This paper describes molecular and functional characterization of a novel dioxygenase enzyme from the aquatic cyanobacterium Anabaena cylindrica. The enzyme showed a strong inhibition by excess of the precursor L‐DOPA. However, its heterologous expression has allowed detecting the formation of the main compounds in the biosynthetic pathway of betalains. The discovery of this novel dioxygenase in the phylum cyanobacteria shows that betalains' formation is more diverse than expected.

Summary
The biosynthesis of betalamic acid, the structural unit of pigments betalains, is performed by enzymes with 4,5‐DOPA‐extradiol‐dioxygenase activity. These enzymes were believed to be limited to plants of the order Caryophyllales and to some fungi. However, the discovery of Gluconacetobacter diazotrophicus as the first betalain‐forming bacterium opened a new field in the search for novel biological systems able to produce betalains. This paper describes molecular and functional characterization of a novel dioxygenase enzyme from the aquatic cyanobacterium Anabaena cylindrica. The enzyme was found to be a homodimer of a polypeptide of 17.8 kDa that, opposite to previous related enzymes, showed a strong inhibition by excess of the precursor L‐DOPA. However, its heterologous expression has allowed detecting the formation of the main compounds in the biosynthetic pathway of betalains. In addition, phylogenetic analysis has shown that this enzyme is not close related to enzymes from plants, fungi or proteobacteria such as G. diazotrophicus. The presence of enzymes that produce these health‐promoting compounds is more diverse than expected. The discovery of this novel dioxygenase in the phylum cyanobacteria expands the presence of betalamic acid‐forming enzymes in organisms of different nature with no apparent relationship among them.
Introduction
Betalains are a class of hydrophilic, nitrogen‐containing pigments present in plants of the order Caryophyllales which lack the most common pigments anthocyanins (Gandía‐Herrero and García‐Carmona, 2013). These two families of pigments are mutually exclusive, and they have never located together in the same plant (Castellanos‐Santiago and Yahia, 2008; Brockington et al., 2011). Betalains have high antioxidant and free radical scavenging activities in vitro (Gandía‐Herrero et al., 2010) and they show a health‐promoting effect in vivo, expanding the lifespan of Caenorhabditis elegans (Guerrero‐Rubio et al., 2019a). Several studies have probed its importance as chemopreventive molecules in different lines of cancer cells (Sreekanth et al., 2007) and a reduction of tumours in mice after their intake (Lechner et al., 2010). In addition, betalain‐rich extracts have a positive activity in humans (Pietrzkowski et al., 2010; Rahimi et al., 2019) which is probably related to the antioxidant and anti‐inflammatory activities of betalain pigments (Vidal et al., 2014).
Betalains are classified into two groups that share betalamic acid as structural and chromophoric unit. This molecule is able to condense with amines or amino acids, producing the yellow pigments betaxanthins, or to condense with cyclo‐DOPA, producing the violet pigments betacyanins. The biosynthesis of betalamic acid is one of the main steps in the betalains’ production (Gandía‐Herrero and García‐Carmona, 2013), and its formation implies the action of the enzyme 4,5‐DOPA‐extradiol‐dioxygenase (4,5‐DODA). The enzyme 4,5‐DODA catalyses the ring cleavage of L‐DOPA, giving rise to 4,5‐seco‐DOPA, an intermediate which spontaneously yields betalamic acid (Fig. 1) (Sasaki et al., 2009). Betalain‐related pigments are also present in some fungi as Amanita (Stintzing and Schliemann, 2007) and Hygrocybe (Babos et al., 2011). In addition, dioxygenase activity similar to the fungi’s enzymes has been detected in Escherichia coli (Gandía‐Herrero and García‐Carmona, 2014). These enzymes produce, besides betalamic acid, muscaflavin, a related pigment produced due to an 2,3‐dioxygenase activity (Girod and Zryd, 1991; Mueller et al., 1997).
Fig. 1.
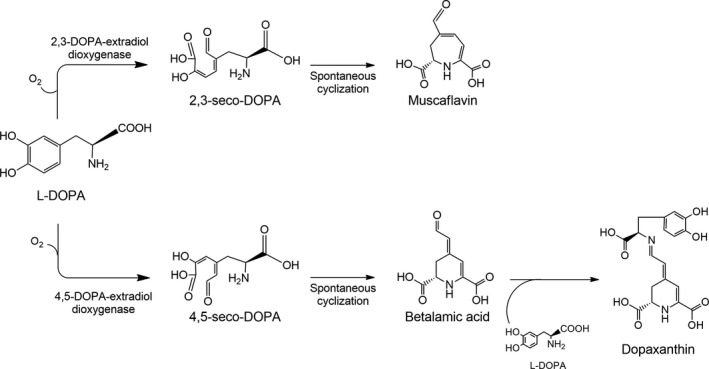
Scheme of activity of DOPA‐extradiol‐dioxygenase (DODA) enzymes. DODA enzymes catalyse the cleavage of L‐DOPA between C2 and C3, yielding 2,3‐seco‐DOPA, or between C4 and C5, yielding 4,5‐seco‐DOPA. Both compounds are intermediates which, by spontaneous cyclization, give rise to muscaflavin and betalamic acid respectively. The condensation of betalamic acid with L‐DOPA yields dopaxanthin.
Our last studies revealed that betalamic acid is not only present in plants of order Caryophyllales and fungi Amanita and Hygrocybe but it is also present in bacteria as Gluconacetobacter diazotrophicus, the first betalain‐producing bacterium described (Contreras‐Llano et al., 2019). G. diazotrophicus contains the smallest DODA enzyme (17.8 kDa) able to produce betalamic acid and muscaflavin. Besides, the high stability of this enzyme has led its heterologous expression in a recombinant system, the fully characterization and time evolution of intermediates 2,3‐seco‐DOPA and 4,5‐seco‐DOPA and the development of biofactories for betalains’ production (Guerrero‐Rubio et al., 2019b).
In the present study, we expand the range of betalain‐producing bacteria describing the cloning, expression, purification, molecular and functional characterization of a DOPA‐extradiol‐dioxygenase enzyme from Anabaena cylindrica, the first cyanobacterium described producing betalamic acid.
Results and discussion
Anabaena cylindrica cultures produce betalamic acid
The discovery of Gluconacetobacter diazotrophicus as the first betalain‐forming bacterium (Contreras‐Llano et al., 2019) has opened a new field in the search for novel biological systems able to produce betalains. Previous phylogenetic analysis showed sequence homology for the dioxygenase enzyme of Gluconacetobacter in several proteobacteria, such as Bradyrhizobium, Komagataeibacter, Mesorhizobium and Inquilinus. An extended phylogenetic analysis performed in this work has shown the presence of homologous proteins not only in proteobacteria genus but also in the phylum cyanobacteria (Fig. S1). Within the sequences found in this analysis, several genera belonging to the order Nostocales, such as Anabaena, Nostoc, Trichormus or Aphanizomenon, showed sequence similarity with the dioxygenase enzyme of G. diazotrophicus. Among them, A. cylindrica, an autotrophic filamentous bacterium with nitrogen‐fixing capabilities, was identified and chosen as a representative species of the phylum cyanobacteria to study the possible betalamic acid‐producing abilities in this group of water organisms. Anabaena cylindrica is a worldwide‐distributed species which is well known for its use in nitrogen‐fixing studies since many decades (Allen and Arnon, 1955). However, there is no evidence on the existence of the betalains pathway in this species and a betalamic acid‐forming activity has never been described. Thanks to the interest in this aquatic microorganism its genome has been completely sequenced (Shih et al., 2013). A. cylindrica cultures enriched with increasing concentrations of L‐DOPA up to its solubility limit at 7.6 mM were employed. Preliminary assays of A. cylindrica cultures supplemented with L‐DOPA as a precursor in the formation of betalamic acid did not produce any yellow pigmentation, but they produced the darkening of the medium. It was hypothesized that this was due to the production of DOPA‐chrome, a product derived from L‐DOPA in the production of melanins by oxidase enzymes. Previous studies have reported that ascorbic acid or an analogous reducing agent is necessary in the biosynthesis pathway to recover the pool of L‐DOPA, thus avoiding the accumulation of DOPA‐chrome (Gandía‐Herrero and García‐Carmona, 2013). The addition of sodium ascorbate 15 mM to the media did not produce the expected results, and the conversion of L‐DOPA to DOPA‐chrome was again evident from the darkening of culture media.
Then, it was considered that this might be due to the presence of an ascorbate peroxidase particularly active in A. cylindrica, demonstrated to be involved in the scavenging of hydrogen peroxide (Miyake et al., 1991). This enzyme is able to capture sodium ascorbate molecules, by reducing them and limiting their bioavailability to revert the conversion of L‐DOPA to DOPA‐chrome. Taking these considerations into account, EDTA 100 mM was also added to the reaction media in order to avoid the reduction of sodium ascorbate. 7‐day cultures of A. cylindrica were supplemented with L‐DOPA at concentrations 0.23, 0.76, 1.90, 3.80 and 7.60 mM, 15 mM sodium ascorbate and 100 mM EDTA. Non‐supplemented cultures were employed as controls. After two days, supernatants were analysed by HPLC‐ESI/TOF/MS and pure pigment dopaxanthin was employed as a standard to check out the activity of a hypothetical DODA enzyme. This betaxanthin is the result of the condensation of betalamic acid with L‐DOPA, and its presence reveals the formation of betalamic acid from L‐DOPA through the presence of a 4,5‐DOPA‐extradiol‐dioxygenase enzyme. Control pigment analysis yielded a retention time of 12.1 min with an exact mass value of 391.1136 m/z (Fig. 2). The same peak was obtained in cultures of A. cylindrica supplemented with 3.8 and 7.6 mM of L‐DOPA but it was not found at lower concentrations of the added substrate neither in its absence.
Fig. 2.
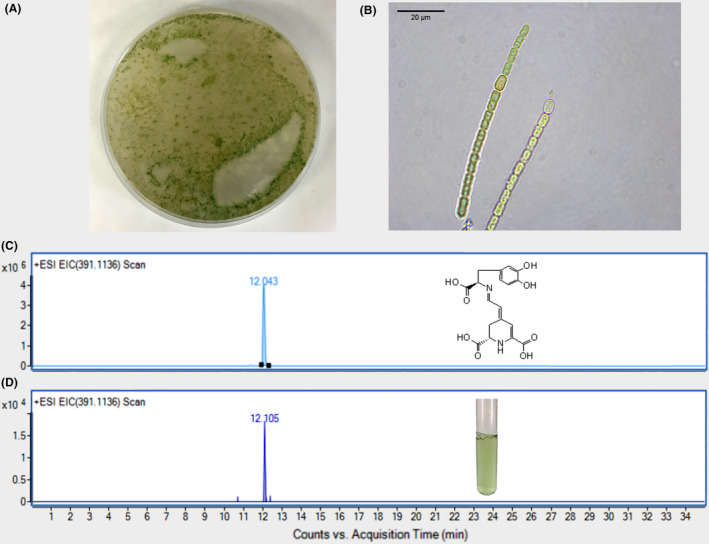
Detection of dopaxanthin in cultures of Anabaena cylindrica supplemented with L‐DOPA.
A. 3‐day plate of BG‐11 medium containing A. cylindrica.
B. Microscopy image of the filamentous distribution of A. cylindrica cells. Scale bar: 20 μM.
C and D. Chromatograms from HPLC‐ESI‐TOF‐MS analysis. Standard dopaxanthin was followed at EIC 391.1136 m/z (C) and the same peak was detected in culture media of A. cylindrica supplemented with L‐DOPA 7.6 mM, 15 mM sodium ascorbate and 100 mM EDTA (D).
Therefore, the existence of a betalamic acid‐forming DODA activity in A. cylindrica has been demonstrated. This metabolism does not seem to be active or relevant in the development of A. cylindrica but it is able to produce betalamic acid when the precursor amino acid is present in the medium. The betalamic acid produced condenses with L‐DOPA to yield its derived betaxanthin, dopaxanthin. A regulatory system for the 4,5‐DOPA‐extradiol‐dioxygenase activity or the biosynthesis of betalains in microorganisms has not been described. However, in A. cylindrica it is here demonstrated that ascorbate peroxidase plays a role in limiting the access of the main enzyme in the pathway to the initial substrate L‐DOPA. Betalamic acid and the final pigment dopaxanthin can only be produced by the cyanobacteria when the activity of the peroxidase enzyme is lowered. This may point out to a yet unknown regulatory system that control expression and activity of betalamic acid‐forming dioxygenases in microorganisms. In plants, expression of 4,5‐DOPA‐extradiol dioxygenase is highly controlled and it only appears in specific organs and developmental stages (Sasaki et al., 2009; Bean et al., 2018).
A. cylindrica 4,5‐DODA sequence, expression and purification
The search for non‐plant L‐DOPA dioxygenases able to produce betalamic acid ‐ and thus to start the biosynthetic pathway of betalains in prokaryotes ‐ has led us to search for sequences related to that described in G. diazotrophicus. Phylogenetic analysis shows a sequence susceptible to produce betalains in the genome of A. cylindrica. The genome of this autotrophic cyanobacterium is deposited in the NCBI database with the access number NZ_AP018166.1 (project PRJDB5665). Its chromosome presents a size of 7.06 Mb where the sequence for the protein WP_015213489.1, corresponding to the 4,5‐DODA sequence, is located with the locus tag CA722_RS27340. The sequence for the protein WP_015213489.1 is accessible from the databases and was here used as a template to obtain a synthetic sequence optimized for expression in E. coli. This optimization introduced mutations which did not change the amino acid sequence coded. The modified sequence was inserted into the multiple cloning site of the expression vector pRSETA, obtaining the recombinant plasmid pRSETA‐AcDODA.
This plasmid was used to transform thermocompetent E. coli BL21 (DE3) cells. For an increased expression of activity in transformed E. coli cultures, optimization conditions of IPTG concentration and temperature of induction was performed using IPTG at final concentrations of 0.1, 0.5 and 1 mM and temperatures of 20 and 25 °C. In all cases, 20 h after induction L‐DOPA 7.6 mM and ascorbate 15 mM were added to the cultures and dopaxanthin formation was evaluated by HPLC. This measure allowed to establish the optimal conditions for the activity, and thus protein expression. The highest dopaxanthin content was achieved after induction with 0.1 mM IPTG at 20 °C, as shown in Fig. S2. These conditions were then employed for protein purification.
The purified enzyme was run on an SDS‐PAGE electrophoresis gel, revealing that the recombinant A. cylindrica DODA was purified to homogeneity after affinity separation. A higher amount of pure protein was obtained when the protein solution was eluted at pH 7.5 (Fig. 3A). The results of the purification process are shown in Table 1, where it can be seen that a purification yield of 75% and a purification fold of 1.6 were obtained, in accordance with the electrophoretic estimation of DODA expression.
Fig. 3.
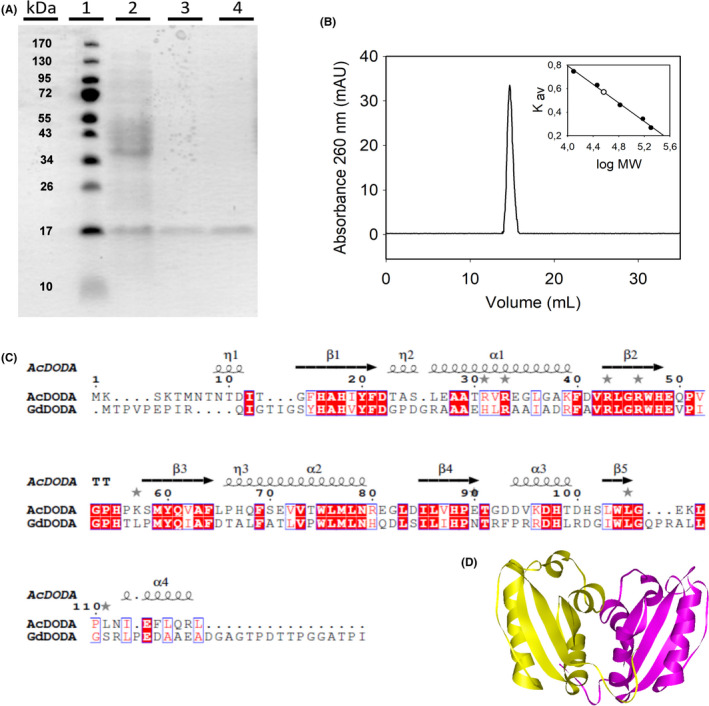
Characterization of DODA enzyme from Anabaena cylindrica.
A. SDS‐PAGE electrophoretic analysis of the expression and purification of AcDODA from E. coli BL21 cells. Lane 1, molecular weight markers; lane 2, soluble protein content of cells harvested 20 h after IPTG induction (0.1 mM); lane 3, eluted protein after affinity chromatography purification; and lane 4, final purified protein obtained after desalting on PD‐10 columns.
B. Analysis of the AcDODA enzyme by gel filtration chromatography. Elution was followed at a wavelength of λ = 260 nm. Inset: Calibration curve and molecular mass determination of the detected dimer.
C. Comparison of DODA sequences from A. cylindrica and G. diazotrophicus. Sequence alignment with structural considerations was performed with Expresso (Armougom et al., 2006). Conserved blocks of amino acids are squared. Strictly conserved residues are shown in red.
D. Structural model for the dimer of DODA enzyme from A. cylindrica by using the template PDB‐ID 2PEB from Nostoc punctiforme. The two monomeric units are shown in yellow and purple.
Table 1.
Expression and purification of Anabaena cylindrica dioxygenase.
| Volume (ml) | Protein (mg ml−1) | Total Prot. (mg) | Activity (µM min−1) a | Specific Activity (µmol min−1 mg−1) | Purif. fold | Yield (%) | |
|---|---|---|---|---|---|---|---|
| Crude extract b | 7.0 | 6.18 | 43.29 | 0.743 | 0.773 | 1.0 | 100 |
| Ni2+ chrom. | 7.0 | 2.82 | 19.7 | 0.594 | 1.264 | 1.6 | 75 |
Activity was determined using 100 µl of protein solution under the assay conditions.
Crude extract was obtained from a cellular paste harvested from a 0.5 l culture.
Molecular characterization
The band corresponding to the recombinant protein obtained by SDS‐PAGE yields an estimated molecular mass of 17 kDa (Fig. 3), consistent with the molecular weight calculated according to the protein sequence (17.8 kDa). The purified protein was analysed by HPLC‐ESI‐MS‐TOF in order to determine its accurate molecular mass. Mass spectra showed a single peak with a molecular mass of 17.806 kDa, which agrees with the above‐mentioned values obtained by SDS‐PAGE and for the theorical molecular weight. To fully characterize the protein, its peptide mass fingerprint was determined by MALDI‐TOF analysis after trypsin digestion. The main peptides identified corresponded to the masses 437.89 m/z [(R)WHEQPVGPHPK(S)], 1052.51 m/z [(R)EGLDILVHPETGDDVKDHTDHSLWLGEK(L)], 621.86 m/z [(K)LPLNIEFLQR(L)], 479.89 m/z [(K)DHTDHSLWLGEK(L)], 868.94 m/z [(R)EGLDILVHPETGDDVK(D)].
The purified protein was subjected to gel filtration chromatography on a Superdex 200 column under native conditions in order to determine whether this DODA enzyme is a monomer or if it forms oligomers. In this sense, different concentrations of purified protein were employed to investigate the possibility of an equilibrium among the monomer and the possible oligomers. A single peak eluting at the same volume was obtained in all cases, with a molecular mass estimated of 32.9 kDa (Fig. 3). This value doubles the expected mass according to the SDS‐PAGE and TOF analyses. Therefore, the results obtained by gel filtration show that the DODA protein of A. cylindrica is a dimer under native conditions.
These results strongly support the sequence homology found by a search performed on the databases for sequences with known structures. The A. cylindrica DODA has 71.3% identity (83.0% similarity) with a putative dioxygenase (PDB entry 2PEB) enzyme characterized from Nostoc punctiforme PCC 73102 (additional information can be found in Appendix S1). X‐ray crystallography at 1.46 Å resolution has demonstrated that this protein is a dimer composed of two monomers of 122 amino acids each. The similarity between these two enzymes agrees with the molecular characterization performed for AcDODA in the present study. In addition, A. cylindrica DODA residues can be assigned to specific secondary motifs also found in the enzyme from the only prokaryote known to produce betalains, G. diazotrophicus, by structurally assisted sequence comparison (Armougom et al., 2006; Robert and Gouet, 2014). Figure 3C shows the structural similitude between both enzymes with respect to the residues involved in the formation of β‐strands and α‐helixes. Taking into account the molecular similarities between the A. cylindrica DODA and G. diazotrophicus enzymes and with the protein crystalized from N. punctiforme, a three‐dimensional model of the novel DODA from A. cylindrica was performed (Biasini et al., 2014; Bienert et al., 2017). The resulting structure can be found in Fig. 3D where a dimer modelled by the bioinformatic techniques is shown. The predicted dimeric protein finds experimental support in Fig. 3B where the multimeric nature of the novel enzyme is unambiguously determined.
Kinetic characterization
The activity of the recombinant DODA enzyme from A. cylindrica was characterized spectrophotometrically by its addition to a reaction medium with the amino acid L‐DOPA as substrate. The addition of the enzyme yielded a yellow coloration with a λmax of 414 nm. Spectral changes were not observed in the absence of the enzyme and, therefore, they were considered proof of the activity of AcDODA. The described activity agrees with the absorbance properties reported for betalamic acid and muscaflavin extracted from plants and fungi (Trezzini and Zrÿb, 1991; Mueller et al., 1997; Gandía‐Herrero et al., 2012) and for those formed in vitro by the enzyme from the fungus A. muscaria (Girod and Zryd, 1991), from the plants B. vulgaris (Gandía‐Herrero and García‐Carmona, 2012) and M. jalapa (Sasaki et al., 2009) and from the bacterium G. diazotrophicus (Contreras‐Llano et al., 2019).
The highest activity for the novel DODA enzyme was determined at pH 7.0 (Fig. 4C). Dependence of the activity rate on substrate concentration was analysed at the optimum pH determined. In these assays, AcDODA enzyme showed a reduction of its activity at high L‐DOPA concentrations, and thus a strong inhibition by excess of L‐DOPA as clearly shown in Fig. 4A. This phenomenon is known as inhibition by excess of substrate. In this kinetic model, the plot of the double‐reciprocal (inverse activity rate as a function of the inverse of L‐DOPA concentration) provides a curve like that shown in Fig. 4B (Segel, 1975). The equation for the obtained curve showed the kinetic parameters estimated as K m = 53 M and V max = 105.8 mM min−1, and a substrate inhibition constant of 31.9 μM. The kinetic mechanism and its corresponding equation are shown in Fig. S3. The values determined are far from those obtained for previously characterized DODA enzymes, which did not show substrate inhibition for L‐DOPA. Previous works reported that values determined for Km were 6.9 mM for B. vulgaris 4,5‐DODA (Gandía‐Herrero and García‐Carmona, 2012), 7.9 mM for E. coli YgiD (Gandía‐Herrero and García‐Carmona, 2014), 3.9 mM for A. Muscaria dioxygenase (Girod and Zryd, 1991) and 1.4 mM for G. diazotrophicus dioxygenase (Contreras‐Llano et al., 2019). Thus, the novel enzyme here described presents a singular behaviour with respect to the activity in the presence of L‐DOPA as substrate.
Fig. 4.
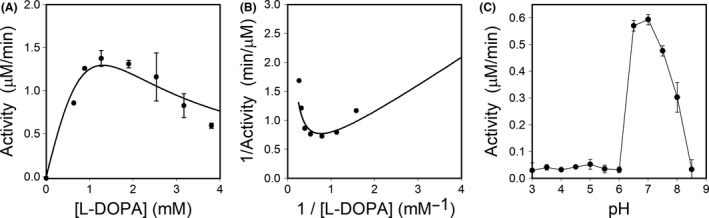
A. cylindrica dioxygenase activity characterization.
A. Enzyme activity at different concentrations of L‐DOPA, measured in 50 mM sodium phosphate buffer pH 7.0.
B. Double‐reciprocal plot showing the inverse activity rate as a function of the inverse of substrate concentration.
C. Effect of pH on the activity of the novel dioxygenase enzyme. Reactions were performed with 2.5 mM L‐DOPA in 50 mM sodium acetate buffer for pH values ranging from 3.5 to 5.0 and in 50 mM sodium phosphate buffer for pH values ranging from 5.5 to 8.5.
Figure 3 shows how AcDODA and GdDODA have a close structural similarity with respect to the residues involved in the formation of β‐strands and α‐helixes. However, the residues that differ between these enzymes could be involved in the kinetic properties of AcDODA. Some of these residues could produce a three‐dimensional conformation that affects the union of L‐DOPA to the active site. Kinetically the meaning of the inhibition by excess of substrate indicates the possibility of the blockage of the catalytic active site by the access of more than one molecule of substrate. Although the biological significance is unclear, this may function as a relevant regulatory mechanism that has not been described in plants and fungi (Reed et al., 2010).
Functional characterization
Escherichia coli (pRSETA‐AcDODA) expressing the novel DODA from A. cylindrica was used to study the evolution of the intermediates of the biosynthetic pathway of betalains and for the biotechnological obtention of their products. The induced expression of the enzyme was performed in LB media under optimal conditions of temperature and IPTG concentration as mentioned above. Then, the reaction was followed for 100 h by the analysis of culture supernatants by HPLC. As Fig. 5 shows, the formation of intermediates 2,3‐ and 4,5‐seco‐DOPA occurs in the first hours of reaction and these reached the maximum concentrations after 7 h. They were detected at λ = 360 nm with retention times of 6.7 and 7.1 min respectively. Betalamic acid was detected at λ = 405 nm with a retention time of 13.8 min and a minor peak was detected with a λmax of 403 nm and a retention time of 15.5 min. This peak corresponds to muscaflavin. HPLC analysis also showed a peak with a λmax of 470 nm and a retention time of 13.2 min, corresponding to dopaxanthin, which is accumulated due to the condensation of betalamic acid and L‐DOPA.
Fig. 5.
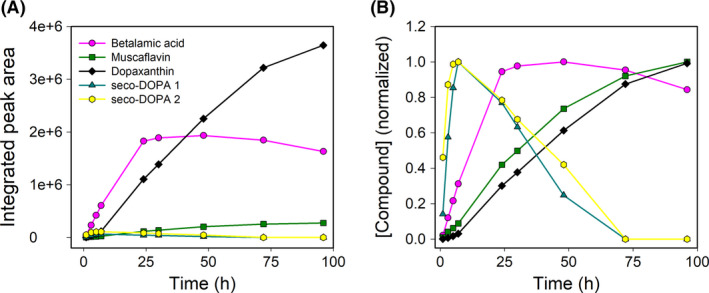
HPLC analysis of the evolution of betalains and intermediates in E. coli (pRSETA‐AcDODA) cultures. Time evolution of intermediates and final compounds in the supernatant of the culture medium followed for 100 h under optimal conditions. Seco‐DOPAs were detected at λmax = 360 nm, betalamic acid and muscaflavin were detected at λmax = 405 nm, and dopaxanthin was detected at λmax = 480 nm. (A) Integrated peak areas obtained from HPLC analysis. (B) Normalized data.
Thus, the DODA enzyme from A. cylindrica presents dual 2,3‐ and 4,5‐DOPA‐extradiol‐dioxygenase activities similar to those described for the dioxygenase enzymes characterized from G. diazotrophicus (Contreras‐Llano et al., 2019), E. coli (Gandía‐Herrero and García‐Carmona, 2014) and A. muscaria (Mueller et al., 1997).
DODA enzyme from A. cylindrica was compared, in terms of sequence homology and phylogeny, with the dioxygenase enzymes previously characterized to produce betalains and with the sequences of potential dioxygenase enzymes related to the G. diazotrophicus sequence. The conserved block of G. diazotrophicus among residues His91 and Asp122 was used for sequence evaluation of the A. cylindrica enzyme (Chang et al., 2015; Li et al., 2015). This block includes one of the three strictly conserved histidines present in the plant enzymes, which is present in GdDODA and in the novel AcDODA as His 101. It also presents acid (Asp100) and hydrophobic (Leu102) residues flanking out the His101, as occurs in the enzymes described from plants (Gandía‐Herrero and García‐Carmona, 2014). Phylogenetic analysis (Fig. 6) of the region conserved showed that there is a compact clade formed by plant sequences and the homologue YgiD protein separated from A. muscaria dioxygenase and another compact clade formed by proteobacteria enzymes. However, A. cylindrica appears as an individual clade apart, showing that although its enzyme produces betalamic acid and muscaflavin, AcDODA is not closely related to these two branches.
Fig. 6.
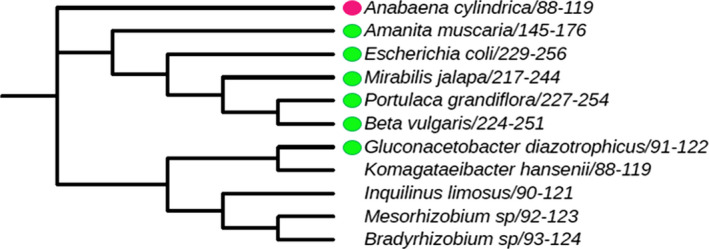
Phylogenetic analysis of characterized betalamic acid‐forming DODAs and related proteobacteria sequences. The analysis was performed with the conserved block of G. diazotrophicus which includes the His 101. This residue is conserved in all characterized 4,5‐DOPA‐dioxygenases known to produce betalamic acid (green dot) and several proteobacteria. The conserved residues were also detected in the novel Anabaena cylindrica enzyme here described to produce dopaxanthin (pink dot), and it conforms an independent clade. Multiple sequence alignment was performed using Clustal Omega, and the phylogenetic tree was inferred by using the phylogeny tool from the ClustalW2 package and plotted using ITOL.
DOPA‐extradiol‐dioxygenase, the key enzyme involved in the betalains’ biosynthetic pathway of Caryophyllales plants, is also present in the cyanobacterium Anabaena cylindrica. The protein sequence has been optimized for its recombinant expression in E. coli, purified to homogeneity and characterized for the first time as a protein of 17.8 kDa which forms dimers under native conditions. Reactions performed in one‐pot experiments show how this enzyme is able to produce betalamic acid and muscaflavin from the amino acid L‐DOPA due to a dual activity 2,3‐ and 4,5‐DOPA‐extradiol‐dioxygenase. This work provides evidence of the presence of DODA enzymes in the phylum cyanobacteria and expands the presence of betalain‐forming capabilities in prokaryotes along with the previously described enzymes from E. coli (Gandía‐Herrero and García‐Carmona, 2014) and G. diazotrophicus (Contreras‐Llano et al., 2019). The sequence analysis of the known enzymes has shown little phylogenetic relationship of the A. cylindrica DODA with those previously described. This seems to indicate that dioxygenase enzymes able to cleave the ring of L‐DOPA to produce betalamic acid may be more diverse and frequent than expected. This opens the possibility of finding novel betalamic acid‐forming dioxygenases in organisms of different nature with no apparent relationship among them.
Experimental procedures
Chemicals, bacterial strains, plasmids and enzymes
Distilled water was purified using a Milli‐Q system (Millipore, Bedford, MA, USA). HPLC‐grade acetonitrile was obtained from Fisher Scientific UK (Leicestershire, UK). E. coli BL21 (DE3) cells and the plasmid pRSETA were obtained from Invitrogen (Waltham, MA, USA). HypperLadderTM 1kb DNA ladder was obtained from Bioline Reagents Ltd (London, UK). Taq DNA polymerase was purchased from Thermo Fisher Scientific Inc. (Waltham, MA, USA). All other chemicals and reagents were obtained from Sigma (St. Louis, MO, USA).
Anabaena cylindrica culture
Anabaena cylindrica was acquired from Spanish Bank of Algae (BEA – Banco Español de Algas, Gran Canaria, Spain). Active growing microorganisms were inoculated in 50 ml cultures of BG‐11 medium (Videau and Cozy, 2019) that contains, per litre, 1.5 g NaNO3, 0.036 g CaCl2‐2H2O, 0.012 g FeNH4‐citrate, 0.001 g Na2‐EDTA, 0.04 g K2HPO4, 0.075 g MgSO4‐7H2O, 0.02 g Na2CO3 and 1 ml of trace metals solution. Components of trace metal solution are, per litre, 2.86 g H3Bo3, 1.81 g MnCl2‐4H2O, 0.222 g ZnSO4‐7H2O, 0.391 g Na2MoO4‐2H2O, 0.079 g CuSO4‐5H2O and 0.0494 g Co(NO3)2. These cultures were maintained seven days at 20 °C with orbital shaken at 150 rpm and light/dark cycles of 14:8 h. Afterwards, cultures media were supplemented with varying concentrations of L‐DOPA (from 0 to 7.6 mM), sodium ascorbate 15 mM and 100 mM EDTA and further incubated at 20 °C. Media were collected after two days and analysed by HPLC‐ESI/TOF/MS by searching for compounds derived from the synthesis of betalamic acid.
Anabaena cylindrica DODA sequence and cloning
The sequence of the protein WP_015213489 from Anabaena cylindrica, deposited at the National Center for Biotechnology Information (NCBI, Bethesda, MD, USA), was used as a template to synthetically obtain the 4,5‐DODA sequence from A. cylindrica. The sequence was enhanced for E. coli expression (Geneart, Regensburg, Germany), and the synthetic DODA gene yielded a 357 bp product that was expressed into the recombinant plasmid pRSETA, which encodes for an additional N‐terminal sequence containing a 6 His‐tag. The new plasmid pRSETA‐AcDODA was transformed into E. coli BL21 (Invitrogen) thermocompetent cells and plated onto LB agar plates containing ampicillin (Amp) 50 μg ml−1. The resulting colonies were then analysed and confirmed by PCR.
PCR amplification was performed using Taq DNA polymerase and the following primers: AcDODA‐F (5´ TGGGGACATATGAAAAGCAAAAC) and AcDODA‐R (5´ GCTGCAGATCTCGAGTTACAGAC). This amplification yielded a 384 bp product, which coincides with the entire DODA synthetic gene plus the additional 6 His‐tag sequence. The plasmid pRSETA‐AcDODA was subsequently used in further experiments.
Expression and purification
The AcDODA protein derived from plasmid pRSETA‐AcDODA was expressed in E. coli BL21 (Invitrogen), grown at 37 °C in LB medium containing Amp 50 μg ml−1 up to an A600 of 0.8–1.2. Induction was performed for 20 h with different concentrations of isopropyl‐1‐thio‐β‐D‐galactopyranoside (IPTG) at different temperatures. Protein production for the following steps was performed in 0.5 l cultures of LB medium under the optimized conditions. In the optimal conditions, cells were harvested by centrifugation and resuspended in sodium phosphate buffer 50 mM, pH 8.0, with 0.3 M sodium chloride. Cell lysis was performed by sonication in a Cole‐Parmer 4710 series ultrasonic homogenizer (Chicago, IL, USA). Recombinant A. cylindrica DODA was purified from E. coli cells by His‐select nickel affinity gel (Sigma) according to the manufacturer’s instructions. After non‐tagged proteins were washed, the His‐tagged protein was eluted with a buffer supplemented with 250 mM imidazole. Immediately after the elution, the protein solution was desalted using PD10 columns (General Electric Healthcare, Milwaukee, USA) and eluted into phosphate buffer 20 mM pH 7.5. Purified protein was quantified using the Bradford protein assay (Bio‐Rad, Hercules, CA, USA; Bradford, 1976), and bovine serum albumin was used as standard to obtain a calibration curve. Samples were analysed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE), by application to 15% polyacrylamide gels and stained using a standard Coomassie Blue method.
Gel filtration
Samples of pure recombinant protein were applied to a Superdex 200 10/300 GL column equilibrated with sodium phosphate buffer 50 mM, pH 7.5, with 150 mM NaCl. The protein was eluted with the same buffer at a flow rate of 0.5 ml min−1. Elutions were performed in an Äkta purifier apparatus (General Electric Healthcare) and monitored at 280 nm. Column calibration was performed with the following protein markers (Sigma): cytochrome C (12.4 kDa), carbonic anhydrase (29 kDa), albumin (66 kDa), alcohol dehydrogenase (150 kDa) and β‐amylase (200 kDa).
MALDI‐TOF‐MS protein analysis
Matrix solution for peptide analyses was α‐cyano‐4‐hydroxycinnamic acid (20 mg ml−1) in ACN/water/TFA (70:30:0.1). The peptide sample was dissolved in 0.1% TFA and mixed with the matrix solution. One microlitre of this mixture was applied to the atmospheric pressure matrix‐assisted laser desorption ionization (AP‐MALDI) target plate and allowed to dry. Experiments were carried out with an Agilent Time of Flight (TOF) Mass Spectrometer (Agilent Technologies, Santa Clara, CA, USA), equipped with an AP‐MALDI Ion Source with an N2 laser (337 nm). Samples were measured in reflectron mode to identify molecular formulas based on precise mass measurements in positive mode. External calibration of the spectrometer was performed with standard peptides from the ProteoMass™ Peptide MALDI‐MS Calibration Kit (Sigma). Data were recorded and processed with Agilent MassHunter Workstation Software. Peptide mass fingerprint determination was carried out using Agilent Spectrum Mill Software. Determination of protein absolute molecular mass was carried out using an HPLC‐ESI‐MS‐TOF system. This system comprises an HPLC Agilent VL 1100 apparatus equipped with an autosampler μ‐well plate and a capillary pump, connected to an Agilent 6100 Mass Spectrometer Time of Flight (MS‐TOF), and an electrospray ionization interface (ESI) was used. The column employed was a Zorbax Poroshell 300SB‐C18, 1 × 75 mm with 5 μm (Agilent Technologies, Santa Clara, CA, USA). The column was operated at 60 °C, and the samples were injected with a flux of 0.2 ml min−1. The protein was eluted with a linear gradient using water/ACN/formic acid (95:4.9:0.1) as solvent A, and water/ACN/formic acid (10:89.9:0.1) as solvent B. A linear gradient was performed for 30 min from 0% to 90% of B. Protein separation was monitored at 210 and 280 nm using a multiple wavelength detector. The mass spectrometer was operated in positive mode in the range of 100‐2200 m/z, using a capillary voltage of 3.5 kV. Nebulizer gas pressure was 30 psi, and drying gas flux was 8 l min−1 at a temperature of 350 °C. External spectrometer calibration was carried out using the ProteoMass™ Peptide MALDI‐MS Calibration Kit (Sigma‐Aldrich, St. Louis, MO, USA). Two different peptides were used as controls (cytochrome C and carbonic anhydrase, Sigma‐Aldrich). All data were recorded and processed through the software Agilent MassHunter Workstation Qualitative Analysis Software (Agilent Technologies), and the intact molecular weight of the protein was obtained using the deconvolution algorithm from this software.
Trypsin digestion
The protein sample was prepared in 100 μl of buffer NH4HCO3 50 mM, pH 8.0, with 0.02% ProteaseMAX™ Surfactant (Promega, Madison, WI, USA). Then, the sample was reduced with DTT 10 mM for 20 min at 56 °C and alkylated with iodoacetamide 50 mM at room temperature in the dark for 20 min. One microgram of proteomics grade trypsin (Promega) was added and the sample was incubated for 4 h at 37 °C. After that, the sample was centrifuged at 15 000 g for 1 min to collect the condensate and the digestion was stopped by adding 0.5% TFA. Peptides were cleaned up with C18 ZipTips (Millipore) and evaporated using an Eppendorf vacuum concentrator model 5301.
HPLC analysis
A Shimadzu LC‐20AD apparatus (Kyoto, Japan) equipped with an SPD‐M20A photodiode array detector was used for analytical HPLC separations performed with a 250 × 4.6 mm Kromasil 100 C‐18 column packed with 5 μm particles (Teknokroma, Barcelona, Spain) (Gandía‐Herrero et al., 2005). A linear gradient was performed using water with 0.05% TFA as solvent A, and acetonitrile with 0.05% TFA as solvent B. The linear gradient was performed for 24 min from 0% B to 35% B with the flow was 1 ml min−1, and the injection volume was 50 μl.
Absorbance spectroscopy
Enzyme activity was determined using a continuous spectrophotometric method by measuring the absorbance at λ = 414 nm due to the presence of betalamic acid and muscaflavin (Girod and Zryd, 1991; Gandía‐Herrero and García‐Carmona, 2012). Unless otherwise stated, the reaction medium contained sodium phosphate buffer 50 mM, pH 7, L‐DOPA 7.6 mM, and sodium ascorbate 100 mM at a final volume of 300 μl. Measurements were performed at 25 °C in 96‐well plates in a Synergy HT plate reader (Bio‐Tek Instruments, Winooski, USA). Betalamic acid solutions of known concentration were employed to calibrate the plate reader detector signal. The molar extinction coefficient of betalamic acid at 424 nm, Ɛ = 24 000 M‐1 cm‐1, was employed for the quantification of betalamic acid (Trezzini and Zrÿb, 1991). Kinetic data analysis was performed by using non‐linear regression fitting with the SigmaPlot Scientific Graphing for Windows version 10.0 (2006, Systat Software, San Jose, CA, USA).
E. coli (pRSETA‐AcDODA) culture conditions
Escherichia coli BL21 cells harbouring pRSETA‐AcDODA were grown in 50 ml LB supplemented with Amp 50 μg ml−1 until reached an O.D.600 0.8–1.0. Then, heterologous expression of the DODA enzyme was performed by addition of 0.1 mM IPTG. The culture was kept at 20 °C for 20 h and then centrifuged 10 min at 7 500 rpm. Supernatant was discarded, and cells were diluted in 50 ml of distilled water containing L‐DOPA 7.6 mM and sodium ascorbate 15 mM. Culture was maintained at 20 °C with orbital shaken for 200 h, and samples were taken at different hours in order to follow the formation of derived products. Samples were centrifuged 1 min at 7 500 rpm, and the supernatant was immediately analysed by HPLC.
Phylogenetic analysis of novel betalamic acid‐forming DODAs
The DODA enzyme from G. diazotrophicus was used as a template to search similar enzymes. The obtention of a neighbour‐joining phylogenetic tree (Altschul et al., 2005; Li et al., 2015) found the DODA enzyme from A. cylindrica by sequence similarity. The relationship of the novel enzymes with betalamic acid‐forming dioxygenases previously reported was performed from the conserved block of G. diazotrophicus which includes the His 101. In all cases, multiple sequence alignment was performed using Clustal Omega from the EMBL‐EBI (European Molecular Biology Laboratory – European Bioinformatics Institute) (Madeira et al., 2019) and the phylogenetic tree was inferred by using the phylogeny tool from the ClustalW2 package of the same platform and plotted using iTOL (Interactive Tree of Life; Letunic and Bork, 2019).
Conflict of interest
The authors declare no competing financial interest.
Supporting information
Fig. S1. Phylogenetic analysis of dioxygenase enzymes related to Gluconacetobacter diazotrophicus (orange). These enzymes correspond to the phyllum proteobacteria (yellow) and cyanobacteria (blue), including Anabaena cylindrica (navy blue).
Fig. S2. Determination of the optimal conditions for the heterologous expression of AcDODA in E. coli cells. A: Comparison of the obtention of the products from the activity of AcDODA and derived compounds in E. coli cultures supplemented with different concentrations of IPTG and maintained under different temperatures. The reactions were followed by HPLC in order to evaluate the presence of the enzyme. B: Macroscopic images of E. coli (pRSETA‐AcDODA) cultures, after 24 hours of reaction, supplemented with 1.0 (left), 0.5 (centre) or 0.1 (right) mM IPTG and maintained at 20 °C (top) or 25 °C (bottom).
Fig. S3. Schematic representation and rate equation for inhibition by excess of substrate.
Appendix S1. Structure modelling of the novel AcDODA enzyme
Acknowledgements
The authors are grateful to Dr. Alejandro Torrecillas (SAI, University of Murcia – Spain) for skilful technical assistance in ESI‐MS/MS, MALDI‐TOF and TOF/Q‐TOF mass spectrometry experiments. This work was supported by ‘Ministerio de Ciencia e Innovación’, project AGL2017‐86526‐P (MCI/AEI/FEDER, UE) and by ‘Programa de Ayudas a Grupos de Excelencia de la Región de Murcia, Fundación Séneca, Agencia de Ciencia y Tecnología de la Región de Murcia’ (Project 19893/GERM/15). M.A.G.‐R. holds a contract financed by MEC‐FSE (Spain).
Microbial Biotechnology (2020) 13(6), 1948–1959
Funding information
This work was supported by ‘Ministerio de Ciencia e Innovación’, project AGL2017‐86526‐P (MCI/AEI/FEDER, UE) and by ‘Programa de Ayudas a Grupos de Excelencia de la Región de Murcia, Fundación Séneca, Agencia de Ciencia y Tecnología de la Región de Murcia’ (Project 19893/GERM/15). M.A.G.‐R. holds a contract financed by MEC‐FSE (Spain).
References
- Allen, M.B. , and Arnon, D.I. (1955) Studies on nitrogen‐fixing blue‐green algae. I. growth and nitrogen fixation by Anabaena cylindrica Lemm. Plant Physiol 30: 366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Wootton, J.C. , Gertz, E.M. , Agarwala, R. , Morgulis, A. , Schaffer, A.A. , and Yu, Y.‐K. (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J 272: 5101–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armougom, F. , Moretti, S. , Poirot, O. , Audic, S. , Dumas, P. , Schaeli, B. , et al (2006) Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D‐Coffee. Nucleic Acids Res 34: W604–W608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babos, M. , Halász, K. , Zagyva, T. , Zöld‐Balogh, A. , Szegő, D. , and Bratek, Z. (2011) Preliminary notes on dual relevance of ITS sequences and pigments in Hygrocybe taxonomy. Persoonia 26: 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean, A. , Sunnadeniya, R. , Akhavan, N. , Campbell, A. , Brown, M. , and Lloyd, A. (2018) Gain‐of‐function mutations in beet DODA2 identify key residues for betalain pigment evolution. New Phytol 219: 287–296. [DOI] [PubMed] [Google Scholar]
- Biasini, M. , Bienert, S. , Waterhouse, A. , Arnold, K. , Studer, G. , Schmidt, T. , et al (2014) SWISS‐MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42: W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert, S. , Waterhouse, A. , De Beer, T.A.P. , Tauriello, G. , Studer, G. , Bordoli, L. , and Schwede, T. (2017) The SWISS‐MODEL Repository‐new features and functionality. Nucleic Acids Res 45: D313–D319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Brockington, S.F. , Walker, R.H. , Glover, B.J. , Soltis, P.S. , and Soltis, D.E. (2011) Complex pigment evolution in the Caryophyllales. New Phytol 190: 854–864. [DOI] [PubMed] [Google Scholar]
- Castellanos‐Santiago, E. , and Yahia, E.M. (2008) Identification and quantification of betalains from the fruits of 10 Mexican prickly pear cultivars by high‐performance liquid chromatography and electrospray ionization mass spectrometry. J Agric Food Chem 56: 5758–5764. [DOI] [PubMed] [Google Scholar]
- Chang, J.M. , Di Tommaso, P. , Lefort, V. , Gascuel, O. , and Notredame, C. (2015) TCS: A web server for multiple sequence alignment evaluation and phylogenetic reconstruction. Nucleic Acids Res 43: W3–W6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras‐Llano, L.E. , Guerrero‐Rubio, M.A. , Lozada‐Ramirez, J.D. , García‐Carmona, F. , and Gandía‐Herrero, F. (2019) First betalain‐producing bacteria break the exclusive presence of the pigments in the plant kingdom. MBio 10: e00345‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandía‐Herrero, F. , and García‐Carmona, F. (2012) Characterization of recombinant Beta vulgaris 4,5‐DOPA‐extradiol‐dioxygenase active in the biosynthesis of betalains. Planta 236: 91–100. [DOI] [PubMed] [Google Scholar]
- Gandía‐Herrero, F. , and García‐Carmona, F. (2013) Biosynthesis of betalains: yellow and violet plant pigments. Trends Plant Sci 18: 334–343. [DOI] [PubMed] [Google Scholar]
- Gandía‐Herrero, F. , and García‐Carmona, F. (2014) Escherichia coli protein YgiD produces the structural unit of plant pigments betalains: characterization of a prokaryotic enzyme with DOPA‐extradiol‐dioxygenase activity. Appl. Microbiol. Biotechnol. 98: 1165–1174. [DOI] [PubMed] [Google Scholar]
- Gandía‐Herrero, F. , García‐Carmona, F. , and Escribano, J. (2005) A novel method using high‐performance liquid chromatography with fluorescence detection for the determination of betaxanthins. J Chromatogr A 1078: 83–89. [DOI] [PubMed] [Google Scholar]
- Gandía‐Herrero, F. , Escribano, J. , and García‐Carmona, F. (2010) Structural implications on color, fluorescence, and antiradical activity in betalains. Planta 232: 449–460. [DOI] [PubMed] [Google Scholar]
- Gandía‐Herrero, F. , Escribano, J. , and García‐Carmona, F. (2012) Purification and antiradical properties of the structural unit of betalains. J Nat Prod 75: 1030–1036. [DOI] [PubMed] [Google Scholar]
- Girod, P.‐A. , and Zryd, J.‐P. (1991) Biogenesis of betalains: Purification and partial characterization of dopa 4,5‐dioxygenase from Amanita muscaria . Phytochemistry 30: 169–174. [Google Scholar]
- Guerrero‐Rubio, M.A. , Hernández‐García, S. , García‐Carmona, F. , and Gandía‐Herrero, F. (2019) Extension of life‐span using a RNAi model and in vivo antioxidant effect of Opuntia fruit extracts and pure betalains in Caenorhabditis elegans . Food Chem 274: 840–847. [DOI] [PubMed] [Google Scholar]
- Guerrero‐Rubio, M.A. , López‐Llorca, R. , Henarejos‐Escudero, P. , García‐Carmona, F. , and Gandía‐Herrero, F. (2019) Scaled‐up biotechnological production of individual betalains in a microbial system. Microb Biotechnol 12: 993–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, J.F. , Wang, L.‐S. , Rocha, C.M. , Larue, B. , Henry, C. , McIntyre, C.M. , et al (2010) Drinking water with red beetroot food color antagonizes esophageal carcinogenesis in N‐nitrosomethylbenzylamine‐treated rats. J Med Food 13: 733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I. , and Bork, P. (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47: W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Cowley, A. , Uludag, M. , Gur, T. , McWilliam, H. , Squizzato, S. , et al (2015) The EMBL‐EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43: W580–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira, F. , Park, Y.M. , Lee, J. , Buso, N. , Gur, T. , Madhusoodanan, N. , et al (2019) The EMBL‐EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47: W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake, C. , Michihata, F. , and Asada, K. (1991) Scavenging of hydrogen peroxide in prokaryotic and eukaryotic algae: acquisition of ascorbate peroxidase during the evolution of cyanobacteria. Plant Cell Physiol 32: 33–43. [Google Scholar]
- Mueller, L.A. , Hinz, U. , and Zryd, J.‐P. (1997) The formation of betalamic acid and muscaflavin by recombinant dopa‐dioxygenase from Amanita . Phytochemistry 44: 567–569. [Google Scholar]
- Pietrzkowski, Z. , Nemzer, B. , Spórna, A. , Stalica, P. , Tresher, W. , Keller, R. , et al (2010) Influence of Betalain rich extract on reduction of discomfort associated with osteoarthritis. New Med 1: 12–17. [Google Scholar]
- Rahimi, P. , Mesbah‐Namin, S.A. , Ostadrahimi, A. , Abedimanesh, S. , Separham, A. , and Asghary Jafarabadi, M. (2019) Effects of betalains on atherogenic risk factors in patients with atherosclerotic cardiovascular disease. Food Funct 10: 8286–8297. [DOI] [PubMed] [Google Scholar]
- Reed, M.C. , Lieb, A. , and Nijhout, H.F. (2010) The biological significance of substrate inhibition: a mechanism with diverse functions. BioEssays 32: 422–429. [DOI] [PubMed] [Google Scholar]
- Robert, X. , and Gouet, P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, N. , Abe, Y. , Goda, Y. , Adachi, T. , Kasahara, K. , and Ozeki, Y. (2009) Detection of DOPA 4,5‐dioxygenase (DOD) activity using recombinant protein prepared from Escherichia coli cells harboring cDNA encoding DOD from Mirabilis jalapa. Plant Cell Physiol 50: 1012–1016. [DOI] [PubMed] [Google Scholar]
- Segel, I.H. (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady State Enzyme Systems. New York, NY, USA: Wiley. [Google Scholar]
- Shih, P.M. , Wu, D. , Latifi, A. , Axen, S.D. , Fewer, D.P. , Talla, E. , et al (2013) Improving the coverage of the cyanobacterial phylum using diversity‐driven genome sequencing. Proc Natl Acad Sci USA 110: 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekanth, D. , Arunasree, M.K. , Roy, K.R. , Chandramohan Reddy, T. , Reddy, G.V. , and Reddanna, P. (2007) Betanin a betacyanin pigment purified from fruits of Opuntia ficus‐indica induces apoptosis in human chronic myeloid leukemia cell line‐K562. Phytomedicine 14: 739–746. [DOI] [PubMed] [Google Scholar]
- Stintzing, F. , and Schliemann, W. (2007) Pigments of fly agaric (Amanita muscaria). Z Naturforsch C J Biosci 62: 779–785. [DOI] [PubMed] [Google Scholar]
- Trezzini, G.F. , and Zrÿb, J.‐P. (1991) Characterization of some natural and semi‐synthetic betaxanthins. Phytochemistry 30: 1901–1903. [Google Scholar]
- Vidal, P.J. , López‐Nicolás, J.M. , Gandía‐Herrero, F. , and García‐Carmona, F. (2014) Inactivation of lipoxygenase and cyclooxygenase by natural betalains and semi‐synthetic analogues. Food Chem 154: 246–254. [DOI] [PubMed] [Google Scholar]
- Videau, P. , and Cozy, L.M. (2019) Anabaena sp. strain PCC 7120: laboratory maintenance, cultivation, and heterocyst induction. Curr Protoc Microbiol 52: e71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Phylogenetic analysis of dioxygenase enzymes related to Gluconacetobacter diazotrophicus (orange). These enzymes correspond to the phyllum proteobacteria (yellow) and cyanobacteria (blue), including Anabaena cylindrica (navy blue).
Fig. S2. Determination of the optimal conditions for the heterologous expression of AcDODA in E. coli cells. A: Comparison of the obtention of the products from the activity of AcDODA and derived compounds in E. coli cultures supplemented with different concentrations of IPTG and maintained under different temperatures. The reactions were followed by HPLC in order to evaluate the presence of the enzyme. B: Macroscopic images of E. coli (pRSETA‐AcDODA) cultures, after 24 hours of reaction, supplemented with 1.0 (left), 0.5 (centre) or 0.1 (right) mM IPTG and maintained at 20 °C (top) or 25 °C (bottom).
Fig. S3. Schematic representation and rate equation for inhibition by excess of substrate.
Appendix S1. Structure modelling of the novel AcDODA enzyme


