Summary
Environmental DNA approaches are increasingly used to detect microorganisms in environmental compartments, including water. They show considerable advantages to study non‐cultivable microorganisms like Bonamia ostreae, a protozoan parasite inducing significant mortality in populations of flat oyster Ostrea edulis. Although B. ostreae development within the host has been well described, questions remain about its behaviour in the environment. As B. ostreae transmission is direct, seawater appears as an interesting target to develop early detection tools and improve our understanding of disease transmission mechanisms. In this context, we have developed an eDNA/eRNA approach allowing detecting and quantifying B. ostreae 18S rDNA/rRNA as well as monitoring its presence in seawater by real‐time PCR. B. ostreae DNA could be detected up to 4 days while RNA could be detected up to 30 days, suggesting a higher sensitivity of the eRNA‐based tool. Additionally, more than 90% of shed parasites were no longer detected after 2 days outside the oysters. By allowing B. ostreae detection in seawater, this approach would not only be useful to monitor the presence of the parasite in oyster production areas but also to evaluate the effect of changing environmental factors on parasite survival and transmission.
An eDNA/eRNA approach was developed, allowing detecting and quantifying Bonamia ostreae 18S rDNA/rRNA as well as monitoring its presence in seawater by real‐time PCR. B. ostreae DNA and RNA could be detected up to 4 and 30 days respectively, suggesting a higher sensitivity of the eRNA based tool. More than 90% of shed parasites were no longer detected after 2 days outside the oysters.
Introduction
The flat oyster Ostrea edulis (Linnaeus, 1758) is a bivalve species native to Europe. The history of its production has been affected by overfishing, environmental degradation and several disease outbreaks (Buestel et al., 2009), including the emergence of bonamiosis in 1979, an epizootic disease caused by the protozoan parasite Bonamia ostreae. This parasite was first described in Ile Tudy (Brittany, France) in the context of flat oyster mortality (Pichot et al., 1980) and has since spread around Europe, Canada (British Columbia) and United States of America (California, Maine and Washington States) (OIE, 2020), mainly through aquaculture‐related movements (Pogoda et al., 2019). More recently, it has also been reported in Ostrea chilensis in New Zealand (Lane et al., 2016). Today, bonamiosis still causes severe losses, particularly in natural populations, that might hamper the different projects of flat oyster restoration currently undertaken in several European countries (Pogoda et al., 2019; NORA, 2020). Considering its impact on natural and farmed bivalves, B. ostreae is notifiable to the World Organisation for Animal Health and the European Union.
Oysters infected with B. ostreae can be found in various ecosystems, from estuaries and intertidal zones to deep coastal waters or lagoons. All life stages of the flat oyster including larvae, spat and adults can be infected by the parasite. However, mortalities mainly affect flat oysters older than 2 years (Culloty and Mulcahy, 1996). New oysters can become infected with B. ostreae throughout the year but prevalence and infection intensity within flat oyster usually show a peak at the end of winter‐early spring (Culloty and Mulcahy, 1996; Arzul et al., 2006; Engelsma et al., 2010).
Bonamia ostreae belongs to the Haplosporida order. It multiplies within haemocyte cytoplasm where it can be observed under light microscopy as small rounded cells measuring between 2 and 5 µm in diameter. Sometimes, the parasite is present extracellularly. Its presence is often associated with intense haemocyte infiltration (Arzul and Carnegie, 2015). Based on an electron microscopy study, a possible life cycle of B. ostreae in which the parasite is suspected to enter and leave its host through gills has been suggested (Montes et al., 1994). This hypothesis is supported by the observation of parasites in gill epithelium of highly infected oysters by histology and in situ hybridization observation (Arzul and Carnegie, 2015).
Bonamia ostreae is not cultivable; however, a protocol allows isolating parasites from highly infected oysters (Mialhe et al., 1988). It is possible to experimentally infect oysters by injecting parasites isolated following this protocol in the adductor muscle or the pericardic cavity of the flat oyster (Hervio et al., 1995). Additionally, it is possible to infect oysters by maintaining them in cohabitation with infected oysters, demonstrating that the parasite can be directly transmitted and does not require intermediate host to complete its life cycle (Culloty et al., 1999; Lallias et al., 2008).
As many other shellfish diseases, bonamiosis is a non‐pathognomonic disease and its diagnosis relies on cytology, histology and PCR‐based approaches. Several PCR assays have been developed allowing the detection of B. ostreae DNA in flat oysters including some conventional (Carnegie et al., 2000; Cochennec et al., 2000) and, more recently, some real‐time PCR tools (Robert et al., 2009; Ramilo et al., 2013). The surveillance of the parasite is carried out by testing flat oysters as recommended by the OIE (OIE, 2020) whereas its detection outside Ostrea edulis had only been investigated for research purposes.
As B. ostreae is suggested to be directly transmitted through water column (Arzul and Carnegie, 2015), understanding its life cycle requires approaches allowing the detection of the parasite and monitoring its presence in seawater. Considering that B. ostreae is not cultivable, environmental DNA (eDNA)‐based approach appears of interest to monitor its presence in seawater. Although presenting some methodological constraints particularly in aquatic environments, eDNA analyses allow rapid, non‐invasive and cost‐efficient biodiversity monitoring (Harper et al., 2019). Most of the currently developed pathogen eDNA studies target bacteria and more particularly human pathogens (Green et al., 2011; Twing et al., 2011; Casanovas‐Massana et al., 2018) but some work have been carried out on marine and freshwater pathogens (Goarant and Merien, 2006; Johnson and Brunner, 2014; Strepparava et al., 2014; Rusch et al., 2018). In the context of shellfish diseases, eDNA studies have revealed new parasite clades and parasite diversity (Ward et al., 2016; Ward et al., 2018), but also to investigate pathogen survival outside its host (Eiler et al., 2006; Friedman et al., 2014) or the infection process (Parizadeh et al., 2018). Nevertheless, DNA detection does not allow discriminating between viable and non‐viable cells. In this context, it can be interesting to focus on RNA detection, as it is suggested to be a better proxy for the presence of metabolically active stages, due to its rapid degradation in the environment (McCarthy et al., 2001; Nocker et al., 2007; Bae and Wuertz, 2009; Pochon et al., 2017). Indeed, being able to discriminate viable and non‐viable cells is fundamental, especially where parasite life cycles are poorly known such as B. ostreae cycle. However, recent studies also suggest that RNA can persist in the environment through the protection within extracellular vesicles (Cristescu, 2019). RNA may not allow a strict distinction between viable and non‐viable cells, that is why it could be preferable to associate both DNA and RNA detection in environmental studies.
In this context, we have developed and combined real‐time PCR‐based approaches to detect B. ostreae 18S rDNA and rRNA in seawater. These tools are useful for detecting and quantifying the parasite in seawater but can also be used to better understand parasite life cycle outside its host.
Results
Real‐time PCR efficiency, limit of detection, limit of quantification and standard curves
Bonamia ostreae DNA detection
The efficiency, limit of detection (LOD) and limit of quantification (LOQ) of the Bonamia ostreae real‐time PCR detection approach were determined from DNA extracted from five 10‐fold dilution series of 1 µm polycarbonate membranes artificially contaminated with 2.5 to 2.5 x 105 purified parasites (per quarter membrane) (see Experimental procedures section for further details).
Real‐time PCR efficiency was 93.47 % (SD = 4.35), and the analysis of parasite dilution series allowed the detection of B. ostreae DNA from 2.5 to 2.5 × 105 parasites (per quarter membrane). However, the lowest tested parasite amount was only detected twice out of the five tests. Considering that the detection limit was between 2.5 and 25 parasites per quarter membrane, DNA extracted from this last condition was diluted in order to get the following equivalent parasite quantities: 15, 7.5 and 2.5 parasites. These dilutions were tested six times. It was possible to detect 15 parasites in the six replicates, 7.5 parasites in five replicates and 2.5 parasites in one replicate. The LOD of the DNA‐based approach was set at 7.5 parasites per quarter membrane as it represents more than 75% of the tested replicates.
The LOQ of the B. ostreae DNA detection approach was also set at 7.5 parasites as it was the last amount of detected parasites in the linear dynamic range of the standard curve.
A total of 30 measures could be associated with cycle threshold (Ct) and the number of detected parasites (N parasites). The data were thus considered as normally distributed. The following linear regression was performed on Ct values obtained by real‐time PCR and logarithm of number of detected parasites, ln(N parasites): Ct DNA = −1.52 × ln(N parasites) + 41.2 (Fig. 1). This regression was associated to a high R 2 = 0.97 meaning that 97% of the variability on the Ct can be explained by the N parasites. Furthermore, the analysis of studentized residuals (Fig. 2 red dots) allowed validating the linear regression. These residuals were homogeneously and continuously distributed around 0 and more than 95% of them (29 over 30) were between −2 and 2. The prediction interval included all except one point, which was close to the limit of detection of the approach.
Fig. 1.
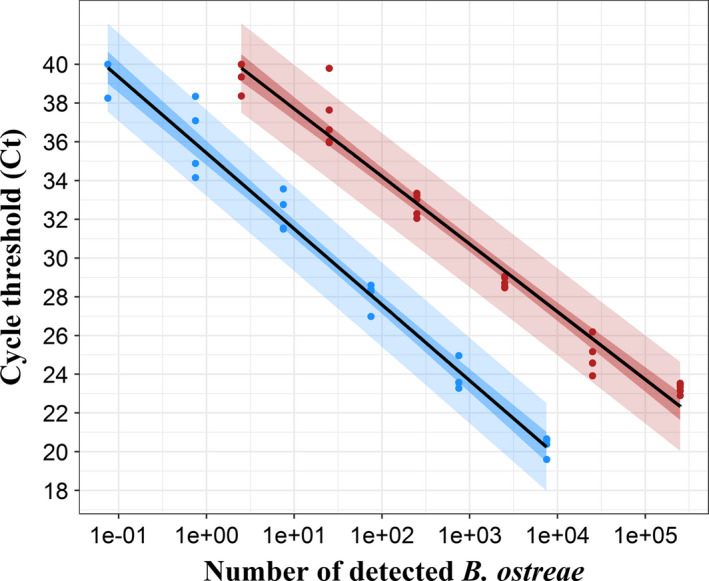
Standard curves established from DNA (red) and RNA (blue) real‐time PCR analyses of Bonamia ostreae dilution series. Ct DNA = −1.52 × ln(N parasites) + 41.2, R 2 = 0.97; Ct RNA = −1.7 × ln(N parasites) + 35.42, R 2 = 0.98. Dots represent experimental data, solid lines represent linear regressions and coloured areas represent 95% confidence (dark) and 95% prediction (light) intervals associated with each regression. For each parasite amount, n = 5 for DNA and n = 4 for RNA.
Fig. 2.
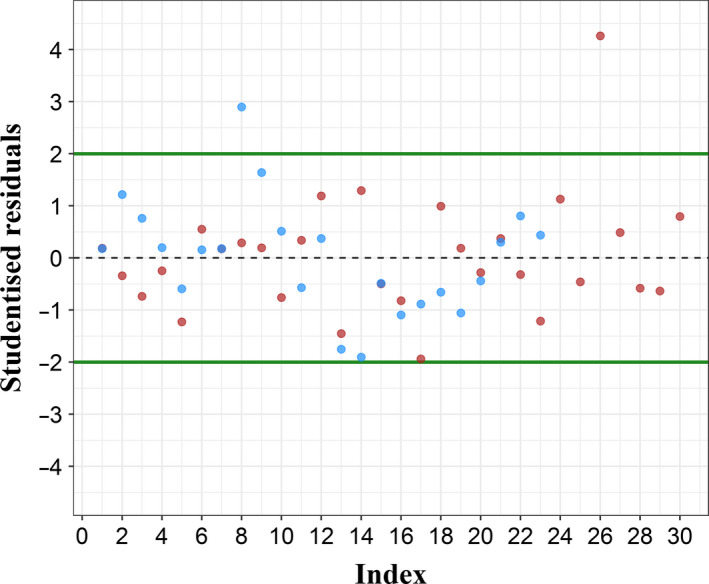
Studentized residuals associated with each regression. Red dots represent studentized residuals associated with DNA standard curve (n = 30). Blue dots represent studentized residuals associated with RNA standard curve (n = 24).
Bonamia ostreae RNA detection
The efficiency, LOD and LOQ of the Bonamia ostreae RNA detection approach were determined by analysing by reverse transcription real‐time PCR RNA extracted from four 10‐fold dilution series of 1 µm polycarbonate membranes artificially contaminated with 7.5 × 10−2 to 7.5 × 106 purified parasites (per quarter membrane) (see Experimental procedures section for further details).
Real‐time PCR efficiency was 81.96 % (SD = 9.07), and the analysis of parasite dilution series allowed the detection of B. ostreae RNA from 7.5 × 10−2 to 7.5 × 103 parasites (per quarter membrane). The lowest tested parasite amount was only detected once whereas the one with 0.75 parasites was detected four times out of four. Thereby, the LOD of the RNA‐based approach was set at 0.75 parasites per quarter membrane.
The LOQ of the B. ostreae RNA detection approach was also set at 0.75 parasites as it was the last amount of detected parasites in the linear dynamic range of the standard curve. Less than 30 measures (n = 24) could be associated with Ct and N parasites. Data normality was checked using Shapiro–Wilk test. Ct data were found normally distributed, contrary to N parasites data, whereas ln(N parasites) data were normally distributed. As for the DNA detection‐based approach, the following linear regression was performed on Ct and ln(N parasites): Ct RNA = −1.7 × ln(N parasites) + 35.42 (Fig. 1). This regression was associated with a high R 2 = 0.98, which means that 98% of the variability on the Ct can be explained by the number of detected parasites. The analysis of studentized residuals (Fig. 2, blue dots) allowed validating the linear regression. These residuals were indeed homogeneously and continuously distributed around 0 and more than 95% of them (23 over 24) were between −2 and 2. The prediction interval included all except one experimental point, which was close to the limit of detection of the approach.
Monitoring of Bonamia ostreae presence in seawater
Bonamia ostreae presence in seawater was monitored from twelve freshly released parasite suspensions obtained by incubating infected flat oysters in UV‐treated seawater filtered at 1 µm during 24 h. After 24 h, oysters were removed and parasite presence in seawater was monitored until one month by real‐time PCR and reverse transcription real‐time PCR (see Experimental procedures section for further details).
At t = 0, depending on the tested parasite suspension, from 4.00 × 103 to 7.02 × 105 parasites and from 1.37 × 103 to 4.30 × 104 parasites were detected per quarter membrane using the DNA‐ and RNA‐based approaches respectively (Table 1). After 96 h, parasite DNA was still detected in 2 of the 3 tested suspensions (3.74 × 101 and 1.06 × 103 parasites), while parasite RNA was still detected in all three tested suspensions (from 6.17 to 6.04 × 101 parasites) (Table 1). After 7 days, parasite DNA was no longer detected, whereas parasite RNA remained detectable in most of the tested suspensions even after 30 days (Table 1). Nevertheless, the amount of detected parasite was at least 100 times lower than the number of parasites detected at the beginning of the experiment.
Table 1.
Number of detected Bonamia ostreae in each tested parasite suspension at different times after removing flat oysters using both DNA and RNA real‐time PCR‐based approaches.
| t = 0 day | t = 1 day | t = 2 days | t = 7 days | t = 15 days | t = 30 days | |
|---|---|---|---|---|---|---|
| DNA | ||||||
| Suspension 1 | 4.00E + 03 | 4.20E + 03 | 7.50E + 00 a | 7.50E + 00 a | 7.50E + 00 a | 7.50E + 00 a |
| Suspension 2 | 1.48E + 05 | 1.22E + 05 | 1.21E + 04 | 7.50E + 00 a | 7.50E + 00 a | 7.50E + 00 a |
| Suspension 3 | 1.54E + 05 | 1.14E + 05 | 9.97E + 00 | 7.50E + 00 a | 7.50E + 00 a | 7.50E + 00 a |
| Suspension 4 | 6.22E + 05 | 1.03E + 05 | 1.58E + 04 | 7.50E + 00 a | 7.50E + 00 a | 7.50E + 00 |
| Suspension 5 | 3.42E + 05 | 1.05E + 05 | 6.08E + 03 | 7.50E + 00 a | 7.50E + 00 a | 7.50E + 00 a |
| Suspension 7 | 7.02E + 05 | 2.83E + 04 | 1.03E + 04 | 7.50E + 00 a | 7.50E + 00 a | 7.50E + 00 a |
| Suspension 8 | 1.55E + 05 | 3.13E + 05 | 2.02E + 04 | 7.50E + 00 a | 7.50E + 00 a | 7.50E + 00 a |
| Suspension 9 | 1.84E + 05 | 4.73E + 05 | 9.10E + 04 | 7.50E + 00 a | 7.50E + 00 a | 7.50E + 00 a |
| RNA | ||||||
| Suspension 1 | 1.25E + 04 | 2.63E + 03 | 1.37E + 02 | 8.28E + 00 | 1.03E + 01 | 1.27E + 01 |
| Suspension 2 | 4.60E + 03 | 2.18E + 04 | 7.20E + 02 | 7.50E−01 a | 4.73E + 01 | 7.50E−01 a |
| Suspension 3 | 8.29E + 03 | 7.50E−01 a | 2.36E + 02 | 7.50E−01 a | 3.52E + 01 | 2.69E + 00 |
| Suspension 4 | 1.10E + 04 | 1.16E + 04 | 4.69E + 02 | 3.00E + 00 | 2.07E + 01 | 4.35E + 00 |
| Suspension 5 | 1.37E + 03 | 7.50E−01 a | 3.66E + 01 | 1.32E + 01 | 1.39E + 00 | 7.74E + 00 |
| Suspension 7 | 4.30E + 04 | 1.62E + 04 | 3.25E + 00 | 7.50E−01 a | 2.38E + 01 | 7.50E−01 a |
| Suspension 8 | 2.35E + 04 | 7.50E−01 a | 4.38E + 03 | 7.50E−01 a | 2.12E + 01 | 3.84E + 03 b |
| Suspension 9 | 2.30E + 03 | 1.34E + 04 | 8.30E + 01 | 9.23E + 00 | 7.50E−01 a | 7.50E−01 a |
| t = 0 h | t = 4 h | t = 8 h | t = 24 h | t = 28 h | t = 32 h | t = 48 h | t = 56 h | t = 72 h | t = 96 h | |
|---|---|---|---|---|---|---|---|---|---|---|
| DNA | ||||||||||
| Suspension 10 | 6.35E + 03 | 5.12E + 03 | 2.55E + 04 | 7.99E + 03 | 3.71E + 03 | 2.10E + 03 | 6.55E + 02 | 3.27E + 02 | 3.01E + 01 | 3.74E + 01 |
| Suspension 11 | 2.26E + 04 | 4.50E + 04 | 2.24E + 04 | 2.82E + 04 | 1.79E + 04 | 1.79E + 04 | 8.18E + 03 | 8.15E + 03 | 1.72E + 03 | 7.50E + 00 |
| Suspension 12 | 2.26E + 04 | 4.19E + 04 | 4.71E + 04 | 1.89E + 04 | 1.65E + 04 | 1.10E + 04 | 7.92E + 03 | 2.28E + 03 | 1.06E + 03 | |
| RNA | ||||||||||
| Suspension 10 | 4.00E + 02 | 1.52E + 03 | 8.42E + 02 | 1.26E + 03 | 1.06E + 03 | 5.95E + 02 | 1.78E + 02 | 5.29E + 01 | 1.11E + 02 | 6.04E + 01 |
| Suspension 11 | 5.14E + 02 | 9.01E + 01 | 1.40E + 02 | 2.16E + 02 | 2.02E + 02 | 2.46E + 01 | 4.93E + 01 | 7.50E−01 a | 5.99E + 01 | 1.82E + 01 |
| Suspension 12 | 1.32E + 02 | 2.24E + 02 | 1.33E + 01 | 6.39E + 01 | 1.51E + 02 | 6.84E + 01 | 2.03E + 01 | 9.91E + 00 | 6.17E + 00 | |
Fixed at the limit of detection.
Considered as outlier and not taken in account in further analysis.
The effect of the initial (t = 0) number of parasites on its subsequent detection (t = 48 h for the nine first suspensions and t = 96 h for the three last suspensions) was tested by computing Spearman’s (rho) and Kendall’s (tau) correlation coefficients, as data were not normally distributed (Shapiro–Wilk test). Correlation was found positive for both DNA (rho = 0.69, P‐value = 0.02, and tau = 0.53, P‐value = 0.03) and RNA detection (rho = 0.39, P‐value = 0.2, and tau = 0.38, P‐value = 0.12), but only marginally significant in the case of DNA.
Median percentages were then computed for each time and for both DNA and RNA detection (Fig. 3, solid lines).
Fig. 3.
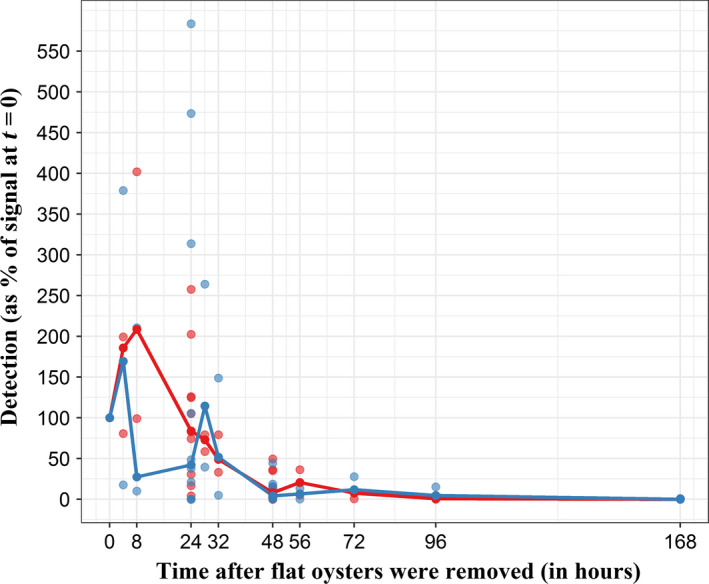
Temporal trend of Bonamia ostreae DNA (red) and RNA (blue) detection after removing flat oysters. Detection on Y‐axis is computed for each sampling time as the ratio between the number of detected parasites at t = τ and the number of detected parasites at t = 0. Dots represent experimental data, and solid lines represent the median detection trend. For 0, 24, 48 and 168 h, n = 11. For other sampling times, n = 3. In order to facilitate reading, t = 168 h is the last experimental data showed on the graph, considering that number of detected parasites at t = 360 h and t = 720 h were almost the same.
Median DNA detection increased from t = 0 to t = 8 h (209% of the initial detection) and then decreased to less than 1% of the initial detection at t = 168 h (7 days). From 28 h, the amount of detected parasites became statistically different from t = 0 (P‐value0−28 = 5.27 × 10−4, Wilcoxon–Mann–Whitney): at 28 h, 73% of the initial parasite quantity was still detected. At t = 48 h, only 8% of the initial amount of parasites remained detectable (P‐value0−48 = 2.55 × 10−5, Wilcoxon–Mann–Whitney).
Median RNA detection increased from t = 0 to t = 4 h (169% of the initial detection), decreased between t = 4 h and t = 8 h (27% of the initial detection) before increasing again from t = 8 h to t = 28 h (114% of the initial detection). Finally, after 28 h it decreased up to about 5% of the initial detection. After 48 h, the amount of detected parasites became statistically different from t = 0 (P‐value0−48 = 2.55 × 10−5, Wilcoxon–Mann–Whitney). At this time, 6% of the initial signal remained detectable.
Bonamia ostreae DNA and RNA detections showed the same evolution over time (Fig. 3). As data were considered as normally distributed (n > 30), correlation between DNA and RNA detection was investigated by computing Pearson’s coefficients. Significantly positive correlation was found (cor = 0.53; P‐value = 4.87 × 10−7 and cor = 0.67; P‐value = 2.08 × 10−11) between DNA and RNA when considering detection percentages and N parasites respectively.
Discussion
Studies and monitoring of shellfish diseases are mostly based on pathogen detection within their hosts (Barbosa Solomieu et al., 2015). Focusing on pathogens inside their bivalve hosts maximizes the chance to detect them. However, understanding pathogen life cycles and epizootiology is also needed to better monitor and manage associated diseases. In this context, environmental DNA (eDNA)‐based approaches could be very useful as they present considerable advantages to study pathogens outside their host, notably the detection of elusive or non‐cultivable organisms (Taberlet et al., 2012; Bass et al., 2015) like Bonamia ostreae.
Therefore, we have developed for the first time an eDNA/eRNA‐based approach allowing B. ostreae detection and quantification in seawater and then used it to monitor parasite presence in seawater until one month after being released.
Bonamia ostreae detection was carried out by real‐time and reverse transcription real‐time PCR amplifying the 18S rDNA and rRNA, respectively, from nucleic acid extracted from water after filtration on a 1 µm pore size membrane. Quantification has been possible thanks to standard curves established from serially diluted parasite suspensions and real‐time PCR targeting B. ostreae DNA has allowed detecting and quantifying from 7.5 to 2.5 × 105 parasites. The assay targeting RNA has allowed detecting and quantifying from 0.75 to 7.5 × 103 parasites.
Few real‐time PCR assays are currently available to quantify bivalve pathogens and most of them target pathogens in bivalve tissues. For example, a real‐time PCR assay was developed, allowing the quantification of B. ostreae DNA from gill tissues with a 6 log dynamic range and a limit of detection of 50 gene copies per reaction (Robert et al., 2009). A similar approach was developed to detect and quantify the protozoan Perkinsus marinus parasite of the American oyster Crassostrea virginica, not only in oyster tissues but also in environmental waters (Audemard et al., 2004). This DNA‐based approach allowed detecting as low as 3.3 × 10−2 cell per 10 µl reaction mixture.
Bonamia ostreae RNA detection limit appeared 10 times lower than DNA one, indicating a higher sensitivity of the RNA‐based approach. While the gene encoding 18S RNA might be present in several copies in the parasite genome, RNA and more particularly ribosomal RNA are expected to be much more abundant, depending notably on the metabolism of the cells (Roberts et al., 2011; Ryan et al., 2012).
As B. ostreae is not cultivable, contaminated seawater suspensions were obtained by maintaining infected flat oysters in UV‐treated seawater. The use of both RNA‐ and DNA‐based real‐time PCR assays has allowed showing that live infected flat oysters were able to shed parasites. This was suggested (Arzul and Carnegie, 2015) but never demonstrated before, most authors assuming that parasites are released from dying or dead infected oysters.
In our conditions, parasite RNA remained detectable up to 30 days while DNA was no longer detected after 7 days spent outside the host. This difference can be explained by the limit of detection of both approaches. As the limit of detection of the RNA‐based PCR assay is 10 times lower than the DNA approach, smaller parasites quantities can be detected using the RNA assay. This observation is also supported by some recent studies suggesting that, contrary to common thinking, eRNA is abundant and persistent in terrestrial and aquatic environment (Cristescu, 2019). Nevertheless, parasite RNA detected 7, 15 and 30 days post‐shedding always represented less than 1% of the initial detected quantity. N parasites at t = 0 and N parasites at the end of the experiment (t = 48 h for the nine first suspensions and t = 96 h for the three last suspensions) showed a positive correlation for both DNA and RNA, but was only marginally significant for DNA. This observation was quite unexpected but can be explained by the rapid decrease of detected parasites (90% of decrease in 2 days). Whatever the initial amount of shed B. ostreae was, most parasites were no longer detected after 2 days spent outside the host, explaining why the correlation between N parasites at the beginning and N parasites at the end of experiment is so weak.
DNA detection does not allow distinguishing between viable and non‐viable cells whereas RNA detection could be a better proxy of metabolically active cells (Blais et al., 1997; Klein and Juneja, 1997; McCarthy et al., 2001; Nocker et al., 2007; Bae and Wuertz, 2009; Pochon et al., 2017). Nevertheless, it has been shown that eDNA persistence is lower in marine waters than in other systems, such as freshwater. Indeed, eDNA fragments persist above detection threshold for only few days in seawater and for several days or weeks in freshwater (Dejean et al., 2011; Thomsen et al., 2012; Sassoubre et al., 2016; Collins et al., 2018), making DNA detection in marine waters a good proxy for the presence of targeted organism. Thus, the combined DNA and RNA detection at a given time was considered as the best indicator of the presence of metabolically active parasites in this study. In our conditions, DNA detection appeared more repeatable than RNA detection with mean coefficients of variation estimated at 0.74 (±0.49) and 1.24 (±0.51) respectively. Therefore, although DNA detection does not inform on the cell status (viable or non‐viable), it still appears as an interesting target to monitor B. ostreae presence in seawater.
The use of both DNA and RNA detection approaches to monitor B. ostreae presence in seawater yielded consistent results. An increase in parasite detection was observed in the first hours post‐shedding (8 h for DNA and 28 h for RNA) and was followed by a slow decrease until the end of the experiment. Although unexpected, the increase in DNA and RNA amounts during the first hours post‐shedding was not statistically different from results at t = 0 (P‐value > 0.1, Wilcoxon–Mann–Whitney). Even if B. ostreae DNA and RNA could still be detected after 7 days, less than 10 % of the initial parasite amount remained detectable 48 h post‐shedding, suggesting that most of B. ostreae remain present for 2 days in seawater. This result is in agreement with previous work carried on this model (Arzul et al., 2009).
Finally in tested conditions, more than 90% of shed parasites were no longer detected after 2 days. However, it was possible to detect B. ostreae DNA and RNA up to 7 and even 30 days respectively. The significance of these detections for transmission is unclear as detecting DNA and RNA does not inform on the capacity of the parasite to infect new oysters. Moreover, we can assume that the parasite presence can be impacted by environmental conditions including temperature and salinity (Arzul et al., 2009) but also by other environmental factors such as light, turbidity or pH and the presence of other microorganisms in seawater.
By efficiently detecting and quantifying B. ostreae DNA and RNA in seawater, the methodology we have established can be used to monitor the presence of the parasite in zones with wild oyster beds and shellfish farms, but also to evaluate the effect of changing environmental factors on the detection and transmission of the parasite.
Experimental procedures
Selection of flat oysters infected with Bonamia ostreae
Flat oysters collected from Bonamia ostreae infected zones in Brittany (France) were sent to the laboratory and maintained in 225 l raceways supplied by natural seawater (15°C, 35 g NaCl l−1) enriched in Skeletonema costatum (1010 cells per hour) for at least five days before being biopsied. A piece of gill was collected from each oyster after maintaining them in a 50 g l−1 MgCl2 anaesthetic solution for at least 4 h (Suquet et al., 2010). After drying tissues on absorbent paper, imprints were made on a glass slide and stained with Hémacolor® (Merck‐Millipore). Gill imprints were screened for the presence of B. ostreae under light microscope and infection intensity was determined according to previously established criteria (Hervio et al., 1995):
Negative: No parasite detected in a whole imprint,
Low infection (+): About 10 parasites observed in a whole imprint,
Moderate infection (++): About one parasite per observation field,
High infection (+++): Several or numerous parasites observed in each microscopic field.
Oysters found infected were selected either to isolate parasites and establish standard curves and detection limits of our methods or monitor the presence of the parasite in seawater.
Real‐time PCR efficiency, limit of detection, limit of quantification and standard curves
Efficiency, LOD and LOQ of both real‐time PCR approaches were established using Bonamia ostreae isolated from highly infected oysters (Mialhe et al., 1988). Isolated parasites were counted using a Malassez‐cell hemocytometer, and suspension concentration was adjusted before performing 1:10 serial dilutions in 0.22 µm filtered seawater (FSW). Each dilution was filtered onto a 1 µm pore size 47 mm diameter polycarbonate membrane (Whatman® Nuclepore™ Track‐Etched Membranes), stored at −80°C until being used for DNA or RNA extraction. A total of 5 separate diluted series from 10 to 106 parasites per membrane were tested in real‐time PCR for DNA detection. A total of 4 separate diluted series from 3 × 10−1 parasites to 3 × 107 parasites per membrane were tested in real‐time PCR for RNA detection.
Monitoring of Bonamia ostreae presence in seawater
The monitoring of Bonamia ostreae presence in seawater was studied using four batches of 7 to 10 flat oysters known to be infected with B. ostreae by examination of gill imprints as described above. Each batch included approximately 1/3 of lightly, 1/3 of moderately and 1/3 of highly infected oysters.
Oysters were maintained for 24 h in 2 l of UV‐treated seawater filtered at 1 µm. After 24 h, oysters were removed, seawater containing freshly released parasites was transferred into a 2 l glass bottle and complemented with streptomycin–penicillin G (final concentration: 2.4512 U ml−1 and 11.0353 U ml−1 respectively) to avoid bacteria proliferation. Nine parasite suspensions were obtained following this approach and were stored at 15°C and exposed to daylight. 100 ml of parasite suspension was collected after mixing suspension at days 0, 1, 2, 7, 15 and 30 (Fig. 4). Considering first obtained results, three additional parasite suspensions were prepared following the same approach to monitor parasite presence on a shorter period. In these trials, 100 ml of parasite suspension was collected after mixing suspension 0 h, 4 h, 8 h, 1 day, 28 h, 32 h, 2 days, 56 h, 3 days and 4 days after removing oysters (Fig. 4).
Fig. 4.
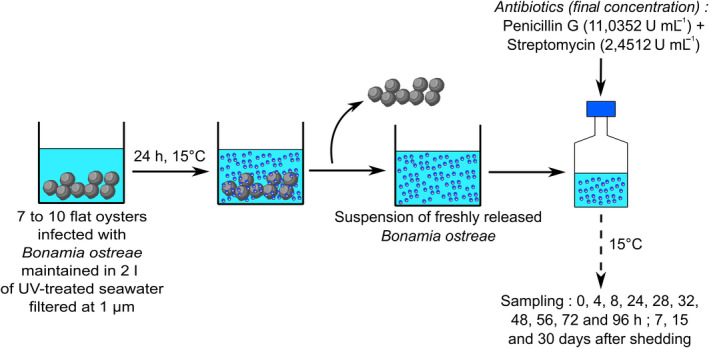
Experimental design used to monitor the presence of Bonamia ostreae outside flat oysters.
Negative controls consisted in 100 ml of seawater collected before immersing flat oysters.
Sample filtration
Seawater samples (100 ml) collected as previously described were pre‐filtered at 20 µm to remove largest particles before being filtered through a 1 µm pore size 47 mm diameter polycarbonate membrane (WhatmanWhatman® Nuclepore™ Track‐Etched Membranes) using a vacuum pump. Each membrane was then cut in four quarters and stored at −80°C until being processed for either DNA (two quarters) or RNA (two quarters) extraction. Quarters for DNA extraction were stored in 1.5 ml Safe‐Lock Tubes (Eppendorf), and quarters for RNA extraction were stored in 1.5 ml DNA LoBind Tubes (Eppendorf). For each sample, one quarter membrane was used for nucleic acid extraction (DNA and RNA), and the other quarter was stored in order to redo analysis if needed.
Nucleic acid extraction
Total DNA was extracted from quarter membranes using the DNeasy® PowerWater® Kit (Qiagen) according to the manufacturer’s protocol with some modifications. After 10 min at 65°C, a mechanical cell lysis was carried out using the Precellys® 24 bead beater (Bertin Technologies) and the following programme: 4 lysis cycles of 20 s at 5000 r.p.m., with 5 s of pause between each cycle. DNA was eluted in 50 µl of the elution solution provided in the kit, and samples were stored at 4°C until being tested by real‐time PCR.
Total RNA was extracted from quarter membranes using the RNeasy® PowerWater® Kit (Qiagen) according to the manufacturer’s protocol with some modifications. Mechanical cell lysis was carried out as described above for DNA extraction. Nucleic acid was eluted in 50 µl of the elution solution provided in the kit. Genomic DNA was removed using the DNase™ Max Kit (Qiagen) following the manufacturer’s recommendations, except that DNase reaction was performed at 37°C for 30 min instead of 20 min. The successful elimination of genomic DNA was checked by testing RNA extracts directly by real‐time PCR targeting Bonamia ostreae 18S rDNA (see below).
cDNAs were synthesized using the Invitrogen™ SuperScript™ III Reverse Transcriptase kit (Thermo Fisher Scientific), following the manufacturer’s instructions. cDNAs were stored at 4°C until being tested by real‐time PCR.
Real‐time PCR
For the detection of Bonamia ostreae 18S rDNA, amplification reactions were carried out as described in the Standard Operating Procedure (SOP) published by European Reference Laboratory for Mollusc Diseases (EURL for Molluscs Diseases, 2020; L. Canier, C. Dubreuil, M. Noyer, D. Serpin, B. Chollet, C. Garcia and I. Arzul, unpublished data). PCR mixture included 12.5 µl 2X Brilliant III Ultra‐Fast QPCR Master Mix (Agilent Technologies); 0.38 µl Bosp2 18S Forward Primer (5′ CAG GAT GCC CTT AGA TGC TC 3′) (20 µM); 0.63 µl Bosp2 18S Reverse Primer (5′ GTA CAA AGG GCA GGG ACG TA 3′) (20 µM); 0.75 µl Bosp2 18S IN Probe (5′ TTG ACC CGG CTT GAC AAG GC 3′) (HEX‐BHQ‐1) (10 µM); 5.75 µl bi‐distilled water; and 5 µl of extracted DNA. The thermal programme was as follows: 95°C for 3 min and 40 cycles of amplification at 95°C for 15 s and 60°C for 1 min.
For the detection of B. ostreae 18S rRNA, PCR mixture included 10 µl 2X Brilliant III Ultra‐Fast SYBR® Green QPCR Master Mix (Agilent Technologies), 2 µl 18S Forward Primer (5′ TCA GCA CTT TTC GAG AAA TCA A 3′) (5 µM), 2 µl 18S Reverse Primer (5′ CCA CCA TGC ATA GAA TCA AGA A 3′) (4 µM), 1 µl bi‐distilled water and 5 µl of undiluted cDNA. The thermal profile was as follows: 95°C for 3 min and 40 cycles of amplification at 95°C for 5 s and 60°C for 20 s. A post‐PCR dissociation curve was run with the following parameters: 95°C for 1 min, 60°C for 30 s and a gradual augmentation from 60°C to 95°C for 40 min (Gervais et al., 2018).
Each sample was analysed in duplicate by real‐time PCR analysis in 96‐microwell plates using the Mx3000p™ thermocycler sequence detector (Stratagene). Positive and negative controls were included in each PCR run. Positive controls consisted in DNA or RNA extracted from known infected samples. Negative controls consisting in 5 µl of bi‐distilled water used in the extraction and real‐time PCR steps were added to each PCR plate.
Data analysis
Data were analysed using R 3.4.2 (2017‐09‐28) (R Core Team, 2017) after computing average Ct from each duplicate. When no amplification was obtained, ‘No Ct’ value was replaced by 40, corresponding to the maximum number of cycles.
Real‐time PCR efficiency, limit of detection and limit of quantification
Efficiency, LOD and LOQ of both DNA and RNA approaches were determined from the analysis of parasite dilution series in real‐time PCR.
After a decimal log conversion of the number of parasites included in the dilution series, PCR efficiency was established for each dilution series (n = 5 and n = 4 for DNA and RNA respectively) using the following formula:
| (1) |
where ‘a’ is the slope of the linear regression performed on Ct ~ log value.
Based on these data, average efficiency and standard deviation were computed for both DNA and RNA approaches.
To be as close as possible to the MIQE guidelines (Bustin et al., 2009), LOD was defined as the lowest detected amount of parasites in at least 75% of tested dilutions (4 out of 5 for the DNA approach or 3 out of 4 for the RNA approach) and LOQ was defined as the last amount of parasites detected in the linear dynamic range of the standard curve.
Standard curves
Standard curves were then established for each real‐time PCR approach by performing a linear regression on semi‐logarithm transformed data obtained from the analysis of parasite dilution series. The equation of the standard curves was the following:
| (2) |
95% confidence and prediction intervals were calculated using predict function in R (Cornillon et al., 2012).
As an output of the regression model, R 2 was used to check the amount of variability of Ct explained by N parasites. Residuals were also analysed graphically to validate the model.
Parasite presence
Parasite presence was measured by testing samples regularly taken from freshly shed parasite suspensions in real‐time PCR.
Number of parasites per quarter membrane was calculated using the following equation (modified from Equation 2):
| (3) |
Parasite presence was then estimated at each time by computing the following ratio:
| (4) |
After testing data normality for each time, non‐parametric statistical tests (Wilcoxon–Mann–Whitney) were performed on presence data to investigate the difference between the amount of detected parasites at t = τ and at t = 0.
Additionally, correlation between the number of detected parasites at the beginning and the end of the experiment as well as between DNA and RNA detection was tested by computing Pearson’s, Spearman’s or Kendall’s correlation coefficients after checking data normality.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by the French Research Institute for Exploitation of the Sea (IFREMER) and Région Nouvelle‐Aquitaine. This work received financial support from the European Union Reference Laboratory for Molluscs Diseases, the French National Research Agency (ANR) project ENVICOPAS (15‐CE35‐0004) and the European project VIVALDI (H2020 n°678589).
Microbial Biotechnology (2020) 13(6), 1807–1818
Funding information
This work was supported by the French Research Institute for Exploitation of the Sea (IFREMER) and Région Nouvelle‐Aquitaine. This work received financial support from the European Union Reference Laboratory for Molluscs Diseases, the French National Research Agency (ANR) project ENVICOPAS (15‐CE35‐0004) and the European project VIVALDI (H2020 n°678589).
References
- Arzul, I. , and Carnegie, R.B. (2015) New perspective on the haplosporidian parasites of molluscs. J Invertebr Pathol 131: 32–42. [DOI] [PubMed] [Google Scholar]
- Arzul, I. , Miossec, L. , Blanchet, E. , Garcia, C. , Francois, C. , and Joly, J.‐P. (2006) Bonamia ostreae and Ostrea edulis: A Stable Host‐parasite System in France? Cairns, Queensland, Australia: Presentation ‐ XI International Symposium for Veterinary Epidemiology and Economics.
- Arzul, I. , Gagnaire, B. , Bond, C. , Chollet, B. , Morga, B. , Ferrand, S. , et al (2009) Effects of temperature and salinity on the survival of Bonamia ostreae, a parasite infecting flat oysters Ostrea edulis. Dis Aquat Organ 85: 67–75. [DOI] [PubMed] [Google Scholar]
- Audemard, C. , Reece, K.S. , and Burreson, E.M. (2004) Real‐Time PCR for detection and quantification of the Protistan parasite Perkinsus marinus in environmental waters. Appl Environ Microbiol 70: 6611–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, S. , and Wuertz, S. (2009) Discrimination of viable and dead fecal Bacteroidales bacteria by Quantitative PCR with Propidium Monoazide. Appl Environ Microbiol 75: 2940–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa Solomieu, V. , Renault, T. , and Travers, M.‐A. (2015) Mass mortality in bivalves and the intricate case of the Pacific oyster, Crassostrea gigas . J Invertebr Pathol 131: 2–10. [DOI] [PubMed] [Google Scholar]
- Bass, D. , Stentiford, G.D. , Littlewood, D.T.J. , and Hartikainen, H. (2015) Diverse applications of environmental DNA methods in parasitology. Trends Parasitol 31: 499–513. [DOI] [PubMed] [Google Scholar]
- Blais, B.W. , Turner, G. , Sooknanan, R. , and Malek, L.T. (1997) A nucleic acid sequence‐based amplification system for detection of Listeria monocytogenes hlyA sequences. Appl Environ Microbiol 63: 310–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buestel, D. , Ropert, M. , Prou, J. , and Goulletquer, P. (2009) History, status, and future of oyster culture in France. J Shellfish Res 28: 813–820. [Google Scholar]
- Bustin, S.A. , Benes, V. , Garson, J.A. , Hellemans, J. , Huggett, J. , Kubista, M. , et al (2009) The MIQE guidelines: minimum information for publication of quantitative real‐time PCR Experiments. Clin Chem 55: 611–622. [DOI] [PubMed] [Google Scholar]
- Carnegie, R.B. , Barber, B.J. , Culloty, S.C. , Figueras, A.J. , and Distel, D.L. (2000) Development of a PCR assay for detection of the oyster pathogen Bonamia ostreae and support for its inclusion in the Haplosporidia. Dis Aquat Organ 42: 199–206. [DOI] [PubMed] [Google Scholar]
- Casanovas‐Massana, A. , Pedra, G.G. , Wunder, E.A. , Diggle, P.J. , Begon, M. , and Ko, A.I. (2018) Quantification of Leptospira interrogans survival in soil and water microcosms. Appl Environ Microbiol 84: e00507‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochennec, N. , Le Roux, F. , Berthe, F. , and Gerard, A. (2000) Detection of Bonamia ostreae based on small subunit ribosomal probe. J Invertebr Pathol 76: 26–32. [DOI] [PubMed] [Google Scholar]
- Collins, R.A. , Wangensteen, O.S. , O'Gorman, E.J. , Mariani, S. , Sims, D.W. , and Genner, M.J. (2018) Persistence of environmental DNA in marine systems. Commun Biol 1: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillon, P.‐A. , Guyader, A. , Husson, F. , Jégou, N. , Josse, J. , Kloareg, M. , et al (2012) Statistiques avec R. Rennes: Presses universitaires de Rennes. [Google Scholar]
- Cristescu, M.E. (2019) Can environmental RNA revolutionize biodiversity science? Trends Ecol Evol 34: 694–697. [DOI] [PubMed] [Google Scholar]
- Culloty, S.C. , and Mulcahy, M.F. (1996) Season‐, age‐, and sex‐related variation in the prevalence of bonamiasis in flat oysters (Ostrea edulis L.) on the south coast of Ireland. Aquaculture 144: 53–63. [Google Scholar]
- Culloty, S.C. , Novoa, B. , Pernas, M. , Longshaw, M. , Mulcahy, M.F. , Feist, S.W. , and Figueras, A. (1999) Susceptibility of a number of bivalve species to the protozoan parasite Bonamia ostreae and their ability to act as vectors for this parasite. Dis Aquat Organ 37: 73–80. [Google Scholar]
- Dejean, T. , Valentini, A. , Duparc, A. , Pellier‐Cuit, S. , Pompanon, F. , Taberlet, P. , and Miaud, C. (2011) Persistence of environmental DNA in freshwater ecosystems. PLoS One 6: e23398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiler, A. , Johansson, M. , and Bertilsson, S. (2006) Environmental influences on Vibrio populations in Northern Temperate and Boreal Coastal Waters (Baltic and Skagerrak Seas). Appl Environ Microbiol 72: 6004–6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsma, M. , Kerkhoff, S. , Roozenburg, I. , Haenen, O. , van Gool, A. , Sistermans, W. , et al (2010) Epidemiology of Bonamia ostreae infecting European flat oysters Ostrea edulis from Lake Grevelingen, The Netherlands. Mar Ecol Prog Ser 409: 131–142. [Google Scholar]
- EURL for Molluscs Diseases (2020) European reference laboratory for molluscs diseases. [WWW Document]https://www.eurl‐mollusc.eu/SOPs.
- Friedman, C.S. , Wight, N. , Crosson, L.M. , White, S.J. , and Strenge, R.M. (2014) Validation of a quantitative PCR assay for detection and quantification of ‘Candidatus Xenohaliotis californiensis’. Dis Aquat Organ 108: 251–259. [DOI] [PubMed] [Google Scholar]
- Gervais, O. , Renault, T. , and Arzul, I. (2018) Molecular and cellular characterization of apoptosis in flat oyster a key mechanisms at the heart of host‐parasite interactions. Sci Rep 8: 12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goarant, C. , and Merien, F. (2006) Quantification of Vibrio penaeicida, the etiological agent of Syndrome 93 in New Caledonian shrimp, by real‐time PCR using SYBR Green I chemistry. J Microbiol Methods 67: 27–35. [DOI] [PubMed] [Google Scholar]
- Green, H.C. , Shanks, O.C. , Sivaganesan, M. , Haugland, R.A. , and Field, K.G. (2011) Differential decay of human faecal Bacteroides in marine and freshwater. Environ Microbiol 13: 3235–3249. [DOI] [PubMed] [Google Scholar]
- Harper, L.R. , Buxton, A.S. , Rees, H.C. , Bruce, K. , Brys, R. , Halfmaerten, D. , et al (2019) Prospects and challenges of environmental DNA (eDNA) monitoring in freshwater ponds. Hydrobiologia 826: 25–41. [Google Scholar]
- Hervio, D. , Bachere, E. , Boulo, V. , Cochennec, N. , Vuillemin, V. , Le Coguic, Y. , et al (1995) Establishment of an experimental infection protocol for the flat oyster, Ostrea edulis, with the intrahaemocytic protozoan parasite, Bonamia ostreae: application in the selection of parasite‐resistant oysters. Aquaculture 132: 183–194. [Google Scholar]
- Johnson, A. , and Brunner, J. (2014) Persistence of an amphibian ranavirus in aquatic communities. Dis Aquat Organ 111: 129–138. [DOI] [PubMed] [Google Scholar]
- Klein, P.G. , and Juneja, V.K. (1997) Sensitive detection of viable Listeria monocytogenes by reverse transcription‐PCR. Appl Environ Microbiol 63: 4441–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallias, D. , Arzul, I. , Heurtebise, S. , Ferrand, S. , Chollet, B. , Robert, M. , et al (2008) Bonamia ostreae ‐induced mortalities in one‐year old European flat oysters Ostrea edulis: experimental infection by cohabitation challenge. Aquat Living Resour 21: 423–439. [Google Scholar]
- Lane, H. , Webb, S. , and Duncan, J. (2016) Bonamia ostreae in the New Zealand oyster Ostrea chilensis: a new host and geographic record for this haplosporidian parasite. Dis Aquat Organ 118: 55–63. [DOI] [PubMed] [Google Scholar]
- McCarthy, A.J. , Saunders, J.R. , and Milner, M.G. (2001) Relationship between nucleic acid ratios and growth in Listeria monocytogenes. Microbiology 147: 2689–2696. [DOI] [PubMed] [Google Scholar]
- Mialhe, E. , Bachère, E. , Chagot, D. , Grizel, H. (1988) ,Isolation and purification of the protozoan Bonamia ostreae (Pichot et al. 1980), a parasite affecting the flat oyster Ostrea edulis L. Aquaculture 71: 293–299. [Google Scholar]
- Montes, J. , Anadón, R. , and Azevedo, C. (1994) A possible life cycle for Bonamia ostreae on the Basis of Electron Microscopy Studies. J Invertebr Pathol 63: 1–6. [Google Scholar]
- Nocker, A. , Sossa, K.E. , and Camper, A.K. (2007) Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J Microbiol Methods 70: 252–260. [DOI] [PubMed] [Google Scholar]
- NORA . (2020). Native Oyster Restoration Alliance [WWW Document]. https://noraeurope.eu/restoration‐projects/.
- OIE . (2020). Manual of diagnostic tests for aquatic animals [WWW Document]. https://www.oie.int/fr/normes/manuel‐aquatique/acces‐en‐ligne/.
- Parizadeh, L. , Tourbiez, D. , Garcia, C. , Haffner, P. , Dégremont, L. , Roux, F.L. , and Travers, M.‐A. (2018) Ecologically realistic model of infection for exploring the host damage caused by Vibrio aestuarianus . Environ Microbiol 20: 4343–4355. [DOI] [PubMed] [Google Scholar]
- Pichot, Y. , Comps, M. , Tige, G. , Grizel, H. , and Rabouin, M.‐A. (1980) Recherches sur Bonamia ostreae gen. n., sp. n., parasite nouveau de l’huître plate Ostrea edulis L. Rev Trav Inst Pêch Marit 43: 131–140. [Google Scholar]
- Pochon, X. , Zaiko, A. , Fletcher, L.M. , Laroche, O. , and Wood, S.A. (2017) Wanted dead or alive? Using metabarcoding of environmental DNA and RNA to distinguish living assemblages for biosecurity applications. PLoS One 12: e0187636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoda, B. , Brown, J. , Hancock, B. , Preston, J. , Pouvreau, S. , Kamermans, P. , et al (2019) The Native Oyster Restoration Alliance (NORA) and the Berlin Oyster Recommendation: bringing back a key ecosystem engineer by developing and supporting best practice in Europe. Aquat Living Resour 32: 13. [Google Scholar]
- R Core Team (2017) R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Ramilo, A. , Navas, J.I. , Villalba, A. , and Abollo, E. (2013) Species‐specific diagnostic assays for Bonamia ostreae and B. exitiosa in European flat oyster Ostrea edulis: conventional, real‐time and multiplex PCR. Dis Aquat Organ 104: 149–161. [DOI] [PubMed] [Google Scholar]
- Robert, M. , Garcia, C. , Chollet, B. , Lopez‐Flores, I. , Ferrand, S. , François, C. , et al (2009) Molecular detection and quantification of the protozoan Bonamia ostreae in the flat oyster, Ostrea edulis . Mol Cell Probes 23: 264–271. [DOI] [PubMed] [Google Scholar]
- Roberts, J.J. , Brandt, S.B. , Fanslow, D. , Ludsin, S.A. , Pothoven, S.A. , Scavia, D. , and Höök, T.O. (2011) Effects of hypoxia on consumption, growth, and RNA:DNA ratios of Young Yellow Perch. Trans Am Fish Soc 140: 1574–1586. [Google Scholar]
- Rusch, J.C. , Hansen, H. , Strand, D.A. , Markussen, T. , Hytterød, S. , and Vrålstad, T. (2018) Catching the fish with the worm: a case study on eDNA detection of the monogenean parasite Gyrodactylus salaris and two of its hosts, Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss). Parasites Vectors 11: 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, D.J. , Sepúlveda, M.S. , Nalepa, T.F. , and Höök, T.O. (2012) Spatial variation in RNA:DNA ratios of Diporeia spp. in the Great Lakes region. J Gt Lakes Res 38: 187–195. [Google Scholar]
- Sassoubre, L.M. , Yamahara, K.M. , Gardner, L.D. , Block, B.A. , and Boehm, A.B. (2016) Quantification of environmental DNA (eDNA) shedding and decay rates for three marine fish. Environ Sci Technol 50: 10456–10464. [DOI] [PubMed] [Google Scholar]
- Strepparava, N. , Wahli, T. , Segner, H. , and Petrini, O. (2014) Detection and quantification of Flavobacterium psychrophilum in water and fish tissue samples by quantitative real time PCR. BMC Microbiol 14: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suquet, M. , Araya, R.G. , Lebrun, L. , Queau, I. , Mingant, C. , and Robert, R. (2010) Anaesthesia and gonad sampling in the European flat oyster (Ostrea edulis). Aquaculture 308: 196–198. [Google Scholar]
- Taberlet, P. , Coissac, E. , Hajibabaei, M. , and Rieseberg, L.H. (2012) Environmental DNA. Mol Ecol 21: 1789–1793. [DOI] [PubMed] [Google Scholar]
- Thomsen, P.F. , Kielgast, J. , Iversen, L.L. , Møller, P.R. , Rasmussen, M. , and Willerslev, E. (2012) Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS One 7: e41732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twing, K.I. , Kirchman, D.L. , and Campbell, B.J. (2011) Temporal study of Helicobacter pylori presence in coastal freshwater, estuary and marine waters. Water Res 45: 1897–1905. [DOI] [PubMed] [Google Scholar]
- Ward, G.M. , Bennett, M. , Bateman, K. , Stentiford, G.D. , Kerr, R. , Feist, S.W. , et al (2016) A new phylogeny and environmental DNA insight into paramyxids: an increasingly important but enigmatic clade of protistan parasites of marine invertebrates. Int J Parasitol 46: 605–619. [DOI] [PubMed] [Google Scholar]
- Ward, G.M. , Neuhauser, S. , Groben, R. , Ciaghi, S. , Berney, C. , Romac, S. , and Bass, D. (2018) Environmental sequencing fills the gap between parasitic Haplosporidians and free‐living Giant Amoebae. J Eukaryot Microbiol 65: 574–586. [DOI] [PMC free article] [PubMed] [Google Scholar]


