1 mg/kg/day PFOA. (C) and (F) are from mice treated with 10 mg/kg/day PFOA. Locations with brown colored spots exampled with arrows are where DNA break happened, indicating unusual cell death.
3.2. Effect of PFOA on Cell proliferation and apoptosis-related genes
To assess whether PFOA exposure causes visible changes in the liver, we quantified liver weight both in PFOA exposed mice and in controls. Experimental results indicate that PFOA significantly increased liver weight in a dose-dependent manner (Fig 3A), possibly due to fatty liver (Fig 1) or increased cell proliferation. To evaluate the effect of PFOA exposure on hepatocyte proliferation in liver, mRNA expression levels were quantified in different cell cycle genes. Significant increase in the expression of cyclin B1 (Ccnb1), cyclin E1 (Ccne1) and cyclin A2 (Ccna2) in PFOA exposed animals equal to or higher than 5 mg/kg/day (Fig 3B) was noted. We also investigated the mRNA expression levels of pro and anti-apoptotic genes (Fig 3C). The expression levels of B cell leukemia/lymphoma 2 (Bcl2) and Bcl2-like 10 (Bcl2l10), which both encode proteins that have shown to suppress cell apoptosis (anti-apoptotic), significantly increased. PFOA exposure did not change the expression of BCL2-associated X protein (Bax), encoding a protein that functions as an apoptotic activator (pro-apoptotic).
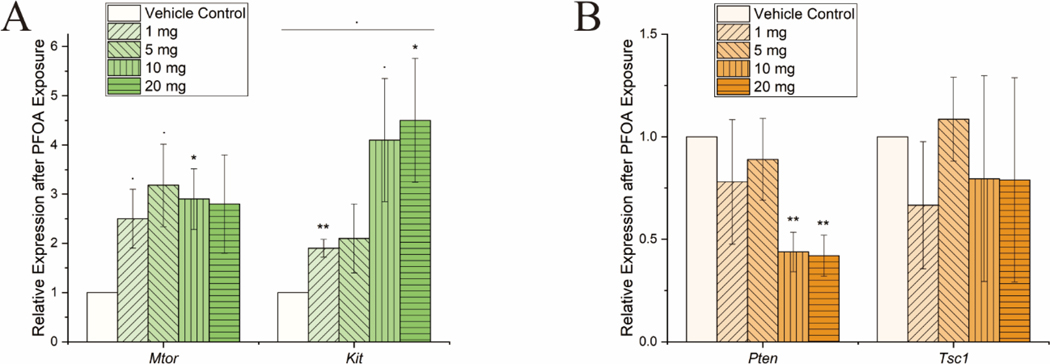
3.3. PFOA exposure activates mTOR signaling pathway
The mechanistic target of rapamycin (mTOR) signaling pathway regulates many metabolic and physiological processes in different organs or tissues. Dysregulation of mTOR signaling has been implicated in many human diseases including fatty liver diseases (Guri et al., 2017; Guo et al., 2018). Since mTOR signaling is involved in lipid biogenesis as well as cell proliferation and survival, the mRNA expression levels of different genes involved in the regulation of mTOR signaling was quantified. A significant increase in both mechanistic target of rapamycin kinase (Mtor) and KIT proto-oncogene receptor tyrosine kinase (Kit) levels was observed with PFOA exposure (Fig 4A). MTOR mediates cellular response to DNA damage and regulates cellular metabolism (Proud, 2004). KIT also plays an essential role in the regulation of cell survival and the activation of mTOR pathway. The mRNA levels of phosphatase and tensin homolog (Pten), a primary inhibitor of mTOR pathway, significantly reduced at PFOA exposure levels 10 mg/kg/day and 20 mg/kg/day. While no significant change in expression of TSC complex subunit (Tsc1), another known inhibitor of mTOR signaling (Fig 3B) was noted.
Figure 4. Activation of mTOR signaling pathway following PFOA exposure.
(A) Histograms demonstrating increased expression of Mtor and Kit with increased PFOA exposure, suggesting activation of mTOR pathway. (B) Histograms also quantifying mRNA levels of inhibitors of mTOR pathway namely Pten and Tsc1. A significant reduction in Pten expression was noted at higher PFOA exposure. The expression levels were normalized with GAPDH. n = 3 for all experimental data. PFOA treatment concentrations are shown as the mass in legends per kg mouse per day. Significance codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1.
3.4. Effect of PFOA on DNA methylation and its regulation
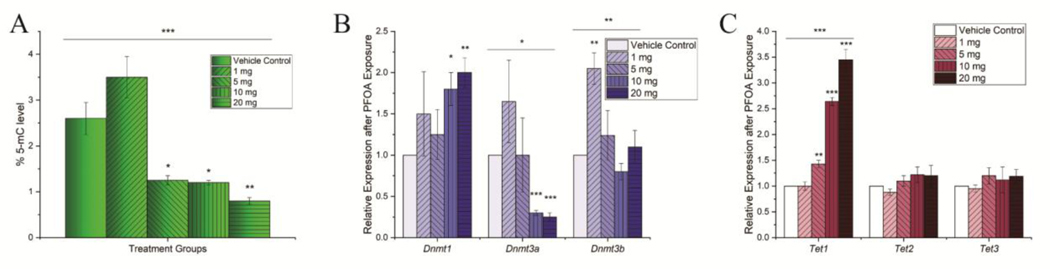
To test this hypothesis that PFOA induces DNA methylation alteration in mouse liver, we quantified DNA methylation levels in the liver. For determining the global DNA methylation levels in mice liver following PFOA exposure, 5-methyl cytosine (5-mC) levels were quantified in liver samples. PFOA greatly induced global hypo-methylation in liver at exposure concentrations of 5, 10, and 20 mg/kg/day (Fig 5A). Next, we quantified the mRNA expression levels of the genes that encode major DNA methylation regulators namely DNMTs and TETs. The PFOA exposure reduced Dnmt3a expression levels at a higher concentration (10mg/kg/day, P<0.00001; 20mg/kg/day, P=0.0002; Fig 5B). The expression of Dnmt1 was also increased significantly at exposure concentrations 10 and 20 mg/kg/day. However, Dnmt3b expression did not change with PFOA exposure in any specific pattern, except it was increased significantly at 1 mg/kg/day PFOA (P = 0.0052; Fig 5B). The PFOA exposure also significantly increased Tet1 expression with no change in Tet2 or Tet3 expression (Fig 5C). Our data showed a concentration dependent effect on the mRNA expression of DNA methylation regulation proteins, especially Dnmt3a and Tet1, and in DNA methylation levels.
Figure 5. PFOA induced epigenetic changes in mice liver.
(A) Histograms demonstrating quantification of 5-mC levels from liver genomic DNA. Increased PFOA concentration induces hypo-methylation (B) Histograms quantifying Dnmts mRNA expression levels. The expression levels of Dnmt1 increased and Dnmt3a decreased significantly with the increased concentration of PFOA (C) Histograms quantifying Tets mRNA expression levels. Tet1 expression levels were elevated dose-dependently. The expression levels were normalized with GAPDH. n = 3 for all experimental data. PFOA treatment concentrations are shown as the mass in legends per kg mouse per day. Significance codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1.
3.5. PFOA exposure induces changes in RNA binding proteins (RBPs) and alternative splicing
Recent study showed that hepatocytes exhibit large-scale transcriptional and post-transcriptional remodeling of the transcriptome after birth (Bhate et al., 2015). Specifically, there is a developmental switch in the mRNA splicing and translation programs, which supports postnatal liver development and maturation. Furthermore, under conditions of liver injury, these programs revert to a fetal-like state to promote proliferation of hepatocytes and restoration of liver function (Bangru et al., 2018). Therefore, we hypothesized that PFOA could alter gene expression of liver cells through directly or indirectly affecting the post-transcriptional regulatory mechanism. These mechanisms are mediated by a class of tissue-specific RNA binding proteins (RBPs) (Lewis et al., 2018; Arif et al., 2017). To explore whether post-transcriptional regulatory programs may be altered within the liver upon exposure to PFOA, we screened various RBPs and measured their relative abundance using western blot analysis (Fig 6A). Subsequently, western blot results with significance levels were presented in Fig 6B. Surprisingly, many RBPs such as polyadenylate-binding protein 2 (PABPN1) and polyadenylate-binding protein 1 (PABPC1) showed significant and dose-dependent reduction in abundance upon treatment with PFOA (Fig 6A, B). These proteins play a major role in the pre-mRNA splicing by binding to the poly(A) tails of transcripts, which in turns influence mRNA translation (Chorghade et al., 2017). We also observed a significant change in the hepatic levels of splicing factors CUGBP Elav-like family member 1 (CELF1), Polypyrimidine tract-binding protein 1 (PTBP1) and Epithelial splicing regulatory protein 2 (ESRP2) following PFOA treatment (Fig 6A, B). Next, we used quantitative real time PCR (qRT-PCR) analysis to determine whether the protein abundance of RBPs was being regulated through a concomitant change in their mRNA levels. Surprisingly, our analysis revealed that the mRNA levels of various RBPs screened did not change significantly (Fig 6C). These data suggest that differences in protein abundance of RBPs following PFOA exposure is post-transcriptional and likely due to changes in translation efficiency of the associated transcript or stability of the protein. To assess changes occurring in alternative splicing, we performed splice assays for various target exons known to change during development (Fig 6D). Interestingly, after PFOA treatment, we found significant changes in exon usage in genes such as Fanci and Hjurp, which are known to play a role in DNA repair (Kato et al., 2007; Lopez-Martinez et al., 2019).
4. Discussion
Liver is a main organ where exogenous chemicals are metabolized and ultimately excreted. Liver cells exposed to high PFOA concentrations could result in liver dysfunction, cell injury, and eventually organ failure. From our experiments, we observed significant dose-dependent increase in liver mass (Fig 3A) and liver cell enlargement (Fig 1, Fig 2), with an increase in PFOA exposure level. Besides the hepatocellular hypertrophy, increased lipid deposits in liver cells were also observed after PFOA exposure (Fig 1), which are characteristic to fatty liver disease. Previous studies have clearly established a correlation between PFOA exposure and increased liver mass, major characteristic feature of the NAFLD (Goldenthal, 1978; Sibinski et al. 1987; Perkins et al., 2004). Though our study is an acute exposure study, results are in line with other studies (Goldenthal, 1978; Sibinski et al. 1987; Perkins et al., 2004).
Our results show increased expression of cell cycle regulation genes Ccnb1, Ccne1, Ccna2, and anti-apoptotic genes Bcl2 and Bcl2l10 (Fig 3B, C). Thus, the increased liver weight is possibly associated with activation of cell cycle genes and repression of apoptotic genes. It is well established that the mTOR pathway affects major eukaryotic signaling networks relevant to cell growth organismal physiology and cellular metabolism (Wiederrecht et al., 1995; Metcalfe et al., 1997). Our results show increase in expression of Mtor and Kit after PFOA exposure (Fig 4A), indicating that mTOR pathway may be activated through the c-kit ligand in PFOA exposed liver. This phenomenon may be further enhanced by subsequent reduction in expression levels of mTOR inhibitor, Pten (Fig 4B). This establishes a relationship between PFOA mediated liver damage, activation of c-kit mediated mTOR pathway and hepatocyte proliferation. Given these results, it is evident that PFOA triggers different molecular pathways.
To determine the mechanisms that caused gene expression changes in liver tissues that were exposed to PFOA, we specifically examined two possible mechanisms, epigenetic changes and alternative splicing. Several recent reports point to the critical role for epigenetics (Jähner et al., 1982; Razin et al., 1984) or changes in splicing (Pihlajamäki et al., 2011; Sen et al., 2013; Elizalde et al., 2014; Sen et al., 2015; Bhate et al., 2015, Cheng et al., 2016; Bangru et al., 2018; Kumar et al., 2019) in various liver diseases thus providing opportunities for the identification of new biomarkers as therapeutic targets.
Few studies have reported on the epigenetic alterations due to PFOA exposure. However, these studies were predominantly carried out in blood samples from population exposed to PFOA toxicants (Guerrero-Preston et al., 2010, Watkins et al., 2014; Kobayashi et al., 2017). One such study has reported on the hypomethylation of specific gene (glutathione-S-transferase Pi) in liver cells in vitro (Tian et al., 2012). Our previous study also showed significant changes in global DNA methylation in vitro with HepG2 cells (Wen et al., 2020). Therefore, it is compelling to investigate epigenetic alterations following PFOA exposure in vivo in mouse liver. Although in vitro studies in human liver L02 cells exhibit no changes in global DNA methylation (Tian et al., 2012), it is evident from our global DNA methylation assay that PFOA induced significant decrease in global DNA methylation levels in liver tissue (Fig 5A). To gain further insights on the alteration in DNA methylation levels, we assessed the mRNA levels of Dnmts and Tets, major regulators of DNA methylation and observed significant changes. PFOA exposure reduced Dnmt3a expression levels at higher concentrations (Fig 5B); thus, affecting de novo DNA methylation. The expression of Dnmt1 increased significantly at higher concentrations (Fig 1B). But without significance in one-way ANOVA test and any specific trend, Dnmt1 changes are less significant. Interestingly, results indicate that increased Dnmt3b levels at low concentration exposure of PFOA may cause DNA hyper-methylation (Fig 5A, B). The PFOA exposure showed a significant dose-dependent increase in Tet1 expression with no change in Tet2 or Tet3 expression (Fig 5C). In essence, our results show a dose-dependent effect of PFOA on the mRNA expression levels of DNA methylation regulation proteins, specifically Dnmt3a and Tet1, and in DNA methylation levels, suggesting that direct exposure to PFOA triggers epigenetic alterations in the liver. Hepatic stellate cell activation is one of the important events in liver fibrosis, wherein, changes in DNA methylation at specific locus of Pten was observed. In particular, Pten gene was hypermethylated leading to decreased expression of Pten (He et al., 2016; Kumar et al., 2018). These prior works suggest possible connection between gene specific DNA methylation changes and Pten mRNA expression. This unusual Pten expression regulation further suggests liver dysfunction upon PFOA exposure.
To date, it is not known whether exposure to organic pollutants such as PFOA leads to alterations in post-transcriptional gene regulatory programs in the liver. In this study, the effects of PFOA on the expression of various RBPs were evaluated. Multiple splicing and factors were noted to show a significant change in their protein (Fig 6A, B) but not mRNA levels (Fig 6C) following PFOA exposure. For instance, both nuclear and cytosolic poly (A) tail binding proteins (PABPC1, PABPN1) showed a dose-dependent reduction in their abundance after PFOA treatment. PABPs act as multifunctional scaffold to form ribonucleoproteins complexes determining the post-transcriptional fate of mRNA (Li et al., 2015). PABPN1 is a nuclear protein that was first discovered for its ability to provide processivity to the poly(A) polymerase and greatly enhance poly(A) tail formation in the nucleus (Mangus et al., 2003). It is also thought to be the primary regulator for controlling the position and length of the poly(A) tail (Goss et al., 2013; Elkon et al., 2013). PABPC1, on the other hand, is primarily cytosolic and stabilizes a “closed loop structure” between the 5’-cap and the 3’-poly(A) tail on most mRNAs (Wells et al., 1998; Kahvejian et al., 2005; Safaee et al., 2012). By linking poly(A) tail to the 5’ end, PABPC1 not only enhances the stability of mRNAs through protection from exonucleases but also promotes their translation through better ribosomal recycling (Coller et al., 1998; Mangus et al., 2003; Goss and Kleiman, 2013). Alterations in PABPN1 and PABPC1 levels following PFOA exposure in the liver are therefore expected to affect mRNA maturation and translation of numerous hepatic transcripts. Consistent with this expectation, we found that changes in protein abundance of various RBPs following PFOA exposure were post-transcriptional and likely driven by changes in translation efficiency of the associated transcript or stability.
Significant reduction in the protein levels of several splicing factors, including ESRP2 was observed after PFOA treatment (Fig 6A, B). ESRP2 is an epithelial splicing regulatory protein that maintains a non-proliferative, mature phenotype of adult hepatocytes (Bhate et al., 2015). ESRP2 is normally absent in fetal livers but begins to be expressed shortly after birth. It is critically important for switching on the adult splicing program for approximately 20% of hepatocyte mRNAs, producing splice variants that encode functional differences in multiple proteins (Bangru et al., 2018). Suppression of ESRP2 following PFOA exposure would result in the expression of fetal mRNA splice variants that might be incompatible with adult hepatic function and therefore promote pathological liver phenotypes. Indeed, after PFOA treatment, we found a significant reduction in the inclusion of developmentally regulated exons within Fanci and Hjurp transcripts (Fig 6D), both of which are implicated in DNA damage and repair response (Kato et al., 2007; Lopez-Martinez et al., 2019). From our TUNEL assay, small brown spots were observed in liver cells from mice treated with 1 mg/kg/day of PFOA and large brown spots were observed in cells from mice treated with 10 mg/kg/day PFOA (Fig 2). Together with the evidence that no detectable spots were observed in control, our results indicated higher DNA damage after PFOA treatment and possible dose-dependent correlation between liver cell DNA damage or apoptosis and PFOA exposure.
5. Conclusion
Our current study has established two major findings, (i) PFOA induces global epigenetic alterations, specifically DNA methylation, in the liver; (ii) PFOA induces tissue-specific changes in RNA binding proteins affecting alternate splicing factors. Our study is also the first to report a dose-dependent effect of PFOA on alternate splicing factor providing new insights on previously unexplored mechanisms of PFOA-mediated hepatotoxicity and pathogenesis. However, additional experiments are needed to further study the epigenetic mechanisms that are influenced by PFOA, as well as the translational regulation of mRNA expressed.
Supplementary Material
Highlights.
Epigenetic alterations and RNA splicing in hepatocellular steatosis were noted
Effect of DNMTs and TETs in liver tissues were evaluated upon PFOA exposure
PFOA exposure alters mTOR expression and decreases pten expression
Tissue specific changes in RNA binding proteins were noted
Changes in alternate splicing factors were identified upon PFOA exposure
Acknowledgements
A.K. is supported by the US National Institute of Health (R01HL126845) and Beckman Fellowship from the Center for Advanced Study at the University of Illinois Urbana-Champaign.
J.I. is supported by UIUC startup grants. A.K. and J.I are also supported by the Planning Grant Award from the Cancer Center at Illinois.
References
- 1.Apelberg BJ, Goldman LR, Calafat AM, Herbstman JB, Kuklenyik Z, Heidler J, Needham LL, Halden RU, Witter FR. Determinants of fetal exposure to polyfluoroalkyl compounds in Baltimore, Maryland. Environ Sci Technol. 2007;41(11):3891–7. [DOI] [PubMed] [Google Scholar]
- 2.Arif W, Datar G, Kalsotra A. Intersections of post-transcriptional gene regulatory mechanisms with intermediary metabolism. Biochim Biophys Acta Gene Regul Mech. 2017;1860(3):349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bangru S, Arif W, Seimetz J, Bhate A, Chen J, Rashan EH, Carstens RP, Anakk S, Kalsotra A. Alternative splicing rewires Hippo signaling pathway in hepatocytes to promote liver regeneration. Nat Struct Mol Biol. 2018;25(10):928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bangru S, Arif W, Seimetz J, Bhate A, Chen J, Rashan EH, Carstens RP, Anakk S, Kalsotra A. Alternative splicing rewires Hippo signaling pathway in hepatocytes to promote liver regeneration. Nat Struct Mol Biol. 2018;25(10):928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banks RE, Smart BE, Tatlow JC. Organofluorine chemistry: Principles and commercial applications. New York (NY): Plenum; 1994: p. 670. [Google Scholar]
- 6.Bhate A, Parker DJ, Bebee TW, Ahn J, Arif W, Rashan EH, Chorghade S, Chau A, Lee JH, Anakk S, Carstens RP, Xiao X, Kalsotra A. ESRP2 controls an adult splicing programme in hepatocytes to support postnatal liver maturation. Nat Commun. 2015;6:8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biegel LB, Hurtt ME, Frame SR, O’Connor JC, Cook JC Mechanisms of extra-hepatic tumor induction by peroxisome proliferators in male cd rats. Toxicol. Sci. 2001;60:44–55. [DOI] [PubMed] [Google Scholar]
- 8.Burris JM, Olsen G, Simpson C, Mandel J. Determination of serum half-lives of several fluorochemicals. 3M Company; Interim Report #2. January 11, 2002. US EPA AR- 226–1086. [Google Scholar]
- 9.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10(11):741–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Y, Luo C, Wu W, Xie Z, Fu X, Feng Y. Liver-Specific Deletion of SRSF2 Caused Acute Liver Failure and Early Death in Mice. Mol Cell Biol. 2016;36(11):1628–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chorghade S, Seimetz J, Emmons R, Yang J, Bresson SM, Lisio M, Parise G, Conrad NK, Kalsotra A. Poly(A) tail length regulates PABPC1 expression to tune translation in the heart. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christopher B, Martin J. 28-Day Oral Toxicity Study with fc-143 in Albino Mice. Industrial bio-test laboratories. Inc, 1977;pp. 8532–10655 (t-1742coc). [Google Scholar]
- 13.Coller JM, Gray NK, Wickens MP. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui L, Zhou QF, Liao CY, Fu JJ, Jiang GB. Studies on the toxicological effects of PFOA and PFOS on rats using histological observation and chemical analysis. Arch Environ Contam Toxicol. 2009;56(2):338–49. [DOI] [PubMed] [Google Scholar]
- 15.Elizalde M, Urtasun R, Azkona M, Latasa MU, Goni S, Garcia-Irigoyen O, Uriarte I, Segura V, Collantes M, Di Scala M, Lujambio A, Prieto J, Avila MA, Berasain C. Splicing regulator SLU7 is essential for maintaining liver homeostasis. J Clin Invest. 2014;124(7):2909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14(7):496–506. [DOI] [PubMed] [Google Scholar]
- 17.Ericson I, Marti-Cid R, Nadal M, van Bavel B, Lindstrom G, Domingo JL. Human exposure to perfluorinated chemicals through the diet: intake of perfluorinated compounds in foods from the Catalan (Spain) Market. J Agric Food Chem 2008;56:1787–1794 [DOI] [PubMed] [Google Scholar]
- 18.Eriksen KT, Raaschou-Nielsen O, Sørensen M, Roursgaard M, Loft S, Møller P. Genotoxic potential of the perfluorinated chemicals PFOA, PFOS, PFBS, PFNA and PFHxA in human HepG2 cells. Mutat Res. 2010;700(1–2):39–43. [DOI] [PubMed] [Google Scholar]
- 19.Freire PF, Martin JP, Herrero O, Peropadre A, de la Pena E, Hazen MJ. In vitro assessment of the cytotoxic and mutagenic potential of perfluorooctanoic acid. Toxicology in Vitro. 2008. August 1;22(5):1228–33. [DOI] [PubMed] [Google Scholar]
- 20.Fletcher T, Galloway TS, Melzer D, Holcroft P, Cipelli R, Pilling LC, Mondal D, Luster M, Harries LW. Associations between PFOA, PFOS and changes in the expression of genes involved in cholesterol metabolism in humans. Environ Int. 2013;57–58:2–10. [DOI] [PubMed] [Google Scholar]
- 21.Goldenthal E. Final Report, Ninety Day Subacute Rat Toxicity Study on Fluorad Fluorochemical FC-143. 1978; International Research and Development Corporation (Study). [Google Scholar]
- 22.Goss DJ, Kleiman FE. Poly(A) binding proteins: are they all created equal? Wiley Interdiscip Rev RNA. 2013;4(2):167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerrero-Preston R, Goldman LR, Brebi-Mieville P, Ili-Gangas C, Lebron C, Witter FR, Apelberg BJ, Hernández-Roystacher M, Jaffe A, Halden RU, Sidransky D. Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics. 2010;5(6):539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo J, Fang W, Chen X, Lin Y, Hu G, Wei J, Zhang X, Yang C, Li J. Upstream stimulating factor 1 suppresses autophagy and hepatic lipid droplet catabolism by activating mTOR. FEBS Lett. 2018;592(16):2725–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guri Y, Colombi M, Dazert E, Hindupur SK, Roszik J, Moes S, Jenoe P, Heim MH, Riezman I, Riezman H, Hall MN. mTORC2 Promotes Tumorigenesis via Lipid Synthesis. Cancer Cell. 2017;32(6):807–823.e12. [DOI] [PubMed] [Google Scholar]
- 26.Hackett JA, Surani MA. Beyond DNA: programming and inheritance of parental methylomes. Cell. 2013;153(4):737–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He L, Gubbins J, Peng Z, Medina V, Fei F, Asahina K, Wang J, Kahn M, Rountree CB, Stiles BL. Activation of hepatic stellate cell in Pten null liver injury model. Fibrogenesis Tissue Repair. 2016;9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hekster FM, Laane RW, de Voogt P. Environmental and toxicity effects of perfluoroalkylated substances. Rev Environ Contam Toxicol. 2003;179:99–121. [DOI] [PubMed] [Google Scholar]
- 29.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011; 333:1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jähner D, Stuhlmann H, Stewart CL, Harbers K, Löhler J, Simon I, Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298(5875):623–8. [DOI] [PubMed] [Google Scholar]
- 31.Johansson N, Fredriksson A, Eriksson P. Neonatal exposure to perfluorooctane sulfonate (PFOS) and perfluorooctanoic acid (PFOA) causes neurobehavioural defects in adult mice. Neurotoxicology. 2008. January 1;29(1):160–9. Johansson, N., Fredriksson, A., Eriksson, P. Neonatal exposure to perfluorooctane sulfonate (PFOS) and perfluorooctanoic acid (PFOA) causes neurobehavioural defects in adult mice. Neurotoxicology. 2008; 29:160–169. [DOI] [PubMed] [Google Scholar]
- 32.Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19(1):104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011; 12(10):715–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67(18):8544–53. [DOI] [PubMed] [Google Scholar]
- 35.Kissa E. Fluorinated surfactants and repellents (2nd edition revised and expanded) (Surfactant science series 97) New York (NY): Marcel Dekker; 2001. p. 640. [Google Scholar]
- 36.Kobayashi S, Azumi K, Goudarzi H, Araki A, Miyashita C, Kobayashi S, Itoh S, Sasaki S, Ishizuka M, Nakazawa H, Ikeno T, Kishi R. Effects of prenatal perfluoroalkyl acid exposure on cord blood IGF2/H19 methylation and ponderal index: The Hokkaido Study. J Expo Sci Environ Epidemiol. 2017;27(3):251–259. [DOI] [PubMed] [Google Scholar]
- 37.Kumar D, Das M, Sauceda C, Ellies LG, Kuo K, Parwal P, Kaur M, Jih L, Bandyopadhyay GK, Burton D, Loomba R, Osborn O, Webster NJ. Degradation of splicing factor SRSF3 contributes to progressive liver disease. J Clin Invest. 2019;130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar P, Raeman R, Chopyk DM, Smith T, Verma K, Liu Y, Anania FA. Adiponectin inhibits hepatic stellate cell activation by targeting the PTEN/AKT pathway. Biochim Biophys Acta Mol Basis Dis. 2018;1864(10):3537–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lake AD, Novak P, Fisher CD, Jackson JP, Hardwick RN, Billheimer DD, Klimecki WT, Cherrington NJ. Analysis of global and absorption, distribution, metabolism, and elimination gene expression in the progressive stages of human nonalcoholic fatty liver disease. Drug Metab Dispos. 2011;39(10):1954–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem. 2015;84:291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis CJ, Pan T, Kalsotra A. RNA modifications and structures cooperate to guide RNA- protein interactions. Nat Rev Mol Cell Biol. 2017;18(3):202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, You B, Hoque M, Zheng D, Luo W, Ji Z, Park JY, Gunderson SI, Kalsotra A, Manley JL, Tian B. Systematic profiling of poly(A)+ transcripts modulated by core 3’ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet. 2015;11(4):e1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang W, Menke AL, Driessen A, Koek GH, Lindeman JH, Stoop R, Havekes LM, Kleemann R, van den Hoek AM. Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PloS one. 2014;9(12). Liang, W., Menke, A. L., Driessen, A., Koek, G. H., Lindeman, J. H., Stoop, R., ... & van den Hoek, A. M. (2014). Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PloS one, 9(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Li J, Zhao Y, Wang Y, Zhang L, Wu Y. The occurrence of perfluorinated alkyl compounds in human milk from different regions of China. Environ Int. 2010;36(5):433–8. [DOI] [PubMed] [Google Scholar]
- 45.Lopez-Martinez D, Kupculak M, Yang D, Yoshikawa Y, Liang CC, Wu R, Gygi SP, Cohn MA. Phosphorylation of FANCD2 Inhibits the FANCD2/FANCI Complex and Suppresses the Fanconi Anemia Pathway in the Absence of DNA Damage. Cell Rep. 2019;27(10):2990–3005.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lyko F, Ramsahoye BH, Kashevsky H, Tudor M, Mastrangelo MA, Orr-Weaver TL, Jaenisch R. Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nat Genet. 1999;23(3):363–6. [DOI] [PubMed] [Google Scholar]
- 47.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4(7):223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin JW, Mabury SA, Solomon K, Muir DCG. Dietary accumulation of perfluorinated acids in juvenile rainbow trout (Oncorhynchus mykiss). Environ Toxicol Chem. 2003;22:189–195 [PubMed] [Google Scholar]
- 49.Metcalfe SM, Canman CE, Milner J, Morris RE, Goldman S, Kastan MB. Rapamycin and p53 act on different pathways to induce G1 arrest in mammalian cells. Oncogene. 1997;15(14):1635–42 [DOI] [PubMed] [Google Scholar]
- 50.Metrick M, Marias A. 28-Day Oral Toxicity Study with FC-143 in Albino Rats. Industrial Bio-Test Laboratories Inc, 1977;pp. 8532–10654 (t-1742coc). [Google Scholar]
- 51.Mondal D, Lopez-Espinosa MJ, Armstrong B, Stein CR, Fletcher T. Relationships of perfluorooctanoate and perfluorooctane sulfonate serum concentrations between mother-child pairs in a population with perfluorooctanoate exposure from drinking water. Environ Health Perspect. 2012;120(5):752–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moylan CA, Pang H, Dellinger A, Suzuki A, Garrett ME, Guy CD, Murphy SK, Ashley-Koch AE, Choi SS, Michelotti GA, Hampton DD, Chen Y, Tillmann HL, Hauser MA, Abdelmalek MF, Diehl AM. Hepatic gene expression profiles differentiate presymptomatic patients with mild versus severe nonalcoholic fatty liver disease. Hepatology. 2014;59(2):471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olsen GW, Burris JM, Ehresman DJ, Froehlich JW, Seacat AM, Butenhoff JL, Zobel LR. Half-life of serum elimination of perfluoro octane sulfonate, perfluoro hexane sulfonate, and perfluorooctanoate in retired fluorochemical production workers. Environ Health Perspect. 2007;115(9):1298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olsen GW, Zobel LR. Assessment of lipid, hepatic, and thyroid parameters with serum perfluorooctanoate (PFOA) concentrations in fluorochemical production workers. International archives of occupational and environmental health. 2007. November 1;81(2):231–46 [DOI] [PubMed] [Google Scholar]
- 55.Perkins RG, Butenhoff JL, Kennedy GL Jr, Palazzolo MJ. 13 - week dietary toxicity study of ammonium perfluorooctanoate (APFO) in male rats. Drug and chemical toxicology. 2004. January 1;27(4):361–78. Perkins, R.G., Butenhoff, J.L., Kennedy, G.L., Palazzolo, M.J. 13-Week dietary toxicity study of ammonium perfluorooctanoate (APFO) in male rats. Drug Chem. Toxicol. 2004;27:361–378. [DOI] [PubMed] [Google Scholar]
- 56.Pihlajamäki J, Lerin C, Itkonen P, Boes T, Floss T, Schroeder J, Dearie F, Crunkhorn S, Burak F, Jimenez-Chillaron JC, Kuulasmaa T, Miettinen P, Park PJ, Nasser I, Zhao Z, Zhang Z, Xu Y, Wurst W, Ren H, Morris AJ, Stamm S, Goldfine AB, Laakso M, Patti ME. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab. 2011;14(2):208–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Proud CG. The multifaceted role of mTOR in cellular stress responses. DNA repair. 2004. August 1;3(8–9):927–34. [DOI] [PubMed] [Google Scholar]
- 58.Olsen GW, & Zobel LR (2007). Assessment of lipid, hepatic, and thyroid parameters with serum perfluorooctanoate (PFOA) concentrations in fluorochemical production workers. International archives of occupational and environmental health, 81(2), 231–246. [DOI] [PubMed] [Google Scholar]; R Core Team, 2019. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/ [Google Scholar]
- 59.Rashid F, Ramakrishnan A, Fields C, Irudayaraj J. Acute PFOA exposure promotes epigenomic alterations in mouse kidney tissues. Toxicology Reports. 2020. January 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Razin A, Webb C, Szyf M, Yisraeli J, Rosenthal A, Naveh-Many T, Sciaky-Gallili N, Cedar H. Variations in DNA methylation during mouse cell differentiation in vivo and in vitro. Proc Natl Acad Sci U S A. 1984;81(8):2275–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Safaee N, Kozlov G, Noronha AM, Xie J, Wilds CJ, Gehring K. Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol Cell. 2012;48(3):375–86. [DOI] [PubMed] [Google Scholar]
- 62.Sen S, Jumaa H, Webster NJ. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nat Commun. 2013;4:1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sen S, Langiewicz M, Jumaa H, Webster NJ. Deletion of serine/arginine-rich splicing factor 3 in hepatocytes predisposes to hepatocellular carcinoma in mice. Hepatology. 2015;61(1):171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shackel NA, Seth D, Haber PS, Gorrell MD, McCaughan GW. The hepatic transcriptome in human liver disease. Comp Hepatol. 2006;5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sibinski L, Allen J, Erickson E. Two Year Oral (Diet) Toxicity/carcinogenicity Study of Fluorochemical fc-143 in Rats. 1987:3M Company; (Riker Exp No. 0281CR0012M3). [Google Scholar]
- 66.Stefani F, Rusconi M, Valsecchi S, Marziali L. Evolutionary ecotoxicology of perfluoralkyl substances (PFASs) inferred from multigenerational exposure: a case study with Chironomus riparius (Diptera, Chironomidae). Aquat Toxicol. 2014;156:41–51. [DOI] [PubMed] [Google Scholar]
- 67.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taylor CK. Fluorinated surfactants in practice In: Karsa D, editor. Design and selection of performance surfactants: Annual surfactants review. New York (NY): John Wiley & Sons; 1999. pp. 271–316. [Google Scholar]
- 69.Tian M, Peng S, Martin FL, Zhang J, Liu L, Wang Z, Dong S, Shen H. Perfluorooctanoic acid induces gene promoter hypermethylation of glutathione-S-transferase Pi in human liver L02 cells. Toxicology. 2012;296(1–3):48–55. [DOI] [PubMed] [Google Scholar]
- 70.Tucker KL, Talbot D, Lee MA, Leonhardt H, Jaenisch R. Complementation of methylation deficiency in embryonic stem cells by a DNA methyltransferase minigene. Proc Natl Acad Sci U S A. 1996;93(23):12920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.US EPA (Environmental Protection Agency). (2001) Analysis of PFOS, FOSA, and PFOA from various food matrices using HPLC electrospray/mass spectrometry. 3 M study conducted by Centre Analytical Laboratories, Inc. 2001. [Google Scholar]
- 72.Vanden Heuvel JP, Kuslikis BI, Van Rafelghem MJ, Peterson RE. Tissue distribution, metabolism, and elimination of perfluorooctanoic acid in male and female rats. J Biochem Toxicol. 1991. Summer;6(2):83–92. [DOI] [PubMed] [Google Scholar]
- 73.Vestergren R, Cousins IT, Trudel D, Wormuth M, Shseringer M. Estimating the contribution of precursor compounds in consumer exposure to PFOS and PFOA. Chemosphere. 2008;73:1617–1624 [DOI] [PubMed] [Google Scholar]
- 74.Wan YJ, Li YY, Xia W, Chen J, Lv ZQ, Zeng HC, Zhang L, Yang WJ, Chen T, Lin Y, Wei J, Xu SQ. Alterations in tumor biomarker GSTP gene methylation patterns induced by prenatal exposure to PFOS. Toxicology. 2010;274(1–3):57–64. [DOI] [PubMed] [Google Scholar]
- 75.Watkins DJ, Wellenius GA, Butler RA, Bartell SM, Fletcher T, Kelsey KT. Associations between serum perfluoroalkyl acids and LINE-1 DNA methylation. Environ Int. 2014;63:71–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2(1):135–40. [DOI] [PubMed] [Google Scholar]
- 77.Wen Y, Mirji N, Irudayaraj J. Epigenetic toxicity of PFOA and GenX in HepG2 cells and their roles in lipid metabolism. Toxicology in Vitro. 2020. February 14:104797. [DOI] [PubMed] [Google Scholar]
- 78.Wiederrecht GJ, Sabers CJ, Brunn GJ, Martin MM, Dumont FJ, Abraham RT. Mechanism of action of rapamycin: new insights into the regulation of Gl-phase progression in eukaryotic cells. Prog Cell Cycle Res. 1995;1:53–71. [DOI] [PubMed] [Google Scholar]
- 79.Yang Q, Xie Y, Depierre J. Effects of peroxisome proliferators on the thymus and spleen of mice. Clin. Exp. Immunol. 2000;122: 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Q, Xie Y, Eriksson AM, Nelson BD, DePierre JW Further evidence for the involvement of inhibition of cell proliferation and development in thymic and splenic atrophy induced by the peroxisome proliferator perfluoroctanoic acid in mice. Biochem. Pharmacol. 2001;62:1133–1140. [DOI] [PubMed] [Google Scholar]
- 81.Ylinen M, Kojo A, Hanhijärvi H, Peura P. Disposition of perfluorooctanoic acid in the rat after single and subchronic administration. Bull Environ Contam Toxicol. 1990;44(1):46–53 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.