Summary
The multi‐enzyme cellulosome complex can mediate the valorization of lignocellulosic biomass into soluble sugars that can serve in the production of biofuels and valuable products. A potent bacterial chassis for the production of active cellulosomes displayed on the cell surface is the bacterium Lactobacillus plantarum, a lactic acid bacterium used in many applications. Here, we developed a methodological pipeline to produce improved designer cellulosomes, using a cell‐consortium approach, whereby the different components self‐assemble on the surface of L. plantarum. The pipeline served as a vehicle to select and optimize the secretion efficiency of potent designer cellulosome enzyme components, to screen for the most efficient enzymatic combinations and to assess attempts to grow the engineered bacterial cells on wheat straw as a sole carbon source. Using this strategy, we were able to improve the secretion efficiency of the selected enzymes and to secrete a fully functional high‐molecular‐weight scaffoldin component. The adaptive laboratory process served to increase significantly the enzymatic activity of the most efficient cell consortium. Internal plasmid re‐arrangement towards a higher enzymatic performance attested for the suitability of the approach, which suggests that this strategy represents an efficient way for microbes to adapt to changing conditions.
Here, we developed a methodological pipeline to produce improved designer cellulosomes, using a cell‐consortium approach, whereby the different components self‐assemble on the surface of L. plantarum. The pipeline served as a vehicle to select and optimize the secretion efficiency of potent designer cellulosome enzyme components, to screen for the most efficient enzymatic combinations and to assess attempts to grow the engineered bacterial cells on wheat straw as a sole carbon source. An adaptive laboratory process served to increase significantly the enzymatic activity of the most efficient cell consortium by internal plasmid rearrangement.

Introduction
Lignocellulosic biomass is the most abundant renewable resource of organic material on Earth (Lamed and Bayer, 1988; Bayer et al., 2008). Therefore, it represents a major portion in the agricultural residues and in the municipal solid waste (Bayer et al., 2007; Fatma et al., 2018). Most of the lignocellulose biomass is composed of sugars that could directly serve in alcoholic fermentations for the production of valuable products. The sugars are arranged in two main polysaccharide groups – cellulose and hemicellulose – that can be efficiently deconstructed by multi‐enzyme complexes termed cellulosomes (Bayer et al., 1983; Bayer et al., 2013; Artzi et al., 2017). These unique extracellular enzymatic complexes are composed of many interacting proteins: that is, the enzymatic subunits and the non‐enzymatic scaffoldins that organize them into the complex. Although cellulosome complexes vary greatly in modular architecture, both between and within the different bacterial species, the molecular basis which determines their assembly comprises the high‐affinity intermodular interactions between a cohesin module and a complementary dockerin module, located on the different interacting components.
The scaffoldins are generally non‐catalytic proteins that integrate the dockerin‐containing enzymes via their repeating cohesin modules. The primary scaffoldin subunit also contains a carbohydrate‐binding module (CBM), which binds the substrate very tightly (Bayer et al., 2004). In general, the primary scaffoldin also bears an anchoring motif that attaches the cellulosome complex to the bacterial cell wall, and it thus mediates between the bacterium and its substrate (Lemaire et al., 1995; Lemaire et al., 1998; Rincon et al., 2005). Cellulosome organization facilitates high levels of synergy among the catalytic subunits via substrate channelling (Moraïs et al., 2011; Zhang, 2011; Wieczorek and Martin, 2012). The proximity of the substrate/cellulosomes/bacterial cell minimizes the diffusion of the hydrolytic products and enzymes, providing the bacterium with a competitive advantage over cellulosome non‐producers (Shoham et al., 1999). The cohesin–dockerin interaction is generally species specific (Pages et al., 1997; Haimovitz et al., 2008), and this feature forms the basis for creation of ‘designer cellulosomes’ – enzymatic complexes in which we can control the position and stoichiometry of the enzymes on the scaffoldin subunit (Bayer et al., 1994; Fierobe et al., 2005; Moraïs et al., 2012). Efficient designer cellulosomes would comprise the three major types of cellulases, that is exoglucanases, endoglucanases and β‐glucosidases, which are responsible for efficient deconstruction of cellulose (Kadam and Demain, 1989; Bayer et al., 1998; Gefen et al., 2012). In addition, it has been shown that incorporation of both xylanases and cellulases into designer cellulosomes further enhances the synergism among the different enzymes and improves degradation of complex cellulosic substrates (Moraïs et al., 2010).
In recent years, the lactic acid bacterium, Lactobacillus plantarum, has been successfully engineered to produce designer cellulosomes in vivo and served both as a tool to obtain insights into intrinsic mechanisms of cellulosomal action and as an emerging platform for biomass‐to‐biofuel conversion (Moraïs et al., 2014; Stern et al., 2017, 2018). The engineered bacterium has proven a suitable producer of cellulases, hemicellulases and cellulosomal components. Indeed, the functionality and stability of the enzymes, the cohesins and the dockerins from various bacterial origins were all demonstrated (Moraïs et al., 2013, 2014; Stern et al., 2018). In addition, the cell‐consortium approach, whereby individual bacterial strains are equipped with single components of the designer cellulosomes, has been shown to be an appropriate strategy to produce functional designer cellulosomes in which the stoichiometry of the components can be controlled (Stern et al., 2018). The resultant secretion of the recombinant dockerin‐bearing enzymes and their self‐assembly onto the chimaeric cohesin‐containing scaffoldin displayed on the bacterial cell surface thus form the final designer cellulosome. In general, fabrication of this type of surface‐displayed enzyme complex would be an attractive strategy for many biotechnological and pharmaceutical applications. L. plantarum is highly suitable for this propose, since it is a generally recognized as safe (GRAS) bacterium that has already been used in many industrial and agricultural applications (Mazzoli et al., 2014).
Adaptive laboratory evolution (ALE) is a useful tool that allows selection of desirable phenotypes through natural evolution in a laboratory environment. In ALE, microbial populations are allowed to grow for extended periods under specified conditions, after which genetic variations that occurred and which improved the phenotypic traits of the culture can be detected. This method has been applied for a variety of applications, and there are increasing perspectives for advancing this technology (Wondraczek et al., 2019). For example, this approach has been successfully demonstrated for xylan utilization in Lactobacillus pentosus, Saccharomyces cerevisiae and Bacillus subtilis (Shen et al., 2012; Zhang et al., 2015; Cubas‐Cano et al., 2019). In another study, Lactobacillus lactis was adapted to grow at 39°C by slowly increasing the growth temperature (Chen et al., 2015). This method is also suitable for complex communities like microbial consortia. In this context, Liu et al. successfully adapted a consortium of three bacterial species for higher biobleaching performance (Liu et al., 2019). Here, we applied this approach in an attempt to adapt an L. plantarum cell consortium to grow on wheat straw.
Lactobacillus plantarum is a well‐characterized lactic acid bacteria (LAB), prevalent in a variety of environmental niches, including some meat, dairy, plant fermentations and in the human gastrointestinal tract (Ahrne et al., 1998; Kleerebezem et al., 2003). This bacterium can grow in the presence of up to 13% (v/v) ethanol in the medium and at pH 3.2 (Alegria et al., 2004). Therefore, it is frequently found in contaminated ethanol fermentations (Limayem et al., 2011; Roach et al., 2013). Furthermore, deconstruction of lignin from plant biomass commonly involves acid pretreatment that needs to be removed (Bayer et al., 2007). In this context, L. plantarum may be added to initial degradation steps due to its production of lactic acid and acid tolerance and would therefore present a potential economic advantage. In addition, L. plantarum can be used in many bioremediation applications, such as for removal of cadmium and lead, which are the two most abundant toxic heavy metals in the environment (Kirillova et al., 2017). L. plantarum is also known as a probiotic, due to its health‐promoting effects in humans, which include cholesterol lowering, diarrhoea prevention and reduction in the symptoms of irritable bowel syndrome (Ducrotte et al., 2012; Bosch et al., 2014; Yang et al., 2014; Seddik et al., 2017). Moreover, several L. plantarum strains are known to produce different antimicrobial compounds, such as organic acids, and antimicrobial peptides, such as bacteriocins, which can be used in food preservation (Zhang et al., 2013; Todorov et al., 2018). In this context, improving the tools for producing and secreting desired products in L. plantarum can prove relevant for many applications.
Here, we used a methodological pipeline to produce improved designer cellulosomes on the surface of L. plantarum. Our pipeline comprises (i) selection of highly active enzymes, (ii) optimized secretion of the selected enzymes and structural cellulosomal proteins (scaffoldins), (iii) selection of the most active enzymatic combinations and (iv) adaptive evolution of the most active enzymatic combination for improvement of growth on wheat straw as a sole carbon source.
Results
Selection of designer cellulosomes components
Lactobacillus plantarum is a mesophilic bacterium that contains 55 glycoside hydrolase (GH) genes, distributed among 18 families. Many of these genes are predicted as β‐glucosidases, and none of them encode for cellulase or xylanase functions according to the CAZy database (Cantarel et al., 2009). Consequently, the bacterium lacks the inherent ability to degrade the major components of the plant fibre (i.e. cellulose and hemicelluloses) but is able to grow on cellobiose as sole carbon source (Stern et al., 2018). The plant cell wall is a complex substrate needing diverse types of enzymes for its breakdown. Therefore, we searched for enzymes with different degradation activities, for example families 5, 9, 10, 11 and 48. GH10 and GH11 families, are associated with xylanase functions, GH5 enzymes can be endoglucanases, xylanases or mannanases, GH48s are usually exoglucanases, while GH9s include exoglucanases, endoglucanases, and processive endoglucanases that can exhibit both endoglucanase and exoglucanase activities.
We performed broad screening of mesophilic enzymes from cellulosomal bacteria, in order to identify the most active enzymes under the growth conditions of L. plantarum (i.e. 37°C and pH below 5). The selected enzymes were chosen from the above GH families that are known to play an important role in cellulosome activity (Moraïs et al., 2016a,b). All of the latter (dockerin‐bearing) enzymes were expressed in E. coli as native enzymes with an appended N‐terminal His‐tag and were purified on a Ni‐NTA column. In total, 11 cellulases and six xylanases were examined (Table S1). The enzymes were tested on model polysaccharidic substrates, carboxymethyl cellulose (CMC) and xylan, and at different pH values (4, 4.5 and 5). The enzymes exhibiting the highest enzymatic activity at low pH, with preference to lower molecular weights, were selected for integration into the designer cellulosome: Clostridium papyrosolvens Cel5, C. papyrosolvens Cel9, C. papyrosolvens Xyn10 and C. papyrosolvens Xyn11 (Fig. S1). For exoglucanase activity, Ruminococcus champanellensis Cel48 was chosen due to its successful expression in E. coli compared to the Cel48s of C. papyrosolvens and R. flavefaciens (data not shown). Additionally, this enzyme exhibited a level of synergism of about twofold when combined with the Cel5 and Cel9 cellulases (Fig. S2).
In order to integrate the selected enzymes into designer cellulosome systems, the catalytic module of each enzyme was fused to a different dockerin module. Cel5 was fused to a dockerin from Archeo g lobus fulgidus (Cel5‐g), Cel9 was fused to a dockerin of B acteroides cellulosolvens (Cel9‐b), Xyn10 was fused to a dockerin of Clostridium t hermocellum (Xyn10‐t), and Xyn11 was fused to a dockerin of A cetivibrio cellulolyticus (Xyn11‐a; Fig. 1). The hydrolytic activities of the chimaeric enzymes were compared to those of wild‐type enzymes and found to be equally active (Stern et al., 2018). For the R. champanellensis exoglucanase Cel48, we created three different chimaeras, with dockerins from B. cellulosolvens, A. cellulolyticus and C. thermocellum that allow us to test different designer cellulosome enzyme combinations. The chimaeric scaffoldin, Scaf‐ATGB, was chosen for this study. It has four cohesins that match the specificities of the above‐described dockerins, a CBM and a cell‐wall‐anchoring sequence (sortase motif; Fig. 1).
Fig. 1.
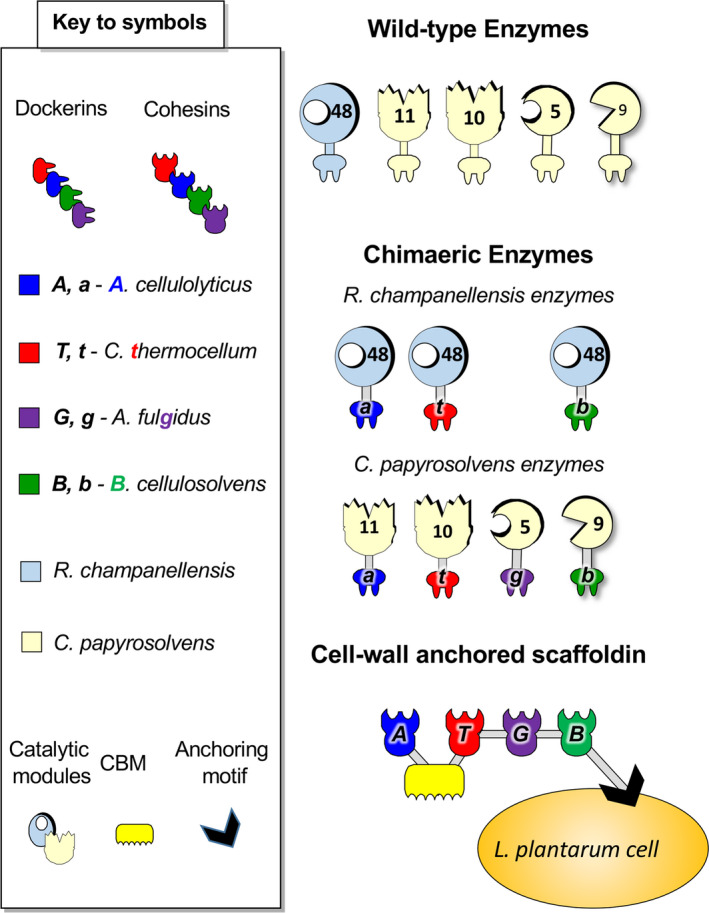
Schematic representation of the recombinant proteins that have been used in this study.
Differential secretion efficiency of heterologous fibrolytic enzymes depends on endogenous signal peptide identity
Lactobacillus plantarum can efficiently secrete heterologous proteins via the pSIP vector system (Sorvig et al., 2003). This is a strictly regulated system based on the bacteriocins sakacin A and sakacin P. One advantage of this system is that the modular design of the pSIP vectors allows easy exchange of the desired elements such as the signal peptide, the gene of interest or the replicon (Sorvig et al., 2003; Mathiesen et al., 2008). The selection of the optimal signal peptide for the desired secreted protein is a crucial step. Using the NucA protein as a reporter gene, the most efficient leader peptide (Lp) of L. plantarum was Lp_3050 (Mathiesen et al., 2009). However, the NucA protein is very small in size (22 kDa), compared to the GH enzymes, and it has been demonstrated that there is high diversity between the secretion efficiency of the same signal peptide with different proteins and hypothesized that smaller proteins are secreted in higher quantities than larger proteins (Mathiesen et al., 2009; Stern et al., 2018). Since the molecular weights of our selected enzymes and scaffoldin vary from 34 to 112 kDa, we wanted to examine the effect of different signal peptides on their secretion efficiency. For this purpose, we selected four different signal peptides originating from secreted L. plantarum proteins with molecular weights higher than 50 kDa but still reported as efficient for NucA protein secretion by Mathiesen et al. (Mathiesen et al., 2009). We also included the best‐performing signal peptide, Lp_3050, according to the previous study (original secreted protein size 11 kDa; Table 1). These five signal peptides were cloned into the pSIP vector fused to the above six designer cellulosome components (enzymes and scaffoldin) to examine variation in their secretion.
Table 1.
Selected signal peptides from the L. plantarum genome.
| Signal peptide | Secreted protein | Secreted protein (kDa) | Secretion efficiency a |
|---|---|---|---|
| 0297 | Extracellular protein, CscC family | 73 | 6 |
| 0373 | Cell‐surface protein precursor, LPXTG‐motif cell‐wall anchor | 125 | 8 |
| 2588 | Cell‐surface adherence protein, collagen‐binding domain, LPXTG‐motif cell‐wall anchor | 57 | 30 |
| 3050 | Extracellular transglycosylase, membrane‐bound | 11 | 1 |
| 3093 | Lysozyme/muramidase, glycoside hydrolase family 25 | 84 | 15 |
Secretion efficiency was according to Mathiesen et al. (2009).
Examination of enzyme secretion
The different plasmids were transformed into electrocompetent L. plantarum cells, and colonies were verified by colony PCR and DNA sequencing. In all cases, the colonies were positive. Carbohydrate‐degradation assays were used to assess enzyme functionality, while Far‐Western blot assays were performed to ensure proper cohesin interaction and the observed size (according to the amino acid sequence) of the secreted protein (Fig. 2). In the Far‐Western blot assay, the detection of the enzymes was verified by interaction of the blotted membrane with purified scaffoldin and antibody against the scaffoldin‐borne CBM. Thus, we can ensure proper binding between the secreted dockerin‐bearing enzymes to their matching cohesin. For all enzymes, a single band of the correct size appeared (Fig. 2, upper panel). The secretion efficiency of the enzymes was also tested by either CMC‐ or xylan‐degradation assay and was in agreement with the Far‐Western results.
Fig. 2.
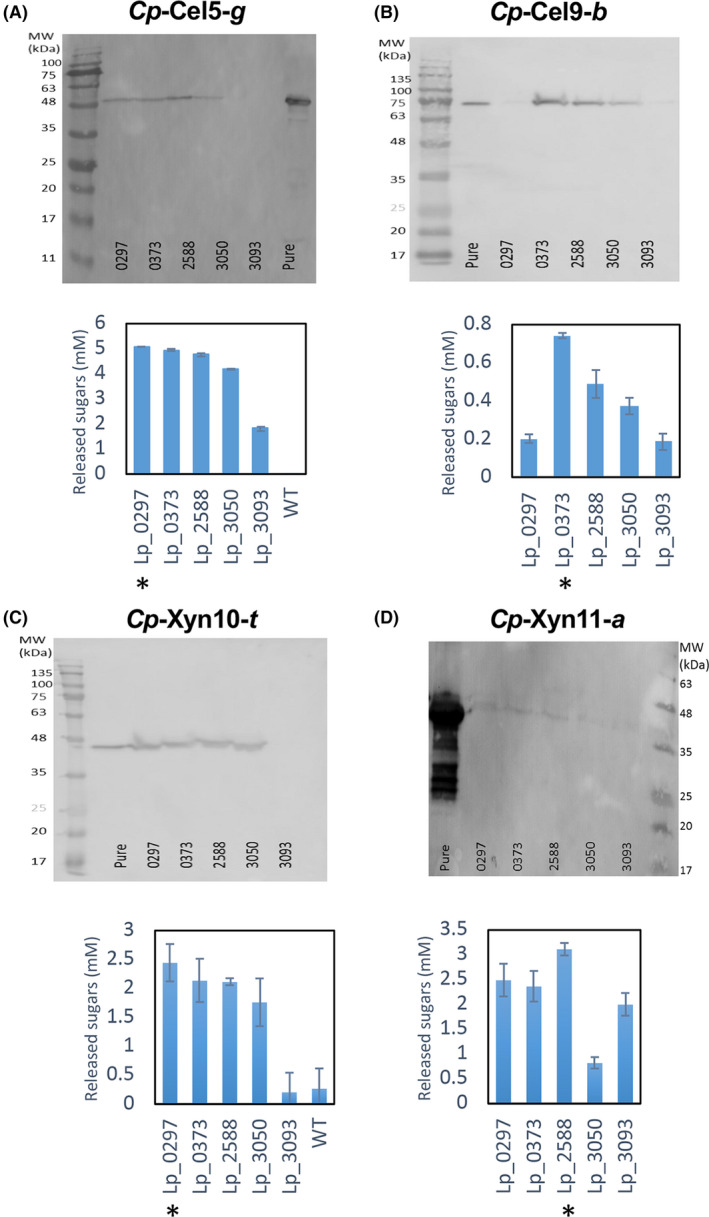
Far‐Western blot analysis and comparative enzymatic activity of the concentrated culture supernatant fractions secreted by five signal peptides from transformed L. plantarum strains. Upper panel, Far‐Western blot; lower panel, enzymatic activity. (A) Secreted Cp‐Cel5‐g, calculated mass 52.4 kDa. (B) Secreted Cp‐Cel9‐b, calculated mass 78.8 kDa. (C) Secreted Cp‐Xyn10‐t, calculated mass 45.8 kDa. (D) Secreted Cp‐Xyn11‐a, calculated mass 34.2 kDa. Concentrated supernatant fluids (30 µl) or purified (pure, see Methods section) recombinant enzyme (1 µg) were loaded onto 12% SDS‐PAGE gels and transferred to a nitrocellulose membrane. For activity assays, concentrated supernatant fluids (30 µl) were incubated with 1% CMC for 2 h or 1% xylan for 1 h at 37°C. Each reaction was performed three times in triplicate, and standard deviations are indicated. The highest performing signal peptide is indicated by an asterisk (*) in the figure for the designated protein.
Interestingly, we observed that the secretion of the orthologous protein depended on the specific signal peptide to which it was fused. Cel5‐g was most highly expressed using signal peptides Lp_0297, Lp_0373 and Lp_2588, while Lp_3093 was much less efficient (Fig. 2A). For Cel9‐b, signal peptide Lp_0373 was the most efficient, Lp_2588 and Lp_3050 exhibited medium levels of secretion, while Lp_0297 and Lp_3093 were very inefficient (Fig. 2B). For Xyn10‐t, the most efficient signal peptide was Lp_0297. Lp_0373, Lp_2588 and Lp_3050 were less efficient while with Lp_3093 negligible levels of activity were detected (Fig. 2C). For Xyn11‐a, Lp_2588 was the most efficient signal peptide, Lp_0297, Lp_0373 and Lp_3093 were less efficient, while only very low levels of activity were detected using Lp_3050 (Fig. 2D).
In the case of Rc Cel48‐b, the activity assays are ineffective, since this enzyme is not active on CMC and its activity on more complex substrates is very low. Therefore, we decided to use signal peptide Lp_2588, based on Western blot assays (data not shown), and the observed performance of this signal peptide for the other proteins. This signal peptide was also appended to the other two forms of this enzyme, Cel48‐a and Cel48‐t. No correlation was observed between the molecular weight of the recombinant protein and its best‐performing signal peptide, which might suggest that more than one secretion mechanism is involved in the secretion of the different enzymes. These results also demonstrated the importance of screening for signal peptide efficiency for each protein, due to the inability to predict.
The best‐performing signal peptide for each protein, as determined by the secretion experiments (see Fig. 2), was used in further experiments. Concentrations of the secreted enzymes in the different cultures were calculated by comparing the concentrated culture supernatant fluids from L. plantarum transformed cells with serial dilutions of the purified recombinant enzymes, by measuring reducing sugar formation on CMC or xylan substrates (Fig. S3). Cellulase concentrations at OD600 = 1 were estimated as 1.54 nM and 11.9 nM for pLp_0297sCel5 and pLp_0373sCel9, respectively. For the xylanases, these values were estimated as 1.12 nM and 0.14 nM for pLp_0297sXyn10 and pLp_2588sXyn11, respectively.
Examination of scaffoldin secretion and anchoring
The scaffoldin module was also expressed using the pSIP plasmid with the addition of a cell‐wall anchor motif (cwa2; Fredriksen et al., 2010), which allows attachment of the scaffoldin to the outer surface of the L. plantarum cells. In order to confirm full expression and functionality of the four cohesin modules of the scaffoldin, an ELISA assay was performed. Despite a previous study reporting the lack of function of two of the cohesins from the Scaf‐ATGB expressed on L. plantarum cell wall (Stern et al., 2018), by examining several signal peptides in the present work, we succeeded in expressing a fully functional scaffoldin. Two of the different signal peptides tested (Lp_0297 and Lp_2588) showed very similar binding profiles for each of the cohesins, indicating high functionality of the cohesins, while with the signal peptides Lp_3093 and Lp_3050, two cohesins (Cohesins G and B) were markedly less active as in Stern et al (Stern et al., 2018; Fig. 3). The negative control (wild‐type cells) displayed negligible or no cohesin‐binding ability. The quantity of the secreted scaffoldin with the plasmid pLp_0297sScafATGB was calculated as 4.6 nM at OD600 = 1, by comparing the binding properties of the pure recombinant proteins (produced in E. coli) to those of the culture pellet from transformed lactobacilli using an ELISA assay (data not shown).
Fig. 3.
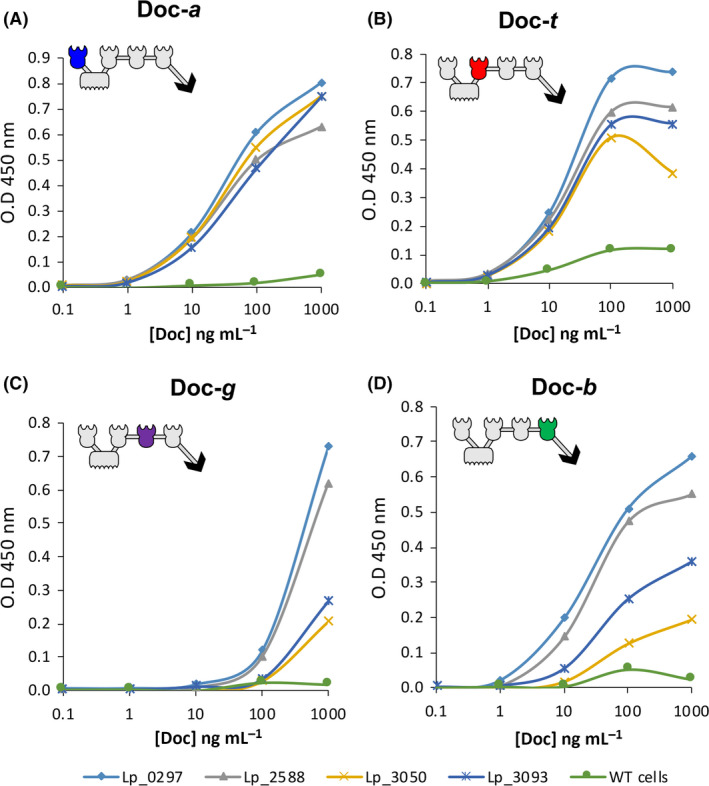
Scaf‐ATGB binding profile using four signal peptides. ELISA interaction assays of (A) Doc‐a (dockerin from A. cellulolyticus), (B) Doc‐t (dockerin from C. thermocellum), (C) Doc‐g (dockerin from A. fulgidus) and (D) Doc‐b (dockerin from B. cellulosolvens). ELISA plates were coated overnight with different amounts of the desired dockerin and then interacted with 100 µl of cell pellet at OD600 = 2. Lp_0297 was selected as signal peptide for expression of Scaf‐ATGB.
Cellulosomal assembly by L. plantarum cell consortia
In previous studies, it was observed that the cellulosome paradigm in L. plantarum was more efficient and stable than the secreted free enzyme approach (Moraïs et al., 2014; Stern et al., 2018). In the present work, we therefore focused on comparing several enzymatic compositions of cell‐wall‐anchored designer cellulosomes (Fig. 4B). Endoglucanase Cp‐Cel5 was the most active enzyme among the tested cellulases (Fig. S1A). Therefore, it was included in all the enzymatic combinations. The cellulosomal assemblies by the different consortia were achieved by mixing equal volumes of overnight cell starter cultures that expressed the desired protein.
Fig. 4.
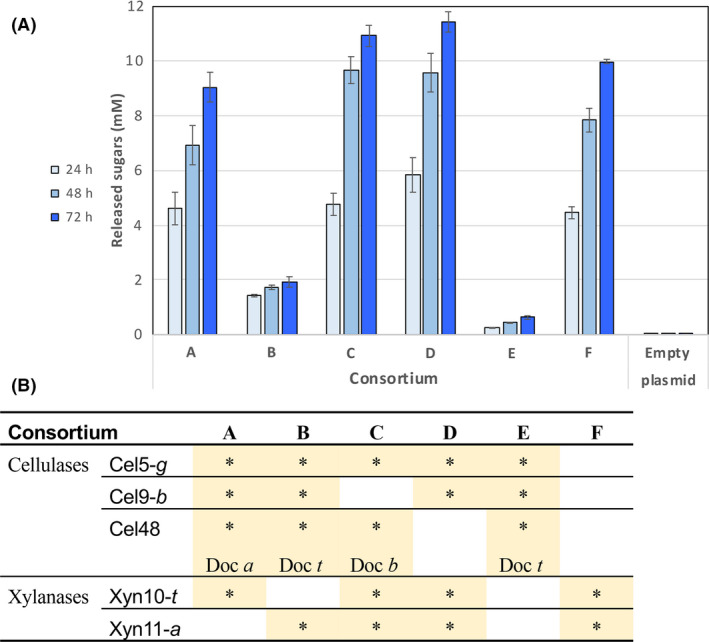
Comparative analysis of wheat straw degradation by different L. plantarum anchored consortia and pLp_empty plasmid cells. (A) Concentrated cell pellet was incubated for 24, 48 and 72 h at 37°C with 40 g l− 1 of pretreated wheat straw. Experiments were conducted three times in triplicate, and standard deviations are as indicated. (B) L. plantarum cell‐ consortium components.
In Figure 4, the enzymatic compositions of each cell consortium and its respective enzymatic activity on wheat straw at three time points are reported. Consortia E and F represent the activities of either the three cellulases alone (E) or the two xylanases (F). While the cellulases are scarcely active on their own in terms of reducing sugar (E), there is an observed synergy between cellulases and xylanases in consortia C and D (both types of enzymes) as opposed to consortium F (xylanases only). Consortia A and B, wherein one xylanase was removed in each, served to highlight the important contribution of Xyn10 (lacking in consortium B) to the enzymatic activity as compared to Xyn11 (lacking in consortium A). The similar degradation levels of consortia C and D, containing either Cel48 or Cel9, indicate the similar contribution of these enzymes. To conclude, we have found that consortium D exhibits the best performance for wheat straw degradation. We therefore selected this consortium for further study.
The ability of two selected consortia, consortium A and consortium D, to utilize the wheat straw substrate as sole carbon source was assessed on a chemically defined medium (CDM; Wegkamp et al., 2010). No growth was obtained for any of the consortia or the wild‐type bacterium after up to 96 h of incubation.
Increase in fibrolytic activity by plasmid rearrangement and selection for lower molecular‐weight enzymes, following consortia growth on plant fibre as a sole carbon source
Here, we applied the ALE approach in an attempt to adapt an L. plantarum cell consortium to grow on wheat straw. For this purpose, we selected cell‐consortium D (comprising two cellulases and two xylanases), which was the most efficient consortium for wheat straw degradation (Fig. 4). Three samples of L. plantarum wild‐type cells and consortium D were tested during the duration of the experiment. For one month, both L. plantarum wild‐type cells and consortium D were grown on poor‐MRS medium (pMRS) supplemented with wheat straw and transferred daily to fresh medium at a dilution of 1:50 (Fig. S4). Each day, the cells were also plated on MRS medium to calculate the colony‐forming units (CFU). The average value of CFU ml−1 was higher in consortia D samples compared to that of the wild‐type samples (1.72 × 107 compared to 1.19 × 107), suggesting some degradation and utilization of the wheat straw. Since pMRS allows low levels of bacterial growth at day 32 (as opposed to CDM), the cells were transferred to CDM + wheat straw (either in liquid medium or in plates) in an attempt to obtain bacterial growth that related only to wheat straw degradation. However, no growth was detected after more than a week.
Interestingly, when we tested the degradation capacity of the consortium cells during the ALE experiment, in all cases we identified an increase in degradation activity of at least one plant fibre polymer. In Figure 5, we observe that at day 1 the three consortium D replicates exhibited equivalent degradation capacities, but at day 32, consortium replicate D2 exhibited an enormous increase in polysaccharide degradation. CMC, xylan‐ and wheat straw‐degrading activities were increased by 3.8‐fold, 4.9‐fold and 4.4‐fold, respectively. All values were normalized with the number of bacterial cells as quantified by RT‐PCR.
Fig. 5.
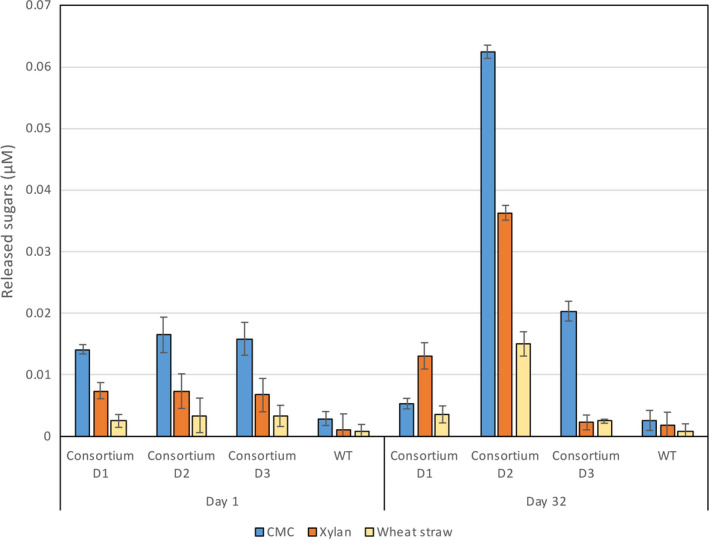
Adaptive laboratory evolution (ALE) of L. plantarum consortium D. Degradation ability of consortium D and wild‐type culture from day 1 and day 32 (following normalization of cell number), after growth for 24 h at 37°C in pMRS + wheat straw medium. Enzymatic activity was tested on 1% CMC, 1% xylan and 40 g l−1 pretreated wheat straw after 24 h at 37°C. Values for reducing sugars were normalized with bacterial cell number. Experiments were conducted four times in triplicate, and standard deviations are as indicated.
Interestingly, we found that the distribution of the different plasmid‐carrying cells was changed tremendously in the three replicates of the experiment, which was reflected as difference in fibrolytic activity. Investigation of plasmid changes during selected days of the experiment was done by PCR using primers that match the pSIP plasmids that were used in this study. Plasmid composition changed during the timecourse of the experiment as can be observed by the distribution of the different PCR products (Fig. 6A). The three consortia replicates were established by the same plasmid composition that varied until day 11 and remained stable until the end of the experiment. Interestingly, after day 11, the two high‐molecular‐weight proteins, the scaffoldin and Cel9, almost completely disappeared in all consortia D samples. Since the PCR results do not reflect quantity, we performed RT‐PCR for the different enzyme genes on day 1, day 16 and day 32 of the developing consortia (Fig. 6B). As anticipated, there was high correlation between the RT‐PCR results and the PCR results. At day 1 the three consortia exhibited the same plasmid profile, while at day 32 they were very different. These results can explain the difference in the activity results between the consortia at day 32 compared to day 1. In consortium D1, more than 80% of the plasmids are of xylanases genes, mainly of Xyn11, which correlates to the slight increase in xylan degradation and decrease in CMC degradation. Similarly, consortium D3 contains ~ 75% of the plasmid for cellulase Cel5 that correlates to the increase in CMC degradation and decrease in xylan degradation. Consortium D2 contains ~ 80% of plasmid Xyn10 and ~ 20% of plasmid Cel5; as a result, we observed marked increase in both xylan and wheat straw degradation. Interestingly, consortium D2 showed substantial improvement while consortium D3 showed only minor improvement in CMC degradation, despite its increased amount of Cel5 plasmid. We therefore tested whether Xyn10 can degrade CMC and discovered that it exhibits some cellulase activity although it is a xylanase (Fig. S5). This finding is very exciting, since only a few Xyn10 enzymes have been reported to have a dual function on cellulose and hemicellulose (Meng et al., 2009; Xue et al., 2015).
Fig. 6.
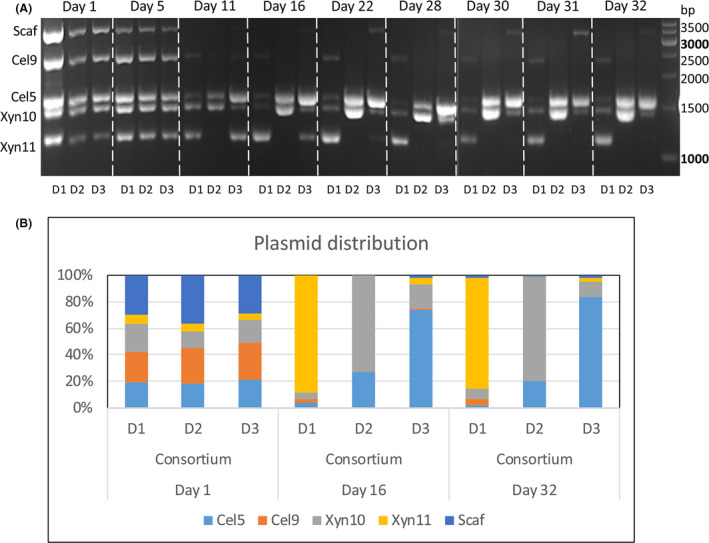
Identification of plasmid arrangements in each consortium D replicate during ALE. (A) DNA was extracted from frozen growth cultures at specific days and was analysed by PCR using primers that fit the pSIP plasmids. The expected sizes of the PCR products are 2334 bp for Cel9, 1596 bp for Cel5, 1443 bp for Xyn10 and 1155 bp for Xyn11. (B) Plasmid distribution of L. plantarum consortia at day 1, day 16 and day 32. The percentage of each plasmid was calculated by dividing plasmid amount by the sum of the total plasmids in the consortium. The copy numbers of each enzyme were determined by RT‐PCR using specific primers for each gene. Experiments were conducted three times in triplicate.
Finally, we checked if the improvement in degradation by consortium D2 at the end of the ALE experiment reflects mainly the changes in plasmid distribution or whether some genetic evolution is also involved. We therefore assembled a reproduced form of consortium D2 (‘pseudo’‐consortium D2), which included the same plasmid distribution as in consortium D2 at day 32. Examination of the relative degradation activity per colony revealed that both consortia have the same degradation profile (Fig. S6). Moreover, sequencing of the GH genes in the plasmids at day 32 confirmed the absence of mutations in any of the genes, although this does not exclude potential genetic modifications at the genome level. Overall, it can thus be concluded from these results that the improvement in degradation ability of consortium D2 at day 32 compared to day 1 is probably due to changes in internal plasmid arrangement and potential genetic modifications in the genome of the microbes.
Discussion
In this study, we have searched for the most suitable and efficient cell‐surface designer cellulosome for production by L. plantarum cells. For this purpose, we established a methodological pipeline, in which several aspects were investigated, including selection of designer cellulosome enzymes, secretion efficiency of the enzymes, optimized enzymatic combination for integration into the mature cell‐surface designer cellulosome and the ability of the cells to grow on wheat straw as a sole carbon source.
Protein secretion efficiency plays a key role in the cell‐consortium approach, whereby each L. plantarum strain expresses and secretes an individual cellulosomal component, which then self‐assemble onto the cell‐surface scaffoldin(s) of one of the strains. The secretion efficiency of heterologous proteins is difficult to predict, since it depends on an optimal combination between the signal peptide, the target protein, and the protein expression and secretion rate (Le Loir et al., 2005; Brockmeier et al., 2006; Mathiesen et al., 2009). The work by Mathiesen et al. (2009) employed the pSIP system to compare the secretion efficiency of the staphylococcal nuclease (NucA) reporter enzymes with 76 signal peptides of L. plantarum. The authors indeed found significant differences between the efficiency of the different signal peptides. Moreover, when they tested the secretion efficiency with the lactobacilli amylase (AmyA), no correlation was observed between the best signal peptides for NucA and the best signal peptides for AmyA. Likewise, similar results were obtained in an extensive work on 173 signal peptides of Bacillus subtilis (Brockmeier et al., 2006). The optimal signal peptide for the cutinase gene was completely different than the one for esterase gene. We therefore examined five different signal peptides originating from proteins with various molecular weights from the genome of L. plantarum to optimize the secretion of our six recombinant proteins. In doing so, we considered whether a connection would exist between molecular weights of the original protein and the ability to secrete heterologous proteins of similar size, but here no correlation was observed. Nevertheless, our results demonstrate that signal peptide screening is an important step towards heterologous protein expression. A previous study reported that Lp_3050 was the most efficient signal peptide for secretion of the low‐molecular‐weight protein, NucA (Mathiesen et al., 2009). However, in this study, while the same signal peptide performed well for Cel5‐g (52.4 kDa) and Xyn10‐t (45.8 kDa) secretion, it was very inefficient for Xyn11‐a (34.2 kDa). In contrast, Lp_2588 was one of the most efficient signal peptides for the majority of the proteins, while it was only of moderate efficiency for NucA (Mathiesen et al., 2009). With this signal peptide screening procedure, we were able to find preferred signal peptides for efficient secretion of high‐molecular‐weight proteins, such as the exoglucanase Cel48‐b (87 kDa) and the scaffoldin Scaf‐ATGB (112 kDa). Moreover, in our previous report (Stern et al., 2018), we were unable to secrete a full functional Scaf‐ATGB by using the original pLp_0373sOFAcwa2 anchoring plasmids (Fredriksen et al., 2010). This was corrected in the current work where satisfactory levels of surface‐attached Scaf‐ATGB were achieved using Lp_2588. These results are important within the context of designer cellulosome research, since many of its components are large proteins, especially the cellulases and scaffoldins.
Secreted proteins are diluted in the environment and are more accessible to proteolytic degradation (Wells and Mercenier, 2008). In contrast, when proteins are anchored to the microbial surface their on‐site concentrations are higher and they are more stable (Moraïs et al., 2014; Stern et al., 2018). Here, we have focused our study on anchored designer cellulosomes, which have proven more efficient in their fibre‐degrading capacity in L. plantarum. In this work, we successfully expressed and displayed large designer cellulosomes on the cell surface of L. plantarum. We thus examined the performance of six consortia that differed in their enzymatic combinations, in order to reveal the contribution of each enzyme in the cellulosomal system. We found that the majority of the degradation of the wheat straw substrate could be attributed to the enzymatic action of the xylanases, particularly by the Xyn10‐producing cells, in the presence or absence of the cellulases. Thus, the degradation products (reducing sugars) are mainly derived from hemicellulose. However, it is possible that some cellulose degradation still occurred, but the products were consumed by the active cells in the pellet. Although hemicellulose degradation products are not consumed by the bacterium (Stern et al., 2018), it is necessary to retain the xylanase activity in the designer cellulosome consortium for removal of the xylan component of the fibre that prevents physical access of the cellulases to the cellulose substrate in the plant cell wall. L. plantarum is able to grow on minimal concentrations of ~ 5.5 mM cellobiose, (Stern et al., 2018), a level which was not attained here in this work. Hence, there remains a need for merging the gap between the low amount of cellobiose obtained by the designer cellulosome machinery and the minimal cellobiose concentration needed for bacterial growth.
The cell‐consortium approach considerably decreases the burden of the cellular machinery of each strain, thereby maximizing their ability to grow and to express the various cellulosomal components. We have used the adaptive laboratory approach for adaptation of the L. plantarum consortium to grow on wheat straw as a sole carbon source. This approach has been successfully demonstrated for xylan utilization in Lactobacillus pentosus, S. cerevisiae and B. subtilis (Shen et al., 2012; Zhang et al., 2015; Cubas‐Cano et al., 2019). The tested consortium was assembled by combination of five L. plantarum strains, where each strain expresses and secretes a different protein, that is Cel5, Cel9, Xyn10, Xyn11 and Scaf‐ATGB. We designed the experiment such that equal amounts of L. plantarum transformed strains were mixed for consortium assembly. We were interested in consortia adaptation of the plasmid copy number that could result in improved degradation ability of the wheat straw substrate. The consortia were grown consistently in the presence of antibiotic, in order to prevent plasmid loss. We noticed that already at day 11, variations in plasmid organization among the three consortia occurred and remained stable during the remainder of the experiment. By the end of the experiment, only three out of the five original plasmids were found in the three consortia. The lost plasmids were those that encoded the high‐molecular‐weight genes, that is the genes for Cel9 (79 kDa) and Scaf‐ATGB (112 kDa), thus suggesting a fitness cost. Interestingly, the scaffoldin itself that does not provide degradation ability to the bacterium was lost, which suggests that in the experimental context only degradation capabilities were selected. Furthermore, the Cel9 enzyme was the least active of the inserted enzymes (Fig. S1), suggesting selection for higher degrading capabilities. The three consortium D replicates evolved into three different consortia that produced three different combinations of enzymes, all of which exhibited an increase in at least one fibrolytic capability. These differences in the three repetitions of consortium D could stem from random genetic drift in the cell population, due to the repetitive sampling step between the transfers. Indeed, it was shown previously that decreasing the population bottleneck (increasing dilution rates between transfers) resulted in lower genetic diversity in bacterial populations (Wein and Dagan, 2019). Consortium D1 produced only xylanase activity, consortium D2 both xylanase and cellulase activity, while consortium D3 produced only cellulase activity. However, only the combination of cellulases and xylanases, as produced by consortium D2, demonstrated improvement in plant fibre‐degradation ability, which emphasizes the importance of combining enzymes with different functions in the designer cellulosome. Moreover, this increase in activity was achieved via plasmid rearmament and not due to changes in gene sequence. We conclude this from the similar levels of performance of consortium D2 and a consortium ('pseudo'‐consortium D2) that had the same plasmid ratio as consortium D2 without going through the adaptive process, which suggests that the improvement observed in consortium D2 over the performance of consortia D1 and D2 may be due to plasmid re‐arrangement at the community level. Although it has previously been suggested that plasmids enable microbes to change gene and enzyme numbers in their cells in a rapid manner, our data suggest a potential strategy for microbes for quick adaptation for gene and enzyme stoichiometry at the community level.
The work presented in this communication further establishes the potency of L. plantarum for assembly of designer cellulosomes for biodegradation of lignocellulosic biomass. The methodological pipeline herein developed enabled the improvement of secretion efficiency of high‐molecular‐weight cellulosomal components as well as the determination of an optimized enzymatic combination. In addition, the adaptive lab experiment served to obtain a cell consortium with improved enzymatic activity. The re‐arrangement of the plasmid ratio by the bacterial community towards higher fibre‐degradation function could not have been predicted and suggest a novel strategy for rapid adaptation of microbes even at the community level.
Potential applications in environmental bioremediation and production of second‐generation biofuels, the development of potent L. plantarum for biomass degradation could serve for assisting fibre‐degrading communities, such as in gut environments. L. plantarum has indeed been demonstrated as one of the most promising LAB strains for carriers of cell‐surface‐displayed vaccines (Kuczkowska et al., 2019). In this context, the proficiency in engineering its bacterial cell surface with high‐molecular‐weight proteins and complexes could be of interest for use as a delivery vehicle of therapeutic compounds such as antigens. Furthermore, expression of cell‐surface scaffoldin will allow the use of multiple antigens that may be beneficial in future clinical applications.
Materials and methods
Cloning
All recombinant proteins employed in this study were first cloned into pET28a plasmids and designed to contain a His‐tag for subsequent purification. The recombinant enzymes Cel5‐g, Cel9‐b, Xyn10‐t and Xyn11‐a were designed and cloned by Stern et al. (2018). The recombinant enzymes Cel48‐a/b/t were obtained by fusing the catalytic module of Ruminococcus champanellensis Cel48 (GenBank GI no. 291544207) to dockerins of Acetivibrio cellulolyticus (ScaB), Bacteroides cellulosolvens (ScaA) and Clostridium thermocellum (Cel48S) as described at Stern et al. (2018).
For expression and secretion in L. plantarum, the glycoside hydrolases were cloned in the modular secretion plasmids pLp_3050sAmy (Mathiesen et al., 2009) by replacing the amylase gene in these plasmids by an appropriately amplified gene fragment, using SalI and HindIII restriction sites. The anchoring scaffoldin, Scaf‐ATGB, was amplified with XhoI and MluI restriction sites (Stern et al., 2018), following which, this fragment was cloned into the pLp_0373sOFAcwa2 anchoring plasmids (Fredriksen et al., 2010). Further change of the signal peptides was achieved by amplifying the Lp_0297, Lp_0373, Lp_2588 and Lp_3093 signal peptides from the L. plantarum WCFS1 genome with restriction sites NcoI and SalI at the ends. Then, each signal peptide was ligated into the cut plasmid pLp_3050sGH or by the restriction‐free method (Unger et al., 2010). Since the pSIP vectors are suitable for both L. plantarum and E. coli, all cloning steps were performed using E. coli XL1 competent cells, which were grown in LB (Luria‐Bertani) medium supplemented with 200 µg ml−1 erythromycin (Ben‐David et al., 2019). The primers that were used in this study are listed in Table S2.
Recombinant protein expression and purification from Escherichia coli
E. coli BL21 (DE3) strain was used for overexpression of the recombinant proteins. Protein expression and purification were performed as described earlier (Moraïs et al., 2016a,b). When needed, proteins were concentrated using Amicon ultraconcentrators (Millipore, Ireland). The purified proteins were stored in 50% (v/v) glycerol at −20°C.
Protein expression in L. plantarum
Electroporation of electrocompetent L. plantarum WCFS1 was conducted according to the protocol of Aukrust and Blom (Aukrust et al., 1995). Freshly inoculated cultures of L. plantarum WCFS1 harbouring a pSIP‐derived expression plasmid were induced at OD600 = 0.3 by adding the inducing peptide for sakacin P production (Caslo Laboratory, Lyngby, Denmark; Eijsink et al., 1996) to a final concentration of 25 ng ml−1, and the cell culture was incubated until OD600 ≈ 1 at 37°C in MRS broth without proteose peptone with 10 μg ml−1 erythromycin (beef extract 10 g l−1, yeast extract 5 g l−1, glucose 20g l−1, polysorbate 80 1 g l−1, ammonium citrate 2 g l−1, sodium acetate 5 g l−1, magnesium sulfate 0.1 g l−1, manganese sulfate 0.05 g l−1, dipotassium phosphate 2 g l−1; Moraïs et al., 2014). For the designer cellulosome strategy, the MRS broth without proteose peptone was used and supplemented with 40 mM CaCl2. A consortium of bacteria producing the different proteins (the cellulases, the xylanases and the scaffoldin) were mixed in equal amounts and induced as described above. Cells expressing empty pLp_plasmid were used to ensure equal amounts of cells in the consortia expressing two and three enzymes. For the secreted proteins, culture supernatant fluids were dialysed overnight in TBS buffer supplemented with 15 mM CaCl2. Afterwards, the supernatant fluids were concentrated to OD600 = 50 by using Amicon centrifugal filters with a 30‐kDa cutoff (Millipore, Molsheim, France). For anchored scaffoldin and designer cellulosomes, cells were washed twice by centrifugation with TBS containing 1% Triton X‐100 and resuspended to eliminate the sugars present in the MRS medium. Finally, the cell pellet was resuspended in TBS at OD600 = 50.
Far‐Western blotting for L. plantarum secreted proteins
Proteins from 50‐times concentrated culture supernatant fluids were separated on SDS‐PAGE gels (12% acrylamide) and transferred to a nitrocellulose membrane using Mini Trans‐Blot cells (Bio‐Rad Laboratories, Israel). All steps were performed at room temperature. Non‐specific protein interactions were blocked by incubating the membrane for 1 h with 5% bovine serum albumin (BSA; prepared in Tris‐buffered saline–Tween 20 [TBS‐T]). The membrane was then interacted with 30 µg of purified Scaf‐ATGB in TBS‐T, containing 1% BSA for 1 h. The membrane was rinsed twice (1 min) with TBS‐T, and rabbit antibody against the scaffoldin CBM (at a dilution of 1:5000; Morag et al., 1995) was incubated with the membrane for 1 h in TBS‐T, containing 1% BSA. The membrane was again rinsed twice (1 min) with TBS‐T and then incubated for 1 h with secondary goat anti‐rabbit antibody labelled with horseradish peroxidase (HRP) [Jackson ImmunoResearch Laboratories (West Grove, PA)], at a dilution of 1:10 000. The membrane was rinsed as described above and then rinsed twice (30 min) with TBS and 1% Triton X‐100. Blots were developed by incubating the membrane for 1 min with equal amounts of ECL kit solutions A and B (Thermo Fisher Scientific, MA, USA). Chemiluminescence was quantified using a luminescent image analyser (ImageQuant LAS 4000 Mini; Danyel Biotech, Israel).
ELISA binding assay
This procedure is based on the protocol developed by Barak et al. (2005). MaxiSorp ELISA plates (Nunc A/S, Roskilde, Denmark) were coated overnight at 4°C with 0.1–1000 ng ml−1 of the desired Xyn‐Doc (100 μl per well) in 0.1 M sodium carbonate (pH 9). The following steps were performed at room temperature with all reagents at a volume of 100 μl per well. The coating solution was discarded and blocking buffer (TBS, containing 10 mM CaCl2, 0.05% Tween 20 and 2% BSA) was added (1 h incubation). The blocking buffer was discarded, and 100 µl of whole bacterial cells at OD600 = 2 of the recombinant Scaf‐ATGB were added. After a 1‐h incubation period, the plates were washed 3 times with wash buffer (blocking buffer without BSA). Primary antibody preparation, rabbit anti‐CBM and secondary antibody preparation, HRP‐labelled anti‐rabbit antibodies were added and washed as described above in the previous section. Then, the plates were washed (4 times) with wash buffer and 100 μl per well TMB + Substrate‐Chromogen (Dako Corp., Carpinteria, CA) were added. Colour formation was terminated upon addition of 1 M H2SO4 (50 μl per well), and the absorbance was measured at 450 nm using a tunable microplate reader (OPTImax, Molecular Devices Corp., Sunnyvale, CA).
Activity assay
Degradation of the lignocellulosic substrate was assayed by mixing pure recombinant enzymes at the desired concentration or a volume of 30 µl of 50‐fold concentrated culture supernatant fractions (as described above) with 100 µl of 2% CMC or 2% xylan in a final volume of 200 µl of 50 mM acetate buffer, pH 5. Samples were incubated at 37°C for 2 h. The reaction was stopped by transferring the tubes to an ice‐water bath followed by centrifugation for 2 min at 14 000 g. The wheat straw assay was conducted in 200 µl of 50 mM acetate buffer, pH 5.0 with 100 µl of 40 g l−1 hypochlorite‐pretreated wheat straw (Moraïs et al., 2012), and 78 µl of cell pellets concentrated 50 fold for 24–72 h at 37°C. All assays were performed in triplicate. Enzymatic activity was determined quantitatively by measuring soluble reducing sugars released from the polysaccharide substrates by the 3,5‐dinitro‐salicylic acid (DNS) method (Miller, 1959; Ghose, 1987). DNS solution (150 µl) was added to 100 µl of sample, and after the reaction mixture was boiled for 10 min, absorbance at 540 nm was measured. Sugar concentrations were determined using a glucose standard curve.
Adaptive laboratory evolution (ALE)
Lactobacillus plantarum cell consortia and wild‐type strains were tested by ALE in triplicate. The assembly of consortium D was done by taking equal amounts of overnight cell starter culture that expressed the desired proteins. Consortia cells and wild‐type cells were washed twice with 0.85 NaCl to remove sugars from the MRS growth medium. Afterwards, 200 µl of washed cells were transferred into 10 ml of poor‐MRS (pMRS; yeast extract 5 g l−1, polysorbate 80 1 g l−1, ammonium citrate 2 g l−1, sodium acetate 5 g l−1, magnesium sulfate 0.1 g l−1, manganese sulfate 0.05 g l−1, dipotassium phosphate 2 g l−1) +2% pretreated wheat straw medium (Moraïs et al., 2012). This medium was prepared in a manner similar to that used to prepare regular MRS but without the addition of glucose, beef extract and peptone. In the pMRS medium, L. plantarum can grow to OD600 of ~ 0.2 compared to OD600 ~ 6 in MRS medium (data not shown). The cell cultures were grown for a period of 32 days, whereby daily aliquots of 200 µl of the overnight cell culture were transferred to a new 10 ml pMRS + 2% wheat straw medium. The remainder of the overnight culture was kept at −80°C after the addition of 20% sterile glycerol. The bacterial colony‐forming units (CFU) of each sample were calculated by plating 10 µl of 1 × 10−7 serial dilutions on MRS plates and counting the number of cells using the ImageJ software. PCR assay was performed by using the forward primer 5ʹ‐GCTGGCTGGCGTAAAGTATGC‐3ʹ and reverse primer 5ʹ‐CAACTGCTGCTTTTTGGCTATCAATC‐3ʹ. The sequence of these primers fits the pLp plasmid backbone, and therefore, all the recombinant genes can be amplified at the same reaction. PCRs were performed using KAPA HiFi HotStart DNA Polymerase KR0370 (KAPA Biosystems) as described in the manual, and DNA samples were purified as described below. For the RT‐PCR and activity assay experiments, a starter culture derived from a frozen sample of the desired day was grown overnight in pMRS + 2% wheat straw medium. On the following day, 200 µl of starters was inoculated into 10 ml of pMRS + 2% wheat straw medium, and the cells were grown for 24 h. Cell aliquots (1 ml) were then taken for RT‐PCR quantification as described below. For activity assays, 78 µl of overnight growing cells (instead of concentrated cell pellets as in Fig. 4) was mixed with CMC, xylan or hypochlorite‐pretreated wheat straw for 24 h. The assays were performed as described above.
Real‐time PCR
DNA preparation was performed as follows: samples (1 ml) were centrifuged for 3 min at 5000 g. The supernatant fraction was then removed, and the cell pellet was washed with TE buffer (10 mM Tris, 1 mM EDTA, brought to pH 8 with HCl). The cells were centrifuged again (3 min at 5000 g), and supernatant fluids were removed. The pellet was resuspended in 100 µl TE buffer and vortexed. Samples were boiled for 15 min at 95°C and then cooled for 15 min on ice. A final centrifuge step was performed at 14 000 g for 5 min, and the supernatant fraction was kept at −20°C until use.
Quantitative real‐time PCR analysis was performed to evaluate the numbers of bacterial cells and the distribution of plasmids in the tested samples. For total bacterial cell number, we used specific primers for the GH39 enzyme of L. plantarum. In addition, we designed unique primers for the different plasmids tested (Table S3). Individual standard curves suitable for quantification of each plasmid were generated by amplifying serial 10‐fold dilutions of quantified gel‐extracted PCR products obtained by amplification of each fragment. The standard curves were obtained using six dilution points and calculated using the QuantStudio™ Real‐Time PCR software. Subsequent quantifications were calculated with the same program using the generated standard curves. RT‐PCR was performed in a 7 µl reaction mixture containing 3.5 µl Biosystems™ Fast SYBR™ Green Master Mix (Thermo Scientific, Massachusetts), 0.14 µl of each primer (10 µM working concentration), 1.82 µl nuclease‐free water and 1.4 µl of DNA. The amplification program was as follows: initial denaturation of 20 s at 95°C and then 40 cycles at 95°C for 2 s, followed by annealing and extension for 20 s at 60°C. To determine the specificity of amplification, a melting curve of PCR products was monitored by slow heating, with fluorescence collection at 1°C increments, from 45 to 99°C.
Construction of ‘pseudo’‐consortium D2
Lactobacillus plantarum cell starter cultures that expressed the Cel5 and Xyn10 proteins were grown overnight at 37°C without shaking in pMRS medium. The two strains were moved to a new tube at a ratio of 20% of Cel5 and 80% of Xyn10. Subsequently, 200 µl of mixed cells was transferred into 10 ml of pMRS + 2% pretreated wheat straw medium and grown overnight at 37°C without shaking. The overnight mixed cell culture was tested for enzymatic activity on CMC, xylan and wheat straw and by RT‐PCR.
Conflict of interest
The authors have no conflict of interest to declare.
Supporting information
Table S1. Enzymes examined in this study.
Table S2. Primers list.
Table S3. Real‐time PCR primers list.
Fig. S1. Comparative enzymatic activity of the selected cellulases and xylanases from different bacteria.
Fig. S2. Synergism between endoglucanases Cel5 and Cel9 with exoglucanase Cel48.
Fig. S3. Quantification of the chimaeric enzymes secreted by L. plantarum.
Fig. S4. Schematic representation of the adaptive laboratory evolution experiment.
Fig. S5. Enzymatic activity of Cel5 and Xyn10 enzymes on CMC and xylan.
Fig. S6. Relative degradation activity on CMC, xylan and wheat straw by consortium D2 at day 32 and “pseudo”‐consortium D2.
Acknowledgements
The research was supported by grants, from the European Research Council (No. 640384) to I.M. and Israel Science Foundation (ISF No. 1947/19) to I.M. and S.M. E.A.B. is the incumbent of the Maynard I. and Elaine Wishner Chair of Bio‐organic Chemistry at the Weizmann Institute of Science.
Microbial Biotechnology (2020) 13(6), 1748–1764
Funding information
The research was supported by grants, from the European Research Council (No. 640384) to I.M. and Israel Science Foundation (ISF No. 1947/19) to I.M. and S.M. E.A.B. is the incumbent of the Maynard I. and Elaine Wishner Chair of Bio‐organic Chemistry at the Weizmann Institute of Science.
References
- Ahrne, S. , Nobaek, S. , Jeppsson, B. , Adlerberth, I. , Wold, A.E. , and Molin, G. (1998) The normal Lactobacillus flora of healthy human rectal and oral mucosa. J Appl Microbiol 85: 88–94. [DOI] [PubMed] [Google Scholar]
- Alegria, E.G. , Lopez, I. , Ruiz, J.I. , Saenz, J. , Fernandez, E. , Zarazaga, M. , et al (2004) High tolerance of wild Lactobacillus plantarum and Oenococcus oeni strains to lyophilisation and stress environmental conditions of acid pH and ethanol. FEMS Microbiol Lett 230: 53–61. [DOI] [PubMed] [Google Scholar]
- Artzi, L. , Bayer, E.A. , and Moraïs, S. (2017) Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nature Rev Microbiol 15: 83–95. [DOI] [PubMed] [Google Scholar]
- Aukrust, T.W. , Brurberg, M.B. , and Nes, I.F. (1995) Transformation of Lactobacillus by electroporation. Methods Mol Biol 47: 201–208. [DOI] [PubMed] [Google Scholar]
- Barak, Y. , Handelsman, T. , Nakar, D. , Mechaly, A. , Lamed, R. , Shoham, Y. , and Bayer, E.A. (2005) Matching fusion‐protein systems for affinity analysis of two interacting families of proteins: the cohesin‐dockerin interaction. J Mol Recognit 18: 491–501. [DOI] [PubMed] [Google Scholar]
- Bayer, E.A. , Kenig, R. , and Lamed, R. (1983) Adherence of Clostridium thermocellum to cellulose. J Bacteriol 156: 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer, E.A. , Morag, E. , and Lamed, R. (1994) The cellulosome–a treasure‐trove for biotechnology. Trends Biotechnol 12: 379–386. [DOI] [PubMed] [Google Scholar]
- Bayer, E.A. , Chanzy, H. , Lamed, R. , and Shoham, Y. (1998) Cellulose, cellulases and cellulosomes. Curr Opin Struct Biol 8: 548–557. [DOI] [PubMed] [Google Scholar]
- Bayer, E.A. , Belaich, J.P. , Shoham, Y. , and Lamed, R. (2004) The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol 58: 521–554. [DOI] [PubMed] [Google Scholar]
- Bayer, E.A. , Lamed, R. , and Himmel, M.E. (2007) The potential of cellulases and cellulosomes for cellulosic waste management. Curr Opin Biotechnol 18: 237–245. [DOI] [PubMed] [Google Scholar]
- Bayer, E.A. , Lamed, R. , White, B.A. , and Flint, H.J. (2008) From cellulosomes to cellulosomics. Chem Rec 8: 364–377. [DOI] [PubMed] [Google Scholar]
- Bayer, E.A. , Shoham, Y. , and Lamed, R. (2013) Lignocellulose‐decomposing bacteria and their enzyme systems In The Prokaryotes: Prokaryotic Physiology and Biochemistry. Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., and Thompson F. (eds). Berlin, Heidelberg: Springer, pp. 215–266. [Google Scholar]
- Ben‐David, Y. , Moraïs, S. , Stern, J. , Mizrahi, I. , and Bayer, E.A. (2019) Cell‐surface display of designer cellulosomes by Lactobacillus plantarum . Methods Enzymol 617: 241–263. [DOI] [PubMed] [Google Scholar]
- Bosch, M. , Fuentes, M.C. , Audivert, S. , Bonachera, M.A. , Peiro, S. , and Cune, J. (2014) Lactobacillus plantarum CECT 7527, 7528 and 7529: probiotic candidates to reduce cholesterol levels. J Sci Food Agric 94: 803–809. [DOI] [PubMed] [Google Scholar]
- Brockmeier, U. , Caspers, M. , Freudl, R. , Jockwer, A. , Noll, T. , and Eggert, T. (2006) Systematic screening of all signal peptides from Bacillus subtilis: a powerful strategy in optimizing heterologous protein secretion in Gram‐positive bacteria. J Mol Biol 362: 393–402. [DOI] [PubMed] [Google Scholar]
- Cantarel, B.L. , Coutinho, P.M. , Rancurel, C. , Bernard, T. , Lombard, V. , and Henrissat, B. (2009) The Carbohydrate‐Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37: D233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Shen, J. , Ingvar Hellgren, L. , Ruhdal Jensen, P. , and Solem, C. (2015) Adaptation of Lactococcus lactis to high growth temperature leads to a dramatic increase in acidification rate. Sci Rep 5: 14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas‐Cano, E. , Gonzalez‐Fernandez, C. , and Tomas‐Pejo, E. (2019) Evolutionary engineering of Lactobacillus pentosus improves lactic acid productivity from xylose‐rich media at low pH. Biores Technol 288: 121540. [DOI] [PubMed] [Google Scholar]
- Ducrotte, P. , Sawant, P. , and Jayanthi, V. (2012) Clinical trial: Lactobacillus plantarum 299v (DSM 9843) improves symptoms of irritable bowel syndrome. World J Gastroenterol 18: 4012–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijsink, V.G.H. , Brurberg, M.B. , Middelhoven, P.H. , and Nes, I.F. (1996) Induction of bacteriocin production in Lactobacillus sakei by a secreted peptide. J Bacteriol 178: 2232–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatma, S. , Hameed, A. , Noman, M. , Ahmed, T. , Shahid, M. , Tariq, M. , et al (2018) Lignocellulosic biomass: a sustainable bioenergy source for the future. Protein Pept Lett 25: 148–163. [DOI] [PubMed] [Google Scholar]
- Fierobe, H.‐P. , Mingardon, F. , Mechaly, A. , Belaich, A. , Rincon, M.T. , Lamed, R. , et al (2005) Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined tri‐functional scaffoldin. J Biol Chem 280: 16325–16334. [DOI] [PubMed] [Google Scholar]
- Fredriksen, L. , Mathiesen, G. , Sioud, M. , and Eijsink, V.G. (2010) Cell wall anchoring of the 37‐kilodalton oncofetal antigen by Lactobacillus plantarum for mucosal cancer vaccine delivery. Appl Environ Microbiol 76: 7359–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefen, G. , Anbar, M. , Morag, E. , Lamed, R. , and Bayer, E.A. (2012) Enhanced cellulose degradation by targeted integration of a cohesin‐fused beta‐glucosidase into the Clostridium thermocellum cellulosome. Proc Natl Acad Sci USA 109: 10298–10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose, T.K. (1987) Measurments of cellulase activity. Pure Appl Chem 59: 257–268. [Google Scholar]
- Haimovitz, R. , Barak, Y. , Morag, E. , Voronov‐Goldman, M. , Shoham, Y. , Lamed, R. , and Bayer, E.A. (2008) Cohesin‐dockerin microarray: diverse specificities between two complementary families of interacting protein modules. Proteomics 8: 968–979. [DOI] [PubMed] [Google Scholar]
- Kadam, S.K. , and Demain, A.L. (1989) Addition of cloned beta‐glucosidase enhances the degradation of crystalline cellulose by the Clostridium thermocellum cellulose complex. Biochem Biophys Res Comm 161: 706–711. [DOI] [PubMed] [Google Scholar]
- Kirillova, A.V. , Danilushkina, A.A. , Irisov, D.S. , Bruslik, N.L. , Fakhrullin, R.F. , Zakharov, Y.A. , et al (2017) Assessment of resistance and bioremediation ability of Lactobacillus strains to lead and cadmium. Int J Microbiol 2017: 9869145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem, M. , Boekhorst, J. , van Kranenburg, R. , Molenaar, D. , Kuipers, O.P. , Leer, R. , et al (2003), Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci USA 100: 1990–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczkowska, K. , Overland, L. , Rocha, S.D.C. , Eijsink, V.G.H. , and Mathiesen, G. (2019) Comparison of eight Lactobacillus species for delivery of surface‐displayed mycobacterial antigen. Vaccine 37: 6371–6379. [DOI] [PubMed] [Google Scholar]
- Lamed, R. , and Bayer, E.A. (1988) The cellulosome of Clostridium thermocellum In Advances in Applied Microbiology. Laskin A.I. (ed). Cambridge, MA: Academic Press, pp. 1–46. [Google Scholar]
- Le Loir, Y. , Azevedo, V. , Oliveira, S.C. , Freitas, D.A. , Miyoshi, A. , Bermudez‐Humaran, L.G. , et al (2005) Protein secretion in Lactococcus lactis : an efficient way to increase the overall heterologous protein production. Microb Cell Fact 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire, M. , Ohayon, H. , Gounon, P. , Fujino, T. , and Beguin, P. (1995) OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S‐layer‐like domains to components of the cell envelope. J Bacteriol 177: 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire, M. , Miras, I. , Gounon, P. , and Beguin, P. (1998) Identification of a region responsible for binding to the cell wall within the S‐layer protein of Clostridium thermocellum . Microbiology 144(Pt 1): 211–217. [DOI] [PubMed] [Google Scholar]
- Limayem, A. , Hanning, I.B. , Muthaiyan, A. , Illeghems, K. , Kim, J.W. , Crandall, P.G. , et al (2011) Alternative antimicrobial compounds to control potential Lactobacillus contamination in bioethanol fermentations. J Environ Sci Health B 46: 709–714. [DOI] [PubMed] [Google Scholar]
- Liu, R. , Chen, Y. , Tian, Z. , Mao, Z. , Cheng, H. , Zhou, H. , and Wang, W. (2019) Enhancing microbial community performance on acid resistance by modified adaptive laboratory evolution. Biores Technol 287: 121416. [DOI] [PubMed] [Google Scholar]
- Mathiesen, G. , Sveen, A. , Piard, J.C. , Axelsson, L. , and Eijsink, V.G.H. (2008) Heterologous protein secretion by Lactobacillus plantarum using homologous signal peptides. J Appl Microbiol 105: 215–226. [DOI] [PubMed] [Google Scholar]
- Mathiesen, G. , Sveen, A. , Brurberg, M.B. , Fredriksen, L. , Axelsson, L. , and Eijsink, V.G.H. (2009) Genome‐wide analysis of signal peptide functionality in Lactobacillus plantarum WCFS1. BMC Genom 10: 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoli, R. , Bosco, F. , Mizrahi, I. , Bayer, E.A. , and Pessione, E. (2014) Towards lactic acid bacteria‐based biorefineries. Biotechnol Adv 32: 1216–1236. [DOI] [PubMed] [Google Scholar]
- Meng, X. , Shao, Z. , Hong, Y. , Lin, L. , Li, C. , and Liu, Z. (2009) A novel pH‐stable, bifunctional xylanase isolated from a deep‐sea microorganism, Demequina sp. JK4. J Microbiol Biotechnol 19: 1077–1084. [PubMed] [Google Scholar]
- Miller, G.L. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Biochem 31: 426–428. [Google Scholar]
- Morag, E. , Lapidot, A. , Govorko, D. , Lamed, R. , Wilchek, M. , Bayer, E.A. , and Shoham, Y. (1995) Expression, purification, and characterization of the cellulose‐binding domain of the scaffoldin subunit from the cellulosome of Clostridium thermocellum . Appl Environ Microbiol 61: 1980–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraïs, S. , Barak, Y. , Caspi, J. , Hadar, Y. , Lamed, R. , Shoham, Y. , et al (2010) Cellulase‐xylanase synergy in designer cellulosomes for enhanced degradation of a complex cellulosic substrate. MBio 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraïs, S. , Barak, Y. , Hadar, Y. , Wilson, D.B. , Shoham, Y. , Lamed, R. , and Bayer, E.A. (2011) Assembly of xylanases into designer cellulosomes promotes efficient hydrolysis of the xylan component of a natural recalcitrant cellulosic substrate. MBio 2: e00233‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraïs, S. , Morag, E. , Barak, Y. , Goldman, D. , Hadar, Y. , Lamed, R. , et al (2012) Deconstruction of lignocellulose into soluble sugars by native and designer cellulosomes. MBio 3: e00508‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraïs, S. , Shterzer, N. , Rozman Grinberg, I. , Mathiesen, G. , Eijsink, V.G. , Axelsson, L. , et al (2013) Establishment of a simple Lactobacillus plantarum cell consortium for cellulase‐xylanase synergistic interactions. Appl Environ Microbiol 79: 5242–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraïs, S. , Shterzer, N. , Lamed, R. , Bayer, E.A. , and Mizrahi, I. (2014) A combined cell‐consortium approach for lignocellulose degradation by specialized Lactobacillus plantarum cells. Biotechnol Biofuels 7: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraïs, S. , Ben David, Y. , Bensoussan, L. , Duncan, S.H. , Koropatkin, N.M. , Martens, E.C. , et al (2016a) Enzymatic profiling of cellulosomal enzymes from the human gut bacterium, Ruminococcus champanellensis, reveals a fine‐tuned system for cohesin‐dockerin recognition. Environ Microbiol 18: 542–556. [DOI] [PubMed] [Google Scholar]
- Moraïs, S. , Cockburn, D.W. , Ben‐David, Y. , Koropatkin, N.M. , Martens, E.C. , Duncan, S.H. , et al (2016b) Lysozyme activity of the Ruminococcus champanellensis cellulosome. Environ Microbiol 18: 5112–5122. [DOI] [PubMed] [Google Scholar]
- Pages, S. , Belaich, A. , Belaich, J.‐P. , Morag, E. , Lamed, R. , Shoham, Y. , and Bayer, E.A. (1997) Species‐specificity of the cohesin‐dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: Prediction of specificity determinants of the dockerin domain. Proteins 29: 517–527. [PubMed] [Google Scholar]
- Rincon, M.T. , Cepeljnik, T. , Martin, J.C. , Lamed, R. , Barak, Y. , Bayer, E.A. , and Flint, H.J. (2005) Unconventional mode of attachment of the Ruminococcus flavefaciens cellulosome to the cell surface. J Bacteriol 187: 7569–7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach, D.R. , Khatibi, P.A. , Bischoff, K.M. , Hughes, S.R. , and Donovan, D.M. (2013) Bacteriophage‐encoded lytic enzymes control growth of contaminating Lactobacillus found in fuel ethanol fermentations. Biotechnol Biofuels 6: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddik, H.A. , Bendali, F. , Gancel, F. , Fliss, I. , Spano, G. , and Drider, D. (2017) Lactobacillus plantarum and its probiotic and food potentialities. Probiot Antimicrob Protein 9: 111–122. [DOI] [PubMed] [Google Scholar]
- Shen, Y. , Chen, X. , Peng, B. , Chen, L. , Hou, J. , and Bao, X. (2012) An efficient xylose‐fermenting recombinant Saccharomyces cerevisiae strain obtained through adaptive evolution and its global transcription profile. Appl Microbiol Biotechnol 96: 1079–1091. [DOI] [PubMed] [Google Scholar]
- Shoham, Y. , Lamed, R. , and Bayer, E.A. (1999) The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol 7: 275–281. [DOI] [PubMed] [Google Scholar]
- Sorvig, E. , Gronqvist, S. , Naterstad, K. , Mathiesen, G. , Eijsink, V.G.H. , and Axelsson, L. (2003) Construction of vectors for inducible gene expression in Lactobacillus sakei and L. plantarum . FEMS Microbiol Lett 229: 119–126. [DOI] [PubMed] [Google Scholar]
- Stern, J. , Artzi, L. , Moraïs, S. , Fontes, C. , and Bayer, E.A. (2017) Carbohydrate depolymerization by intricate cellulosomal systems. Methods Mol Biol 1588: 93–116. [DOI] [PubMed] [Google Scholar]
- Stern, J. , Moraïs, S. , Ben‐David, Y. , Salama, R. , Shamshoum, M. , Lamed, R. , et al (2018) Assembly of synthetic functional cellulosomal structures onto the cell surface of lactobacillus plantarum, a potent member of the gut microbiome. Appl Environ Microbiol 84: e00282‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov, S.D. , de Paula, O.A.L. , Camargo, A.C. , Lopes, D.A. , and Nero, L.A. (2018) Combined effect of bacteriocin produced by Lactobacillus plantarum ST8SH and vancomycin, propolis or EDTA for controlling biofilm development by Listeria monocytogenes. Rev Argent Microbiol 50: 48–55. [DOI] [PubMed] [Google Scholar]
- Unger, T. , Jacobovitch, Y. , Dantes, A. , Bernheim, R. , and Peleg, Y. (2010) Applications of the Restriction Free (RF) cloning procedure for molecular manipulations and protein expression. J Struct Biol 172: 34–44. [DOI] [PubMed] [Google Scholar]
- Wegkamp, A. , Teusink, B. , de Vos, W.M. , and Smid, E.J. (2010) Development of a minimal growth medium for Lactobacillus plantarum . Lett Appl Microbiol 50: 57–64. [DOI] [PubMed] [Google Scholar]
- Wein, T. , and Dagan, T. (2019) The effect of population bottleneck size and selective regime on genetic diversity and evolvability in bacteria. Genome Biol Evol 11: 3283–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, J.M. , and Mercenier, A. (2008) Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria, Nature reviews . Microbiology 6: 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek, A.S. , and Martin, V.J. (2012) Effects of synthetic cohesin‐containing scaffold protein architecture on binding dockerin‐enzyme fusions on the surface of Lactococcus lactis . Microb Cell Fact 11: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondraczek, L. , Pohnert, G. , Schacher, F.H. , Kohler, A. , Gottschaldt, M. , Schubert, U.S. , et al (2019) Artificial microbial arenas: materials for observing and manipulating microbial consortia. Adv Mater 31: e1900284. [DOI] [PubMed] [Google Scholar]
- Xue, X. , Wang, R. , Tu, T. , Shi, P. , Ma, R. , Luo, H. , et al (2015) The N‐Terminal GH10 domain of a multimodular protein from Caldicellulosiruptor bescii is a versatile xylanase/beta‐glucanase that can degrade crystalline cellulose. Appl Environ Microbiol 81: 3823–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, K.M. , Jiang, Z.Y. , Zheng, C.T. , Wang, L. , and Yang, X.F. (2014) Effect of Lactobacillus plantarum on diarrhea and intestinal barrier function of young piglets challenged with enterotoxigenic Escherichia coli K88. J Anim Sci 92: 1496–1503. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.H. (2011) Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol Adv 29: 715–725. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Liu, L. , Hao, Y. , Zhong, S. , Liu, H. , Han, T. , and Xie, Y. (2013) Isolation and partial characterization of a bacteriocin produced by Lactobacillus plantarum BM‐1 isolated from a traditionally fermented Chinese meat product. Microbiol Immunol 57: 746–755. [DOI] [PubMed] [Google Scholar]
- Zhang, B. , Li, N. , Wang, Z. , Tang, Y.J. , Chen, T. , and Zhao, X. (2015) Inverse metabolic engineering of Bacillus subtilis for xylose utilization based on adaptive evolution and whole‐genome sequencing. Appl Microbiol Biotechnol 99: 885–896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Enzymes examined in this study.
Table S2. Primers list.
Table S3. Real‐time PCR primers list.
Fig. S1. Comparative enzymatic activity of the selected cellulases and xylanases from different bacteria.
Fig. S2. Synergism between endoglucanases Cel5 and Cel9 with exoglucanase Cel48.
Fig. S3. Quantification of the chimaeric enzymes secreted by L. plantarum.
Fig. S4. Schematic representation of the adaptive laboratory evolution experiment.
Fig. S5. Enzymatic activity of Cel5 and Xyn10 enzymes on CMC and xylan.
Fig. S6. Relative degradation activity on CMC, xylan and wheat straw by consortium D2 at day 32 and “pseudo”‐consortium D2.


