Abstract
Long noncoding RNAs (lncRNAs), genomic “dark matter,” are deeply involved in diverse biological processes. The lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) is a highly participatory lncRNA; however, its roles in gastric cancer (GC) remain largely unexplored. Here, we demonstrated that the expression of NEAT1 was significantly increased and negatively correlated with prognosis in GC. Subsequent experiments confirmed that KLF5 can induce NEAT1 expression by binding to the NEAT1 promoter region. Further experiments revealed that NEAT1 silencing significantly suppressed cell proliferation both in vitro and in vivo and induced apoptosis. We used mRNA sequencing (mRNA-seq) to identify the preferentially affected genes linked to cell proliferation in cells with NEAT1 knockdown. Mechanistically, NEAT1 bound BRG1 (SMARCA4) directly, modulating H3K27me3 and H3K4me3 in the GADD45A promoter to regulate GADD45A-dependent G2/M cell cycle progression. In addition, BRG1 was significantly upregulated and correlated with outcomes in GC; moreover, it promoted cell proliferation both in vitro and in vivo. Taken together, our data support the importance of NEAT1 in promoting GC tumorigenesis and indicate that NEAT1 might be a diagnostic and therapeutic target in GC.
Keywords: NEAT1, tumorigenesis, prognosis, gastric cancer, epigenetics
Graphical Abstract

Ma and colleagues demonstrated the functions and mechanism of lncRNA NEAT1 in gastric cancer tumorigenesis. NEAT1, which was induced by KLF5, promoted cell proliferation and inhibited apoptosis. Mechanistically, NEAT1 bound BRG1 directly, modulating histone methylation modification to regulate GADD45A-dependent G2/M cell cycle progression.
Introduction
Gastric cancer (GC) is one of the most common gastrointestinal tract cancers. It is a leading cause of cancer-related mortality worldwide and resulted in 782,685 deaths in 2018.1 Because of the lack of specific symptoms, most patients present at the time of diagnosis with lymph node and distant metastasis, resulting in poor prognosis.2,3 Therefore, the identification of unambiguous molecular signatures that may serve as sensitive diagnostic biomarkers and therapeutic targets is essential.
Early studies on cancer hallmarks in GC focused on the protein-coding genome dramatically expanded the understanding of cancer-related molecular mechanisms.4 However, breakthroughs in high-throughput microarray and next-generation sequencing technologies have revealed a diverse cast of noncoding genomic regions. Messenger RNAs (mRNAs) that code for proteins account for < 2% of all sequences, and noncoding RNAs are proposed to drive important cancer phenotypes.5 Among the divergent groups of non-protein-coding sequences are long noncoding RNAs (lncRNAs), which are defined as sequences with a length of > 200 and act through interactions with DNA, RNA, or proteins. lncRNAs can serve as signals in cis, participating in regulating nearby gene expression and chromatin architecture, or can be involved in distant molecular regulation in trans.6 Many studies have shown that aberrantly expressed lncRNAs may act as oncogenes or tumor suppressors in every major cancer type. Our previous studies showed that a variety of lncRNAs are involved in the development of GC.7, 8, 9, 10, 11, 12 For example, KLF5 and MYC–LINC00346–miR-34a-5p crosstalk promotes the proliferation and suppresses the apoptosis of GC cells,7 and components of the LINC01234–miR-204-5P–CBFB axis cooperate in mediating GC tumorigenesis.8
The lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) is encoded by the NEAT1 gene, which is located on chromosome 11q13.1 and has two isoforms: NEAT1_1 (3.7 kb) and NEAT1_2 (23 kb).13 Both isoforms localize to paraspeckles and are an essential component of subnuclear bodies. Upregulated expression of NEAT1 and the oncogenic properties of this lncRNA have been reported in various types of solid tumors.14 In breast cancer, NEAT1 can function as a competing endogenous RNA (ceRNA) and contribute to tumor progression. Octamer-binding transcription factor 4 (Oct4) upregulates NEAT1 expression by binding its promoter region, resulting in tumor progression in non-small cell lung cancer (NSCLC). The Oct4–NEAT1–MALAT1 axis is an independent biomarker for unfavorable prognosis in NSCLC patients.15 Furthermore, NEAT1 is directly targeted by the tumor suppressor p53.16 Thus, NEAT1 could serve as a prognostic biomarker and target for therapeutic interventions. However, the role and molecular mechanisms of NEAT1 in GC are not well known.
In this study, we found that the lncRNA NEAT1, which exhibits aberrant expression in GC tissues, is associated with GC progression and poor prognosis in GC patients. Knockdown of NEAT1 inhibited GC cell proliferation both in vitro and in vivo. KLF5 was found to bind to the promoter region of NEAT1 and modulate its expression. Importantly, NEAT1 negatively regulated growth arrest and DNA damage-inducible 45 alpha (GADD45A), a tumor suppressor protein. Mechanistically, NEAT1 acted as a scaffold for BRG1, recruiting it to install H3K27me3 and H3K4me3 in the promoter region of GADD45A. Taken together, our data indicate that NEAT1 is a promising therapeutic biomarker in GC.
Results
NEAT1 Was Upregulated in GC and Associated with Poor Prognosis
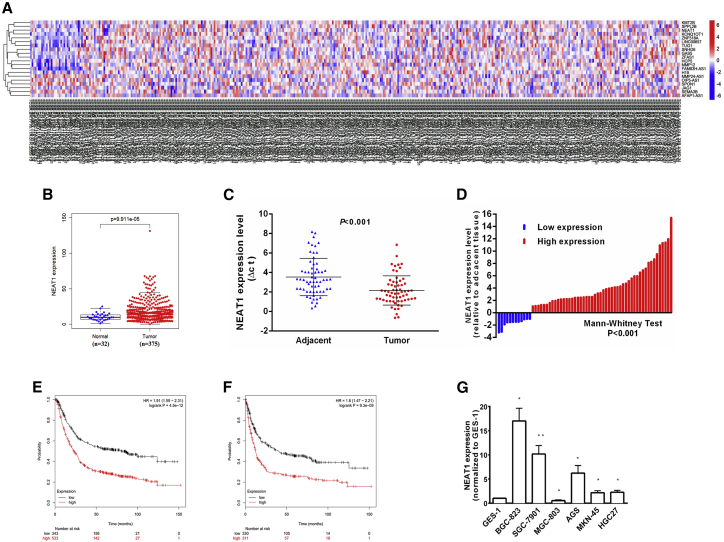
NEAT1 has two isoforms with similar transcription start sites. We selected NEAT1_1 (3.7 kb) for further study. We analyzed the expression of the lncRNA NEAT1 in 32 normal tissues and 375 tumor tissues of patients with GC in The Cancer Genome Atlas (TCGA). NEAT1 expression was obviously upregulated in tumor tissues compared to normal tissues (Figures 1A and B). We also assessed NEAT1 expression in 62 GC cancer tissues and paired adjacent normal tissues and found that NEAT1 expression was significantly higher in these cancer tissues than in the paired adjacent normal tissues (p < 0.001) (Figure 1C). With a 2-fold change in the expression level as the cutoff, 50 of the 62 GC cancer samples showed upregulation of NEAT1, and 12 showed downregulation relative to normal tissues (Figure 1D).
Figure 1.
NEAT1 Expression Is Upregulated in Gastric Cancer Tissues and Associated with Poor Prognosis
(A, B) Relative expression of NEAT1 in gastric cancer tissues compared with normal tissue was analyzed using TCGA data. (C) NEAT1 expression was analyzed by qRT-PCR in gastric cancer tissues and corresponding adjacent nontumor tissues, and the data were presented as the ΔCt value (n = 62). (D) Data are represented as fold change (tumor/normal). (E, F) Kaplan-Meier analysis of OS and FP (first time to progression) according to according to NEAT1 expression levels. (G) qRT-PCR analysis of NEAT1 expression in the normal gastric epithelium cell line (GES-1) and gastric cancer cells. Error bars, mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
To validate the clinical significance of NEAT1, we explored the correlations between NEAT1 expression and clinical features. As shown in Table 1, NEAT1 expression was correlated with lymph node metastasis (p = 0.046), regional lymph node metastasis (p = 0.017), and TNM stage (p = 0.005). Online Kaplan-Meier analysis (http://kmplot.com/) showed that in GC patients, higher levels of NEAT1 correlated with a shorter overall survival (OS) time and first time to progression (FP) (Figures 1E and F). The median OS time was 23.2 months in the high-expression cohort and 80.7 months in the low-expression cohort, while the median FPs were 12.2 months and 40.2 months, respectively. These results suggested that NEAT1 might play an oncogenic role in GC. NEAT1 expression was assessed in GC cell lines and normal gastric mucosal epithelial cells. NEAT1 was significantly overexpressed in BGC-823, SGC-7901, AGS, MKN-45, and HGC27 cells and underexpressed in MGC-803 cells (Figure 1G). BGC-823 and SGC-7901 exhibited the highest expression levels and were selected for studies to investigate the biological functions of NEAT1. Taken together, these results indicated that NEAT1 is upregulated in GC cell lines and GC tissues and is a potential prognostic biomarker in GC.
Table 1.
Correlation between NEAT1 Expression and Clinicopathological Features of GC (n = 62)
| Clinical Parameter | NEAT1 |
Chi-Square Test p value | |
|---|---|---|---|
| Low-Expression Cases (n = 12) | High-Expression Cases (n = 50) | ||
| Age, years | 0.092 | ||
| <50 | 9 | 24 | |
| >50 | 3 | 26 | |
| Gender | 0.767 | ||
| Male | 8 | 38 | |
| Female | 4 | 12 | |
| Location | 0.410 | ||
| Distal | 5 | 27 | |
| Middle | 6 | 15 | |
| Proximal | 1 | 8 | |
| Size | 0.049a | ||
| >5 cm | 5 | 38 | |
| <5 cm | 7 | 12 | |
| Histologic differentiation | 0.991 | ||
| Well | 1 | 3 | |
| Moderately | 3 | 14 | |
| Poorly | 7 | 29 | |
| Undifferentiated | 1 | 4 | |
| Invasion depth | 0.137 | ||
| T1 | 3 | 7 | |
| T2 | 6 | 12 | |
| T3 | 2 | 19 | |
| T4 | 1 | 12 | |
| TNM stages | 0.005a | ||
| I | 6 | 5 | |
| II | 4 | 13 | |
| III | 2 | 27 | |
| IV | 0 | 5 | |
| Lymphatic metastasis | 0.046a | ||
| Yes | 4 | 34 | |
| No | 8 | 16 | |
| Regional lymph nodes | 0.017a | ||
| pN0 | 7 | 15 | |
| pN1 | 4 | 6 | |
| pN2 | 1 | 11 | |
| pN3 | 0 | 18 | |
| Distant metastasis | 0.573 | ||
| Yes | 0 | 5 | |
| No | 12 | 45 | |
Indicates p < 0.05.
KLF5 Regulated the Expression of NEAT1
To explore the mechanism of NEAT1 overexpression in GC, we used the online database JASPAR (http://jaspar.genereg.net/) to predict the potential binding sites in the promoter region of NEAT1. The transcription factor (TF) KLF5 was found to bind to the NEAT1 promoter region. Data indicate that amplification of KLF5, GATA4, and GATA6 in GC appears to be mutually exclusive. KLF5/GATA4/GATA6 may regulate the transcription of target genes and affect tumor cell biological behavior.17 The results of the TCGA database analysis showed that KLF5 expression was positively correlated with that of NEAT1 (Figure 2A). We knocked down KLF5 expression in BGC-823 and SGC-7901 cells with small interfering RNA (siRNA) targeting KLF5 (si-KLF5). The results of both qRT-PCR and western blotting showed that the expression levels of KLF5 and NEAT1 were significantly decreased (Figures S1A and S1C; Figure 2B). By contrast, transfection of the pcDNA-KLF5 overexpression vector increased NEAT1 expression in GC cell lines (Figure 2C; Figures S1B and S1C). The chromatin immunoprecipitation (ChIP) assay results confirmed that KLF5 can bind to the NEAT1 promoter region; its binding capacity was approximately 42 times that of negative control immunoglobulin G (IgG). The binding capacity of KLF5 decreased significantly, to only approximately 1/3 of that in the control group, after KLF5 was silenced (Figure 2D). The bioinformatics tool (JASPAR) predicted five KLF5 binding sites within the NEAT1 promoter, and these sites were confirmed using a luciferase reporter assay (Figures 2E and 2F). Hence, we concluded that KLF5 binds to the promoter of NEAT1 and activates its transcription.
Figure 2.
KLF5, and NEAT1 Are Amplified in GC Tissues, and KLF5 Upregulates NEAT1 Expression in GC Cells
(A) Relationship between NEAT1 expression and KLF5 mRNA levels in TCGA dataset. (B, C) NEAT1 mRNA expression in cells transfected with KLF siRNAs or overexpression vector. Error bars, mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (D) ChIP assay demonstrated endogenous KLF5 binding to the NEAT1 gene promoter. (E) Dual-luciferase reporter assay was performed by cotransfection of the NEAT1 promoter fragment (pGL3-NEAT1-F) or NEAT1 promoter constructs with deletions in different putative binding elements for KLF5. (F) The observed KLF5-mediated expression of luciferase activity was abolished by mutation of NEAT1-MT mutation type.
NEAT1 Promoted GC Cell Proliferation
To determine the biological function of NEAT1 in GC, we used antisense oligonucleotides (ASOs) and small hairpin RNA (shRNA) plasmids to silence NEAT1 and pcDNA3.1 plasmids to overexpress NEAT1. ASO- and shRNA plasmid-mediated knockdown and pcDNA3.1 plasmid-mediated overexpression were confirmed in both BGC-823 and SGC-7901 cells (Figures S1D–S1F). Knockdown of NEAT1 expression significantly inhibited GC cell proliferation compared with that of control cells, as demonstrated by Cell Counting Kit-8 (CCK8) proliferation assays (Figures 3A and 3B). By contrast, overexpression of NEAT1 promoted GC cell growth. Similar results were found in colony formation assays (Figures 3C and 3D). To explore the role of NEAT1 in vivo, BGC-823 cells stably transfected with sh-NEAT1 or control vector were inoculated into nude mice (7 mice per experimental group). Eighteen days after subcutaneous injection, the tumors formed in the sh-NEAT1 group were substantially smaller and had a lower average weight than those in the empty vector group (Figures 3E–3G). These results indicated that NEAT1 may mediate the tumorigenic behavior of GC cells in vivo and in vitro.
Figure 3.
Effects of NEAT1 on Gastric Cancer Cell Proliferation In Vitro
(A, B) CCK8 assays were used to determine the viability of NEAT1 ASO#1 and ASO#2 transfected or pcDNA-NEAT1 transfected gastric cancer cells. Error bars, mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (C, D) Colony formation assays were performed to determine the proliferation. Error bars, mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (E–G) The stable NEAT1 knockdown gastric cancer cells were used for the in vivo assays. The tumors from two groups of nude mice were shown and tumor growth curves were measured and shown after the injection of cells. The tumor volume was calculated every 3 days. Tumor weights from two groups are represented. The data represent the mean ± SD from three independent experiments. Error bars, mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001..
GADD45A Was the Target of NEAT1
To investigate whether NEAT1 promotes GC cell proliferation by altering the cell cycle, we performed flow cytometric analysis to evaluate the effect of NEAT1 downregulation. Compared with control cells, BGC-823 and SGC-7901 cells transfected with NEAT1 ASOs exhibited dramatically increased apoptosis. The results also revealed that NEAT1 knockdown significantly increased the proportion of cells in G2/M phase, with a lower proportion in S phase, indicating that the regulation of DNA damage repair might be affected by NEAT1 silencing, thus promoting apoptosis and suppressing proliferation (Figures 4A and 4B). By contrast, NEAT1 overexpression increased the number of cells in S phase (Figure 4C). All multicellular organisms have mechanisms of host defense against genotoxic stress, and these mechanisms involve cell division, differentiation, and death. Abnormalities in cell cycle progression or apoptosis can cause an imbalance in cell proliferation and death, resulting in tumorigenesis.18 Overall progression of the cell cycle is controlled by two checkpoints at the G1/S and G2/M boundaries, which are the checkpoints for DNA replication and mitotic cell division, respectively.19 DNA damage may block cell cycle progression through these boundaries and trigger apoptosis.20,21 Upon DNA damage, the formation of DNA double-strand breaks (DSBs) is the characteristic for cancer cells. DNA-DSBs are always followed by the phosphorylation of H2AX histone, and the new phosphorylated protein is called γ-H2AX, which starts the DNA repair process.22,23 Thus, γ-H2AX is a key marker for DNA-DSBs. We performed immunofluorescence assays to investigate γ-H2AX immunoreactivity in GC cells treated with NEAT1 knockdown. The number of γ-H2AX immunoreactivity cells was significantly higher in both BGC-823 and SGC-7901 cells treated with NEAT1 knockdown than the contrast cells (Figure 4D).
Figure 4.
Effect of NEAT1 on Gastric Cancer Cell Apoptosis and Cell Cycle In Vitro and In Vivo
(A) Flow cytometry was used to detect the apoptotic rates of cells. Error bars, mean ± SD. NS, no significance; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (B) Flow cytometry showing significant decreases or increases in the proportion of cells in S or G2/M phase, respectively, when NEAT1 was silenced in SGC-7901 and BGC-823 cells. Error bars, mean ± SD. NS, no significance; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (C) Flow cytometry showing significant increases or decreases in the proportion of cells in S or G2/M phase, respectively, when NEAT1 was overexpressed in SGC-7901 and BGC-823 cells. Error bars, mean ± SD. NS, no significance; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (D) Immunofluorescence staining results showing the level of γ-H2AX in BGC-823 and SGC-7901 cells with or without NEAT1 knockdown. Green: γ-H2AX staining, blue: DAPI staining. (E) Heat map of altered genes in silenced cells compared with control cells. (F) Changes in protein levels of GADD45A by western blot in cells with NEAT1. (G) GADD45A protein levels in tumor tissues from sh-NEAT1 or negative control gastric cancer cells were determined by IHC.
To gain insights into the molecular mechanism by which the lncRNA NEAT1 promotes the proliferation of GC cells, we assessed the mRNA expression profiles of BGC-823 cells after knockdown of NEAT1. A set of 274 mRNAs showed a ≥ 2-fold increase in expression in NEAT1-depleted cells; NEAT1 knockdown also reduced the abundance (by ≥ 2-fold) of 557 genes (Figure 4E). Gene ontology (GO) analysis showed that these genes were significantly associated with the mitogen-activated protein kinase (MAPK) signaling pathway, supporting the role of NEAT1 in cell proliferation and the cell cycle (Figure S2A). Differentially expressed genes related to the MAPK signaling pathway, including KLF4, GADD45A, GDF15, IER3, IRAK2, AIFM2, FGF21, FGFR2, and PDGFB, were selected and confirmed by qRT-PCR (Figure S2B). GADD45A is associated with inhibiting cell cycle progression through the G2/M checkpoint and repairing DNA damage.24 GADD45A levels were significantly decreased in NEAT1-overexpressing cells, as determined by qRT-PCR and western blotting. NEAT1 knockdown increased the GADD45A protein expression level (Figure S2C; Figure 4F). Consistent with these findings, immunohistochemical staining analysis of samples from the NEAT1-depleted xenograft mouse model indicated that Ki-67 expression was decreased and GADD45A expression was increased (Figure 4G). These results suggested that NEAT1 may promote GC tumorigenesis by inhibiting GADD45A.
NEAT1 Directly Bound to BRG1 and Recruited it to Epigenetically Silence GADD45A
Next, we explored the mechanism by which lncRNA NEAT1 impairs GADD45A expression. Accumulating studies have indicated that the subcellular distribution of lncRNAs determines their function. Fluorescence in situ hybridization (FISH) and nuclear-cytoplasmic fractionation assays showed that NEAT1 is a nuclear-enriched lncRNA (Figure 5A; Figure S3A). Thus, NEAT1 may interact with nuclear molecules or proteins, controlling transcriptional activity. We found by using the RNA-Protein Interaction Prediction database (RPISeq, http://pridb.gdcb.iastate.edu/RPISeq) that the lncRNA NEAT1 might interact with BRG1 and EZH2. The results of RNA immunoprecipitation (RIP) assays showed that NEAT1 can bind to BRG1 but not to EZH2 (Figures 5B and 5C). The physical interaction of NEAT1 and BRG1 was validated by western blot analysis using the proteins retrieved from RNA pulldown assays (Figure 5D). We also found that NEAT1 cannot influence the expression of BRG1 (Figure S3B), indicating that NEAT1 could contribute to GC proliferation via the synergistic effect of BRG1. BRG1 is an active subunit of the SWI/SNF chromatin remodeling complex. SWI/SNF mediates chromatin remodeling and promotes transcription. Analysis of TCGA datasets indicated that SWI/SNF genes are frequently mutated and are intriguing contributors to cancer. BRG1 can function as a transactivator and bind to the promoter of epithelial-mesenchymal transition (EMT)-related genes, resulting in tumor progression and GC cell metastasis.25 Silencing BRG1 increased but overexpressing BRG1 decreased the GADD45A protein level (Figure 5E; Figures S3C and S3D). The results of ChIP assays confirmed that knockout of either NEAT1 or BRG1 decreased the binding capacity of BRG1 in the promoter region of GADD45A. Considering that histone modifications play a vital role in BRG1-mediated control of gene expression, we investigated whether either NEAT1 or BRG1 modulates the level of H3K27AC, H3K4me3, and H3K27me3. As shown in Figure 5G, silencing either NEAT1 or BRG1 increased the H3K4me3 level and decreased the H3K27me3 level. We hypothesized that NEAT1 may inhibit GADD45A expression by recruiting BRG1 to install H3K27me3 and H3K4me3 in the promoter region of GADD45A. The results of ChIP assays showed that knockdown of either NEAT1 or BRG1 reduced the enrichment of H3K27me3 and increased that of H3K4me3 in the promoter region of GADD45A (Figures 5H and 5I). These results illustrated that NEAT1 can epigenetically decrease the expression of GADD45A by directly interacting with BRG1 and recruiting it to the promoter region of GADD45A in GC.
Figure 5.
Regulation Relationship between NEAT1 and GADD45A
(A) FISH analysis of the location of NEAT1 (green) in nuclear fractions (blue) of SGC-7901 and BGC-823 cells. (B) RNA-Protein Interaction Prediction (RPISeq) showed high interaction probabilities of BRG1, EZH2 and NEAT1. (C) RIP experiments were performed in BGC-823 cells, and the coprecipitated RNA was subjected to qRT-PCR for NEAT1. The fold enrichment of NEAT1 is relative to its matching IgG control. (D) RNA pull-down proved BRG1 combined with NEAT1. (E) GADD45A expression in SGC-7901 and BGC-823 cells transfected with control siRNA, BRG1 siRNA, or pcDNA-BRG1. (F) ChIP-qPCR of GADD45A promoter region after knockdown BRG1 or NEAT1 in BGC-823 and SGC-7901 cells. Antibody enrichment was quantified relative to the amount of input DNA. Antibody directed against IgG was used as a negative control. ∗p < 0.05; ∗∗p < 0.01. (G) H3K27AC, H3K4me3, H3K27me3 protein level in BGC-823 cells following knockdown of NEAT1 or inhibition of BRG1. (H, I) ChIP-qPCR of H3K4me3 and H3K27me3 of the promoter region of GADD45A genes locus after knockdown of NEAT1 or siRNA treatment targeting BRG1. Antibody enrichment was quantified relative to the amount of input DNA. Antibody directed against IgG was used as a negative control. Error bars, mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
BRG1 Promoted GC Cell Proliferation
According to TCGA database analysis, the expression levels of BRG1 are significantly increased in GC tissues compared with normal mucosa (Figures 6A and 6B). qRT-PCR analysis was performed to investigate BRG1 expression in 62 pairs of GC tumor tissues and adjacent normal tissues (Figure S4A). We divided the patients into two groups based on the BRG1 expression level (fold change ≥ 2) and found that BRG1 was overexpressed in 50 of the 62 patients (Figure 6C). We explored the correlation between the BRG1 expression level and the clinicopathological features of patients with GC. In general, BRG1 downregulation was associated with a lower TNM stage, a smaller tumor size, and less extensive regional lymph node metastasis (Table 2). Furthermore, BRG1 overexpression was associated with poor survival, as determined by using an online Kaplan-Meier analysis tool (http://kmplot.com/) (Figures 6D and 6E). The median OS time was 24.6 months in the high-BRG1-expression cohort and 65 months in the low-BRG1-expression cohort, while the corresponding median FPs were 13.1 months and 34.67 months. We performed gain- and loss-of-function experiments to study the biological function of BRG1. We analyzed the expression of BRG1 in GC cell lines and found that BGC-823 and SGC-7901 cells exhibited higher expression levels than GES-1 cells (Figure S4B). Thus, we selected BGC-823 and SGC-7901 cells for further investigation. GC cells were transfected with siRNAs or shRNAs to silence BRG1 or with the BRG1-pcDNA3.1 vector to overexpress BRG1, and the knockdown and overexpression efficiencies were evaluated by qRT-PCR (Figures S4C–S4E). Knockdown of BRG1 expression significantly suppressed cancer cell proliferation and colony formation, whereas BRG1 overexpression had the opposite effects (Figures 6F and 6G). To further demonstrate the role of BRG1 in vivo, xenograft models were established with BRG1 wild-type (WT) or BRG1 knockdown cells. Compared with the tumors from control mice, the tumors from mice injected with sh-BRG1-transfected cells were obviously smaller and lighter (Figure 6H; Figure S4F). Moreover, the results of immunohistochemistry (IHC) indicated that when BRG1 was depleted, Ki-67 staining was significantly reduced, while GADD45A expression was increased. Given that the BRG1-mediated sustained inactivation of GADD45A was vital to NEAT1 expression in GC, a series of rescue experiments was performed. The CCK8 assay results indicated that silencing either NEAT1 or BRG1 inhibited the proliferation of GC cells and that knockdown of GADD45A could partially alleviate this inhibition (Figures 7A and 7B). The results of colony formation assays demonstrated that knockdown of either NEAT1 or BRG1 reduced cell colony formation, while suppression of GADD45A expression rescued this ability (Figures 7C and 7D).
Figure 6.
Effects of BRG1 on Gastric Cancer Cell Proliferation In Vitro and In Vivo
(A, B) Relative expression of BRG1 in gastric cancer tissues compared with normal tissue was analyzed using TCGA data. (C) Data are represented as fold change (tumor/normal). (D, E) Kaplan-Meier analysis of OS and FP (first time to progression) according to according to BRG1 expression levels. (F) CCK8 assays were used to determine the viability of si-BRG1 or pcDNA-BRG1 transfected gastric cancer cells. (G) Colony formation assays were performed to determine the proliferation. Error bars, mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.(H) The stable BRG1 knockdown gastric cancer cells were used for the in vivo assays. The tumors from two groups of nude mice were shown. (I) Ki-67 and GADD45A protein levels in tumor tissues from sh-BRG1 or negative control gastric cancer cells were determined by IHC.
Table 2.
Correlation between BRG1 Expression and Clinicopathological Features of GC (n = 62)
| Clinical Parameter | BRG1 |
Chi-Square Test p value | |
|---|---|---|---|
| Low-Expression Cases (n = 12) | High-Expression Cases (n = 50) | ||
| Age, years | 0.693 | ||
| <50 | 7 | 26 | |
| >50 | 5 | 24 | |
| Gender | 0.943 | ||
| Male | 9 | 37 | |
| Female | 3 | 13 | |
| Location | 0.403 | ||
| Distal | 5 | 27 | |
| Middle | 6 | 15 | |
| Proximal | 1 | 8 | |
| Size | 0.049a | ||
| >5 cm | 5 | 38 | |
| <5 cm | 7 | 12 | |
| Histologic differentiation | 0.934 | ||
| Well | 1 | 3 | |
| Moderately | 4 | 13 | |
| Poorly | 6 | 30 | |
| Undifferentiated | 1 | 4 | |
| Invasion depth | 0.414 | ||
| T1 | 4 | 6 | |
| T2 | 3 | 15 | |
| T3 | 3 | 18 | |
| T4 | 2 | 11 | |
| TNM stages | 0.001a | ||
| I | 6 | 5 | |
| II | 5 | 12 | |
| III | 0 | 29 | |
| IV | 1 | 4 | |
| Lymphatic metastasis | 0.001a | ||
| Yes | 10 | 36 | |
| No | 2 | 14 | |
| Regional lymph nodes | 0.004a | ||
| pN0 | 9 | 13 | |
| pN1 | 2 | 8 | |
| pN2 | 0 | 12 | |
| pN3 | 1 | 17 | |
| Distant metastasis | 0.970 | ||
| Yes | 1 | 4 | |
| No | 11 | 46 | |
Indicates p < 0.05.
Figure 7.
NEAT1 Promotes GC Cell Proliferation and Invasion Partly by Modulating GADD45A
(A) CCK8 assays showed that knockdown of GADD45A partly reversed NEAT1-mediated proliferation. Error bars, mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (B) CCK8 assays showed that knockdown of GADD45A partly reversed BRG1-mediated proliferation. The data represent the mean SD from three independent experiments. Error bars, mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (C) Colony formation assays showed that knockdown of GADD45A partly reversed NEAT1-mediated proliferation. Error bars, mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (D) Colony formation assays showed that knockdown of GADD45A partly reversed BRG1-mediated proliferation. Error bars, mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. (E) Association analysis of the relationship between NETA1 and GADD45A expression levels, in 42 paired gastric cancer tissues. (F) Association analysis of the relationship between BRG1 and GADD45A expression levels in 42 paired gastric cancer tissues. (G) Representative images of BRG1, GADD45A, and Ki-67 protein expression detected by IHC analysis in tumor tissues and normal tissues. (H) Proposed model in which NEAT1 mediates the proliferation of GC.
Next, we measured the expression levels of NEAT1, BRG1, and GADD45A in 42 pairs of GC tissues. GADD45A expression levels were negatively correlated with those of NEAT1 (R2 = −0.226) and BRG1 (R2 = −0.259) (Figures 7E and 7F). Consistent with these results, immunohistochemical staining analysis of NEAT1, BRG1, and GADD45A in GC tissues indicated that patients with high BRG1 expression exhibited lower GADD45A and Ki-67 expression than patients with low BRG1 expression (Figure 7G). Collectively, these findings suggested that NEAT1 might exert oncogenic effects on GC progression by interacting with BRG1 to modulate histone methylation of the GADD45A promoter (Figure 7H).
Discussion
An increasing number of studies have revealed the multiple roles of lncRNAs in the initiation and progression of malignancies. However, the exact mechanisms underlying the abnormal expression and target regulation of lncRNAs remain elusive. In this study, we investigated the role of the lncRNA NEAT1 in GC progression and explored the underlying molecular mechanisms. Our results showed that NEAT1 was significantly upregulated in GC tissues compared with adjacent normal tissues and was associated with poor prognosis. NEAT1 overexpression facilitated GC cell proliferation. Moreover, we demonstrated that KLF5 bound to the NEAT1 promoter. Further experiments showed that NEAT1 inhibited GADD45A expression in GC cells and xenografts. In addition, BRG1 was found to play a critical role in epigenetic repression of GADD45A expression by inducing histone methylation of the GADD45A promoter.
NEAT1 is considered an important regulatory factor in several tumors. In prostate cancer, the transcription factor CDC5L is a target of NEAT1.26 NEAT1 represses ARGN expression, which is essential for DNA damage repair and induces cell cycle arrest, by recruiting CDC5L to the ARGN promoter. In osteosarcoma, NEAT1 interacts with the transcriptional repression complex G9a–DNMT1–Snail and induces EMT, resulting in cell migration and metastasis.27
However, the regulatory patterns that mediate NEAT1 expression and the mechanisms by which it modulates global gene expression have not been well characterized in GC. Dysregulated lncRNAs have consistently been found to be regulated by genome-level alterations. We found a positive correlation between the KLF5 transcription factor and NEAT1 by analyzing TCGA data. The results of ChIP and luciferase reporter assays confirmed high enrichment of KLF5 in the promoter of NEAT1. KLF5 and MYC can bind to the lnc00346 promoter to positively regulate lncRNA transcription and facilitate GC tumorigenesis.7 In bladder cancer, KLF5 acts as a positive regulator of cell proliferation by accelerating G1/S cell cycle progression.28 We proved that KLF5 can activate NEAT1 expression through an interaction with its promoter.
To identify putative NEAT-targeted mRNAs possibly related to the cell cycle and apoptosis, we performed gene expression analysis after NEAT1 knockout. GADD45A was confirmed as a target of NEAT1. GADD45A expression is induced by cytotoxic chemotherapy in a DNA damage-dependent manner,29 and GADD45A is involved in many types of cancers through the maintenance of genomic stability, control of the G2/M checkpoint, and apoptosis.30 Consistent with the results of our study, GADD45A has been reported to be downregulated in GC and to act as a tumor suppressor.31
We also found that NEAT1 recruits BRG1 to methylate the GADD45A promoter region. The SWI/SNF complex contains one of two ATPase subunits—BRM or BRG1—to provide ATP to maintain chromatin architecture or regulate gene expression.32,33 lncRNAs have recently been reported to exert functions via their interaction with the SWI/SNF complex.34 A recent report showed that lncTCF7 recruits the nucleosome remodeling complex SWI/SNF to the TCF7 promoter.35 lncTCF7 triggers TCF7 expression to promote liver cancer stem cell (CSC) self-renewal through the activation of WNT signaling. The lncRNA SchLAP1 inhibits the expression of SNF5, a core component of the SWI/SNF complex that normally functions as a tumor suppressor, leading to prostate cancer progression.36 A well-acknowledged role for BRG1 in gene transcription is its capacity to link nucleosome-modifying enzymes to their target promoters. In addition, BRG1 can interact directly with sequence-specific noncoding RNAs and chromatin remodeling enzymes to integrate into a wide range of cellular contexts. BRG1 endows lung cancer cells with proliferation and migration abilities by activating KDM3A transcription. BRG1 and KDM3A form a complex to remove H3K9 dimethylation from the CCNB1 and LTBP2 promoters. BRG1 knockdown decreases lung cancer malignancy.37 The lncRNA MALAT1 binds BRG1 to epigenetically promote hepatocellular carcinoma cell proliferation and invasion.38 In this study, the results of RNA pulldown and RIP assays confirmed that NEAT1 interacts with BRG1. The ChIP data demonstrated that NEAT1 controls the availability of GADD45A by recruiting BRG1 to increase H3K27me3 enrichment and decrease H3K4me3 enrichment in the GADD45A promoter.
NEAT1 often functions as a ceRNA to sponge microRNAs (miRNAs) in a variety of cancers.39 Cytoplasmic lncRNAs can interact with their target microRNAs or modulate mRNA translation at the posttranscriptional level; however, NEAT1 is a nuclear lncRNA in GC cells. In the nucleus, lncRNAs participate primarily in epigenetic and transcriptional processes.6 We previously identified direct binding between NEAT1 and argonaute-2 (AGO2), a member of the RNA-induced silencing complex (RISC) (data not shown). This interaction with AGO2 indicates that NEAT1 may also play a role in the biogenesis of miRNAs and their transport to the cytoplasm.
From a therapeutic perspective, NEAT1 offers a potential target. However, much remains to be done to achieve its clinical translation. Anticancer drugs inhibit cell growth, partially by blocking DNA synthesis and the formation of both single-strand and double-strand breaks in DNA.40 NEAT1 exerts its effects mostly through the inhibition of DNA damage repair and deregulation of cell cycle progression. However, NEAT1 is also essential for paraspeckle integrity. Silencing NEAT1 may damage normal cells; hence, selective targeting is necessary.
Antibody-drug conjugates (ADCs) are a new approach to selectively deliver cytotoxic anticancer agents to tumors while minimizing the exposure of normal tissues to the drug.41,42 Antigens that are highly expressed in cancer are ideal targets for this therapeutic strategy. Conjugating NEAT1 inhibitors with tumor-specific antibodies is anticipated to improve drug efficiency and decrease side effects.
Preferential reliance on aerobic glycolysis, also termed the “Warburg effect,” fuels cancer cell growth and proliferation.43 Accelerated aerobic glycolysis is recognized to distinguish cancer cells from most normal cells.44 This distinction has been applied to tumor imaging through assessment of altered uptake of the radiolabeled glucose analog [18F] fluoro-2-deoxyglucose (FDG). Continued investigations of cancers have elucidated several metabolic markers, mostly glucose transporters (Glut) and glycolytic enzymes, that are preferentially used in cancer cells.45 Hence, compounds that can be recognized by these markers are suitable for the delivery of NEAT1 inhibitors to targeted cancer cells.
In summary, we identified that the lncRNA NEAT1, whose expression was dependent on KLF5, promoted GC cell proliferation and was correlated with poor prognosis. The NEAT1-BRG1 complex markedly inhibited GADD45A expression by increasing local H3K27me3 modification and decreasing H3K4me3 modification. The lncRNA NEAT1–BRG1–GADD45A axis may thus serve as a promising therapeutic target in GC.
Materials and Methods
Tissue Samples
We obtained 62 paired GC and adjacent nontumor (normal) tissues from patients who were diagnosed with GC based on histopathologic evaluation and had undergone surgery at the First Affiliated Hospital of Nanjing Medical University (Nanjing, China) between 2010 and 2016. None of the patients received chemotherapy or radiotherapy prior to surgery. Among them, there were 11 cases in TNM stage I, 17 cases in TNM stage II, 29 cases in TNM stage III, and 5 cases in TNM stage IV. There were 38 cases with lymphatic metastasis and 5 cases with distant metastasis. All collected tissue samples were immediately snap frozen in liquid nitrogen and stored at −80°C until required. The histologic grade was assessed according to the eighth edition of the International Union against Cancer/American Joint Committee on Cancer tumor-node-metastasis (TNM) staging system. All research complied with the principles of the Declaration of Helsinki. The study was approved by the Research Ethics Committee of the First Affiliated Hospital of Nanjing Medical University, and informed consent was obtained.
Cell Culture
Six human GC cell lines (BGC-823, SGC-7901, AGS, HGC-27, MGC-803, MKN-45) and a normal gastric epithelial cell line (GES-1) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). BGC-823 and MGC-803 cells were cultured in RPMI-1640 medium; SGC-7901, MKN-45, and HGC-27 cells were cultured in DMEM; and AGS cells were cultured in F12 medium. All media were supplemented with 10% fetal bovine serum (FBS) (GIBCO, USA), 100 U/mL penicillin, and 100 mg/mL streptomycin (Invitrogen, USA) in humidified air at 37°C with 5% CO2. All cell lines were authenticated by short tandem repeat DNA profiling.
RNA Extraction and qRT-PCR Assays
Total RNA was extracted from tissues or cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA (1 μg) was reverse transcribed in a final volume of 20 μL using random primers under standard conditions for the Prime Script RT Reagent Kit (Takara). Real-time PCR analyses were performed with SYBR Premix Ex Taq (Takara). The expression levels were normalized to those of GAPDH. Primers were synthesized by Invitrogen. The specific primers used are described in Table S1. qRT-PCR and data collection were conducted in an ABI 7900HT real-time PCR system (Applied Biosystems). Our qRT-PCR data were analyzed and expressed relative to threshold cycle (Ct) values and were then converted to fold-change values.
Cell Transfection
GC cells were transfected with siRNAs and plasmid vectors using Lipofectamine 3000 (Invitrogen) in accordance with the manufacturer’s protocol. The NEAT1 ASOs (NEAT1-ASO#1, #2) and scrambled negative control ASOs were purchased from Exiqon. The BRG1 and GADD45A siRNAs and the scrambled negative control siRNA (si-NC) were provided by Invitrogen. The nucleotide sequences are listed in Table S1. Complementary DNA (cDNA) of the human BRG1 and NEAT1 transcripts and shRNA sequences directed against these transcripts were cloned into the pCDNA3.1 and PLKO vectors. Plasmid vectors for transfection were prepared using DNA Midiprep Kits (QIAGEN) and transfected into cells. Forty-eight hours post transfection, cells were harvested for qRT-PCR or western blot analysis.
Cell Proliferation Assays
Details of the CCK8 and colony formation assay protocols are available in a previously published manuscript.8
Subcellular Fractionation
The nuclear and cytosolic fractions were separated using a PARIS Kit (Invitrogen, USA) according to the manufacturer’s instructions.
RNA Pulldown Assays
RNA pulldown assays were performed with a Pierce Magnetic RNA-Protein Pull-Down Kit according to the manufacturer’s instructions (Thermo, USA). Details of the RNA-protein pulldown assay protocols are available in a previously published manuscript.12
RIP Assays
RIP assays were performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) according to the manufacturer’s instructions. The antibodies used for RIP assays of BRG1 were purchased from Abcam (shown in Table S2). Details of the RIP assay protocol are available in a previously published manuscript.12
Flow Cytometric Analysis
Details of flow cytometric analysis protocols are available in a previously published manuscript.8
ChIP Assays
ChIP assays were performed using a ChIP Assay Kit (Millipore, USA) according to the manufacturer’s instructions as described previously. The primer sequences are shown in Table S1. Details of the ChIP assay protocols are available in a previously published manuscript.12
Western Blot Analysis
Protein extraction and quantification were performed as previously reported.12 The antibodies used are described in Table S2.
Luciferase Reporter Assays
Luciferase assays were performed using a luciferase assay kit (Promega) based on the manufacturer’s protocol. The cDNA fragment containing a WT or mutant NEAT1 fragment was subcloned into the pGL3-Basic luciferase reporter vector (Promega, USA) downstream of the luciferase gene. Human HEK293T cells (1 × 105) cultured in a 24-well plate were cotransfected with 150 ng of either empty vector or KLF5 and with 50 ng of the firefly luciferase reporter vector containing the WT or mutant NEAT1 fragment using Lipofectamine 3000 (Invitrogen). Forty-eight hours after transfection, the luciferase assay was performed using a Dual Luciferase Kit (Promega, USA). The relative firefly luciferase activities were normalized to those of Renilla luciferase. Transfections were repeated in triplicate.
FISH
RNA FISH probes were designed and synthesized by Bogu (Shanghai, China). FISH assays were performed as described previously.8
RNA Transcriptome Sequencing
RNA transcriptome sequencing assays were performed as described previously12 and provided by NovelBio (Shanghai, China).
IHC
Immunohistochemical detection of Ki-67, BRG1, and GADD45A was performed according to a previously described method.12
Animal Experiments
All protocols were approved by the Committee on the Ethics of Animal Experiments of Nanjing Medical University. All experimental procedures involving animals were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals published by the NIH (Bethesda, MD). For the tumorigenicity studies, BGC-823 cells were stably transfected with control shRNA or sh-NEAT1 (3 × 106). Lentiviruses carrying negative control shRNA, sh-NEAT1, sh-BRG1, and empty vector (negative control) were purchased from GenePharma (Shanghai, China). GC cells were subcutaneously injected into either axillary region of male BALB/c nude mice (4–5 weeks old). Tumor volumes and weights were measured every 3 days, and tumor volumes were calculated using the following equation: V = 0.5 ∗ D ∗ d2 (V, volume; D, longest diameter; d, diameter perpendicular to the longest diameter). At 15 days after injection, mice were euthanized, and the subcutaneous growth of each tumor was examined. The primary tumors were cut and subjected to hematoxylin and eosin (H&E) and immunohistochemical staining as described previously.12
Immunofluorescence
Cells were seeded in a cell slide, then washed with Phosphate Buffered Saline (PBST) after 24 h and fixed with 4% paraformaldehyde at 4°C for 30 min followed by 0.5% Triton X-100 20 min, and then incubated with blocking buffer (PBST containing 1% BSA) for 2 h. Next, cells were incubated with primary antibodies targeting γ-H2AX (phospho S139) (1:250 dilution) at 4°C overnight. Washed with PBST, cells were incubated with secondary antibodies (anti-rabbit Alexa 448-conjugated antibodies; 1:200 dilution; Beyotime Biotechnology) for 2 h at room temperature. Nuclei were stained with Hoechst (1:1000 dilution) for 2 min at room temperature. Cells were analyzed immediately with a confocal microscopy after adding antifade mounting medium (Zeiss).
Statistical Analysis
The significance of differences between groups was assessed by a paired, two-tailed Student’s t test, a Wilcoxon test, or a χ2 test. Univariate and multivariate Cox proportional hazards models were used to determine the effects of variables on survival. The Kaplan-Meier method was utilized to analyze the FP and OS. Spearman correlation analysis was used to evaluate correlations between NEAT1, BRG1, and other clinical factors. All statistical analyses were performed using SPSS 20.0 software, and a p value of < 0.05 was considered to indicate statistical significance. The tumor marker prognostic analysis was performed following the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) guidelines.
Author Contributions
P.M., Y.P., F.Y., and Y.F. contributed to designing and organizing the experiments, carrying out the data analysis, and writing the manuscript. W. Liu, C.Z., T.Y., M.X., X.J., X.W., C.S., W. Li, and T.X. contributed to laboratory measurements and data analysis. Y.S. and P.M. contributed to conceiving the ideas, supervising the study, and writing the manuscript.
Conflict of Interest Statement
The authors declare no competing interests.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81802381; No. 81772475; No. 81672896), the Priority Academic Program Development of Jiangsu Higher Education Institutions (JX10231801), projects supported by Nanjing Medical University (NMUC2018005B, JX102GSP201727) and the National Key Research and Development Program: The Key Technology of Palliative Care and Nursing for Cancer Patients (2017YFC1309201). We thank American Journal Experts (https://www.aje.com/) for providing professional English polish.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.09.003.
Supplemental Information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Saka M., Morita S., Fukagawa T., Katai H. Present and future status of gastric cancer surgery. Jpn. J. Clin. Oncol. 2011;41:307–313. doi: 10.1093/jjco/hyq240. [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E., Ducreux M. Colorectal and gastric cancer in 2015: The development of new agents and molecular classifications. Nat. Rev. Clin. Oncol. 2016;13:69–70. doi: 10.1038/nrclinonc.2015.217. [DOI] [PubMed] [Google Scholar]
- 4.Long Y., Wang X., Youmans D.T., Cech T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017;3:eaao2110. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopp F., Mendell J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitt A.M., Chang H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu T.P., Ma P., Wang W.Y., Shuai Y., Wang Y.F., Yu T., Xia R., Shu Y.Q. KLF5 and MYC modulated LINC00346 contributes to gastric cancer progression through acting as a competing endogeous RNA and indicates poor outcome. Cell Death Differ. 2019;26:2179–2193. doi: 10.1038/s41418-018-0236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Chen Z., Yu S., Nie F., Yan S., Ma P., Chen Q., Wei C., Fu H., Xu T. Long Noncoding RNA LINC01234 Functions as a Competing Endogenous RNA to Regulate CBFB Expression by Sponging miR-204-5p in Gastric Cancer. Clin. Cancer Res. 2018;24:2002–2014. doi: 10.1158/1078-0432.CCR-17-2376. [DOI] [PubMed] [Google Scholar]
- 9.Xu T.P., Wang Y.F., Xiong W.L., Ma P., Wang W.Y., Chen W.M., Huang M.D., Xia R., Wang R., Zhang E.B. E2F1 induces TINCR transcriptional activity and accelerates gastric cancer progression via activation of TINCR/STAU1/CDKN2B signaling axis. Cell Death Dis. 2017;8:e2837. doi: 10.1038/cddis.2017.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu T.P., Wang W.Y., Ma P., Shuai Y., Zhao K., Wang Y.F., Li W., Xia R., Chen W.M., Zhang E.B., Shu Y.Q. Upregulation of the long noncoding RNA FOXD2-AS1 promotes carcinogenesis by epigenetically silencing EphB3 through EZH2 and LSD1, and predicts poor prognosis in gastric cancer. Oncogene. 2018;37:5020–5036. doi: 10.1038/s41388-018-0308-y. [DOI] [PubMed] [Google Scholar]
- 11.Ma P., Pan Y., Li W., Sun C., Liu J., Xu T., Shu Y. Extracellular vesicles-mediated noncoding RNAs transfer in cancer. J. Hematol. Oncol. 2017;10:57. doi: 10.1186/s13045-017-0426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu T.P., Liu X.X., Xia R., Yin L., Kong R., Chen W.M., Huang M.D., Shu Y.Q. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 13.Lin Y., Schmidt B.F., Bruchez M.P., McManus C.J. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 2018;46:3742–3752. doi: 10.1093/nar/gky046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y., Wang Y., Fu X., Lu Z. Long non-coding RNA NEAT1 promoted ovarian cancer cells’ metastasis through regulation of miR-382-3p/ROCK1 axial. Cancer Sci. 2018;109:2188–2198. doi: 10.1111/cas.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klec C., Prinz F., Pichler M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol. Oncol. 2019;13:46–60. doi: 10.1002/1878-0261.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adriaens C., Standaert L., Barra J., Latil M., Verfaillie A., Kalev P., Boeckx B., Wijnhoven P.W., Radaelli E., Vermi W. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 17.Chia N.Y., Deng N., Das K., Huang D., Hu L., Zhu Y., Lim K.H., Lee M.H., Wu J., Sam X.X. Regulatory crosstalk between lineage-survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development. Gut. 2015;64:707–719. doi: 10.1136/gutjnl-2013-306596. [DOI] [PubMed] [Google Scholar]
- 18.Vaux D.L., Strasser A. The molecular biology of apoptosis. Proc. Natl. Acad. Sci. USA. 1996;93:2239–2244. doi: 10.1073/pnas.93.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He H., Wang C., Dai Q., Li F., Bergholz J., Li Z., Li Q., Xiao Z.X. p53 and p73 Regulate Apoptosis but Not Cell-Cycle Progression in Mouse Embryonic Stem Cells upon DNA Damage and Differentiation. Stem Cell Reports. 2016;7:1087–1098. doi: 10.1016/j.stemcr.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Connor L., Huang D.C., O’Reilly L.A., Strasser A. Apoptosis and cell division. Curr. Opin. Cell Biol. 2000;12:257–263. doi: 10.1016/s0955-0674(99)00084-8. [DOI] [PubMed] [Google Scholar]
- 21.Kaina B. DNA damage-triggered apoptosis: critical role of DNA repair, double-strand breaks, cell proliferation and signaling. Biochem. Pharmacol. 2003;66:1547–1554. doi: 10.1016/s0006-2952(03)00510-0. [DOI] [PubMed] [Google Scholar]
- 22.Sedelnikova O.A., Bonner W.M. GammaH2AX in cancer cells: a potential biomarker for cancer diagnostics, prediction and recurrence. Cell Cycle. 2006;5:2909–2913. doi: 10.4161/cc.5.24.3569. [DOI] [PubMed] [Google Scholar]
- 23.Kuo L.J., Yang L.X. Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo. 2008;22:305–309. [PubMed] [Google Scholar]
- 24.Tong T., Ji J., Jin S., Li X., Fan W., Song Y., Wang M., Liu Z., Wu M., Zhan Q. Gadd45a expression induces Bim dissociation from the cytoskeleton and translocation to mitochondria. Mol. Cell. Biol. 2005;25:4488–4500. doi: 10.1128/MCB.25.11.4488-4500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang L.Y., Zhao J., Chen H., Wan L., Inuzuka H., Guo J., Fu X., Zhai Y., Lu Z., Wang X. SCFFBW7-mediated degradation of Brg1 suppresses gastric cancer metastasis. Nat. Commun. 2018;9:3569. doi: 10.1038/s41467-018-06038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., Wang X., Song W., Xu H., Huang R., Wang Y., Zhao W., Xiao Z., Yang X. Oncogenic Properties of NEAT1 in Prostate Cancer Cells Depend on the CDC5L-AGRN Transcriptional Regulation Circuit. Cancer Res. 2018;78:4138–4149. doi: 10.1158/0008-5472.CAN-18-0688. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Cheng C. Long noncoding RNA NEAT1 promotes the metastasis of osteosarcoma via interaction with the G9a-DNMT1-Snail complex. Am. J. Cancer Res. 2018;8:81–90. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Pattison J.M., Posternak V., Cole M.D. Transcription Factor KLF5 Binds a Cyclin E1 Polymorphic Intronic Enhancer to Confer Increased Bladder Cancer Risk. Mol. Cancer Res. 2016;14:1078–1086. doi: 10.1158/1541-7786.MCR-16-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pietrasik S., Zajac G., Morawiec J., Soszynski M., Fila M., Blasiak J. Interplay between BRCA1 and GADD45A and Its Potential for Nucleotide Excision Repair in Breast Cancer Pathogenesis. Int. J. Mol. Sci. 2020;21:870. doi: 10.3390/ijms21030870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cannell I.G., Merrick K.A., Morandell S., Zhu C.Q., Braun C.J., Grant R.A., Cameron E.R., Tsao M.S., Hemann M.T., Yaffe M.B. A Pleiotropic RNA-Binding Protein Controls Distinct Cell Cycle Checkpoints to Drive Resistance of p53-Defective Tumors to Chemotherapy. Cancer Cell. 2015;28:831. doi: 10.1016/j.ccell.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Duan C., Zhang B., Deng C., Cao Y., Zhou F., Wu L., Chen M., Shen S., Xu G., Zhang S. Piperlongumine induces gastric cancer cell apoptosis and G2/M cell cycle arrest both in vitro and in vivo. Tumour Biol. 2016;37:10793–10804. doi: 10.1007/s13277-016-4792-9. [DOI] [PubMed] [Google Scholar]
- 32.Peterson C.L., Tamkun J.W. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem. Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 33.Wilson B.G., Roberts C.W. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 34.Jégu T., Blum R., Cochrane J.C., Yang L., Wang C.Y., Gilles M.E., Colognori D., Szanto A., Marr S.K., Kingston R.E., Lee J.T. Xist RNA antagonizes the SWI/SNF chromatin remodeler BRG1 on the inactive X chromosome. Nat. Struct. Mol. Biol. 2019;26:96–109. doi: 10.1038/s41594-018-0176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y., He L., Du Y., Zhu P., Huang G., Luo J., Yan X., Ye B., Li C., Xia P. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Prensner J.R., Iyer M.K., Sahu A., Asangani I.A., Cao Q., Patel L., Vergara I.A., Davicioni E., Erho N., Ghadessi M. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z., Xia J., Fang M., Xu Y. Epigenetic regulation of lung cancer cell proliferation and migration by the chromatin remodeling protein BRG1. Oncogenesis. 2019;8:66. doi: 10.1038/s41389-019-0174-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang M., Wang H., Hu X., Cao X. lncRNA MALAT1 binds chromatin remodeling subunit BRG1 to epigenetically promote inflammation-related hepatocellular carcinoma progression. OncoImmunology. 2018;8:e1518628. doi: 10.1080/2162402X.2018.1518628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong C.M., Tsang F.H., Ng I.O. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 2018;15:137–151. doi: 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- 40.Harte R.J., Matthews J.C., O’Reilly S.M., Tilsley D.W., Osman S., Brown G., Luthra S.J., Brady F., Jones T., Price P.M. Tumor, normal tissue, and plasma pharmacokinetic studies of fluorouracil biomodulation with N-phosphonacetyl-L-aspartate, folinic acid, and interferon alfa. J. Clin. Oncol. 1999;17:1580–1588. doi: 10.1200/JCO.1999.17.5.1580. [DOI] [PubMed] [Google Scholar]
- 41.Chau C.H., Steeg P.S., Figg W.D. Antibody-drug conjugates for cancer. Lancet. 2019;394:793–804. doi: 10.1016/S0140-6736(19)31774-X. [DOI] [PubMed] [Google Scholar]
- 42.Bargh J.D., Isidro-Llobet A., Parker J.S., Spring D.R. Cleavable linkers in antibody-drug conjugates. Chem. Soc. Rev. 2019;48:4361–4374. doi: 10.1039/c8cs00676h. [DOI] [PubMed] [Google Scholar]
- 43.Ganapathy-Kanniappan S. Molecular intricacies of aerobic glycolysis in cancer: current insights into the classic metabolic phenotype. Crit. Rev. Biochem. Mol. Biol. 2018;53:667–682. doi: 10.1080/10409238.2018.1556578. [DOI] [PubMed] [Google Scholar]
- 44.Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat. Rev. Cancer. 2016;16:635–649. doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lipstein M.R., Pal I., Bates S.E., Deng C. Metabolic symbiosis in cancer and its therapeutic implication. Semin. Oncol. 2017;44:233–234. doi: 10.1053/j.seminoncol.2017.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.