Abstract
The gasdermin (GSDM) family consists of gasdermin A (GSDMA), B (GSDMB), C (GSDMC), D (GSDMD), E or DNFA5 (GSDME), and DFNB59 in human. Expressed in the skin, gastrointestinal tract, and various immune cells, GSDMs mediate homeostasis and inflammation upon activation by caspases and unknown proteases. In particular, GSDMD is activated by inflammasome-activated caspases-1/−4/−5/−11 as well as a caspase-8-mediated pathway during Yersinia infection. These caspases cleave GSDMD to release its functional N-terminal fragment (GSDMD-NT) from its auto-inhibitory C-terminal fragment (GSDMD-CT). GSDMD-NTs bind to acid lipids in mammalian cell membranes and bacterial membranes, oligomerize, and insert into the membranes to form large transmembrane pores. Consequently, cellular contents including inflammatory cytokines are released and cells can undergo pyroptosis, a highly inflammatory form of cell death. In this chapter, we summarize recent research findings and present experimental procedures to obtain pure recombinant GSDMs for biochemical studies. We highlight a liposome-based assay that yields robust fluorescence signals for characterizing GSDM activities in vitro and may be applicable to other pore-forming proteins and ion channels in general.
Keywords: Gasdermin, pore-forming protein, caspases, liposome, pyroptosis, inflammation, immunity
1. Introduction to the gasdermin family
1.1. Pyroptosis, inflammasomes, and gasdermins
Eukaryotic cells can undergo various types of programmed cell death when triggered by certain metabolic events, pathogens, and environmental factors. In many types of cells including myeloid and epithelial cells in the immune system, supramolecular complexes known as inflammasomes can sense pathogens and danger to initiate pyroptosis, a form of inflammatory cell death that features cell swelling, lysis, and release of pro-inflammatory cytokines including IL-1β and IL-18 (Martinon, Burns, and Tschopp 2002; Zhang et al. 2018; Bergsbaken, Fink, and Cookson 2009; Schroder and Tschopp 2010). While this inflammatory response has a protective role in recruiting immune cells to remove infected cells, its dysregulation can also cause pathology such as inflammatory bowel disease, gout, and sepsis (Jorgensen and Miao 2015; Martinon et al. 2006; Maloy and Powrie 2011).
Inflammasomes are categorized based on their activating signals and caspases involved. Canonical inflammasomes recognize various infection and danger signals including flagellin, K+ efflux, and cytosolic DNA to activate caspase-1 (Franchi et al. 2009; Petrilli et al. 2007; Zhao et al. 2011; Hornung et al. 2009; Lu et al. 2014). On the other hand, the assembly of the non-canonical inflammasome is induced by lipopolysaccharides (LPS) and oxidized lipids and results in the activation of murine caspase-11 and human caspase-4 and −5 (Kayagaki et al. 2013; Shi et al. 2014; Zanoni et al. 2016). The pore-forming protein gasdermin D (GSDMD) was recently identified as a substrate of these inflammasome-activated caspases, which cleave GSDMD into a functional N-terminal fragment (NT) and an autoinhibitory C-terminal fragment (CT) (Kayagaki et al. 2015; Shi et al. 2015). GSDMD-NTs subsequently translocate to the inner leaflet of mammalian cell membranes and bacterial membranes and bind acid lipids phosphatidylserine (PS), such as phosphatidylinositol phosphates (PIPs), phosphatidic acid (PA), and cardiolipin (CL). Upon lipid binding, GSDMD-NTs oligomerize and carry out membrane insertion to form transmembrane (TM) pores (Ding et al. 2016; Liu et al. 2016; Aglietti et al. 2016).
Although GSDMD pores were discovered in the context of pyroptosis, they also play crucial roles in NETosis (Sollberger et al. 2018; Chen et al. 2018) and can also be present in living cells (Evavold et al. 2018), which may have evolved mechanisms to survive GSDMD pores. For example, caspase-3 and −7 can degrade the pore-forming GSDMD-NT into smaller, non-functional fragments (Taabazuing, Okondo, and Bachovchin 2017; Orning et al. 2018). In addition, GSDMD-perforated membranes may be shed from cells via an ESCRT-dependent mechanism (Ruhl et al. 2018). It has also been discovered recently that caspase-8-mediated cleavage of GSDMD occurs under Yersinia infection (Orning et al. 2018; Sarhan et al. 2018), challenging the notion that GSDMD activation is only carried out by inflammatory caspases.
The mechanistic elucidation of GSDMD kindled investigations into the GSDM protein family. In humans, the family consists of GSDMA, GSDMB, GSDMC, GSDMD, GSDME/DFNA5, and DFNB59; in mice, there are ten GSDMs including three isoforms of GSDMA (GSDMA1–3) and four of GSDMC (GSDMC1–4), and GSDMB is absent. With the exception of DFNB59, all GSDMs adopt a two-domain architecture, with GSDM-NT as the pore-forming fragment and GSDM-CT autoinhibitory. Functionally, it has been reported that the mouse GSDMA3 (mGSDMA3) can modulate mitochondrial homeostasis (Lin et al. 2015), while GSDME is activated by caspase-3 and serves as a mediator of pyroptosis and a tumor suppressor (Rogers et al. 2017; Wang et al. 2017). Knowledge is lacking on the physiological roles of most GSDMs and the stimuli and proteases that activate them.
1.2. Pore-forming mechanism of gasdermins
While GSDMD-NT drives pore formation in cells, full-length (FL) GSDMD lacks pore-forming activity and cytotoxicity due to the autoinhibitory function of GSDMD-CT (Ding et al. 2016; Liu et al. 2016; Aglietti et al. 2016). The structural basis for the autoinhibition in GSDMs has been elucidated by the crystal structure of FL mGSDMA3, a mouse homolog of human GSDMA (Ding et al. 2016). In comparison to GSDM-CTs, which are almost exclusively α-helical, mGSDMA3-NT contains an extended β-sheet formed by nine strands (β3-β11) in addition to several α-helices (α1-α4). In the auto-inhibited inactive state, mGSDMA3-CT folds back onto mGSDMA3-NT to form extensive contacts through hydrophobic, hydrogen bonding and charged interactions, leaving the NT-CT linker accessible to cleavage by proteases. Human and mouse GSDMD-CT and human GSDMB-CT are structurally similar to mGSDMA3-CT, suggesting a conserved auto-inhibitory mechanism across the GSDM family (Liu et al. 2018; Kuang et al. 2017).
The structural transition from autoinhibition to the mature GSDM TM pore has been illustrated by the cryo-electron microscopy (cryo-EM) structure of the mGSDMA3 pore, a 26- to 28-subunit mGSDMA3-NT oligomer with a molecular weight of approximately 0.8 MDa (Ruan et al. 2018). The pore features an anti-parallel β-barrel, with each subunit contributing two β-hairpins or four β-strands, as the transmembrane (TM) region and a soluble rim formed by a globular domain of each subunit. The inner diameter of the pore is around 180 Å, large enough to allow the passage of inflammatory cytokines such as IL-1β. Structural alignment of pore-form mGSDMA3-NT to its auto-inhibited state shows that the globular domain remains largely unaltered whereas drastic conformational changes occur at the TM region. More specifically, the first β-hairpin is formed by the β3-β4-β5 region in auto-inhibited mGSDMA3-NT, and the second β-hairpin by the β7-α4-β8 region that is in close contact with mGSDMA3-CT. Adjacent to the inner leaflet of the bilayer membrane, the positively charged α1 helix of mGSDMA3-NT directly binds the negatively charged head of acid lipid CL. The α1 helix is masked by mGSDMA3-CT in the auto-inhibited conformation, consistent with the observation that FL GSDMs do not bind lipids and form pores (Ding et al. 2016; Liu et al. 2016; Aglietti et al. 2016; Ruan et al. 2018).
2. Generation of recombinant gasdermins and proteases
2.1. Molecular cloning, expression, and purification of gasdermins from E. coli
To avoid the issues that GSDM-NTs are toxic to cells and tend to form aggregates in lipidic environments even in the presence of GSDM-CTs, we recommend expressing and purifying FL GSDMs. We found that FL GSDMs can be yielded in considerable quantities, about 10 mg pure protein from 1 L culture of E. coli, as monomers when they are appended a solubility tag such as a maltose-binding protein (MBP) or a small ubiquitin-like modifier (SUMO). The following steps describe the detailed procedure for cloning, expressing, and purifying recombinant human GSDMD (hGSDMD) for in vitro experiments (Ruan et al. 2018).
Gasdermin cloning and expression
Following standard restriction enzyme cloning protocols, clone the full-length hGSDMD sequence into the pDB.His.MBP vector with a tobacco etch virus (TEV)-cleavable N-terminal 6xHis-MBP tag. We call this construct 6xHis-MBP-hGSDMD. For mGSDMA3, clone the full-length sequence into a pET28a vector with an N-terminal 6xHis-SUMO tag. As the protease that cleaves and activates mGSDMA3 is unknown, insert a human rhinovirus (HRV) 3C protease cleavage site (LEVLFQGP) after residue E262 using standard site-directed mutagenesis methods such as overlap extension PCR. We call this construct 6xHis-SUMO-mGSDMA3–3C.
Using 6xHis-MBP-hGSDMD and 6xHis-SUMO-mGSDMA3–3C as template vectors, introduce mutations using the QuikChange Mutagenesis kit (Agilent Technologies).
Verify all cloning results by sequencing. Transform the 6xHis-MBP-hGSDMD and 6xHis-SUMO-mGSDMA3–3C plasmids into BL21 (DE3) E. coli competent cells and grow on LB plates containing kanamycin (50 μg/mL) overnight at 37 °C.
Pick a single colony into 10 mL LB medium containing kanamycin (50 μg/mL) and culture at 37 °C under shaking at 210 rpm until O.D.600 reaches 0.6–1.0.
Induce protein expression by adding 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) to the culture. Lower the temperature to 18 °C and continue growing the cells for 18 h. The expression level can be checked on SDS-PAGE by comparing uninduced and induced cells.
Harvest the bacterial cells by centrifugation at 3,500 rpm for 30 min and transfer the pellet into a 50 mL tube. Wash with pellet by resuspending it in PBS, redoing centrifugation, and discarding the supernatant. Freeze the pellet with liquid nitrogen and store at −80 °C for future use.
Gasdermin purification
Fully thaw and resuspend a pellet in 10 mL of lysis buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 5 mM imidazole, and 2 mM β-mercaptoethanol (2ME) per gram of pellet.
Ultrasonicate the pellet suspension for 10 min on an ice-water bath to fully lyse the cells.
Transfer the lysate to a centrifuge tube and spin at 15000 rpm for 30 min at 4 °C. Collect the supernatant, which contains most of the recombinant GSDMs.
Equilibrate Ni-NTA beads (Qiagen) with lysis buffer. For 1 L of bacterial culture, we recommend using 3 mL of Ni-NTA beads. The following steps are based on 1 L of culture and the quantities of reagents can be scaled accordingly.
Incubate the equilibrated Ni-NTA beads with the supernatant at 4 °C for 1 h.
Load the supernatant-bead mixture onto a glass Econo-Column and collect flow-through fractions.
Wash the beads with at least 20 column volumes (CVs) of wash buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 20 mM imidazole, and 2 mM 2ME). Collect wash fractions.
Elute recombinant GSDMs off the column with 5 CVs of elution buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 300 mM imidazole, and 2 mM 2ME). Collect elution fractions. Analyze the supernatant, flow-through, wash, and elution fractions using SDS-PAGE. The elution fractions should contain recombinant GSDMs at around 100 kD for 6xHis-MBP-hGSDMD and around 70 kD for 6xHis-SUMO-mGSDMA3–3C.
Pool elution fractions containing pure recombinant GSDMs. Spin the mixture at 15,000 rpm for 10 min at 4 °C to remove insoluble aggregates. Load the sample into a Superdex 200 (10/300) size exclusion column equilibrated with gel filtration buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl). Pool and save monomer peak fractions. This step removes soluble aggregates that hinder complete enzymatic cleavage in the following step.
Add TEV to the pool to cleavage the MBP tag off the recombinant hGSDMD, or add ULP1 to cleave the SUMO tag off the recombinant mGSDMA3. The protease to GSDM concentration ratio should be around 1:50. We observe nearly complete cleavage after 24 h at 4 °C. TEV, ULP1, and the 3C protease used for cleaving engineered mGSDMA3, are available commercially or can be generated using bacterial expression systems and straightforward Ni-NTA purification procedures. We recommend using TEV and ULP1 tagged with 6xHis for easier removal of these enzymes later.
After the 24-h cleavage, incubate the reaction mixture with 1 mL of Ni-NTA beads equilibrated with gel filtration buffer for 1 h. Then, Load the mixture onto a glass Econo-Column and collect flow-through fractions. MBP, SUMO, ULP1 and TEV will be bound to the beads and stay in the column due to the presence of 6xHis tags.
Pool the flow-through fractions containing GSDMs without MBP or SUMO, which should show a molecular weight of around 50 kD on SDS-PAGE. Spin the mixture at 15,000 rpm for 10 min at 4 °C to remove insoluble aggregates. Load the sample into a Superdex 200 (10/300) size exclusion column equilibrated with gel filtration buffer. Pool and concentrate monomer peak fractions to 5 μM. Aliquot the proteins, freeze with liquid nitrogen, and store at −80 °C for future use.
2.2. Overproduction of caspase-11 using insect cells
Caspase-11, an upstream activating protease for GSDMD, can be overexpressed in insect cell systems and purified for in vitro cleavage of hGSDMD. We employed baculovirus-infected Sf9 cells to generate caspase-11 following the established protocols for the Bac-to-Bac Baculovirus Expression System (Invitrogen), briefly described as following.
Full-length caspase-11 sequence was cloned into the pFastBac-HTa vector with a TEV-cleavable N-terminal 6xHis tag. Verify results by sequencing.
Transform the pFastBac-HTa-caspase-11 plasmid into E. coli DH10Bac and the bacteria cultured on an LB plate containing kanamycin (50 μg/ml), gentamycin (7 μg/ml), tetracycline (10 μg/ml), Bluo-gal (100 μg/ml), and IPTG (40 μg/ml) for 48 h at 37 °C.
Pick a white colony and grow in a miniprep LB culture containing kanamycin (50 μg/ml), gentamycin (7 μg/ml), and tetracycline (10 μg/ml) for 12 h at 37 °C under shaking at 210 rpm.
Extract bacmids from the miniprep culture using the isopropanol precipitation method.
Transfect Sf9 insect cells with the bacmid to generate P1, P2, and P3 baculoviruses following the manufacturer’s instructions.
Culture 1 L of Sf9 cells to a density of 2–3 million cells/ml. Then, infect the cells with 20 mL of P3 viruses and culture the infected cells for 48 h at 27 °C.
Collect the cells by spinning the culture at 2,500 rpm for 15 min at 4 °C. Wash the pellet with PBS. Freeze with liquid nitrogen and store at −80 °C for future use.
Purify 6xHis-caspase-11 following the same protocol as for 6xHis-MBP-hGSDMD and 6xHis-SUMO-mGSDMA3. Eluate the 6xHis-caspase-11 from the Ni-NTA resin, concentrate to 2 μM, and aliquot for storage. Note that caspase-11 undergoes autoproteolysis to generate active fragments, and therefore storage at 4 °C over time can enhance its enzymatic activity (Yang, Chang, and Baltimore 1998).
3. Liposome leakage assay
3.1. Liposome formulation
GSDM-NTs bind acidic lipids, including PS, PIPs, and CL, via charge-charge interactions. Liposomes that contain these acidic lipids can recruit GSDM-NTs, which form pores and enable the exchange of materials across the liposomal membranes. Based on this principle, we designed a liposome leakage assay that monitors GSDM activities in vitro (Ruan et al. 2018). Briefly, we encapsulate terbium ions (Tb3+) into the liposomes and provide dipicolinic acid (DPA) outside the liposomes. GSDM pore formation on the liposomal membranes induces the leakage of Tb3+ into the external buffer containing DPA, leading to a fluorogenic reaction between Tb3+ and DPA. The following steps describe the process to generate liposomes for the leakage assay. The lipids (Avanti Polar Lipids) we used were dissolved in chloroform to 25 mg/ml, upon which the calculations below are based.
Mix 80 μL phophatidylcholine (PC), 128 μL phosphatidylethanolamine (PE), and 64 μL CL in a glass tube. Note that other lipid combinations can be used provided that acidic lipids such as PIPs, PS, and CL are included.
Evaporate the chloroform under a steady stream of nitrogen gas while rotating the tube to create a lipid film on the glass surface.
Using a vortex mixer, suspend the lipid film in 1 mL buffer A (20 mM HEPES at pH 7.4, 150 mM NaCl, 50 mM sodium citrate, and 15 mM TbCl3) to generate the liposomes.
Push the liposome suspension through a 100-nm Whatman® Nuclepore™ Track-Etched Membrane 30 times using an extruder to obtain liposomes more homogeneous in size.
Equilibrate a Superose 6 (10/300 GL) gel filtration column with buffer B (20 mM HEPES at pH 7.4, 150 mM NaCl). Load the liposomes onto the column and collect 5 mL of void fractions to produce a stock of PC-PE-CL liposomes at 1.6 mM lipid concentration.
3.2. In vitro pore formation, assay protocol, and data interpretation
The leakage of Tb3+ from GSDM-perforated liposomes can be monitored by an increase in fluorescence intensity when Tb3+ binds DPA in the external buffer (Fig. 1A). Below is a protocol for the liposome leakage assay using purified hGSDMD and caspase-11 as an example. The combination of purified mGSDMA3–3C and the 3C protease will yield similar results.
Fig. 1. A liposome leakage assay for monitoring gasdermin activity.
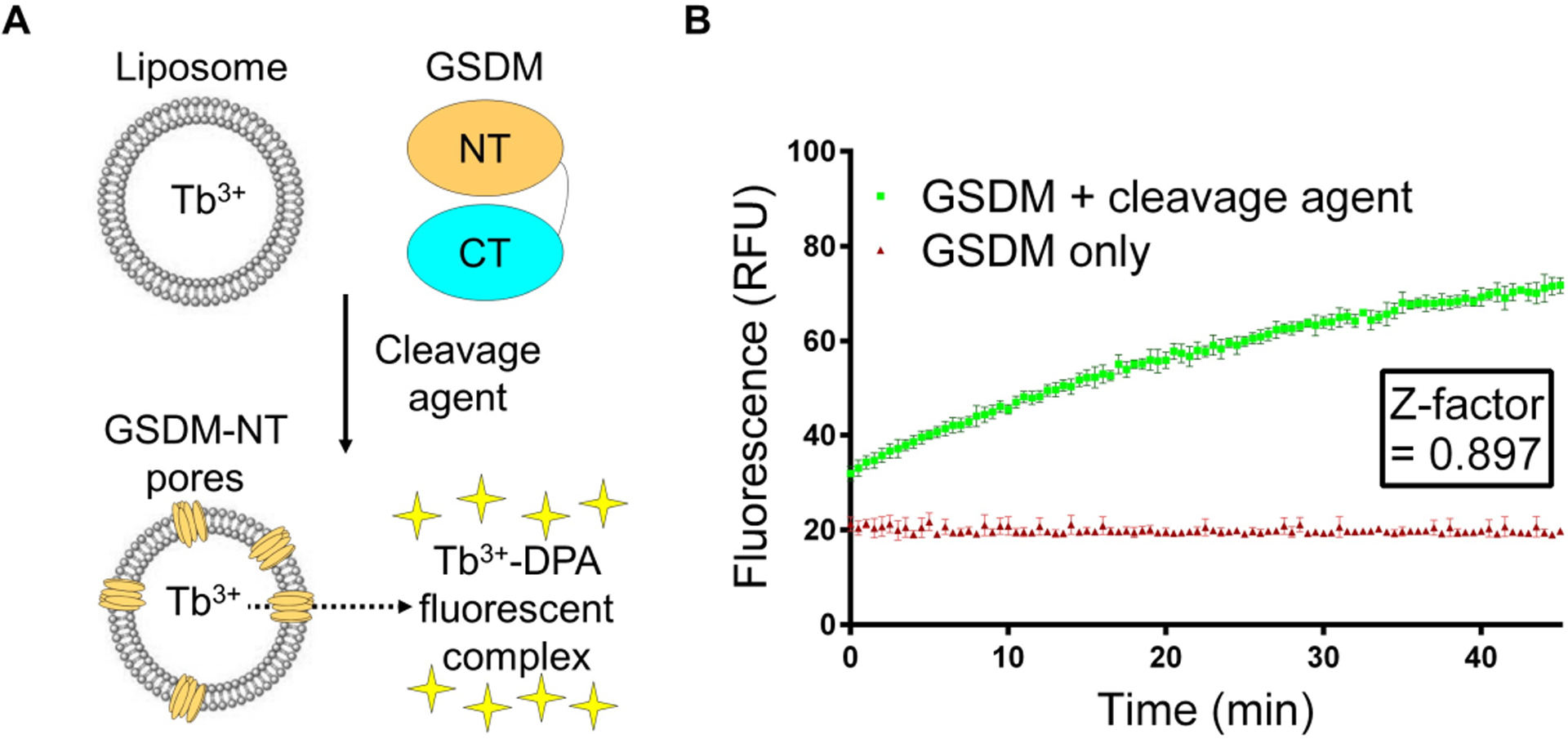
(A) A scheme of the liposomal assay. (B) Statistical analysis using the Z-factor. Data are plotted as mean ± standard deviation.
Dilute the PC-PE-CL liposomes with buffer C (20 mM HEPES at pH 7.4, 150 mM NaCl, and 50 μM DPA) to a final lipid concentration of 50 μM for the leakage assay.
In a 50 uL reaction system, add 30 μL liposomes (final lipid concentration: 30 μM), 5 μL hGSDMD (final concentration: 0.5 μM), 5 uL caspase-11 (final concentration: 0.2 μM). To set up a negative control, replace the caspase-11 with buffer C.
Mix the reactions well using a pipette, and transfer the reactions to Corning 3820 384-well assay plate.
Using a Molecular Devices SpectraMax M5 plate reader, continuously record fluorescence at 545 nm after excitation at 276 nm for 45 min at 30 s intervals.
The fluorescence data can be plotted with respect to time. A robust increase in fluorescence, which indicates active GSDM pore formation, is expected when both GSDM and the cleavage agent are present, in comparison to no or little fluorescence in the absence of the cleavage agent. To quantify the separation between the positive signal and the negative control, we employ a statistical measure known as the Z-factor, which is frequently used in high-throughput screening (Zhang 1999). The Z-factor is calculated based on the means (μ) and standard deviations (σ) of the experimental (exp) and control groups (ctr) as follows.
A Z-factor of less than 0.5 indicates an insignificant separation between the two groups, whereas a Z-factor between 0.5 and 1 suggests significant separation. Using the assay protocol above, we are able to reproducibly achieve a Z-factor of around 0.9 (Fig. 1B).
4. Conclusions and discussion
GSDMs represent a new family of β-barrel pore-forming proteins that play vital roles in innate immunity and cell death. While the functions and activation mechanisms of GSDMD and GSDME have been more extensively studied, the physiological significance and the pathways that regulate the generation of other GSDM-NTs remain largely elusive. In addition to aiding the identification of new upstream activating enzymes for GSDMs, the liposome leakage assay that monitors GSDM activities in vitro can also characterize the effects of GSDM mutations, some of which are related to human diseases. Similar liposomal assays have been employed to study other membrane proteins including pore-forming proteins and ion channels (Wang et al. 2014; Faudry, Perdu, and Attree 2013; Posson et al. 2018), suggesting the plasticity and wide applicability of this liposome-based method.
Acknowledgments
This work was supported by US NIH grants DP1HD087988, R01A139914, and R01Al124491 to H.W., a Charles A. King Trust Postdoctoral Fellowship to J.R., and an Albert J. Ryan Fellowship to S.X..
Abbreviations
- GSDM
Gasdermin
- FL
Full-length
- NT
N-terminal fragment
- CT
C-terminal fragment
- TM
Transmembrane
- TEV
Tobacco Etch Virus protease
- 3C
Human Rhinovirus 3C protease
- CL
Cardiolipin
- PE
Phosphatidylethanolamine
- PS
Phosphatidylserine
- PC
Phosphatidylcholine
- PIP
Phosphatidylinositol phosphate
- Cryo-EM
Cryo-electron microscopy
- 2ME
β-mercaptoethanol
- TbCl3
Terbium(III) chloride
- DPA
Dipicolinic acid
- PBS
Phosphate buffered saline
References
- Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, Ciferri C, Dixit VM, and Dueber EC. 2016. ‘GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes’, Proc Natl Acad Sci U S A, 113: 7858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken T, Fink SL, and Cookson BT. 2009. ‘Pyroptosis: host cell death and inflammation’, Nature Reviews Microbiology, 7: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KW, Monteleone M, Boucher D, Sollberger G, Ramnath D, Condon ND, von Pein JB, Broz P, Sweet MJ, and Schroder K. 2018. ‘Noncanonical inflammasome signaling elicits gasdermin D-dependent neutrophil extracellular traps’, Sci Immunol, 3. [DOI] [PubMed] [Google Scholar]
- Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, and Shao F. 2016. ‘Pore-forming activity and structural autoinhibition of the gasdermin family’, Nature, 535: 111–6. [DOI] [PubMed] [Google Scholar]
- Evavold CL, Ruan J, Tan Y, Xia S, Wu H, and Kagan JC. 2018. ‘The Pore-Forming Protein Gasdermin D Regulates Interleukin-1 Secretion from Living Macrophages’, Immunity, 48: 35–44 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faudry E, Perdu C, and Attree I. 2013. ‘Pore formation by T3SS translocators: liposome leakage assay’, Methods Mol Biol, 966: 173–85. [DOI] [PubMed] [Google Scholar]
- Franchi L, Eigenbrod T, Munoz-Planillo R, and Nunez G. 2009. ‘The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis’, Nat Immunol, 10: 241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, and Fitzgerald KA. 2009. ‘AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC’, Nature, 458: 514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I, and Miao EA. 2015. ‘Pyroptotic cell death defends against intracellular pathogens’, Immunol Rev, 265: 130–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, and Dixit VM. 2015. ‘Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling’, Nature, 526: 666–71. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, Forsberg LS, Carlson RW, and Dixit VM. 2013. ‘Noncanonical inflammasome activation by intracellular LPS independent of TLR4’, Science, 341: 1246–9. [DOI] [PubMed] [Google Scholar]
- Kuang SY, Zheng J, Yang H, Li SH, Duan SY, Shen YF, Ji CN, Gan JH, Xu XW, and Li JX. 2017. ‘Structure insight of GSDMD reveals the basis of GSDMD autoinhibition in cell pyroptosis’, Proceedings of the National Academy of Sciences of the United States of America, 114: 10642–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PH, Lin HY, Kuo CC, and Yang LT. 2015. ‘N-terminal functional domain of Gasdermin A3 regulates mitochondrial homeostasis via mitochondrial targeting’, Journal of Biomedical Science, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, and Lieberman J. 2016. ‘Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores’, Nature, 535: 153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang C, Rathkey JK, Yang J, Dubyak GR, Abbott DW, and Xiao TS. 2018. ‘Structures of the Gasdermin D C-Terminal Domains Reveal Mechanisms of Autoinhibition’, Structure, 26: 778–84 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, and Egelman EH. 2014. ‘Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes’, Cell, 156: 1193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy KJ, and Powrie F. 2011. ‘Intestinal homeostasis and its breakdown in inflammatory bowel disease’, Nature, 474: 298–306. [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, and Tschopp J. 2002. ‘The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta’, Mol Cell, 10: 417–26. [DOI] [PubMed] [Google Scholar]
- Martinon F, Petrilli V, Mayor A, Tardivel A, and Tschopp J. 2006. ‘Gout-associated uric acid crystals activate the NALP3 inflammasome’, Nature, 440: 237–41. [DOI] [PubMed] [Google Scholar]
- Orning P, Weng D, Starheim K, Ratner D, Best Z, Lee B, Brooks A, Xia SY, Wu H, Kelliher MA, Berger SB, Gough PJ, Bertin J, Proulx MM, Goguen JD, Kayagaki N, Fitzgerald KA, and Lien E. 2018. ‘Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death’, Science, 362: 1064.-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrilli V, Papin S, Dostert C, Mayor A, Martinon F, and Tschopp J. 2007. ‘Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration’, Cell Death Differ, 14: 1583–9. [DOI] [PubMed] [Google Scholar]
- Posson DJ, Rusinova R, Andersen OS, and Nimigean CM. 2018. ‘Stopped-Flow Fluorometric Ion Flux Assay for Ligand-Gated Ion Channel Studies’, Methods Mol Biol, 1684: 223–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, and Alnemri ES. 2017. ‘Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death’, Nat Commun, 8: 14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Xia S, Liu X, Lieberman J, and Wu H. 2018. ‘Cryo-EM structure of the gasdermin A3 membrane pore’, Nature, 557: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl S, Shkarina K, Demarco B, Heilig R, Santos JC, and Broz P. 2018. ‘ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation’, Science, 362: 956–60. [DOI] [PubMed] [Google Scholar]
- Sarhan J, Liu BC, Muendlein HI, Li P, Nilson R, Tang AY, Rongvaux A, Bunnell SC, Shao F, Green DR, and Poltorak A. 2018. ‘Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection’, Proceedings of the National Academy of Sciences of the United States of America, 115: E10888–E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, and Tschopp J. 2010. ‘The inflammasomes’, Cell, 140: 821–32. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, and Shao F. 2015. ‘Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death’, Nature, 526: 660–5. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, and Shao F. 2014. ‘Inflammatory caspases are innate immune receptors for intracellular LPS’, Nature, 514: 187–92. [DOI] [PubMed] [Google Scholar]
- Sollberger G, Choidas A, Burn GL, Habenberger P, Di Lucrezia R, Kordes S, Menninger S, Eickhoff J, Nussbaumer P, Klebl B, Kruger R, Herzig A, and Zychlinsky A. 2018. ‘Gasdermin D plays a vital role in the generation of neutrophil extracellular traps’, Sci Immunol, 3. [DOI] [PubMed] [Google Scholar]
- Taabazuing CY, Okondo MC, and Bachovchin DA. 2017. ‘Pyroptosis and Apoptosis Pathways Engage in Bidirectional Crosstalk in Monocytes and Macrophages’, Cell Chem Biol, 24: 507–14 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF, Wang FS, and Wang X. 2014. ‘Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3’, Mol Cell, 54: 133–46. [DOI] [PubMed] [Google Scholar]
- Wang YP, Gao WQ, Shi XY, Ding JJ, Liu W, He HB, Wang K, and Shao F. 2017. ‘Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin’, Nature, 547: 99.-+. [DOI] [PubMed] [Google Scholar]
- Yang X, Chang HY, and Baltimore D. 1998. ‘Autoproteolytic activation of pro-caspases by oligomerization’, Mol Cell, 1: 319–25. [DOI] [PubMed] [Google Scholar]
- Zanoni I, Tan Y, Di Gioia M, Broggi A, Ruan J, Shi J, Donado CA, Shao F, Wu H, Springstead JR, and Kagan JC. 2016. ‘An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells’, Science, 352: 1232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD and Oldenburg KR 1999. ‘A simple statistical parameter for use in evaluation and validation of high throughput screening assays’, Journal of biomolecular screening, 4: 67–73. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen X, Gueydan C, and Han J. 2018. ‘Plasma membrane changes during programmed cell deaths’, Cell Res, 28: 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, and Shao F. 2011. ‘The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus’, Nature, 477: 596–600. [DOI] [PubMed] [Google Scholar]


