Abstract
Adipogenic differentiation is the process by which preadipocytes become mature adipocytes, cells that store energy and regulate metabolic homeostasis. During differentiation, neutral lipids that accumulate in adipocytes can be detected using stains and used as an index of cell differentiation. However, imaging tools for evaluating intracellular lipid droplets remain at their infancy. Nutrition, stress, or chemical exposure can dysregulate adipogenic differentiation and lipid metabolism. Therefore, the aims of this study were to develop an accurate, standardized approach to quantify lipid droplet size of mature adipocytes and a clustering approach to analyze the total lipid content per adipocyte. For the lipid droplet analysis, we used two approaches, the free online computer software of reference, ImageJ, and another free online computer software, CellProfiler. For ImageJ, we used an already developed macro designed to identify particles and quantify their area, and for CellProfiler, we developed a new analysis pipeline. Our results show that CellProfiler is able to accurately identify a greater number of lipid droplets compared to ImageJ. A clustering analysis is also possible using CellProfiler which allows for the quantification of total lipid content per individual adipocyte to provide insight into single-cell responsiveness to adipogenic stimuli. CellProfiler streamlines the lipid droplet phenotypic analysis of adipocytes compared to more traditional analysis methods. In conclusion, this novel image analysis tool can provide with a more precise evaluation of lipid droplet and adipogenesis dysregulation, a critical need in the understanding of metabolic disorders.
Keywords: adipocyte, lipid droplet, image segmentation, quantitative analyses
Introduction
Adipogenesis is the process by which mesenchymal stromal cells or committed preadipocytes differentiate into mature adipocytes. This is a tightly controlled process that may be altered by several factors, including nutrition, stress, as well as environmental factors (Ghaben and Scherer 2019). Understanding how these factors can alter adipogenesis can help provide insights into strategies to prevent adipose tissue accumulation resulting in metabolic disorders and/or obesity (Ghaben and Scherer 2019). Studies to understand adipogenesis often use the preadipocyte mouse cell line 3T3-L1. Preadipocytes are induced to differentiate in vitro for 8 to 12 days during which cells are exposed to a differentiation cocktail that may contain insulin, peroxisome proliferator-activated receptor gamma (PPARγ) agonists, and/or 3-isobutyl-1-methylxanthine (IMBX), among others (Zhao et al. 2019). During the differentiation process, cells begin to accumulate lipid droplets leading to the formation of mature adipocytes with a specific transcriptomic profile, including increased PPARγ, FABP4, and adiponectin expression (Lee JE et al. 2019).
Current techniques to detect intracellular lipid accumulation after differentiation include the use of dyes that will stain for neutral triglycerides and lipids. While there are over 50 different probes to detect lipid droplets (Fam et al. 2018), two of the most common probes are Oil Red O, a lysochrome-diazo dye that stains lipids with a red color at 518 nm, and BODIPY493/503, an organoboron fluorescent dye. BODIPY493/503 is more selective for lipid droplet detection, as the poor solubility of Oil Red O can damage or result in the fusion of lipid droplets during the ethanol/isopropanol staining step (Fukumoto and Fujimoto 2002). After lipid staining, visualization of lipid droplets is most routinely carried out by transmission light microscopy or fluorescence microscopy, with fluorescence probes being more sensitive to detection (Klymchenko 2017). Methods to quantify intracellular lipid accumulation after differentiation range from visual observation of stain (most often Oil Red O or Nile Red) on a culture plate (Prieto-Echague et al. 2017; Diep et al. 2018) or digital quantification (for Oil Red O or BODIPY) (Park et al. 2017; Pu and Veiga-Lopez 2017). Current analytical methods use basic software that allows for deconvolution of images based on color detection to quantify total lipid content. The ImageJ free software is one of the most commonly used image analysis platforms for lipid content evaluation. Although ImageJ allows the development of macros to automate a series of ImageJ commands for complex analyses, standard components of the software do not allow for proper identification of individual lipid droplets. In recent years, software programs that allow for the automated analyses of images have also been used to evaluate lipid content. CellProfiler is an open-access software that is designed for high-throughput modular analysis and quantification of data from biological images allowing for great flexibility in automated particle analyses. CellProfiler’s point-and-click interface makes the software user-friendly, so that users can create protocols for image analyses, called “pipelines”, to measure their phenotype of interest. These pipelines can be shared and downloaded in .cppipe or .cpproj formats within an ever-growing community with over 3,000 users on its online forum (https://github.com/CellProfiler/CellProfiler) (McQuin et al. 2018). Even though CellProfiler has been used to quantify lipid droplets (Majithia et al. 2014; Kristof et al. 2016; Campos et al. 2018), the development of a CellProfiler pipeline that quantifies lipid droplets has not yet been described so that a standardized pipeline can be used by the community.
Lipid droplets have also been involved in the regulation of cellular lipid metabolism, membrane formation, intracellular trafficking, protein-protein interactions and define mitochondrial bioenergetics (Blom et al. 2011; Onal et al. 2017; Benador et al. 2018). In addition, lipid droplets of different sizes are enriched for different lipids, with large droplets enriched for triacylglycerol and smaller with cholesteryl esters (Chanderbhan et al. 1982; McIntosh et al. 2010). In recent years, it has become evident that defects in lipid droplet biogenesis, their size, and changes in proteins present in the surface of lipid droplets (perilipins) are associated with pathophysiological conditions, including obesity, fatty liver disease, and atherosclerosis, type 2 diabetes, Alzheimer’s disease, cardiovascular disease, and cancer (Onal et al. 2017; Dalhaimer 2019), many of which have been associated with chemical exposures (Al-Eryani et al. 2015; Deierlein et al. 2017; Heindel et al. 2017; Lee YM et al. 2018; Dallio et al. 2019). Adipogenesis and thus, lipid droplet formation and accumulation can be dysregulated in the presence of environmental chemical exposures (Schmidt et al. 2012; Biemann et al. 2014; Boucher et al. 2016; Pu et al. 2017). Therefore, understanding specific lipid droplet dysregulations can have implications for disorders such as diabetes, non-alcoholic fatty liver disease, atherosclerosis and obesity (Onal et al. 2017; Dalhaimer 2019). Thus, the first aim of this study was to develop a systematic analysis for, not only total intracellular lipid content, but also individual lipid droplet detection. We analyzed lipid content using the already developed ImageJ macro, “Analyze Particles”, and a newly developed CellProfiler pipeline. During adipogenesis, individual cells can respond differently to adipogenic stimuli. This can result in a mixed population of cells, with some that have fully differentiated into mature adipocytes displaying large intracellular lipid droplets and others that have not undergone full differentiation and bear smaller lipid droplets. Image analyses that allow for the identification of individual cell lipid content is the first step towards the understanding of single-cell responsiveness to adipogenic stimuli, a poorly understood phenomenon. Therefore, the second aim of this study was to develop an automated analysis that will allow for the quantification of intracellular lipid content within a single cell.
Material and Methods
Cell Culture and Adipocyte Differentiation
The commercially available 3T3-L1 murine preadipocyte cell line was used as a model of adipogenic differentiation. Cells (American Type Culture Collection, ATCC, Manassas, VA, USA; within passage 6) were grown in basal medium in a twenty-four well plate until confluency. Basal medium consisted of DMEM/F12 medium and was supplemented with 1 % penicillin-streptomycin, 10 mM HEPES, and 10 % fetal bovine serum (Corning, Manassas, VA, USA). Cells were then allowed to grow for an additional 2 days before cells were induced to differentiate, as previously described (Pu and Veiga-Lopez 2017). In brief, 3T3-L1 cells were exposed for 2 days to differentiation medium consisting of basal medium supplemented with 33 μM biotin, 17 mM pantothenate, 1 μg/ml insulin, 1 μM dexamethasone, and 0.5 mM IBMX. Thereafter, cells were cultured for 8 additional days in basal differentiation medium supplemented with 1μg/ml insulin. Media was replaced every 48 hours. After 10 days of differentiation, cells were fixed with 10% formalin for 15 min at room temperature until further immunostaining.
Immunofluorescence and Imaging
Lipid droplets in differentiated adipocytes were identified with a BODIPY493/503 stain (D3922, Invitrogen, Carlsbad, CA, USA) and the nuclei were identified with a DAPI stain following the manufacturer’s protocol. Formalin-fixed 3T3-L1 cells were washed three times in DPBS and incubated with BODIPY stain working solution (1 μg/ml in DPBS) for 20 min. Cells were then washed three times with DPBS and incubated with DAPI (1 μg/ml in DPBS) for 5 min before being washed again with DPBS. Cells were imaged using a Nikon Eclipse Ti-U fluorescence microscope (Tokyo, Japan) with a high performance EMCCD & CCD camera (Tucson, AZ, USA) on a 10x objective lens (images were 1,940 × 1,460 pixels in size). FITC and DAPI channels using two fluorescence channels at 519 nm and 461 nm were used to capture BODIPY stained lipid droplets and DAPI stained nuclei, respectively, using the NIS-Elements imaging software (AR 4.40.00, Nikon, Melville, NY, USA). A total of 50 systematic random images were captured for further analyses. A subset of 12 representative images were used for further analyses, 6 with low and 6 with high lipid droplet density (See Supplemental Figures 6–17).
ImageJ Software:Lipid Analysis
Lipid analysis was conducted using the open-source ImageJ software (https://imagej.net/Fiji/Downloads). The ImageJ step-by-step protocol (for MacOS) used in this study for image analysis is shown in Supplemental Figure 1. First, individual color images were imported into ImageJ by selecting the “File” option followed by the dropdown “Open” option (Supplemental Figure 1A). Next, the uploaded color images were converted into an 8-bit grayscale image by selecting the “Image”, Type”, and “8-bit” options as shown in Supplemental Figure 1B. Then, a threshold defined by the range of minimum and maximum values of the RGB color (16 and 255 arbitrary units) was selected to convert the grayscale image to a binary image by selecting the options “Image”, “Adjust”, and Threshold” (Supplemental Figure 1C). (Supplemental Figure 1D). After all parameters from the “Threshold” option were setup, “Set” was selected. The “Analyze Particles” macro was run by first selecting the “Analyze” option and then the macro “Analyze Particles” (Supplemental Figure 1E). Thereafter, maximum and minimum values for the size and circularity of the objects (size: 0 to ∞ and circularity: 0.00 to 1.00) was selected (Supplemental Figure 1F). Once all parameters from the “Analyze Particles” macro were setup, “OK” was selected to run the automated analysis. A separate “Results” window provided the area of each particle identified and a separate “Summary” window provided the output variables: total number of identified particles, the total positive lipid area (in pixels), the average size of each particle (in pixels), and the percent of positive lipid area per image.
CellProfiler Software:Lipid Analysis
Lipid analysis was also conducted using CellProfiler (version 3.1.8; https://cellprofiler.org/releases/). A step-by-step protocol (for MacOS) for image analysis using CellProfiler is shown in Supplemental Figure 2. CellProfiler’s workspace interface displays four basic modules (Supplemental Figure 2A Left Panel: “Images”, “Metadata”, “NamesAndTypes”, and “Groups”) related to uploading and sorting of images. The “Images” module was used to first drag and drop either individual images or folders of images to be analyzed (see “Drop files and folders here”). In this study, images were uploaded as .png files, but CellProfiler supports various image file formats. Then, the “NamesAndTypes” module was used to sort between BODIPY and DAPI stained images based on the file names. After uploading and sorting images, different modules related to the analysis of images and output of data were added to the pipeline using the “Adjust modules +” option (Supplemental Figure 2A, arrow). After selecting the “Adjust modules” option, modules were added from a side window (Supplemental Figure 2B). The pipeline panel shown in Supplemental Figure 2C lists the image analysis modules that were used to identify and quantify lipid droplets in a BODIPY stained image which included two “ColorToGray” modules, two “IdentifyPrimaryObject” modules, followed by “MeasureObjectIntensity”, “FilterObjects”, “MeasureObjectSizeShape”, and “ExportToSpreadsheet”. This pipeline allows for the measurement of the following variables from each lipid droplet: area, mean and median radius, eccentricity, perimeter, compactness, Euler number, form factor, minor and major axis length, minimum and maximum Feret diameter, and solidity (see definitions for these terms below). This pipeline, annotated with notes to understand the function of each module, is provided as Supplemental File 1 (CP_ Pipeline_Lipid Droplet Analysis).
Lipid Droplet Measurements
The CellProfiler software defines the lipid droplets output variables as follows. The area is defined by the number of pixels within a lipid droplet. The mean and median radius provide a measurement of the width of each droplet in pixels. Eccentricity is a measurement of how closely each object resembles a circle as it is the ratio of the distance between the foci of an ellipse and its major axis length. The perimeter is the total number of pixels around the boundary of an object. Compactness refers to the mean square distance of the object’s pixels from the center of the object divided by the area, meaning that a filled circle will have a compactness of 1, while objects with holes in them will have a value greater than one. Euler number is the number of objects in the region minus the number of holes in those objects. Form factor is calculated as 4 × π × area/perimeter2 and will have a value of one for a perfectly circular object. Major and minor axis length refer to the length (pixels) of the major and minor axes of an ellipse. Minimum and maximum Feret diameter (pixels) is the distance between two parallel lines tangent on either side of the object and refer to the smallest and largest possible diameters in a specific lipid droplet. Solidity is the proportion of the pixels in the convex hull that are also in the object.
Manual Lipid Analysis
To provide a comparison between imaging analyses, lipid content was also manually analyzed on three adipocytes from a sample image containing a high density of mature adipocytes. To measure the area of each lipid droplet, the already outlined lipid droplets were again outlined in ImageJ using the “Freehand selection”. After each droplet had been manually outlined with the “Freehand selection”, the area of that individual droplet was determined using the “Analyze” and “Measure” options from the dropdown menu on ImageJ. This was repeated for each lipid droplet in the three images. To compare the manually acquired measurements to the ImageJ and CellProfiler automated analyses, all three images were processed using the ImageJ “Analyze Particles” macro (Supplemental Figure 3B, 3E, 3H) and the CellProfiler pipeline (Supplemental Figure 3C, 3F, 3I) as described above (see “ImageJ Software: Lipid Analysis” and “CellProfiler Software: Lipid Analysis”).
CellProfiler Software:Clustering Analysis to Identify Differentiated Adipocytes
CellProfiler can not only automatically identify lipid droplets, but additional modules can also be added so that lipid droplets pertaining to the same differentiated adipocyte can be clustered together (Supplemental Figure 2D). The following modules were added to cluster lipid droplets: “SplitOrMergeObjects”, “ConvertObjectsToImage”, “IdentifyPrimaryObjects”, “FilterObjects”, “OverlayObjects”, “MeasureObjectSizeShape”, and “SaveImages”. This pipeline is provided as Supplemental File 2 (CP_ Pipeline_Lipid Droplet Clustering Analysis).
Statistics
All data are presented as mean ± SEM. Comparisons between the two groups (low- and high-density images) were analyzed by an independent T-test using PASW Statistics for Windows release 18.0.1. Logarithmic transformation was applied to account for normality of data. Differences were considered significant at P<0.05.
Results
ImageJ Software:Lipid Analysis
When manually analyzing each image with the ImageJ “Analyze Particles” macro, original images containing BODIPY stained lipid droplets (Figure 1A) were first converted to grayscale images (Figure 1B). Then, grayscale images were converted to binary (black and white) images (Figure 1C). Thresholding was done based on the signal intensity of the images to be analyzed in order to filter out any background signal. It was also a required step to generate binary (black and white) images (Figure 1C) because only binary images can be run by the “Analyze Particles” ImageJ macro. In Figure 1C, the black area represents the total positive lipid area and the white area represents the area of the image that does not contain lipid content. From the binary image, lipid particles were identified using the “Analyze Particles” macro. Even though ImageJ allows the user to select the maximum and minimum size and circularity of objects to be identified so that anything that is not an object of interest can be excluded from the analysis, these parameters were kept at their original settings in this study (size: 0 to ∞ and circularity: 0.00 to 1.00). The parameters were kept at this setting since the positive lipid area was already identified in the binary image. After running the analysis, ImageJ generated a new image displaying the particles that were identified (Figure 1D) and a separate “Results” window that provided the area of each particle identified. ImageJ also provided a separate “Summary” window which displayed the output variables (see Methods section).
Figure 1. Sample image processing using ImageJ “Analyze Particles”.
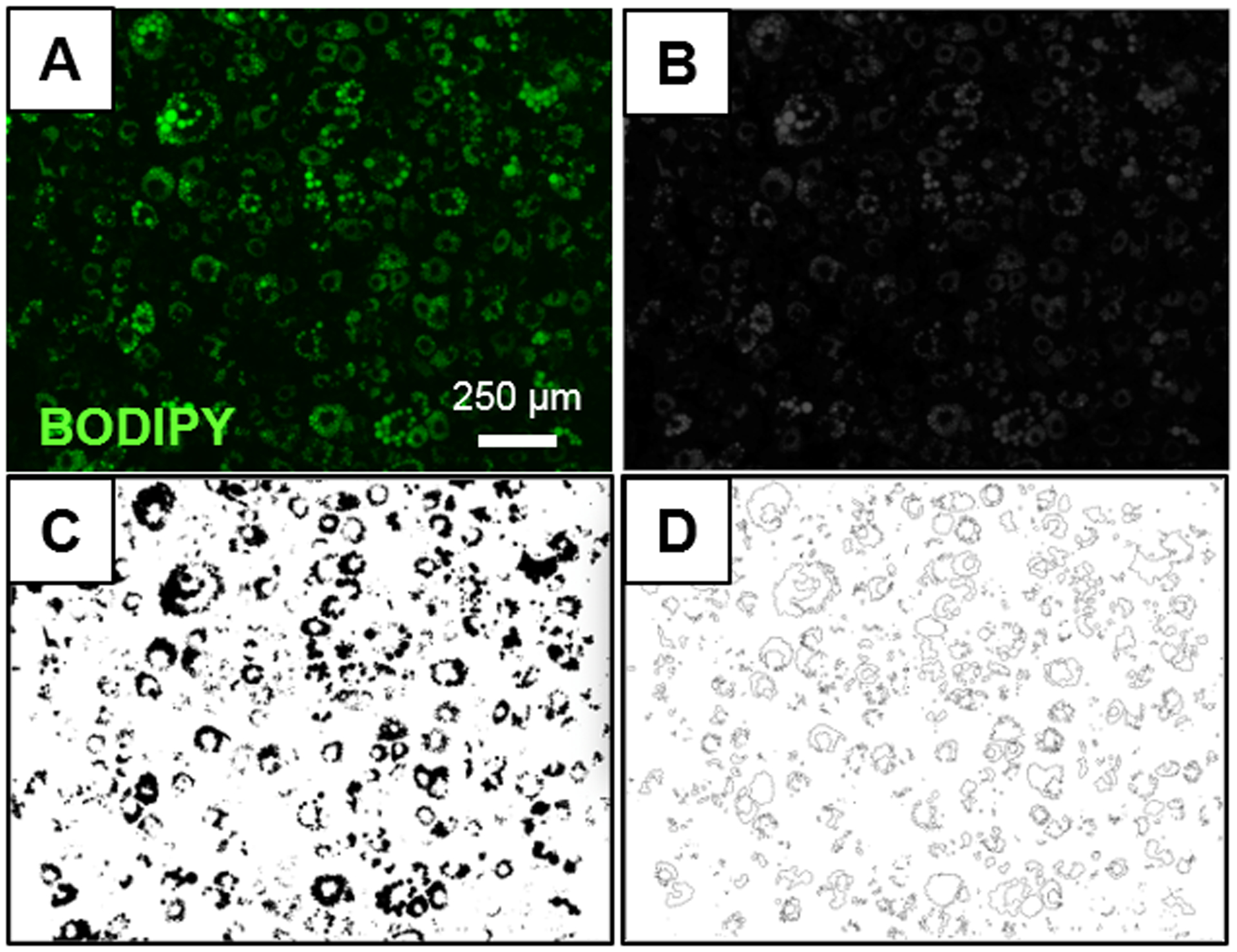
(A) Original image of differentiated adipocytes with a high density of BODIPY stained lipid droplets. (B) Grayscale image after conversion of the original image to an 8-bit image. (C) Binary image after applying an intensity threshold to the 8-bit image with ImageJ. (D) Lipid content identified by “Analyze Particles” macro in ImageJ.
CellProfiler Software:Lipid Analysis
After images were uploaded onto the CellProfiler software using the “Images” module, BODIPY and DAPI stained images were sorted into two different groups using the “NamesAndTypes” module. Then, two “ColorToGray” modules were used to convert each of the BODIPY and DAPI color images (Figure 2A) to grayscale images (Figure 2B). The first “IdentifyPrimaryObjects” module was used to count the number of nuclei in each DAPI image (Figure 2C) based on the size, shape, and pixel intensity of the nuclei (minimum diameter: 8; maximum diameter: 80-pixel units). The second “IdentifyPrimaryObjects” module was then used to count the number of lipid droplets in each BODIPY stained image also based on the size, shape, and pixel intensity of the lipid droplets with the output shown in Figure 2D (minimum diameter: 6; maximum diameter: 80-pixel units). This second “IdentifyPrimaryObjects” module can be used for altering the intensity detection threshold. Examples of different threshold correction factors (0.95 – 1.10) and their corresponding lipid droplet identification outputs are shown in Supplemental Figure 4. For instance, a 7.6% increase in the number of lipid droplets was observed when the threshold was lowered by 0.1 arbitrary units. An additional “FilterObjects” module was included to ensure that only the strongest signals were quantified, while less intense signals were excluded from the final lipid droplet analysis (minimum intensity: 0.15; maximum intensity:1 arbitrary units). The “MeasureObjectSizeShape” module measured the output variables described in the Lipid Droplet Measurements Methods section. These data were then exported onto a .csv file using the “ExportToSpreadsheet” module. This module also provided a read out of the total number of nuclei per each image.
Figure 2. Sample image processing using CellProfiler.
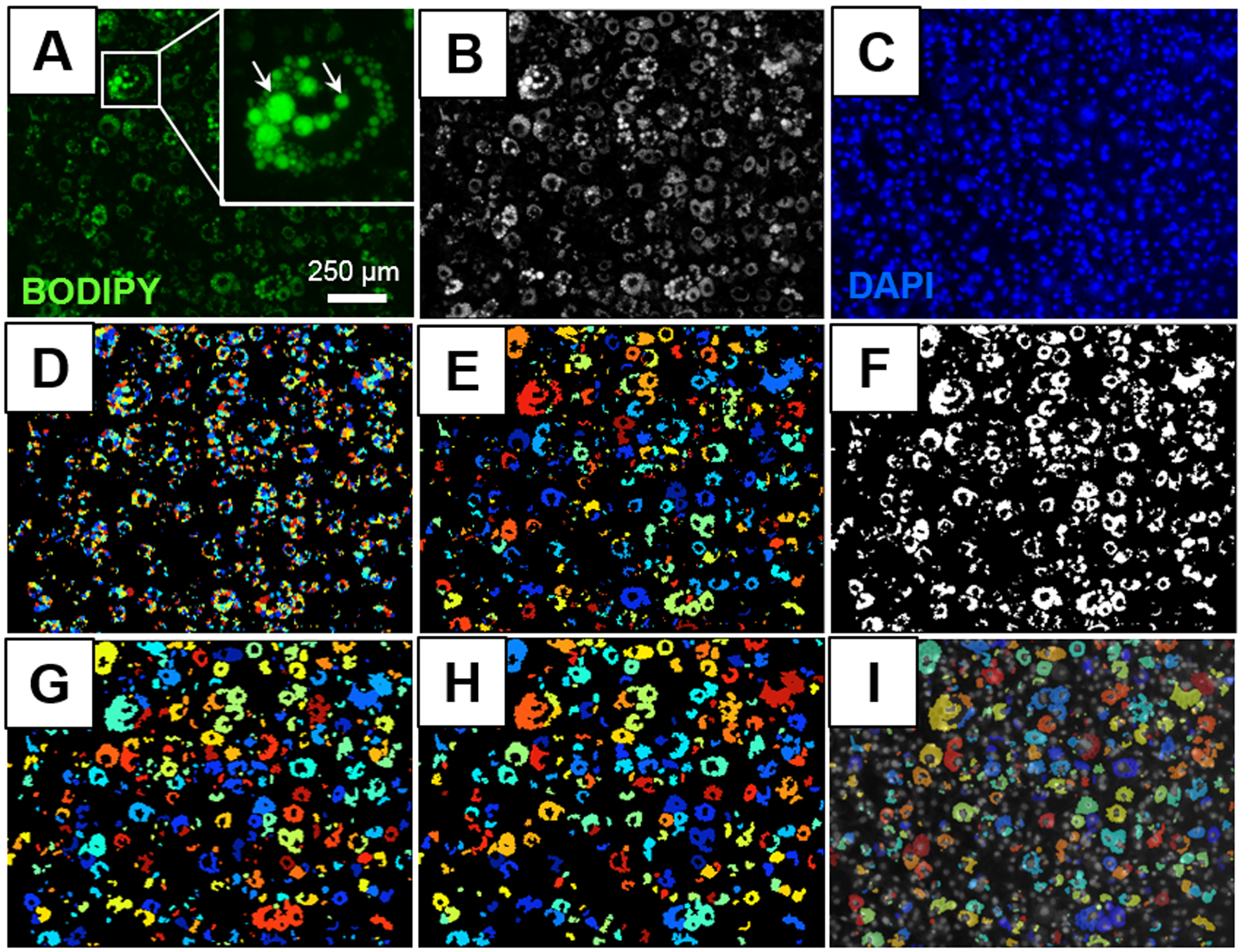
(A) Original image of differentiated adipocytes with a high density of BODIPY stained lipid droplets. Image also contains a magnified picture of a differentiated adipocyte to model the diversity in lipid droplet sizes within a single cell. (B) Conversion of original image to grayscale using the “ColorToGray” module. (C) Corresponding image of DAPI stained nuclei. (D) Identified lipid droplets using the “IdentifyPrimaryObjects” and “FilterObjects” modules. (E) Touching lipid droplets were grouped together using the “SplitOrMergeObjects” module. (F) Identified objects from the “SplitOrMergeObjects” module were converted to a binary image using the “ConvertObjectsToImage” module. (G) Adipocytes were identified by size, shape, and intensity from the binary image using the “IdentifyPrimaryObjects” module. (H) Objects below the area threshold were excluded from the final analysis using the “FilterObjects” module. (I) Differentiated adipocytes were overlaid with corresponding nuclei using the “OverlayObjects” module.
Comparison of Lipid Droplet Identification using ImageJ and CellProfiler
A set of 12 images were analyzed using both, the ImageJ “Analyze Particles” macro, and the CellProfiler pipeline shown in Supplemental Figures 1 and 2, respectively (images used are in Supplemental Figures 6). As a first example, two of these images with low- (Figure 3A) and high-lipid density (Figure 3D) were used to assess the utility of the two analytical methods. ImageJ identified lipid content from a binary image that was made by applying an intensity threshold based on the size and circularity of the particles of interest. The CellProfiler pipeline identified lipid particles by their average diameter, pixel intensity and shape (Table 1). Both ImageJ (Figure 3B, 3E) and CellProfiler (Figure 3C, 3F) could calculate the percent positive lipid area in an image once the lipid content had been identified. CellProfiler identified a greater amount of lipid content in both the low- and high-density pictures with 2.9 % and 17.4 % positive lipid area (Figure 3C, 3F) compared to ImageJ which identified 2.0 % and 13.5 % positive lipid area respectively (Figure 3B, 3E). When evaluating individual lipid droplet results, ImageJ displayed the area of each lipid particle in a new “Results” window and CellProfiler provided the data for individual lipid droplet area in a .csv file. The lipid droplet sizes from the “Results” window and the .csv file were used to generate histograms for both the images with low- (Figure 3G) and high-lipid content (Figure 3H) and to compare the lipid droplet size distribution between analytical methods. The majority of the lipid droplets identified by CellProfiler were ~100 pixels vs. 1–2 pixels when using ImageJ (Figure 3G, 3H). The distribution of the lipid particles’ area identified by ImageJ showed a positive skewness compared to that of CellProfiler. This skewed distribution was also evident in the maximum lipid droplet area, with ImageJ displaying a maximum area of 13,426 pixels in the high-density image and 7,276 pixels in the low-density image compared to 1,065 and 713 in CellProfiler, respectively (Supplemental Figure 5).
Figure 3. Comparing lipid area identified by ImageJ and CellProfiler in low vs. high lipid droplet content images.
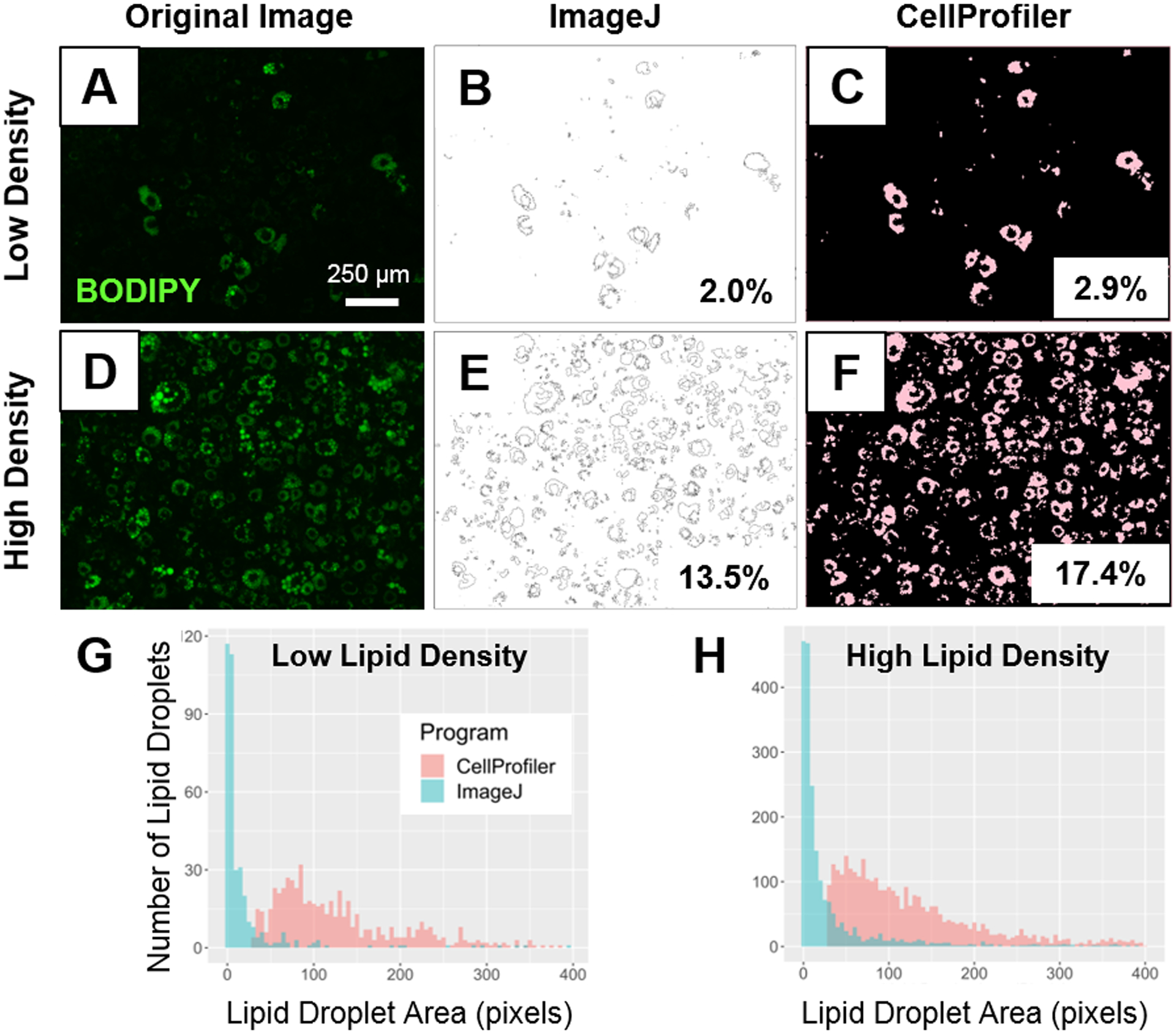
Top: Original image with low density lipid content (A) and corresponding processed images with identified percent positive lipid area identified by ImageJ (B) and CellProfiler (C). Original image with high density lipid content (D) and corresponding processed images with identified percent positive lipid area identified by ImageJ (E) and CellProfiler (F). Bottom: Comparison of the individual lipid droplet area quantification between ImageJ and CellProfiler (G, H). Lipid droplet area distribution in a sample image with low (G) and high (H) density of lipid droplets in CellProfiler (red) and ImageJ (blue).
Table 1.
ImageJ vs. CellProfiler imaging comparative summary.
| ImageJ “Analyze Particles” | CellProfiler Pipeline | |
|---|---|---|
| Analysis Type | Manual | Automatic |
| Identification | Done by size and circularity | Done by pixel intensity, size, and shape |
| Result | Wider range of lipid particle sizes | Greater number of lipid droplets with more consistent area |
| Clustering Analysis | No | Yes |
We further validated the differences between the ImageJ and CellProfiler analyses by analyzing additional images (6 with high- and 6 with low-lipid content; Supplemental Figure 6). CellProfiler identified a greater number of lipid droplets compared to ImageJ in both low- and high-density images (Table 2). Mean lipid droplet area was significantly different between analytical methods within lipid density images. A large disparity in lipid droplet distribution (percentiles and median values) was also observed in ImageJ vs. CellProfiler-processed images, independent of the amount of lipid droplet density. While both programs provided a list of the area of each individual lipid particle in each image, only the CellProfiler analysis provided additional measurements (see “Lipid Droplet Measurements” in Methods section). To evaluate the accuracy of lipid droplet detection using ImageJ and Cell Profiler, manual identification was also conducted in three images displaying single adipocytes (Supplemental Figure 3). CellProfiler was able to identify a greater number of lipid droplets in each adipocyte than ImageJ compared to the manual identification. CellProfiler was also able to identify lipid droplets that were closer to the area of the manually measured lipid droplets than ImageJ by an order of magnitude.
Table 2.
Lipid droplet measurements analyzed with ImageJ and CellProfiler from images with low and high lipid density.
| Lipid Droplets Parameters | Low Density Images (n = 5) | High Density Images (n = 5) | |||
|---|---|---|---|---|---|
| ImageJ | CellProfiler | ImageJ | CellProfiler | ||
| Total Number | 1,564 | 3,379 | 6,642 | 12,686 | |
| Lipid Droplet Area (px) | Mean (± SEM) | 293 ± 37b | 154 ± 2b | 286 ± 19a | 158 ± 1b |
| Median | 8 | 113 | 9 | 112 | |
| Mode | 1 | 46 | 1 | 37 | |
| Minimum | 1 | 29 | 1 | 29 | |
| Maximum | 31,107 | 1,432 | 44,172 | 1,777 | |
| 25th Percentile | 2 | 67 | 3 | 66 | |
| 50th Percentile | 8 | 113 | 9 | 112 | |
| 75th Percentile | 35 | 193 | 45 | 199 | |
| Mean Radius (px) | ND | 2.21 | ND | 2.23 | |
| Mean Eccentricity | ND | 0.66 | ND | 0.65 | |
For this analysis, lipid droplets from all 5 images were combined. Mean radius (pixels) and eccentricity were measured by CellProfiler, but could not be determined using the “Analyze Particles” macro on ImageJ. a ≠ b denotes P < 0.05 within image densities and between analysis programs. Diff: difference, ND: not detectable, and n/a: not available.
CellProfiler Software:Clustering Analysis
In order to cluster lipid droplets together by adipocyte, modules (Supplemental Figure 2D) were added onto the pipeline shown in Supplemental Figure 2C. First, the “SplitOrMergeObjects” module was added onto the pipeline to merge any touching lipid droplets together to identify them as one object (Figure 2E). Then, those objects were converted to a binary image with the “ConvertObjectsToImage” module (Figure 2F). This conversion was done so that the differentiated adipocytes could be more accurately identified with the “IdentifyPrimaryObjects” module which distinguishes objects based on their size (minimum diameter: 80; maximum diameter: 500-pixel units) and can de-clump objects by their shape (Figure 2G). After differentiated adipocytes were initially identified with “IdentifyPrimaryObjects”, objects with an area below 400 pixels were excluded from the final analysis with the “FilterObjects” module to remove any background signal as seen in Figure 2H. The differentiated adipocytes were overlaid on the original DAPI image with the “OverlayObjects” module (Figure 2I) and they were measured with “MeasureObjectSizeShape”. Finally, the new overlaid images were saved with “SaveImages” and the measurements of the differentiated adipocytes including their individual area, radius, and location were exported on a .csv file with the “ExportToSpreadsheet” module. By adding these additional modules, CellProfiler can be used to compare the number of differentiated adipocytes to the number of nuclei to calculate the percent of preadipocytes that differentiated. As an example, an image with low lipid density (Figure 4A) and an image with high lipid density (Figure 4D) were used to demonstrate how the identified lipid droplets (Figure 4B, 4E) were clustered together to identify the differentiated adipocytes (Figure 4C, 4F).
Figure 4. Clustering Analysis by CellProfiler.
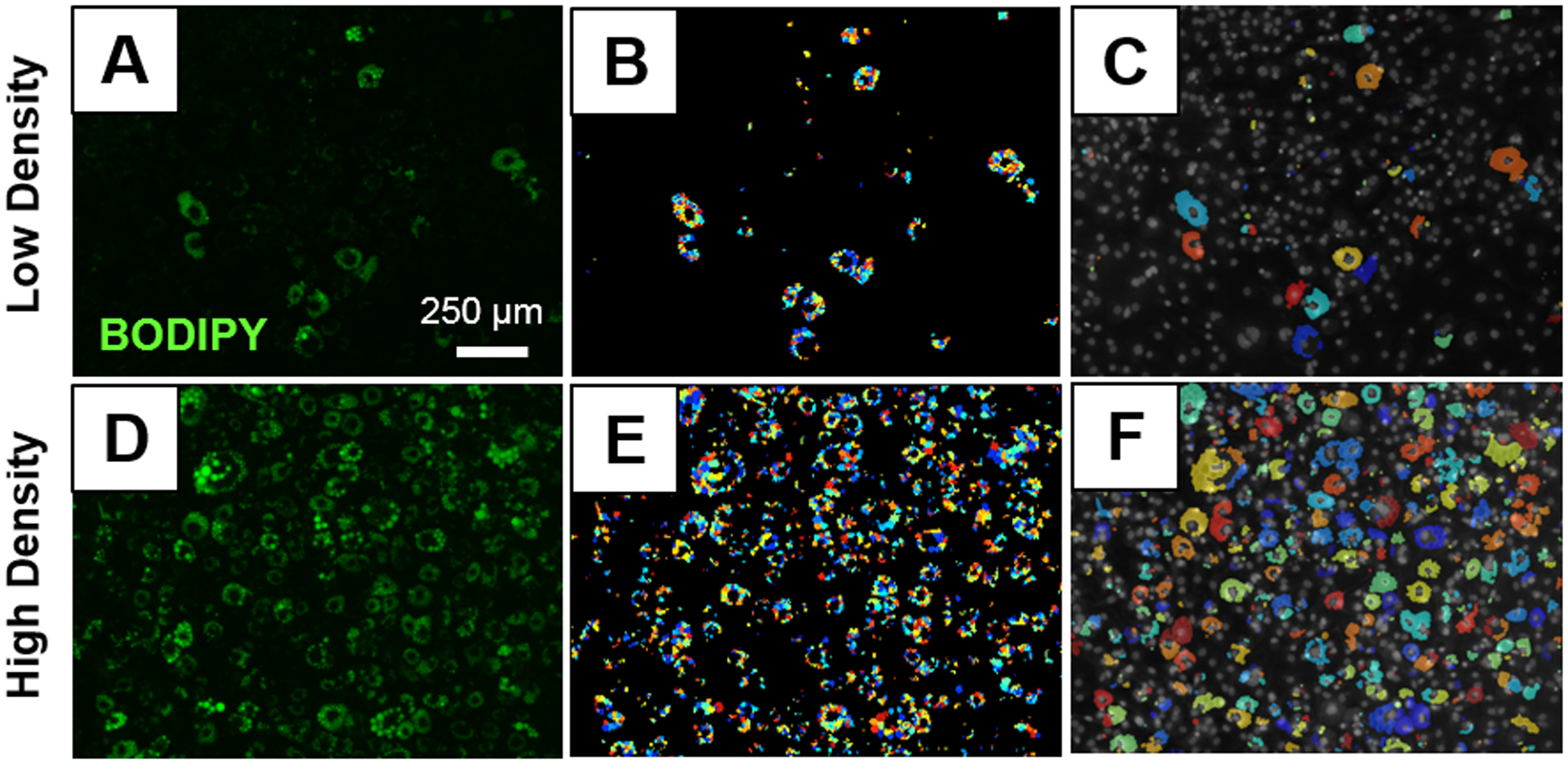
Original BODIPY stained images with low (A) and high (D) lipid content. Individual lipid droplets identified on low (B) and high (E) lipid density images. Clusters of adipocytes identified on low (C) and high (F) density images.
Discussion
In this study we have developed a CellProfiler pipeline to automatically analyze double-stained images with BODIPY (lipid stain) and DAPI (nuclei stain) that allow for 1) the identification of individual lipid droplets and 2) the clustering of lipid droplets associated with a specific adipocyte. Altogether, this novel quantification pipeline allows for a streamlined and accurate identification of lipid droplets that is not possible with the widely used ImageJ program as summarized in Table 1. The CellProfiler clustering analysis of lipid droplets allows for the quantification of lipid content per adipocyte which can provide insight to how individual cells respond to adipogenic stimuli. Importantly, this automated analysis of individual lipid droplets and lipid content per cell can be a useful tool in understanding how cells respond to, for instance, environmental chemicals that have been shown to affect adipogenic differentiation (Helies-Toussaint et al. 2014; Boucher et al. 2016; Pu et al. 2017).
ImageJ is a widely used open-access software for the analysis of images including the quantification of lipid droplet area (Exner et al. 2019; Lv et al. 2019). CellProfiler is another open-access image analysis software that has been utilized for a variety of analyses to identify cellular components and phenotypes including muscle histopathology (Lau et al. 2018), identification of macrophages (Roeper et al. 2017), immunohistochemical analysis (Tollemar et al. 2018), and more recently, 3D images of preimplantational embryos (McQuin et al. 2018). While CellProfiler program has also been used to quantify lipid droplets (Majithia et al. 2014; Kristof et al. 2016; Campos et al. 2018) and adipose spheroid accumulation (Klingelhutz et al. 2018), the development of a CellProfiler pipeline that quantifies lipid droplets has not yet been described. CellProfiler can help streamline the process of analyzing lipid droplets while also yielding accurate data. Because CellProfiler allows for the automated analysis of a set of images, it can greatly reduce the amount of time it would take to analyze a set of images using ImageJ and also help reduce errors when analyzing images individually.
In the CellProfiler pipeline shown in Supplemental Figure 2C, the “IdentifyPrimaryObjects” module identifies lipid droplets according to pixel intensity, average lipid droplet size, and by their shape. When setting the parameters in this module, we observed that slight modifications in the threshold correction factor can affect the number of lipid droplets that are identified in the image (Supplemental Figure 4). This threshold correction factor was also found to be critical in other image analyses, like in the proper identification of muscle fibers (Lau et al. 2018). Overall, the CellProfiler “IdentifyPrimaryObjects” module leads to an accurate identification of lipid droplets because it is able to distinguish between droplets based on pixel intensity which cannot be done with the ImageJ “Analyze Particle” macro.
Specific information regarding the size of individual lipid droplets is becoming increasingly important because lipid droplet size can correlate with adipocyte maturity (Onal et al. 2017), but also with insulin resistance and obesity (Liu et al. 2016; Covington et al. 2017; Nielsen et al. 2017). Ultimately, differences in lipid droplets can provide insights as to how factors (stress, nutritional, environmental, or chemical) may affect adipogenic differentiation. This type of ‘between-droplet’ identification by pixel intensity is not possible with the “Analyze Particles” macro because each image has to first be converted into a binary image (Figure 1C) prior to running the macro, which results in an image that has lost the underlying pixel gradient information. Using ImageJ, the only option to modify the parameters for identifying lipid particles is by their size and circularity, which can range from 0 to ∞ and from 0 to 1, respectively. When kept at those parameters, all the positive lipid area from the binary image is classified as a lipid particle without distinguishing between individual droplets (see Figures 1D). If those parameters were to be arbitrarily modified to be more stringent (for example: size: 0 to 750 pixels and circularity: 0.25 to 1), a smaller percent of positive lipid area will be identified by the “Analyze Particles” macro. In this study, the parameters were kept at the original settings (size: 0 to ∞ and circularity: 0 to 1) so that any positive lipid content was not excluded from the analysis. Other studies have compared the “Analyze Particles” ImageJ macro to CellProfiler when identifying nuclei and found the flexibility of the CellProfiler modules to be more conducive to analyzing larger and more complex images than ImageJ (Roeper et al. 2017). Specifically, the CellProfiler modules “IdentifyPrimaryObjects” and “MeasureObjectSizeShape” that were used to identify the nuclei in human macrophages and mouse vascular smooth muscle cells revealed that, the thresholding from the ImageJ “Analyze Particles” macro can lead to the incorrect separation of small particles from the larger nucleus (Roeper et al. 2017).
When comparing the individual lipid droplets identified by each program, CellProfiler was able to identify 216 % and 191 % more lipid droplets than ImageJ in both, low- and high-lipid density sample images, respectively (Table 2). This is because the “Analyze Particles” macro was unable to distinguish between touching lipid droplets in a binary image, meaning that any touching lipid droplets were classified as one large particle instead of several, smaller particles. These large particles also explain why the maximum size of the lipid particles identified by ImageJ were significantly larger than those identified by CellProfiler in both, high- and low-density sample images (Table 2). This is the main analytical limitation when using the ImageJ software. Because CellProfiler identifies lipid droplets based on their pixel intensity, it can distinguish between touching lipid droplets since the lipid droplet borders have lower pixel intensities. This ability to distinguish touching lipid droplets results in more accurate identification of lipid droplets. Lipid droplets with more consistent sizes are reflected in a smaller range in area, with majority of the lipid droplets being ~100 pixels compared to 1 – 2 pixels when using ImageJ. These small particles identified by ImageJ resulted in a left-skewed lipid droplet size distribution plot (Figure 3G and 3H). This is due to the fact that particles ranging in size from 0 to ∞ were included in the analysis, a parameter that was kept at this setting because an intensity threshold was already applied when creating a binary image in ImageJ.
In this study, we have demonstrated that CellProfiler can accurately measure lipid droplet size and it can provide additional measurements like lipid droplet location, mean and median radius, eccentricity, perimeter, compactness, Euler number, form factor, minor and major axis length, minimum and maximum Feret diameter, and solidity. Even though macro development is possible with the ImageJ software to allow for more complex and specific image analyses than that of the “Analyze Particles” macro, developing new macros requires a deep understanding of complex programming language. The availability of modules to add onto an analysis pipeline makes CellProfiler user-friendly for a broad audience as it does not require programming knowledge (Carpenter et al. 2006). It is important to note that the accuracy of CellProfiler lipid droplet detection using our novel pipeline is not fully accurate, especially for small lipid droplets. This is particularly noticeable when very small lipid droplets are in direct contact with a large lipid droplet (Supplemental Figure 3A and C, arrows) or when two small lipid droplets with similar intensity are close to each other (Supplemental Figure 3G and I, arrows). In our experience, the major limitation for small lipid droplet detection is the resolution of the original image used for analysis. Additionally, CellProfiler frequently updates its algorithms, high-throughput capabilities and user interface to make the software more accurate and user-friendly (Kamentsky et al. 2011). Therefore, future available algorithms may aid in improving small lipid droplet detection.
Because tools that automate single-cell analyses of heterogeneous populations of adipocytes remain at their infancy, lipid accumulation is most often quantified by measuring the amount of staining across an entire population of cells (Regnier et al. 2015; Boucher et al. 2016). Using the CellProfiler program, we were able to develop a pipeline to cluster lipid droplets together within a single cell to identify the lipid content per adipocyte (Supplemental Figure 2D). Altogether, we were able to successfully identify, not only the number of differentiated adipocytes, but also the number of lipid droplets per adipocyte and the total lipid area per cell. Being able to quantify these parameters is a significant advancement over current image analysis methods. This clustering analysis allows for the quantification of single-cell responsiveness across a heterogeneous population and can thus be useful in identifying cells with different responsiveness to adipogenic stimuli or with different adipogenic potential (Sowa et al. 2013; Boschi et al. 2014; Regnier et al. 2015). Additionally, this pipeline can also be used to calculate the fraction of cells in a given population that differentiated into mature adipocytes by comparing the total number of differentiated adipocytes to the total number of nuclei. This novel tool can specifically be used study cellular responses to environmental chemical exposures which have been shown to alter adipogenic differentiation (Helies-Toussaint et al. 2014; Boucher et al. 2016; Pu et al. 2017).
In conclusion, these novel pipelines in CellProfiler allow for the accurate identification and quantification of individual lipid droplets and lipid content per adipocyte. By gaining a greater understanding in how different adipogenic stimuli affects lipid accumulation in individual cells as well as larger populations of adipocytes, we are steps closer to developing strategies to prevent adipose accumulation in humans that can lead to obesity or other metabolic disorders (Ghaben and Scherer 2019).
Supplementary Material
Funding:
Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institute of Health (1K22ES026208 and R01ES027863 to A.V-L.), Michigan State University (MSU) AgBioResearch and the United States Department of Agriculture (USDA) National Institute of Food and Agriculture. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest: The authors report no declarations of interest.
References
- Al-Eryani L, Wahlang B, Falkner KC, Guardiola JJ, Clair HB, Prough RA, Cave M. 2015. Identification of Environmental Chemicals Associated with the Development of Toxicant-associated Fatty Liver Disease in Rodents. Toxicologic pathology. 43(4):482–497. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benador IY, Veliova M, Mahdaviani K, Petcherski A, Wikstrom JD, Assali EA, Acin-Perez R, Shum M, Oliveira MF, Cinti S et al. 2018. Mitochondria Bound to Lipid Droplets Have Unique Bioenergetics, Composition, and Dynamics that Support Lipid Droplet Expansion. Cell Metab. 27(4):869–885 e866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemann R, Fischer B, Navarrete Santos A. 2014. Adipogenic effects of a combination of the endocrine-disrupting compounds bisphenol A, diethylhexylphthalate, and tributyltin. Obesity facts. 7(1):48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom T, Somerharju P, Ikonen E. 2011. Synthesis and biosynthetic trafficking of membrane lipids. Cold Spring Harb Perspect Biol. 3(8):a004713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschi F, Rizzatti V, Zamboni M, Sbarbati A. 2014. Lipid droplets fusion in adipocyte differentiated 3T3-L1 cells: a Monte Carlo simulation. Exp Cell Res. 321(2):201–208. [DOI] [PubMed] [Google Scholar]
- Boucher JG, Ahmed S, Atlas E. 2016. Bisphenol S Induces Adipogenesis in Primary Human Preadipocytes From Female Donors. Endocrinology. 157(4):1397–1407. [DOI] [PubMed] [Google Scholar]
- Campos V, Rappaz B, Kuttler F, Turcatti G, Naveiras O. 2018. High-throughput, nonperturbing quantification of lipid droplets with digital holographic microscopy. J Lipid Res. 59(7):1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7(10):R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanderbhan R, Noland BJ, Scallen TJ, Vahouny GV. 1982. Sterol carrier protein2. Delivery of cholesterol from adrenal lipid droplets to mitochondria for pregnenolone synthesis. J Biol Chem. 257(15):8928–8934. [PubMed] [Google Scholar]
- Covington JD, Johannsen DL, Coen PM, Burk DH, Obanda DN, Ebenezer PJ, Tam CS, Goodpaster BH, Ravussin E, Bajpeyi S. 2017. Intramyocellular Lipid Droplet Size Rather Than Total Lipid Content is Related to Insulin Sensitivity After 8 Weeks of Overfeeding. Obesity (Silver Spring). 25(12):2079–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhaimer P 2019. Lipid Droplets in Disease. Cells. 8(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallio M, Diano N, Masarone M, Gravina AG, Patane V, Romeo M, Di Sarno R, Errico S, Nicolucci C, Abenavoli L et al. 2019. Chemical Effect of Bisphenol A on Non-Alcoholic Fatty Liver Disease. Int J Env Res Pub He. 16(17). English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deierlein AL, Rock S, Park S. 2017. Persistent Endocrine-Disrupting Chemicals and Fatty Liver Disease. Curr Environ Health Rep. 4(4):439–449. [DOI] [PubMed] [Google Scholar]
- Diep DTV, Hong K, Khun T, Zheng M, Ul-Haq A, Jun HS, Kim YB, Chun KH. 2018. Anti-adipogenic effects of KD025 (SLx-2119), a ROCK2-specific inhibitor, in 3T3-L1 cells. Sci Rep. 8(1):2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner T, Beretta CA, Gao Q, Afting C, Romero-Brey I, Bartenschlager R, Fehring L, Poppelreuther M, Fullekrug J. 2019. Lipid droplet quantification based on iterative image processing. J Lipid Res. 60(7):1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fam TK, Klymchenko AS, Collot M. 2018. Recent Advances in Fluorescent Probes for Lipid Droplets. Materials (Basel). 11(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto S, Fujimoto T. 2002. Deformation of lipid droplets in fixed samples. Histochem Cell Biol. 118(5):423–428. [DOI] [PubMed] [Google Scholar]
- Ghaben AL, Scherer PE. 2019. Adipogenesis and metabolic health. Nat Rev Mol Cell Biol. 20(4):242–258. [DOI] [PubMed] [Google Scholar]
- Heindel JJ, Blumberg B, Cave M, Machtinger R, Mantovani A, Mendez MA, Nadal A, Palanza P, Panzica G, Sargis R et al. 2017. Metabolism disrupting chemicals and metabolic disorders. Reproductive toxicology. 68:3–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helies-Toussaint C, Peyre L, Costanzo C, Chagnon MC, Rahmani R. 2014. Is bisphenol S a safe substitute for bisphenol A in terms of metabolic function? An in vitro study. Toxicol Appl Pharmacol. 280(2):224–235. [DOI] [PubMed] [Google Scholar]
- Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. 2011. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 27(8):1179–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingelhutz AJ, Gourronc FA, Chaly A, Wadkins DA, Burand AJ, Markan KR, Idiga SO, Wu M, Potthoff MJ, Ankrum JA. 2018. Scaffold-free generation of uniform adipose spheroids for metabolism research and drug discovery. Sci Rep. 8(1):523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymchenko AS. 2017. Solvatochromic and Fluorogenic Dyes as Environment-Sensitive Probes: Design and Biological Applications. Acc Chem Res. 50(2):366–375. [DOI] [PubMed] [Google Scholar]
- Kristof E, Doan-Xuan QM, Sarvari AK, Klusoczki A, Fischer-Posovszky P, Wabitsch M, Bacso Z, Bai P, Balajthy Z, Fesus L. 2016. Clozapine modifies the differentiation program of human adipocytes inducing browning. Transl Psychiatry. 6(11):e963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau YS, Xu L, Gao Y, Han R. 2018. Automated muscle histopathology analysis using CellProfiler. Skelet Muscle. 8(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Schmidt H, Lai B, Ge K. 2019. Transcriptional and Epigenomic Regulation of Adipogenesis. Mol Cell Biol. 39(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YM, Jacobs DR, Lee DH. 2018. Persistent Organic Pollutants and Type 2 Diabetes: A Critical Review of Review Articles. Front Endocrinol. 9 English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Takahashi Y, Desai N, Zhang J, Serfass JM, Shi YG, Lynch CJ, Wang HG. 2016. Bif-1 deficiency impairs lipid homeostasis and causes obesity accompanied by insulin resistance. Sci Rep. 6:20453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X, Liu J, Qin Y, Liu Y, Jin M, Dai J, Chua BT, Yang H, Li P. 2019. Identification of gene products that control lipid droplet size in yeast using a high-throughput quantitative image analysis. Biochim Biophys Acta Mol Cell Biol Lipids. 1864(2):113–127. [DOI] [PubMed] [Google Scholar]
- Majithia AR, Flannick J, Shahinian P, Guo M, Bray MA, Fontanillas P, Gabriel SB, Go TDC, Project NJFAS, Consortium STD et al. 2014. Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes. Proc Natl Acad Sci U S A. 111(36):13127–13132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh AL, Storey SM, Atshaves BP. 2010. Intracellular lipid droplets contain dynamic pools of sphingomyelin: ADRP binds phospholipids with high affinity. Lipids. 45(6):465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D et al. 2018. CellProfiler 3.0: Next-generation image processing for biology. PLoS biology. 16(7):e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Christensen AE, Nellemann B, Christensen B. 2017. Lipid droplet size and location in human skeletal muscle fibers are associated with insulin sensitivity. Am J Physiol Endocrinol Metab. 313(6):E721–E730. [DOI] [PubMed] [Google Scholar]
- Onal G, Kutlu O, Gozuacik D, Dokmeci Emre S. 2017. Lipid Droplets in Health and Disease. Lipids Health Dis. 16(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YK, Obiang-Obounou BW, Lee J, Lee TY, Bae MA, Hwang KS, Lee KB, Choi JS, Jang BC. 2017. Anti-Adipogenic Effects on 3T3-L1 Cells and Zebrafish by Tanshinone IIA. Int J Mol Sci. 18(10). English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Echague V, Lodh S, Colman L, Bobba N, Santos L, Katsanis N, Escande C, Zaghloul NA, Badano JL. 2017. BBS4 regulates the expression and secretion of FSTL1, a protein that participates in ciliogenesis and the differentiation of 3T3-L1. Sci Rep-Uk. 7 English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y, Gingrich J, Steibel JP, Veiga-Lopez A. 2017. Sex-specific modulation of fetal adipogenesis by gestational bisphenol A and bisphenol S exposure. Endocrinology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y, Veiga-Lopez A. 2017. PPARgamma agonist through the terminal differentiation phase is essential for adipogenic differentiation of fetal ovine preadipocytes. Cell Mol Biol Lett. 22:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier SM, El-Hashani E, Kamau W, Zhang X, Massad NL, Sargis RM. 2015. Tributyltin differentially promotes development of a phenotypically distinct adipocyte. Obesity (Silver Spring). 23(9):1864–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeper M, Braun-Dullaeus RC, Weinert S. 2017. Semiautomatic High-Content Analysis of Complex Images from Cocultures of Vascular Smooth Muscle Cells and Macrophages: A CellProfiler Showcase. SLAS Discov. 22(7):837–847. [DOI] [PubMed] [Google Scholar]
- Schmidt JS, Schaedlich K, Fiandanese N, Pocar P, Fischer B. 2012. Effects of di(2-ethylhexyl) phthalate (DEHP) on female fertility and adipogenesis in C3H/N mice. Environ Health Perspect. 120(8):1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa Y, Imura T, Numajiri T, Takeda K, Mabuchi Y, Matsuzaki Y, Nishino K. 2013. Adipose stromal cells contain phenotypically distinct adipogenic progenitors derived from neural crest. PLoS One. 8(12):e84206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollemar V, Tudzarovski N, Boberg E, Tornqvist Andren A, Al-Adili A, Le Blanc K, Garming Legert K, Bottai M, Warfvinge G, Sugars RV. 2018. Quantitative chromogenic immunohistochemical image analysis in cellprofiler software. Cytometry A. 93(10):1051–1059. [DOI] [PubMed] [Google Scholar]
- Zhao X, Hu H, Wang C, Bai L, Wang Y, Wang W, Wang J. 2019. A comparison of methods for effective differentiation of the frozen-thawed 3T3-L1 cells. Anal Biochem. 568:57–64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


