Ulcerative colitis is a chronic colon inflammatory disease, the probiotics might alleviate UC symptoms by modulating the gut mucosal microbiota, our research has provided new insights into the mechanism of symptom alleviation in UC by applying probiotics‐based adjunctive treatment.

Summary
This was a pilot study aiming to evaluate the effects of probiotics as adjunctive treatment for ulcerative colitis (UC). Twenty‐five active patients with UC were assigned to the probiotic (n = 12) and placebo (n = 13) groups. The probiotic group received mesalazine (60 mg kg−1 day−1) and oral probiotics (containing Lactobacillus casei Zhang, Lactobacillus plantarum P‐8 and Bifidobacterium animalis subsp. lactis V9) twice daily for 12 weeks, while the placebo group received the same amounts of mesalazine and placebo. The clinical outcomes were assessed. The gut mucosal microbiota was profiled by PacBio single‐molecule, real‐time (SMRT) sequencing of the full‐length 16S rRNA of biopsy samples obtained by colonoscopy. A significantly greater magnitude of reduction was observed in the UC disease activity index (UCDAI) in the probiotic group compared with the placebo group (P = 0.043), accompanying by a higher remission rate (91.67% for probiotic‐receivers versus 69.23% for placebo‐receivers, P = 0.034). The probiotics could protect from diminishing of the microbiota diversity and richness. Moreover, the gut mucosal microbiota of the probiotic‐receivers had significantly more beneficial bacteria like Eubacterium ramulus (P < 0.05), Pediococcus pentosaceus (P < 0.05), Bacteroides fragilis (P = 0.02) and Weissella cibaria (P = 0.04). Additionally, the relative abundances of the beneficial bacteria correlated significantly but negatively with the UCDAI score, suggesting that the probiotics might alleviate UC symptoms by modulating the gut mucosal microbiota. Our research has provided new insights into the mechanism of symptom alleviation in UC by applying probiotic‐based adjunctive treatment.
Introduction
Ulcerative colitis (UC) is a chronic subtype of inflammatory bowel disease (IBD) that is characterized by bloody diarrhoea and abdominal pain (Ni et al., 2017). It is of great interest to improve the treatment strategy for UC. Anti‐inflammatory drugs, including mesalazine (5‐aminosalicylic acid), corticosteroids and azathioprine, are traditionally used for treating UC (Schwartz and Cohen, 2008). However, these standard drugs are not effective for all patients with UC, and some of them cause serious side effects, for example, leucopenia, infection, pancreatitis and increased risk of malignancy (Wilson et al., 2010). Hence, alternative treatments or adjuvant standard treatments are welcomed.
The pathogenesis of UC is still not entirely clear, but recent clinical evidence suggests close links between UC and gut dysbiosis. The inflammation in UC may be associated with certain gut bacteria and their influence to the overall intestinal microbiota. Since the gut microbiota plays an important role in regulating the host immunity, gut dysbiosis might cause host immune dysregulation and thus trigger chronic inflammatory disorders like UC (Miele et al., 2009; Matthes et al., 2010; Nishida et al., 2018). In general, patients with IBD have a greater number of pro‐inflammatory bacteria on their gut mucosa, particularly some members of the family Enterobacteriaceae (Swidsinski et al., 2005). On the other hand, some conflicting results have been reported. Previous studies found increased abundances of bifidobacteria and Faecalibacterium prausnitzii in the ileal mucosal samples of patients with IBD (Willing et al., 2010; Hill et al., 2014).
In recent years, treatment strategies targeting patients' gut microbiota have been suggested for the management of UC (Qi et al., 2018). Probiotics are live microorganisms that, when administered in adequate amounts, confer a beneficial effect to the host (Hill et al., 2014). Some studies have shown that the administration of probiotic preparations helped induce and maintain remission in adults with UC, for example, Escherichia coli Nissle 1917 (Matthes et al., 2010), some Bifidobacterium (B.) and Lactobacillus (L.) strains (Ganji‐Arjenaki and Rafieian‐Kopaei, 2018), VSL#3 (a mixture of eight probiotic bacteria, namely L. paracasei, L. plantarum, L. acidophilus, L. delbrueckii subsp. bulgaricus, B. longum, B. breve, B. infantis and Streptococcus thermophilus) (Miele et al., 2009; Tursi et al., 2010). However, even though many studies have reported encouraging symptom alleviation effects in patients with UC after probiotic consumption, few reports have further explored the mechanism behind, particularly the role of patients' gut microbiota in UC manifestations and remission (Tursi et al., 2005; Van Gossum, 2007).
This pilot study assessed the effects of the probiotic mix Probio‐Fit® (a highly concentrated probiotic product that contained L. casei Zhang, L. plantarum P‐8 and B. animalis subsp. lactis V9) as adjunctive treatment for UC when used with standard regimen. These probiotic strains have shown desirable in vivo effects in previous studies. For example, L. casei Zhang suppressed the serum level of tumour necrosis factor‐alpha while enhanced secretory immunoglobulin‐A and interleukin 2 in mice (Ya et al., 2008). The L. plantarum P‐8 strain reduced inflammation in adults of different ages (Wang et al., 2014). The B. animalis subsp. lactis V9 strain is a beneficial bacterium isolated from the gut microbiota of a healthy Mongolian child (Sun et al., 2010). Probio‐Fit® has been shown to improve diarrhoea in dogs by improving their gut microbiota composition and function (Xu et al., 2019). The primary efficacy outcomes of the current trial were the UC disease activity index (UCDAI) and clinical remission; meanwhile, changes in the gut mucosal microbiota of patients were monitored before and after the treatment.
Results
Participant flow and baseline characteristics
The participant flow and the patients’ baseline characteristics are illustrated in Figure 1 and Table 1, respectively. A total of 30 patients with UC (13 males and 17 females; average age of 44, range = 27–64) were enrolled at the start of the trial. However, five patients were disqualified based on exclusions. A total of 25 subjects accomplished the clinical trial. They were assigned to two groups (probiotic group, n = 12; placebo group, n = 13; Table 1). The gut mucosal samples of 18 individuals were collected at study entry and at week 12 (Fig. 1). Most patients suffered from rectal (14 patients) and left colon (8 patients) inflammation, while two patients suffered from pancolitis and one had inflammation at the sigmoid colon (Table 1).
Figure 1.
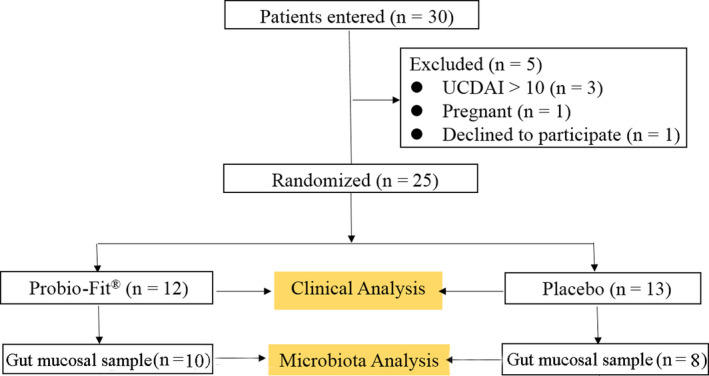
Study flow chart (ulcerative colitis disease activity index, UCDAI).
Table 1.
Baseline characteristics of patients.
| Characteristics | Placebo group | Probiotic group | P (Pearson’s chi‐square test) |
|---|---|---|---|
| Average age (range) | 47.9 (27–64) | 45.8 (28–63) | 0.512 |
| Gender (male: female) | 9:4 | 1:11 | 0.002 |
| Location of colon inflammation (number of patients) | |||
| Sigmoid colon | 1 | 0 | 0.626 |
| Left side | 5 | 3 | |
| Rectum | 6 | 8 | |
| Entire intestinal colon lining | 1 | 1 | |
Clinical outcome assessment at week 12
The clinical outcomes of patients were assessed at week 12. The clinical improvement was more obvious for the probiotic group (nine, two and one patients attained complete remission, partial remission and of no noticeable efficacy, respectively), while three, six and four patients attained complete remission, partial remission and lack of clinical efficacy for the placebo group (P = 0.034; Table 2). Thus, the overall remission rate was 91.67% for the probiotic group (versus 69.23% for the placebo group). Moreover, the probiotic‐receivers showed more significant reduction in stool frequency (P = 0.0035; Fig. 2A), UCDAI score (P = 0.043; Fig. 2B) and rectal bleeding rate (Pearson’s chi‐square test, P = 0.035; Table 2). Particularly, a higher number of probiotic‐receiving patients showed ≥ twofold reduction in UCDAI score at week 12, and none of the probiotic‐receivers showed increased UCDAI score at week 12. In contrast, one placebo‐receiver showed a steady UCDAI score, while another patient had an increased UCDAI score (Fig. 2C and D). Changes in these outcome parameters supported a significantly higher clinical efficacy of using probiotics as adjunctive therapy for UC.
Table 2.
Clinical outcomes after 12‐week treatment.
| Groups | Complete remission | Partial remission | Lack of efficacy | P (Pearson’s chi‐square test) | Rectal bleeding | P (Pearson’s chi‐square test) |
|---|---|---|---|---|---|---|
| Placebo | 3 | 6 | 4 | 0.034 | 6 | 0.035 |
| Probiotics | 9 | 2 | 1 | 1 |
Figure 2.
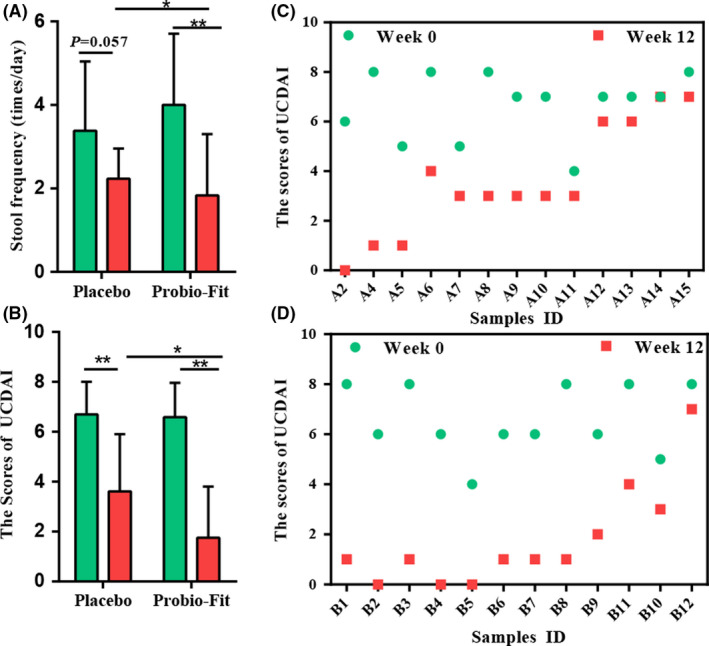
Effects of treatments on clinical indicator. (A) The effects of probiotics on the stool frequency. (B) The ulcerative colitis disease activity index (UCDAI). (C) Changes in UCDAI in individuals in the placebo group (n = 13). (D) Probiotic group (n = 12). The height of the bars represents the mean value, and error bars represent standard deviation. Significant differences are indicated by asterisks (*P < 0.05 and **P < 0.01, respectively; Pearson’s chi‐square test).
Treatment‐directed modulation of gut mucosal microbiota diversity and structure
The diversity and richness of UC gut mucosal microbiota were evaluated by the Shannon diversity index and Chao1 index, respectively (Fig. 3A and B). No significant differences (P > 0.05) were detected between the placebo and probiotic groups at week 0 and week 12 for both indexes. However, numerical decreases were observed in the microbiota diversity and richness of the placebo (received only mesalazine together with dextrin) but not the probiotic group (Fig. 3A and B). These results suggested that administering probiotics could help protect from the reduction in the mucosal microbiota diversity and richness caused by mesalazine treatment.
Figure 3.
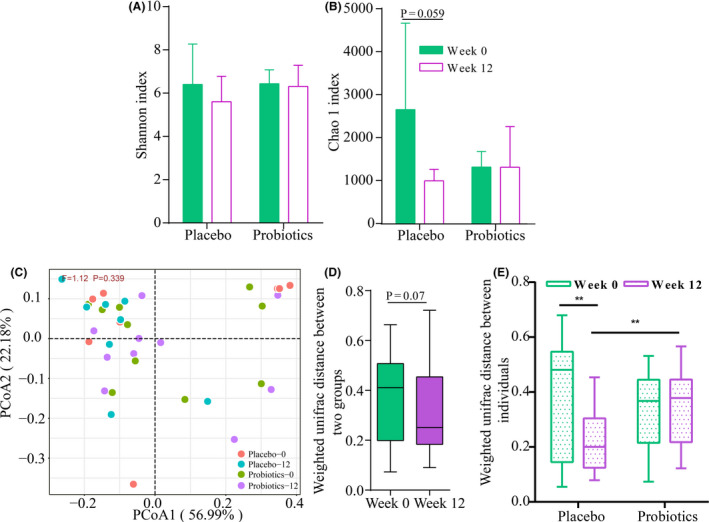
Effects of treatments on the ulcerative colitis (UC) gut mucosal microbiota. (A) Shannon index. (B) Chao1 index at week 0 and week 12. (C) Principal coordinates analysis (PCoA; weighted UniFrac distance) of UC gut mucosal microbiota of the two groups at week 0 and week 12. F‐value and P‐value on the PCoA score plots represent the difference of two groups calculated by permutational multivariate analysis of variance (PERMANOVA). (D) Difference in the weighted UniFrac distance between the two treatment groups at week 0 and week 12. (E) Difference in the weighted UniFrac distance among individuals within groups at week 0 and week 12 (*P < 0.05, **P < 0.01 and ***P < 0.001, respectively; Mann–Whitney test).
To explore how the UC gut mucosal microbiota responded to the probiotic administration, principal coordinates analysis (PCoA) was performed based on the weighted UniFrac distance. No obvious treatment‐based clustering pattern was seen on the PCoA score plot, which was confirmed by an insignificant difference among the four subgroups (F = 1.12, P = 0.339, PERMANOVA), suggesting that the probiotic supplementation did not produce a marked shift in the UC gut mucosal microbiota community (Fig. 3C). However, interestingly, although not significant, the weighted UniFrac distances between the gut mucosal microbiota of the two treatment groups were shorter at week 12 than week 0 (Fig. 3D). Additionally, the weighted UniFrac distances among the placebo‐receivers but not the probiotic‐receivers decreased significantly from week 0 to week 12 (P = 0.0039). Moreover, at week 12, the weighted UniFrac distance was significantly higher for the probiotic group compared with the placebo group (P = 0.0019), even though no significant difference was observed between the two groups at the baseline level (Fig. 3E).
Treatment‐directed changes in UC gut mucosal microbiota composition
To investigate treatment‐directed changes in the UC gut mucosal microbiota composition, the microbial taxonomic profiles of biopsy samples obtained at weeks 0 and 12 were investigated. The relative abundance of some genera markedly increased in the placebo group at week 12 compared with week 0 (P < 0.05), such as Streptococcus, Propionibacterium and Sphingomonas. On the other hand, for the probiotic group, the relative abundance of the genera Leuconostoc (P = 0.06) and Weissella (P = 0.08) increased, accompanied by the decrease in Pseudoflavonifractor at week 12 (P < 0.05; Fig. 4A). Additionally, at week 12, significantly more Weissella, Ruminococcus, Eubacterium, Blautia and Pediococcus were detected in the probiotic group compared with the placebo group, while significantly less Lactococcus and Meiothermus were found in the probiotic group compared with the placebo group (P < 0.05; Fig. 4B).
Figure 4.
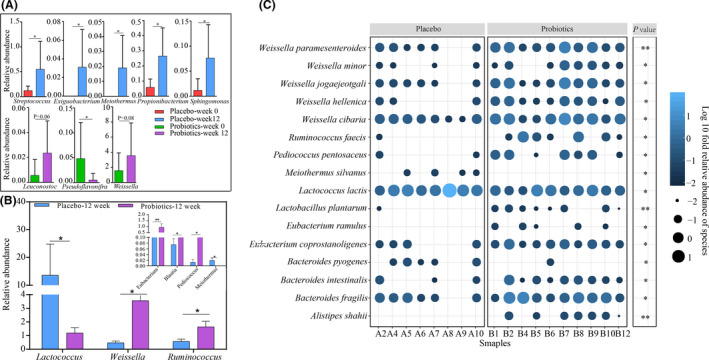
Effects of treatments on the gut mucosal microbiota composition of patients with ulcerative colitis. (A) Differential abundant bacteria identified in the two treatment groups at the genus level, comparison between two time points of the same group; differential abundant (B) genus and (C) species identified between the placebo and probiotic groups at week 12. *P < 0.05, **P < 0.01 and ***P < 0.001, respectively; Mann–Whitney test.
At the species level, 16 significant differential abundant species were found between the placebo and probiotic groups at week 12 (P < 0.05; Fig. 4C). The species Alistipes shahii, Eubacterium ramulus, L. plantarum, Pediococcus pentosaceus and Ruminococcus faecis were detected exclusively in the probiotic group, and their relative abundances increased significantly at week 12 in the probiotic group but not the control group (Fig. S1). Moreover, some other species of the probiotic group also increased significantly at week 12, including Bacteroides fragilis (P = 0.02), Bacteroides intestinalis (P = 0.04), Eubacterium coprostanoligenes (P = 0.02), Weissella cibaria (P = 0.04) and Weissella paramesenteroides (P = 0.004). In contrast, the species, Bacteroides pyogenes, Lactococcus lactis and Meiothermus silvanus, decreased significantly at week 12, and their relative abundances were significantly lower than the placebo group at week 12 (P < 0.05; Fig. 4C, Fig. S1).
Correlation between differential gut mucosal microbes and UCDAI
To identify how the changes in the gut mucosal microbes were associated with the symptom improvement (measured by the UCDAI score), a correlation network was constructed using Spearman’s rank correlation analysis (Fig. 5). Negative correlations were found between most of the differentially enriched bacterial species identified at week 12 of the probiotic group, such as members of the genera Pediococcus (Pediococcus pentosaceus) and Weissella (Weissella cibaria, Weissella paramesenteroides, Weissella jogaejeotgali). The relative abundances of the Weissella species showed correlated negatively with the UCDAI score, suggesting that these taxa might help relieve the symptoms of UC (Table 2).
Figure 5.
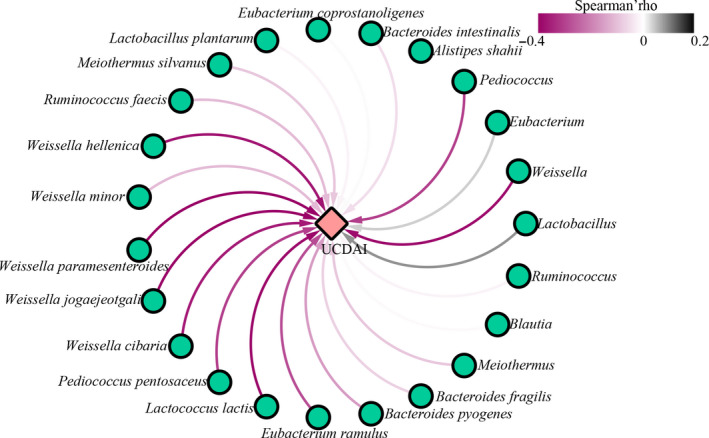
Spearman's correlation network plot of differential bacteria and ulcerative colitis disease activity index (UCDAI). The green circles and red diamond represent significantly modulated bacteria and UCDAI, respectively. Significant correlations between bacterial species and UCDAI are connected by curved lines. The line colour represents the correlation strength as illustrated by the colour scale of Spearman's rho, ranking between 0.2 and −0.4. A value greater than zero indicates a positive relationship, and vice versa.
Discussion
Ulcerative colitis is a chronic colon inflammatory disease, and it is commonly managed by administrating anti‐inflammatory medications like mesalazine. Although multiple contributory factors, including genetics, host inflammation and immunity, gut microbiota, and other potential environmental conditions, have been suggested to promote the development of UC, the precise aetiology and pathogenesis remain inconclusive. In recent years, much evidence has suggested a strong link between the host intestinal microbiota and UC pathogenesis, and providing the fact that the gut microbiota plays a key role in regulating the host immunity, UC pathogenesis could be relating to dysregulated interactions between the gut mucosal microbiota and host immunity (Guarner, 2008; Manichanh et al., 2012). Hence, apart from using conventional anti‐inflammatory medications, modulation of the intestinal microbiota has become a new target for adjunctive treatment (Qi et al., 2018).
Our results showed that significantly more pronounced beneficial effects and potential reduction in colon inflammation were observed in patients with UC who received the adjunctive treatment (mesalazine plus probiotics) compared with the placebo group. The probiotic‐receivers reported significantly lower UCDAI scores compared with subjects in the placebo group, and there was also more striking improvement in the remission ratio, stool frequency and rectal bleeding (P < 0.05). The efficacy of probiotics in managing active UC has previously been reported. For example, probiotics, such as VSL#3, Escherichia coli Nissle 1917 (Willing et al., 2010; Hill et al., 2014; Qi et al., 2018), and Saccharomyces boulardii (Guslandi et al., 2003), have been shown to induce clinical remission in patients with UC. However, the effectiveness of probiotics in maintaining clinical remission in patients with UC seems to be different. For example, a published clinical study observed no significant difference in clinical remission in patients with UC receiving Probio‐TecAB‐25 (a probiotic mix that contained L. acidophilus La‐5, and B. animalis subsp. lactis BB‐12) or placebo material (Wildt et al., 2011). One main reason for performing the current work was that many probiotics have shown strain‐specific effects in their health‐promoting effects. Thus, it would be necessary to perform independent clinical trials to characterize the specific beneficial function for each strain. The current results have shown for the first time the symptom alleviation effects and improvement of quality of life in patients with UC after the consumption of the applied probiotic mix.
Furthermore, since the gut mucosal microbiota might play role in the pathogenesis of UC, the present study monitored the changes in the gut mucosal microbiota upon the 12‐week treatment of mesalazine or mesalazine plus probiotics. Our data showed non‐significant reduction in the UC gut mucosal microbiota richness (represented by the Chao1 index) for both groups at week 12. However, interestingly, a higher magnitude of decrease was observed in the Chao1 index of the placebo group (P = 0.059) compared with the probiotic group (P = 0.54). The gut microbiota of patients with IBD was characterized by a diminished biodiversity owing to a shifted balance between commensal and potentially pathogenic microorganisms (Mosca et al., 2016). On the other hand, a previous study also reported that mesalazine treatment could reduce the mucosal bacteria of patients with UC (Swidsinski et al., 2005). Thus, the difference in the extent of reduction in gut mucosal microbial richness could be a beneficial effect of adjunctive treatment with probiotics in maintaining a relatively stable microbial richness.
Then, the overall structure of UC mucosal microbiota before and after the 12‐week treatment was analysed. The results of PCoA (weighted UniFrac distance) showed that the symptom alleviation effects were not due to drastic shift in the gut microbial structure. Yet, comparing with the placebo‐receivers, the average weighted UniFrac distance was significantly higher among the probiotic‐receivers with a greater within‐group variation at week 12. The increase in the UniFrac distance after the 12‐week probiotic treatment might be deemed as a desirable effect, as a recent study reported that the remission of UC patients was accompanied by an increase in the Bray–Curtis distance (Kinchen et al., 2018), another beta‐diversity index similar to the UniFrac distance. On the other hand, since the gut microbiota, its response and its resilience towards external stimuli are known to vary largely between individuals (Dethlefsen and Relman, 2011; Lichtman et al., 2016), the observed differences and changes could also be natural variability among individuals or responses driven by mesalazine treatment with/without probiotics. The intake of mesalazine has previously been reported to modulate the intestinal mucosal microbiota (Schirmer et al., 2018).
The PacBio SMRT technology is advantageous over other sequencing platforms in producing long sequence reads; thus, it is capable of describing microbiota profiles at a high taxonomic resolution when in combined use in sequencing full‐length metagenomic 16S rRNA genes (Toma et al., 2014). This study firstly applied such technology in profiling the gut mucosal microbiota composition to the species level. Several known butyrate‐producers that belonged to the Clostridium cluster XIV group, namely Ruminococcus (P < 0.05), Blautia (P < 0.05) and Eubacterium (P < 0.01), were significantly enriched in the probiotic group compared with the placebo group at week 12. Meanwhile, significantly more Weissella sequences were detected in the probiotic group at week 12 (P < 0.05). Weissella is a new genus of lactic acid bacteria that has been shown to suppress lipopolysaccharide‐induced pro‐inflammatory stress and confer other beneficial effects in mice (Singh et al., 2018). Our data showed that the relative abundance of Weissella correlated negatively and significantly with the UCDAI, suggesting a possible role of this genus in regulating the intestinal mucosal immunity of patients with UC. At the species level, five Weissella species were significantly enriched in the probiotic group at week 12. Among these differential abundant species, Weissella cibaria has been reported to enhance the activity of natural killer cells (Lee et al., 2018). Bacteroides fragilis was another species that was significantly enriched in the probiotic group at week 12. Generally, Bacteroides fragilis are classified into non‐toxigenic and toxigenic strains. Non‐toxigenic Bacteroides fragilis seem to exert beneficial effects to the host. They have been shown to induce interleukin 10 expression that helped protect against induced colitis (Chiu et al., 2014). Moreover, Bacteroides fragilis have been shown to promote immune system maturation and suppress abnormal inflammation, possibly via altering subjects’ intestinal microbiota structure (Chan et al., 2019; Fan et al., 2019). Bacteroides fragilis could also inhibit pathogenic bacteria. Thus, non‐toxigenic Bacteroides fragilis have been suggested as candidate probiotics (Li et al., 2017). In contrast, the enterotoxigenic Bacteroides fragilis strains could secrete the Bacteroides fragilis toxin (BFT), which induced severe colonic inflammation and tumour development in mice (Rabizadeh et al., 2007). The taxonomic resolution of 16S rRNA microbiota profiling in the current work was not fine enough to distinguish between non‐toxigenic and toxigenic strains of Bacteroides fragilis; thus, further work has to be done to confirm if the increased portion of Bacteroides fragilis was beneficial or potential harmful.
In addition, the adjunctive treatment with probiotics also induced the appearance of some beneficial bacteria in most samples, such as Eubacterium ramulus, L. plantarum and Ruminococcus faecis, which might have been present in an undetectable level at week 0. Eubacterium ramulus are human intestinal physiologic bacteria that have been demonstrated to degrade flavonoids (Braune et al., 2001). The probiotic mix used in this work contained L. plantarum; therefore, the increase in the relative abundance of this species could either be due to gut colonization of the ingested L. plantarum or a stimulatory effect of growth of gut lactobacilli. Lactobacillus plantarum are considered as probiotic bacteria that confer numerous beneficial effects to the host, such as anti‐inflammation (Gao et al., 2017). Overall, the probiotics modulated subjects’ gut mucosal microbiota, particularly enhanced the beneficial bacterial subpopulation, which helped reduce localized inflammation.
Although some clinical trials have shown a high efficacy of managing mild‐to‐moderate UC with probiotics (Willing et al., 2010; Hill et al., 2014; Qi et al., 2018), a previous study reported no significant clinical effect and time to relapse in UC patients compared with placebo (Matsuoka et al., 2018). The discrepancy could be due to strain‐specific effect of the applied probiotics. Among the reports that found clinical efficacy of administering probiotics, only few studies have successfully linked the clinical improvements with specific changes in the mucosal microbiota related to probiotic administration. Thus, the current work not only serves as additional evidence demonstrating the beneficial effect of ingesting probiotics in UC but also deciphers part of the physiologic mechanism by performed in‐depth analysis to identify changes in patients’ gut mucosal microbiota and their correlation with symptom alleviation.
In conclusion, the present study demonstrated that the application of probiotics in conjunction with mesalazine markedly increased the clinical efficacy of treatment for active UC compared with using the regular regimen alone. The improvement of clinical symptoms was accompanied by modest changes in patients' gut mucosal microbiota composition. This work has provided further insights into a potential mechanism of adjunctive treatment of UC by probiotic supplementation. However, a larger sample size would be desirable for further confirming the clinical efficacy of the current treatment. Future studies should also attempt to elucidate the treatment‐induced clinical effects from the perspective of gut microbial metabolism.
Experimental procedures
Ethics
All subjects provided written informed consent prior to the start of the study. The Ethical Committees of Inner Mongolia Agricultural University and the Affiliated Hospital of Inner Mongolia Medical University approved the study. The study was registered in the Chinese Clinical Trial Registry (chiCTR‐IPR‐17010306).
Patients
Since this work was a pilot study that aimed to study the effect of Probio‐Fit® in managing UC, power calculation was not employed. However, the number of patients recruited in this work was referenced to some similar studies published previously, in which 18–34 patients participated (Guslandi et al., 2003; Bibiloni et al., 2005; Furrie et al., 2005). Thus, a total of 30 patients who expressed willingness in complying to this work were recruited. They suffered from active UC for over three months between December 2016 and July 2017, and they visited the Department of Gastroenterology of the Affiliated Hospital of Inner Mongolia Medical University, Huhhot, China.
The severity of UC was assessed by the UCDAI score; that is, the total scores of four variables listed in Table 3 (D'Haens et al., 2007). Inclusion criteria: (i) age ≥ 18 years; (ii) mild‐to‐moderate UC patients (a UCDAI score ranging from three to ten); (iii) UC involving at least the rectosigmoid region. Exclusion criteria: a UCDAI score greater than ten; intake of additional medication, such as steroids, antibiotics and probiotics within four weeks before this study; pregnant or lactating women; smokers; individuals suffering from known cardiovascular diseases and renal insufficiency, malignant tumours, infectious disease and other digestive tract diseases, diabetes mellitus or autoimmune diseases; individuals that had previous abdominal surgical history. Five of the recruited patients were excluded (Fig. 1). The subjects were asked not to change from their usual diet during the course of the clinical trial except that they were requested to avoid from spicy, greasy and irritating diets.
Table 3.
Ulcerative colitis disease activity index (UCDAI).
| Score | Stool frequency | Rectal bleeding | Mucosal appearance | Disease activity rated by physicians |
|---|---|---|---|---|
| 0 | Normal | None | Normal | Normal |
| 1 | 1–2 Stools/day | Streaks of blood | Mild friability | Mild |
| 2 | 3–4 Stools/day | Obvious blood | Moderate friability | Moderate |
| 3 | > 4 Stools/day | Mostly blood | Exudation, spontaneous bleeding | Severe |
Study design
This was a prospective 12‐week randomized, double‐blind, placebo‐controlled study. A total of twenty‐five UC patients met the inclusion criteria; and they were randomly assigned to the probiotic (n = 12) or placebo (n = 13) group, respectively. All patients received the standard regimen (mesalazine at a dose of 60 mg kg−1 day−1). Additionally, the probiotic group received two sachets of probiotic product (Probio‐Fit®) per day orally, while the control group received two sachets of placebo material daily. Probio‐Fit® was available in individually sealed plastic sachets (Beijing Scitop Bio‐tech Shareholding, Beijing, China). Each sachet contained dextrin as the excipient and a mixture of three bacterial strains, that is, 3 × 109 CFU g−1 of L. casei Zhang, 3 × 109 CFU g−1 of L. plantarum P‐8, and 4 × 109 CFU g−1 of B. animalis subsp. lactis V9 (a total delivery of 2 × 1010 CFU g−1 per day). The placebo sachets appeared identical to the probiotic sachets, and they contained only dextrin.
Samples collection and clinical parameters
A complete physical examination, colonoscopy and histology of biopsy samples were performed before (week 0) and after (week 12) the course of treatment. Before colonoscopy, all patients fasted overnight and took 137.15 g of oral polyethylene glycol electrolyte powder (Shu Tai Shen Pharmaceutical, Beijing, China) for stool purging. For each biopsy sampling, three pieces of mucosal tissue were taken at the junction of the rectum and sigmoid colon with biopsy forceps during colonoscopy (colonoscope model: Pentax EC38‐I10F) after rinsing the intestinal mucosal surface thoroughly. The mucosa samples were quickly frozen in liquid nitrogen before transferring to −80°C refrigerator for storage. Only 18 sets of mucosal samples could be included in the microbiota analysis (ten and eight for the probiotic and placebo groups, respectively), as two patients disagreed with the biopsy sampling at week 0 while another five patients did not undergo the biopsy sampling at week 12.
Patients’ clinical symptoms (rating of symptom severity by physicians), endoscopic appearance, stool frequency, conditions of rectal bleeding, and UCDAI score were observed and recorded at week 0 and week 12. Each variable was graded on a scale of 0 (normal) to 3 (most severe). The UCDAI score ranged from 0 to 12 (most severe). Moreover, complete remission was defined as UCDAI score ≤ 2 (with scores of 0 point for both stool frequency and rectal bleeding severity; and at least a one‐point reduction in the mucosal appearance score from baseline). Partial remission was defined as UCDAI score ≤ 4 (with the sum of stool frequency and rectal bleeding severity scores of ≤ 1). A UCDAI score of ≥ 5 was considered as lack of efficacy.
Genomic DNA extraction, Polymerase Chain Reaction (PCR) and 16S rRNA gene sequencing
Microbiota analysis was performed on the gut mucosal samples of 18 patients taken at weeks 0 and 12. The sample genomic DNA was extracted with the QIAGEN DNA Stool Mini‐Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The quality of genomic DNA was assessed by agarose gel electrophoresis and spectrophotometric analysis. Samples with DNA concentration > 100 ng μl−1 and an optical density ratio (260 nm to 280 nm) between 1.8 and 2.0 were used for PCR.
The full‐length 16S rRNA genes were amplified from the extracted DNA by PCR using the forward 27F (5ʹ‐AGAGTTTGATCMTGGCTCAG‐3ʹ) and reverse 1492R (5ʹ‐ACCTTGTTACGACTT‐3ʹ) primers, with the introduction of a set of 16‐base barcodes for each sample (Mosher et al., 2013). The PCR program was as follows: 95°C for 4 min; 30 cycles of 95°C for 30 s, 60°C for 45 s and 72°C for 30 s with a final extension at 72°C for 5 min (Liu et al., 2015).
The barcoded‐amplicons were sequenced by PacBio single‐molecule, real‐time (SMRT) sequencing technology. Briefly, the 16S rRNA gene amplicons were used for constructing DNA libraries with the Pacific Biosciences SMRT Bell™ template prep kit 1.0. The sequencing reaction was performed with P6‐C4 chemistry on a PacBio RS II instrument (Pacific BioSciences of California, USA) following the guidelines of the manufacturer (Mosher et al., 2013).
Bioinformatics analyses
The protocol RS_ReadsOfinsert.1 in the SMRT Portal (version 2.7) was employed for extracting sequence information from the raw data. Raw sequences were strictly filtered to form a high‐quality sequence dataset, according to the criteria: (i) minimum full passes of up to 5; (ii) minimum predicted accuracy of 90; (iii) an insert read length of 1400 to 1800 bp. The filtered reads were then barcode‐sorted into different samples, followed by trimming the barcode and primer sequences.
The extracted high‐quality sequences were analysed by the Quantitative Insights into Microbial Ecology (QIIME) package (version 1.7) (Caporaso et al., 2010b). Briefly, a sequence from each cluster was chosen as the representative sequence to be aligned by pynast (Caporaso et al., 2010a) and uclust (Edgar, 2010) under 100% clustering of sequence identity. The representative sequences were classified into operational taxonomic units (OTUs) under the threshold of 97% identity using uclust (Haas et al., 2011). The OTUs within the dataset were assigned to the lowest taxonomic level using the Ribosomal Database Project II (RDP‐II, Release 11.5) and Greengenes (version 13.8) databases at a minimum bootstrap threshold of 80% (DeSantis et al., 2006; Cole et al., 2007). The de novo taxonomic tree constructed by the representative chimera‐checked OTU set with fasttree was used to assess the alpha‐ and beta‐diversity (Price et al., 2009).
To compare the sequencing depth and alpha‐diversity between samples, the Shannon–Wiener index and the number of observed species of each sample were calculated. The beta‐diversity, reflecting the microbiota community structure, was assessed by PCoA of the weighted UniFrac distance derived from the phylogenetic tree (Lozupone and Knight, 2005).
Data statement
Raw sequence data have been deposited to the MG‐RAST database under the project number mgp90220.
Statistical analyses
Statistical analyses were performed using R packages (http://www.r‐project.org/). Mann–Whitney test was used to assess differences in the UCDAI score, stool frequency and abundances of taxa between groups/time points. The severity of rectal bleeding and clinical efficacy between different groups were compared using chi‐square test (adjusted by Pearson’s chi‐square test). A P‐value < 0.05 was considered to be statistically significant for all statistical comparison. The Benjamini–Yekutieli method was used to control for multiple testing in assessing differences in microbiota composition between groups (Benjamini and Yekutieli, 2001). Graphs were generated by graphpad prism 6 and R packages. Correlations between gut mucosal bacteria and clinical parameters were calculated by using the Spearman's rank correlation coefficient in R package.
Conflict of interests
The authors declare that they have no conflict of interest.
Supporting information
Fig. S1. Relative abundances of some gut mucosal bacterial species of patients with ulcerative colitis at weeks 0 and 12. Error bars represent standard deviation, *P < 0.05 and **P < 0.01, respectively; Mann–Whitney test. P‐values are shown in cases where a marginal difference in relative abundance was found.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [Grant number 31720103911] and the Inner Mongolia Science & Technology Major Projects [Grant number ZDZX2018018].
Microbial Biotechnology (2020) 13(6), 2032–2043
Funding information
This work was supported by the National Natural Science Foundation of China [Grant number 31720103911] and the Inner Mongolia Science & Technology Major Projects [Grant number ZDZX2018018].
Contributor Information
XiaoFeng Zhou, Email: doctorzxf@126.com.
Heping Zhang, Email: hepingdd@vip.sina.com.
References
- Benjamini, Y. , and Yekutieli, D. (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188. [Google Scholar]
- Bibiloni, R. , Fedorak, R.N. , Tannock, G.W. , Madsen, K.L. , Gionchetti, P. , Campieri, M. , et al (2005) VSL#3 probiotic‐mixture induces remission in patients with active ulcerative colitis. Am J Gastroenterol 100: 1539–1546. [DOI] [PubMed] [Google Scholar]
- Braune, A. , Gutschow, M. , Engst, W. , and Blaut, M. (2001) Degradation of quercetin and luteolin by Eubacterium ramulus . Appl Environ Microbiol 67: 5558–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J.G. , Bittinger, K. , Bushman, F.D. , DeSantis, T.Z. , Andersen, G.L. , and Knight, R. (2010a) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J.G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F.D. , Costello, E.K. , et al (2010b) QIIME allows analysis of high‐throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J.L. , Wu, S. , Geis, A.L. , Chan, G.V. , Gomes, T.A.M. , Beck, S.E. , et al (2019) Non‐toxigenic Bacteroides fragilis (NTBF) administration reduces bacteria‐driven chronic colitis and tumor development independent of polysaccharide A. Mucosal Immunol 12: 164–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, C.C. , Ching, Y.H. , Wang, Y.C. , Liu, J.Y. , Li, Y.P. , Huang, Y.T. , et al (2014) Monocolonization of Germ‐Free Mice with Bacteroides fragilis protects against Dextran sulfate sodium‐induced acute colitis. Biomed Res Int 2014: 675786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, J.R. , Chai, B. , Farris, R.J. , Wang, Q. , Kulam‐Syed‐Mohideen, A.S. , McGarrell, D.M. , et al (2007) The ribosomal database project (RDP‐II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35: D169–D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis, T.Z. , Hugenholtz, P. , Larsen, N. , Rojas, M. , Brodie, E.L. , Keller, K. , et al (2006) Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen, L. , and Relman, D.A. (2011) Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA 108: 4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Haens, G. , Sandborn, W.J. , Feagan, B.G. , Geboes, K. , Hanauer, S.B. , Irvine, E.J. , et al (2007) A review of activity indices and efficacy end points for clinical trials of medical therapy in adults with ulcerative colitis. Gastroenterology 132: 763–786. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Fan, H. , Chen, Z. , Lin, R. , Liu, Y. , Wu, X. , Puthiyakunnon, S. , et al (2019) Bacteroides fragilis Strain ZY‐312 Defense against Cronobacter sakazakii‐Induced Necrotizing Enterocolitis In Vitro and in a Neonatal Rat Model. Msystems 4(4): e00305–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrie, E. , Macfarlane, S. , Kennedy, A. , Cummings, J.H. , Walsh, S.V. , O'Neil, D.A. , et al (2005) Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut 54: 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji‐Arjenaki, M. , and Rafieian‐Kopaei, M. (2018) Probiotics are a good choice in remission of inflammatory bowel diseases: a meta analysis and systematic review. J Cell Physiol 233: 2091–2103. [DOI] [PubMed] [Google Scholar]
- Gao, P. , Chen, M. , Zheng, S. , Wang, L. , Shi, H. , Su, X. , et al (2017) Feed‐additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken. Microbiome 5: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner, F. (2008) What is the role of the enteric commensal flora in IBD? Inflamm Bowel Dis 14: S83–S84. [DOI] [PubMed] [Google Scholar]
- Guslandi, M. , Giollo, P. , and Testoni, P.A. (2003) A pilot trial of Saccharomyces boulardii in ulcerative colitis. Eur J Gastroen Hepat 15: 697–698. [DOI] [PubMed] [Google Scholar]
- Haas, B.J. , Gevers, D. , Earl, A.M. , Feldgarden, M. , Ward, D.V. , Giannoukos, G. , et al (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454‐pyrosequenced PCR amplicons. Genome Res 21: 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, C. , Guarner, F. , Reid, G. , Gibson, G.R. , Merenstein, D.J. , Pot, B. , et al (2014) The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastro Hepat 11: 506–514. [DOI] [PubMed] [Google Scholar]
- Kinchen, J. , Chen, H.H. , Parikh, K. , Antanaviciute, A. , Jagielowicz, M. , Fawkner‐Corbett, D. , et al (2018) Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell 175: 372‐+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.J. , Lee, A. , Yoo, H.J. , Kim, M. , Noh, G.M. , and Lee, J.H. (2018) Supplementation with the probiotic strain Weissella cibaria JW15 enhances natural killer cell activity in nondiabetic subjects. J Funct Foods 48: 153–158. [Google Scholar]
- Li, Z. , Deng, H. , Zhou, Y. , Tan, Y. , Wang, X. , Han, Y. , et al (2017) Bioluminescence imaging to Track Bacteroides fragilis inhibition of Vibrio parahaemolyticus infection in mice. Front Cell Infect Microbiol 7: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman, J.S. , Ferreyra, J.A. , Ng, K.M. , Smits, S.A. , Sonnenburg, J.L. , and Elias, J.E. (2016) Host‐microbiota interactions in the pathogenesis of antibiotic‐associated diseases. Cell Rep 14: 1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W.J. , Zheng, Y. , Kwok, L.Y. , Sun, Z.H. , Zhang, J.C. , Guo, Z. , et al (2015) High‐throughput sequencing for the detection of the bacterial and fungal diversity in Mongolian naturally fermented cow's milk in Russia. BMC Microbiol 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone, C. , and Knight, R. (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh, C. , Borruel, N. , Casellas, F. , and Guarner, F. (2012) The gut microbiota in IBD. Nat Rev Gastro Hepat 9: 599–608. [DOI] [PubMed] [Google Scholar]
- Matsuoka, K. , Uemura, Y. , Kanai, T. , Kunisaki, R. , Suzuki, Y. , Yokoyama, K. , et al (2018) Efficacy of Bifidobacterium breve fermented milk in maintaining remission of ulcerative colitis. Dig Dis Sci 63: 1910–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes, H. , Krummenerl, T. , Giensch, M. , Wolff, C. , and Schulze, J. (2010) Clinical trial: probiotic treatment of acute distal ulcerative colitis with rectally administered Escherichia coli Nissle 1917 (EcN). BMC Complement Altern Med 10: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele, E. , Pascarella, F. , Giannetti, E. , Quaglietta, L. , Baldassano, R.N. , and Staiano, A. (2009) Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis. Am J Gastroenterol 104: 437–443. [DOI] [PubMed] [Google Scholar]
- Mosca, A. , Leclerc, M. , and Hugot, J.P. (2016) Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front Microbiol 7: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher, J.J. , Bernberg, E.L. , Shevchenko, O. , Kan, J. , and Kaplan, L.A. (2013) Efficacy of a 3rd generation high‐throughput sequencing platform for analyses of 16S rRNA genes from environmental samples. J Microbiol Meth 95: 175–181. [DOI] [PubMed] [Google Scholar]
- Ni, J. , Wu, G.D. , Albenberg, L. , and Tomov, V.T. (2017) Gut microbiota and IBD: causation or correlation? Nature Rev. Gastroenterol Hepatol 14: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, A. , Inoue, R. , Inatomi, O. , Bamba, S. , Naito, Y. , and Andoh, A. (2018) Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol 11: 1–10. [DOI] [PubMed] [Google Scholar]
- Price, M.N. , Dehal, P.S. , and Arkin, A.P. (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Q. , Liu, Y.N. , Jin, X.M. , Zhang, L.S. , Wang, C. , Bao, C.H. , et al (2018) Moxibustion treatment modulates the gut microbiota and immune function in a dextran sulphate sodium‐induced colitis rat model. World J Gastroenterol 24: 3130–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabizadeh, S. , Rhee, K.J. , Wu, S. , Huso, D. , Gan, C.M. , Golub, J.E. , et al (2007) Enterotoxigenic bacteroides fragilis: a potential instigator of colitis. Inflamm Bowel Dis 13: 1475–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer, M. , Denson, L. , Vlamakis, H. , Franzosa, E.A. , Thomas, S. , Gotman, N.M. , et al (2018) Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe 24: 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, M. , and Cohen, R. (2008) Optimizing conventional therapy for inflammatory bowel disease. Curr Gastroenterol Rep 10: 585. [DOI] [PubMed] [Google Scholar]
- Singh, S. , Bhatia, R. , Singh, A. , Singh, P. , Kaur, R. , Khare, P. , et al (2018) Probiotic attributes and prevention of LPS‐induced pro‐inflammatory stress in RAW264.7 macrophages and human intestinal epithelial cell line (Caco‐2) by newly isolated Weissella cibaria strains. Food Funct 9: 1254–1264. [DOI] [PubMed] [Google Scholar]
- Sun, Z.H. , Chen, X. , Wang, J.C. , Gao, P.F. , Zhou, Z.M. , Ren, Y. , et al (2010) Complete genome sequence of probiotic Bifidobacterium animalis subsp lactis Strain V9. J Bacteriol 192: 4080–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski, A. , Weber, J. , Loening‐Baucke, V. , Hale, L.P. , and Lochs, H. (2005) Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol 43: 3380–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma, I. , Siegel, M.O. , Keiser, J. , Yakovleva, A. , Kim, A. , Davenport, L. , et al (2014) Single‐molecule long‐read 16S sequencing to characterize the lung microbiome from mechanically ventilated patients with suspected pneumonia. J Clin Microbiol 52: 3913–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursi, A. , Brandimarte, G. , Giorgetti, G.M. , Forti, G. , Modeo, M.E. , Elisei, W. , et al (2005) Low‐dose balsalazide plus high‐potency probiotic preparation is more effective than balsalazide alone or mesalazine in the treatment of acute mild‐to‐moderate ulcerative colitis. Gastroenterology 128: A17. [PubMed] [Google Scholar]
- Tursi, A. , Brandimarte, G. , Papa, A. , Giglio, A. , Elisei, W. , Giorgetti, G.M. , et al (2010) Treatment of relapsing mild‐to‐moderate ulcerative colitis with the probiotic VSL#3 as adjunctive to a standard pharmaceutical treatment: a double‐blind, randomized, placebo‐controlled study. Am J Gastroenterol 105: 2218–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gossum, A. (2007) Probiotics and inflammatory bowel diseases (IBD). Nutr Clin Metab 21: 81–84. [Google Scholar]
- Wang, L. , Zhang, J. , Guo, Z. , Kwok, L. , Ma, C. , Zhang, W. , et al (2014) Effect of oral consumption of probiotic Lactobacillus planatarum P‐8 on fecal microbiota, SIgA, SCFAs, and TBAs of adults of different ages. Nutrition 30: 776–783. [DOI] [PubMed] [Google Scholar]
- Wildt, S. , Nordgaard, I. , Hansen, U. , Brockmann, E. , and Rumessen, J.J. (2011) A randomised double‐blind placebo‐controlled trial with Lactobacillus acidophilus La‐5 and Bifidobacterium animalis subsp lactis BB‐12 for maintenance of remission in ulcerative colitis. J Crohns Colitis 5: 115–121. [DOI] [PubMed] [Google Scholar]
- Willing, B.P. , Dicksved, J. , Halfvarson, J. , Andersson, A.F. , Lucio, M. , Zheng, Z. , et al (2010) A Pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139: 1844–1854.e1. [DOI] [PubMed] [Google Scholar]
- Wilson, D.C. , Thomas, A.G. , Croft, N.M. , Newby, E. , Akobeng, A.K. , Sawczenko, A. , et al (2010) Systematic review of the evidence base for the medical treatment of paediatric inflammatory bowel disease. J Pediatr Gastr Nutr 50: S14–S34. [DOI] [PubMed] [Google Scholar]
- Xu, H. , Zhao, F. , Hou, Q. , Huang, W. , Liu, Y. , Zhang, H. , et al (2019) Metagenomic analysis revealed beneficial effects of probiotics in improving the composition and function of the gut microbiota in dogs with diarrhoea. Food Funct 10: 2618–2629. [DOI] [PubMed] [Google Scholar]
- Ya, T. , Zhang, Q.J. , Chu, F.L. , Merritt, J. , Bilige, M.H. , Sun, T.S. , et al (2008) Immunological evaluation of Lactobacillus casei Zhang: a newly isolated strain from koumiss in Inner Mongolia, China. Bmc Immunol 9: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Relative abundances of some gut mucosal bacterial species of patients with ulcerative colitis at weeks 0 and 12. Error bars represent standard deviation, *P < 0.05 and **P < 0.01, respectively; Mann–Whitney test. P‐values are shown in cases where a marginal difference in relative abundance was found.


