Abstract
Recent studies have increasingly shown that the chemical modification of mRNA plays an important role in the regulation of gene expression. N7-methylguanosine (m7G) is a type of positively-charged mRNA modification that plays an essential role for efficient gene expression and cell viability. However, the research on m7G has received little attention to date. Bioinformatics tools can be applied as auxiliary methods to identify m7G sites in transcriptomes. In this study, we develop a novel interpretable machine learning-based approach termed XG-m7G for the differentiation of m7G sites using the XGBoost algorithm and six different types of sequence-encoding schemes. Both 10-fold and jackknife cross-validation tests indicate that XG-m7G outperforms iRNA-m7G. Moreover, using the powerful SHAP algorithm, this new framework also provides desirable interpretations of the model performance and highlights the most important features for identifying m7G sites. XG-m7G is anticipated to serve as a useful tool and guide for researchers in their future studies of mRNA modification sites.
Keywords: N7-Methylguanosine, m7G, prediction, XGBoost, machine learning, SHAP, model interpretation, feature selection, ENAC, SCPseDNC
Graphical Abstract

N7-methylguanosine modification of mRNA plays a key role in the regulation of gene expression. In this study, we developed a computational model termed XG-m7G based on XGBoost and SHAP to identify N7-methylguanosine sites from sequence information. This will help the discovery of N7-methylguansine and its genetic function.
Introduction
Precise regulation of gene expression is vital for the growth and development of organisms in both physiological and pathological processes.1 Post-translational modification of mRNA was recently found to regulate gene expression. For instance, N7-methylguanosine (m7G) is one of the most important mRNA modifications that can be formed during mRNA capping.2,3 Subsequent experiments have proven that m7G is indispensable for several types of gene processing, including RNA splicing, polyadenylation, and mRNA stability.4, 5, 6, 7, 8 With the development of new experimental technology, such as next-generation sequencing (NGS) and immunoprecipitation sequencing (MeRIP-seq), research data on mRNA translation has rapidly increased.9 To date, MODOMICS is a database of RNA modifications that covers more than 160 types of modified ribonucleotides,10 and these data provide an opportunity to build bioinformatics tools that can identify m7G sites. Using the support vector machine (SVM) classifier, Chen et al.8 proposed the first m7G prediction model, iRNA-m7G, by fusing three kinds of features. iRNA-m7G achieved a sensitivity (Sn) of 89.07%, a specificity (Sp) of 90.69%, and a Matthew’s correlation coefficient (MCC) of 0.8 on the jackknife test. In light of the importance of m7G function, we think that it is necessary to further enhance the model performance on m7G sites identification.
Herein, we propose a novel predictor for identifying m7G sites, termed XG-m7G, which applies the extreme gradient boosting (XGBoost) algorithm as the classifier. XG-m7G utilized six types of feature encoders, including binary encoding, composition of k-spaced nucleic acid pairs (CKSNAP), enhanced nucleic acid composition (ENAC), nucleotide chemical property (NCP), nucleotide density (ND), and the series correlation pseudo-dinucleotide composition (SCPseDNC), as its inputs. XGBoost is applied as a classification algorithm to train the model and test its performance. Then, the unified framework SHAP (Shapley additive explanations) is used to interpret predictions,39 rank the feature importance, identify which features are most important, and further select the optimal feature sets. Our benchmarking experiments show that XG-m7G achieved an MCC of 0.825 and 0.839 on 10-fold cross-validation and jackknife tests, respectively, both of which are superior to that of an existing unique model iRNA-m7G.
Results
In this study, we proposed a novel model, XG-m7G, for identifying m7G sites efficiently and accurately from RNA sequences. The performance of XG-m7G was compared with the latest m7G sitesidentification model iRNA-m7G by using both 10-fold cross-validation and jackknife tests. These results indicate that our model outperformed iRNA-m7G in terms of several major metrics, including Sn, Sp, accuracy (Acc), MCC, and area under the receiver operating characteristic (ROC) curve (AUC). Then, we explored the most important features based on a model interpretation method, that is, SHAP, and verified their contribution for identifying m7G sites. In addition, we constructed a web server, which allows interested users to both use our model to identify m7G sites and train their specific models based on their own datasets expediently.
Discussion
XGBoost Outperforms State-of-the-Art Algorithms in m7G Site Prediction
To find the best-performing classification algorithm, four state-of-the-art classifiers, i.e., k-nearest neighbor (KNN),11 SVM,12 logistic regression (LR),13 and random forest (RF),14 were used to predict m7G sites alongside XGBoost. For each classifier, the important parameters were searched and selected according to the prediction results on 10-fold cross-validation. More specifically, we optimized the neighbors k of KNN; kernel function, C, and gamma of SVM; penalty and solver for LR; and n_estimators of RF. For XGBoost, there were four parameters that need to be considered, which were n_estimators ∈ [10, 100, 1,000], max_depth ∈ [3, 5, 7], learning_rate ∈ [0.1, 0.2, 0.3], and gamma ∈ [0.001, 0.01, 0.1]. The final parameter settings of the machine learning algorithms are provided in Table 1.
Table 1.
Parameter Settings of XGBoost, KNN, SVM, LR, and RF
| XGBoost | KNN | SVM | LR | RF |
|---|---|---|---|---|
| n_estimators = 1,000, max_depth = 3, learning_rate = 0.2, gamma = 0.001 | k_neighbors = 4 | kernel = “rbf”, C = 10, gamma = 0.02 | penalty = “l1”, C = 10, solver = “liblinear” | n_estimators = 10 |
To minimize the potential effect of randomness on the experimental results, we conducted the 10-fold cross-validation on the five different classifiers by running 100 rounds. Subsequently, the average value of each evaluation metric was calculated and compared. As shown in Figure 1, XGBoost displayed the best performance according to Sp, Acc, MCC, and AUC; however, for the Sn, XGBoost was lower than KNN by about 1.44%. For clarity, we have also listed the win-draw-loss results in Table 2. For each algorithm (KNN, SVM, LR, and RF), “win” means the number of times XGBoost outperformed it, “draw” represents an equivalent performance between the two, and “loss” represents XGBoost being worse. Moreover, the significance of the difference in the prediction results was analyzed by using a Student’s t test. As shown in Table 2, all p values were far less than 0.01, indicating that there was a statistically significant difference between XGBoost and the other four algorithms.
Figure 1.
Average Results of Five Classification Algorithms after 10-Fold Cross Validation Running for 100 Rounds
Table 2.
“Win-Draw-Loss” Results for XGBoost Compared with Other Classifiers
| Classifier | KNN | SVM | LR | RF |
|---|---|---|---|---|
| Sn | 3-0-97 | 96-0-4 | 100-0-0 | 100-0-0 |
| p value | 5.6103E−34 | 6.6038E−32 | 4.0955E−59 | 1.3241E−80 |
| Sp | 100-0-0 | 100-0-0 | 100-0-0 | 100-0-0 |
| p value | 3.5946E−95 | 2.2062E−40 | 6.5396E−71 | 7.9065E−58 |
| Acc | 100-0-0 | 100-0-0 | 100-0-0 | 100-0-0 |
| p value | 8.8436E−72 | 2.1478E−47 | 5.2734E−79 | 4.2097E−84 |
| MCC | 100-0-0 | 100-0-0 | 100-0-0 | 100-0-0 |
| p value | 8.4909E−71 | 6.9919E−79 | 4.2602E−84 | 2.9426E−47 |
| AUC | 100-0-0 | 100-0-0 | 100-0-0 | 100-0-0 |
| p value | 1.6011E−131 | 1.5336E−70 | 6.9470E−89 | 1.3920E−112 |
In addition, for each classifier, the largest AUC achieved in the 100 rounds of 10-fold cross-validation was recorded, and its corresponding ROC curve is shown in Figure 2. Again, XGBoost outperformed the other four algorithms on m7G site prediction, and it achieved the highest AUC of 0.965.
Figure 2.
ROC Curves of the Five Classification Algorithms
The Effect of Feature Encoding on Model Prediction
In this study, six different feature-encoding schemes were used to generate the feature vectors. The performance of each type of feature is listed in Table S1. Afterward, we used the SHAP algorithm to characterize feature importance and assess feature behavior in our samples. For convenience, we named all features as follows: binary-1, binary-2, …, binary-164; CKSNAP-165, CKSNAP-166, …, CKSNAP-212, …; and SCPseDNC-537, SCPseDNC-538, …, SCPseDNC-672. According to Equation 15, SHAP values were calculated and the top 20 features for all samples are plotted in Figure 3.
Figure 3.
Top 20 Features Sorted by SHAP
In Figure 3A, each row represents a feature, and each point is the SHAP value of an instance. Redder sample points indicate that the value of the feature is larger, and bluer sample points indicate that the value of the feature is smaller; the abscissae represent the SHAP values. For clarity, the area in which feature NCP-466 is located has been magnified in Figure 3B. If the SHAP value is positive, this means that the feature drives the predictions toward m7G sites and has a positive effect; if negative, the feature drives the predictions toward non-m7G sites and has a negative effect. We observed that when features such as NCP-432 and NCP-438 take high SHAP values, the model is driven toward positive m7G prediction. Conversely, when these features take low SHAP values, the model is driven toward non-m7G prediction. We also observed that the features SCPseDNC-546, SCPseDNC-543, SCPseDNC-567, and SCPseDNC-579 only promote the prediction of non-m7G sites, while other features, such as SCPseDNC-541, SCPseDNC-538, SCPseDNC-599, and SCPseDNC-549, only promote the prediction of m7G sites by the model.
In order to further explore the contribution of the top 20 features sorted by SHAP to the model performance, we retrained the model without these features and evaluated the performance of the resulting model on jackknife tests. Figure 4 shows the comparison with and without these top 20 features. We also found that selecting a different number of features would influence the model performance. Therefore, to further improve our model performance, we took the mean value of the absolute values of SHAP for each feature to rank the importance of features. Afterward, we designed and implemented a series of experiments. The feature ranking results were exported and are listed in the Table S2. In addition to the k-fold cross-validation test, the jackknife test is also frequently used as a cross-validation method in statistical prediction.15 For a given benchmark dataset, the jackknife test can always yield a unique outcome.15 Next, we kept the top 50, 100, 150, 200, 250, 300, 350, 400, 450, 500, 550, and 600 features according to the sorting to find the best feature combination by performing jackknife tests. All of the performance results are listed in Table S3. We found that the model achieved the overall best performance when the feature dimension was reduced to 150. To illustrate the effectiveness of feature selection, we provided the performance comparison results of the models trained using the original features and the optimal features on jackknife tests in Table S4 and Figure 4. The results also showed that the model trained using the selected optimal features by SHAP clearly achieved an improve predictive performance compared with the model trained using all of the original features.
Figure 4.
Comparison of Models Trained Using Different Dimensional Features
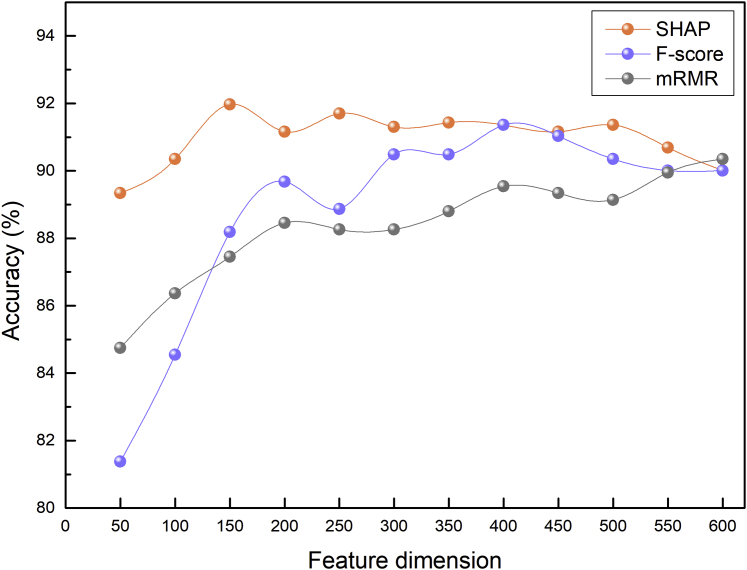
In addition, we also conducted the same feature ranking and selection processes using the F-score and minimum redundancy-maximum relevance (mRMR), which has been extensively adopted to reduce the feature dimension in the fields of bioinformatics and computational biology.16,17 Figure 5 provides the performance comparison of these three feature selection technologies. As can be seen, the SHAP curve was above the F-score curve and the mRMR curve. More specifically, after sorting by the F-score, the combination of the top 400 features reached the best Acc of 91.36%. In comparison, after the ranking by mRMR, the combination of the top 600 features reached the best Acc of 90.35%. These results indicate the effectiveness of SHAP for identifying m7G sites.
Figure 5.
Comparison of SHAP, F-score, and mRMR Dimensionality Reduction Methods on Jackknife Tests
Comparison with iRNA-m7G
To the best of our knowledge, iRNA-m7G is the only model established for searching m7G sites in RNA sequences. Therefore, we compared the performance of XG-m7G with iRNA-m7G by a 10-fold cross-validation test. Table 3 lists the performance comparison of XG-m7G and iRNA-m7G in terms of Sn, Sp, Acc, MCC, and AUC values. Clearly, our proposed XG-m7G method achieved a better performance than iRNA-m7G in terms of four evaluation metrics. Specifically, XG-m7G achieved an improvement of 2.82% and 1.41% for Sn and Acc, respectively. The MCC and AUC of XG-m7G were 0.025 and 0.026, respectively, higher than those of iRNA-m7G. Consequently, a jackknife test was also applied to estimate the performance of XG-m7G. XG-m7G obtained Sn of 92.17%, Sp of 91.77%, Acc of 91.97%, MCC of 0.839, and AUC of 0.972, demonstrating its superior performance to iRNA-m7G. Altogether, these results confirm that our proposed model XG-m7G outperforms iRNA-m7G in identifying m7G sites.
Table 3.
Predictive Performance Comparison of XG-m7G with iRNA-m7G by 10-Fold Cross-Validation and Jackknife Test
| Cross-Validation Test | Methods | Sn (%) | Sp (%) | Acc (%) | MCC | AUC |
|---|---|---|---|---|---|---|
| 10-Fold | iRNA-m7G | 88.66 | 90.96 | 89.81 | 0.800 | 0.946 |
| XG-m7G | 91.48 | 90.96 | 91.22 | 0.825 | 0.972 | |
| Jackknife | iRNA-m7G | 89.07 | 90.69 | 89.88 | 0.800 | – |
| XG-m7G | 92.17 | 91.77 | 91.97 | 0.839 | 0.972 |
Implementation of the XG-m7G Web Server
We developed a web server for XG-m7G to perform convenient prediction and analyses of distinctive m7G sites, which is freely available at http://flagship.erc.monash.edu/XG-m7G/. The XG-m7G server was implemented using PHP, HTML, CSS, JavaScript, and Python running Apache2 and configured in the Linux environment on an eight-core server machine with 32 gigabytes (GB) of memory and three hard disks with a total of 1.25 terabytes (TB) of memory. The server requires users to paste the sequences or upload a text file in the FASTA format as the input. Figure 6A illustrates an instance of the prediction steps. As shown, the prediction results are output in probability ranking and they are capable of being viewed in ascending or descending order. Furthermore, we designed a practical function “train model” to allow users to train their own models with their training data as shown in Figure 6B. The computational time needed for the testing task is determined by the number and total length of sequences provided. For 100 sequences with 41 nt residues each, the training task will be accomplished in a few seconds. We hope that this function can provide a theoretical and useful reference for interested researchers.
Figure 6.
Instructions for Using the XG-m7G Web Server
(A) Example of the “prediction” function of the XG-m7G web server. (B) Example of the “train model” function of the XG-m7G web server
Materials and Methods
Overall Framework
The overall framework of XG-m7G is illustrated in Figure 7. As shown, there were five major steps in the development of XG-m7G. First, we collected benchmark datasets for m7G sites. Second, these sequences were transformed into numeric vectors by several feature-encoding methods. Third, we analyzed and interpreted the feature effect with SHAP on XGBoost, and then the most important features were selected and identified. Fourth, we evaluated the model performance and, lastly, we developed a web server for XG-m7G.
Figure 7.
Overall Framework of XG-m7G
Benchmark Datasets
In this study, the benchmark datasets for training and evaluating our XG-m7G model were collected from Chen et al.8 Chen et al. obtained 801 m7G site-containing sequences by mapping the 801 base-resolution m7G sites in human HeLa and HepG2 cells that had been detected by Zhang et al.9 These sequences, with the m7G sites in the center of the sequences, were extracted to 41 bp, with 20 bp upstream and 20 bp downstream. CD-HIT18 was then used to remove redundant sequences19 using a threshold of 80%, after which 741 positive samples containing m7G sites were retained. The negative samples were first collected from those 41-bp-long sequences with the intermediate guanosine that have not been detected as m7G sites (namely non-m7G site) by the MeRIP-seq method. In order to further avoid the potential problem of low Sn caused by the unbalanced data, 741 such sequences containing non-m7G sites with the sequence similarity of less than 80% were selected to constitute the final negative sample dataset.
Feature Extraction
One of the critical steps is to encode each RNA fragment as a numerical vector. In this study, six types of feature encoding schemes were applied to establish the predictor, including binary encoding, CKSNAP, ENAC, NCP, ND, and SCPseDNC. These encoding schemes have been extensively used for identification of pseudouridine sites,20 prediction of citrullination sites,21 and prediction of m6A sites22,23 with demonstrated performance. It is noteworthy that all features used in this study are available and can be calculated using BioSeq-Analysis2.024 and iLearn.25 A detailed description of these feature-encoding schemes is provided in the following sections.
Binary Encoding
In the binary feature encoding scheme, each nucleotide is represented by a four-dimensional binary vector, e.g., A is coded as (1, 0, 0, 0), C is coded as (0, 1, 0, 0), G is coded as (0, 0, 1, 0), and U is coded as (0, 0, 0, 1). Therefore, each sample has a total of 164 binary features.
CKSNAP
The CKSNAP encoding scheme represents the frequencies of nucleotide pairs separated by k residues. The CKSNAP feature contains 16 values corresponding to pairs of nucleic acids: {AA, AC, AG, ..., UG, UU}. Taking k = 1 as an example, CKSNAP can be given as follows:
| (1) |
where ∗ represents any nucleotide of A, C, G, and U, represents the number of nucleic acid pairs X∗Y that occur in the sequence, and represents the total number of one-spaced nucleic acid pairs in the sequence. In this study, k = 0, 1, and 2, and the corresponding dimension of CKSNAP features was 48.
ENAC
ENAC encoding calculates the nucleic acid composition based on a fixed-length window, which continuously slides from the 5′ to 3′ terminus of each nucleotide sequence.25 This method is usually applied to encode nucleotide sequences of equal length. ENAC can be calculated as follows:
| (2) |
where S represents the size of the sliding window, represents the number of nucleic acids t in the sliding window r,, and . In this study, the size of the sliding window was 2, and the corresponding feature dimension was 160.
NCP and ND
Each nucleotide of A, C, G, and U has a different chemical structure and chemical binding feature. The NCP encoding scheme incorporates the chemical properties of these four types of nucleotides into the representation. The nucleotides can be classified into three different groups26 as follows:
| (3) |
where
| (4) |
Based on these three chemical properties, the nucleotide A is mapped to the numeric vector , C is mapped to , G is mapped to , and U is mapped to .
ND is also termed accumulated nucleotide frequency (ANF),25 which integrates the nucleotide frequency information and the distribution of each nucleotide in the RNA sequence. The density of any nucleotide at the position i in an RNA sequence can be given by the following formula:
| (5) |
where is the length of the sliding substring concerned, while l is the corresponding locator’s sequence position.
The NCP and ND encodings are often used together to take into account both chemical property and long-range sequence order information.20 As an example, the sequence fragment ACGCGGAUUA can be represented as {(1, 1, 1, 1), (0, 1, 0, 0.5), (1, 0, 0, 0.33), (0, 1, 0, 0.5), (1, 0, 0, 0.4), (1, 0, 0, 0.5), (1, 1, 1, 0.29), (0, 0, 1, 0.125), (0, 0, 1, 0.22), (1, 1, 1, 0.3)}. Each sample has 164 NCP and ND features.
SCPseDNC
The SCPseDNC encoding27 is defined as follows:
| (6) |
where
| (7) |
where for is the normalized occurrence frequency of the i-th dinucleotide in the sequence, w is the weight factor ranging from 0 to 1, and Λ is the number of physicochemical indices. Six indices (i.e., Rise (RNA), Roll (RNA), Shift (RNA), Slide (RNA), Tilt (RNA), Twist (RNA)) were set as the default indices for RNA sequences. () is the j-tier correlation factor, defined as:
| (8) |
Where λ represents the highest counted rank (or tier) of the correlation along the nucleotide sequence, and the correlation function is:
| (9) |
where , and is the numerical value of the u-th (u = 1,2, …,) physicochemical index of the dinucleotide at position i, and represents the corresponding value of the dinucleotide at position j. By setting = 20 and w = 0.9, we generated a 136-dimensional vector.
Machine Learning Algorithm
XGBoost28,29 is a type of optimized distributed gradient boosting algorithm. The principle of the XGBoost algorithm can be summarized as follows:
Assume a training dataset of the size n, where denotes an m-dimensional feature vector with the corresponding (output) category :
| (10) |
where K represents the number of trees, represents the score that is associated with the model’s k-th tree, and F denotes the space of scoring functions available for all boosting trees.
Differing from another tree-based algorithm, GBDT (gradient boosting decision tree), XGBoost uses the second-order Taylor expansion to approximate the loss function, and it is more efficient in avoiding the over-fitting issue mainly by adding regularization terms to the objective function. For more details about XGBoost, please refer to Chen and Guestrin.25 In recent years, XGBoost has been extensively utilized in bioinformatics and computational biology for addressing a range of challenging tasks, such as pseudouridine site identification,30 on-target activity prediction of single guide RNAs (sgRNAs),31 recognition of internal ribosome entry sites,32 and so on. In this study, we applied XGBoost to develop the m7G prediction model. Our results demonstrated that XGBoost achieved a better predictive performance than did other machine learning algorithms for m7G site prediction.
Evaluation Metrics
To evaluate the prediction performance of XG-m7G we used four metrics, that is, Sn, Sp, Acc, and MCC, which have previously been used to assess the performance of predictors in other studies.33,34 We also used ROC curves,35, 36, 37, 38 which plot the true-positive rate against the false-positive rate, and AUC to further assess the model performance. Sn, Sp, Acc, and MCC are defined as follows:
| (11) |
| (12) |
| (13) |
| (14) |
where represents the total number of m7G site-containing sequences, represents the total number of non-m7G sequences, represents the number of m7G site-containing sequences incorrectly predicted as non-m7G sequences, and represents the number of non-m7G sequences incorrectly predicted as m7G site-containing sequences.
Notably, these metrics are also used in the existing method iRNA-m7G for identification of m7G sites. Therefore, it is convenient to make a fair and credible comparison of XG-m7G and iRNA-m7G.
SHAP
SHAP is a unified framework for interpreting predictions, proposed in 2017 as the only consistent and locally accurate feature attribution method based on expectations.39 This technique can interpret feature importance scores from complex training models, and it provides an interpretable prediction for a test sample. SHAP values have been proposed as a unified measure of feature importance, as they assign an importance value () to each feature representing the effect of including that feature in model prediction. In cooperative game theory, SHAP values can be computed as follows:
| (15) |
where represents the set of all features and represents all feature subsets obtained from after removing the feature. Then, two models, and , are retrained, and predictions of these two models are compared to the current input , where represents the values of the input features in the set . To estimate from differences, the SHAP approach approximates the Shapley value by either performing Shapley sampling or Shapley quantitative influence.
Author Contributions
C.J., J.S., and F.L. conceived the initial idea and designed the methodology. F.L. and Y.B. implemented the algorithm, conducted the experiments, and processed the results. F.L., Y.B., D.X., and Z.G. developed the web server. All authors drafted, revised, and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by Fundamental Research Funds for the Central Universities (3132020170 and 3132019323) and the National Natural Science Foundation of Liaoning Province (20180550307). This work was also supported by the National Health and Medical Research Council of Australia (NHMRC) (1144652 and 1127948), the Australian Research Council (ARC) (LP110200333 and DP120104460), and by a Major Inter-Disciplinary Research (IDR) project awarded by Monash University.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.08.022.
Contributor Information
Fuyi Li, Email: fuyi.li@monash.edu.
Cangzhi Jia, Email: cangzhijia@dlmu.edu.cn.
Jiangning Song, Email: jiangning.song@monash.edu.
Supplemental Information
References
- 1.Chmielowska-Bąk J., Arasimowicz-Jelonek M., Deckert J. In search of the mRNA modification landscape in plants. BMC Plant Biol. 2019;19:421. doi: 10.1186/s12870-019-2033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowling V.H. Regulation of mRNA cap methylation. Biochem. J. 2009;425:295–302. doi: 10.1042/BJ20091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furuichi Y. Discovery of m7G-cap in eukaryotic mRNAs. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 2015;91:394–409. doi: 10.2183/pjab.91.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindstrom D.L., Squazzo S.L., Muster N., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., 3rd, Hartzog G.A. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drummond D.R., Armstrong J., Colman A. The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res. 1985;13:7375–7394. doi: 10.1093/nar/13.20.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis J.D., Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- 7.Murthy K.G., Park P., Manley J.L. A nuclear micrococcal-sensitive, ATP-dependent exoribonuclease degrades uncapped but not capped RNA substrates. Nucleic Acids Res. 1991;19:2685–2692. doi: 10.1093/nar/19.10.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W., Feng P., Song X., Lv H., Lin H. iRNA-m7G: identifying N7-methylguanosine sites by fusing multiple features. Mol. Ther. Nucleic Acids. 2019;18:269–274. doi: 10.1016/j.omtn.2019.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L.S., Liu C., Ma H., Dai Q., Sun H.L., Luo G., Zhang Z., Zhang L., Hu L., Dong X., He C. Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol. Cell. 2019;74:1304–1316.e8. doi: 10.1016/j.molcel.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai Y., Huang T., Hu L., Shi X., Xie L., Li Y. Prediction of lysine ubiquitination with mRMR feature selection and analysis. Amino Acids. 2012;42:1387–1395. doi: 10.1007/s00726-011-0835-0. [DOI] [PubMed] [Google Scholar]
- 12.Meng C., Jin S., Wang L., Guo F., Zou Q. AOPs-SVM: a sequence-based classifier of antioxidant proteins using a support vector machine. Front. Bioeng. Biotechnol. 2019;7:224. doi: 10.3389/fbioe.2019.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F., Li C., Marquez-Lago T.T., Leier A., Akutsu T., Purcell A.W., Ian Smith A., Lithgow T., Daly R.J., Song J., Chou K.C. Quokka: a comprehensive tool for rapid and accurate prediction of kinase family-specific phosphorylation sites in the human proteome. Bioinformatics. 2018;34:4223–4231. doi: 10.1093/bioinformatics/bty522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lv Z., Jin S., Ding H., Zou Q. A random forest sub-Golgi protein classifier optimized via dipeptide and amino acid composition features. Front. Bioeng. Biotechnol. 2019;7:215. doi: 10.3389/fbioe.2019.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou K.C., Zhang C.T. Prediction of protein structural classes. Crit. Rev. Biochem. Mol. Biol. 1995;30:275–349. doi: 10.3109/10409239509083488. [DOI] [PubMed] [Google Scholar]
- 16.Bi Y., Jin D., Jia C.Z. EnsemPseU: identifying pseudouridine sites with an ensemble approach. IEEE Access. 2020;8:79376–79382. [Google Scholar]
- 17.Jia C., Bi Y., Chen J., Leier A., Li F., Song J. PASSION: an ensemble neural network approach for identifying the binding sites of RBPs on circRNAs. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa522. Published online May 19, 2020. [DOI] [PubMed] [Google Scholar]
- 18.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou Q., Lin G., Jiang X., Liu X., Zeng X. Sequence clustering in bioinformatics: an empirical study. Brief. Bioinform. 2020;21:1–10. doi: 10.1093/bib/bby090. [DOI] [PubMed] [Google Scholar]
- 20.Liu K., Chen W., Lin H. XG-PseU: an eXtreme Gradient Boosting based method for identifying pseudouridine sites. Mol. Genet. Genomics. 2020;295:13–21. doi: 10.1007/s00438-019-01600-9. [DOI] [PubMed] [Google Scholar]
- 21.Ju Z., Wang S.Y. Prediction of citrullination sites by incorporating k-spaced amino acid pairs into Chou’s general pseudo amino acid composition. Gene. 2018;664:78–83. doi: 10.1016/j.gene.2018.04.055. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y., Zeng P., Li Y.H., Zhang Z.D., Cui Q.H. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y., He N., Chen Y., Chen Z., Li L. BERMP: a cross-species classifier for predicting m6A sites by integrating a deep learning algorithm and a random forest approach. Int. J. Biol. Sci. 2018;14:1669–1677. doi: 10.7150/ijbs.27819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B., Gao X., Zhang H. BioSeq-Analysis2.0: an updated platform for analyzing DNA, RNA and protein sequences at sequence level and residue level based on machine learning approaches. Nucleic Acids Res. 2019;47:e127. doi: 10.1093/nar/gkz740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z., Zhao P., Li F., Marquez-Lago T.T., Leier A., Revote J., Zhu Y., Powell D.R., Akutsu T., Webb G.I. iLearn: an integrated platform and meta-learner for feature engineering, machine-learning analysis and modeling of DNA, RNA and protein sequence data. Brief. Bioinform. 2020;21:1047–1057. doi: 10.1093/bib/bbz041. [DOI] [PubMed] [Google Scholar]
- 26.Chen W., Tang H., Ye J., Lin H., Chou K.C. iRNA-PseU: identifying RNA pseudouridine sites. Mol. Ther. Nucleic Acids. 2016;5:e332. doi: 10.1038/mtna.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen W., Lei T.Y., Jin D.C., Lin H., Chou K.C. PseKNC: a flexible web server for generating pseudo K-tuple nucleotide composition. Anal. Biochem. 2014;456:53–60. doi: 10.1016/j.ab.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Chen T.Q., Guestrin C. 2016. XGBoost: a scalable tree boosting system. In KDD ’16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; pp. 785–794. [Google Scholar]
- 29.Li Y., Niu M., Zou Q. ELM-MHC: an improved MHC identification method with extreme learning machine algorithm. J. Proteome Res. 2019;18:1392–1401. doi: 10.1021/acs.jproteome.9b00012. [DOI] [PubMed] [Google Scholar]
- 30.Liu K., Chen W., Lin H. XG-PseU: an eXtreme gradient boosting based method for identifying pseudouridine sites. Mol. Genet. Genomics. 2020;295:13–21. doi: 10.1007/s00438-019-01600-9. [DOI] [PubMed] [Google Scholar]
- 31.Liu B., Luo Z., He J. sgRNA-PSM: predict sgRNAs on-target activity based on position-specific mismatch. Mol. Ther. Nucleic Acids. 2020;20:323–330. doi: 10.1016/j.omtn.2020.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J., Gribskov M. IRESpy: an XGBoost model for prediction of internal ribosome entry sites. BMC Bioinformatics. 2019;20:409. doi: 10.1186/s12859-019-2999-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lv H., Zhang Z.-M., Li S.-H., Tan J.-X., Chen W., Lin H. Evaluation of different computational methods on 5-methylcytosine sites identification. Brief Bioinform. 2020;21:982–995. doi: 10.1093/bib/bbz048. [DOI] [PubMed] [Google Scholar]
- 34.Zhang M., Li F., Marquez-Lago T.T., Leier A., Fan C., Kwoh C.K., Chou K.C., Song J., Jia C. MULTiPly: a novel multi-layer predictor for discovering general and specific types of promoters. Bioinformatics. 2019;35:2957–2965. doi: 10.1093/bioinformatics/btz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding Y., Tang J., Guo F. Identification of drug-side effect association via multiple information integration with centered kernel alignment. Neurocomputing. 2019;325:211–224. [Google Scholar]
- 36.Li F., Wang Y., Li C., Marquez-Lago T.T., Leier A., Rawlings N.D., Haffari G., Revote J., Akutsu T., Chou K.C. Twenty years of bioinformatics research for protease-specific substrate and cleavage site prediction: a comprehensive revisit and benchmarking of existing methods. Brief. Bioinform. 2019;20:2150–2166. doi: 10.1093/bib/bby077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li F., Chen J., Leier A., Marquez-Lago T., Liu Q., Wang Y., Revote J., Smith A.I., Akutsu T., Webb G.I. DeepCleave: a deep learning predictor for caspase and matrix metalloprotease substrates and cleavage sites. Bioinformatics. 2020;36:1057–1065. doi: 10.1093/bioinformatics/btz721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li F., Zhang Y., Purcell A.W., Webb G.I., Chou K.-C., Lithgow T., Li C., Song J. Positive-unlabelled learning of glycosylation sites in the human proteome. BMC Bioinformatics. 2019;20:112. doi: 10.1186/s12859-019-2700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lundberg S.M., Lee S.I. Advances in Neural Information Processing Systems. MIT Press; Cambridge: 2017. A unified approach to interpreting model predictions; pp. 4765–4774. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.